ABSTRACT
Dormancy is a protective state in which diverse bacteria, including Mycobacterium tuberculosis, Staphylococcus aureus, Treponema pallidum (syphilis), and Borrelia burgdorferi (Lyme disease), curtail metabolic activity to survive external stresses, including antibiotics. Evidence suggests dormancy consists of a continuum of interrelated states, including viable but nonculturable (VBNC) and persistence states. VBNC and persistence contribute to antibiotic tolerance, reemergence from latent infections, and even quorum sensing and biofilm formation. Previous studies indicate that the protein mechanisms regulating persistence and VBNC states are not well understood. We have queried the VBNC state of Micrococcus luteus NCTC 2665 (MI-2665) by quantitative proteomics combining gel electrophoresis, high-performance liquid chromatography, and tandem mass spectrometry to elucidate some of these mechanisms. MI-2665 is a nonpathogenic actinobacterium containing a small (2.5-Mb), high-GC-content genome which exhibits a well-defined VBNC state induced by nutrient deprivation. The MI-2665 VBNC state demonstrated a loss of protein diversity accompanied by increased levels of 18 proteins that are conserved across actinobacteria, 14 of which have not been previously identified in VNBC. These proteins implicate an anaplerotic strategy in the transition to VBNC, including changes in the glyoxylate shunt, redox and amino acid metabolism, and ribosomal regulatory processes. Our data suggest that MI-2665 is a viable model for dissecting the protein mechanisms underlying the VBNC stress response and provide the first protein-level signature of this state. We expect that this protein signature will enable future studies deciphering the protein mechanisms of dormancy and identify novel therapeutic strategies effective against antibiotic-tolerant bacterial infections.
IMPORTANCE Dormancy is a protective state enabling bacteria to survive antibiotics, starvation, and the immune system. Dormancy is comprised of different states, including persistent and viable but nonculturable (VBNC) states that contribute to the spread of bacterial infections. Therefore, it is imperative to identify how bacteria utilize these different dormancy states to survive antibiotic treatment. The objective of our research is to eliminate dormancy as a route to antibiotic tolerance by understanding the proteins that control dormancy in Micrococcus luteus NCTC 2665. This bacterium has unique advantages for studying dormancy, including a small genome and a well-defined and reproducible VBNC state. Our experiments implicate four previously identified and 14 novel proteins upregulated in VBNC that may regulate this critical survival mechanism.
KEYWORDS: antibiotic tolerance, proteomics, stress response, systems biology
INTRODUCTION
Dormancy is a remarkable stress response in which bacteria curtail metabolic activity to survive potentially fatal external stresses. Dormancy has broad implications for many pathogenic bacteria, including Mycobacterium tuberculosis, Staphylococcus aureus, Treponema pallidum (syphilis), and Borrelia burgdorferi (Lyme disease) (1, 2). Additionally, stress responses appear to have importance in bacterial life processes like quorum sensing and biofilm formation (3, 4, 5).
The recognized importance of dormancy does not provide a simple definition for this stress state. Recent evidence suggests that dormancy consists of a continuum that spans bacterial stress responses, including the recognized viable but nonculturable (VBNC) and persistence states (6). Persister cells have been identified as a “small subpopulation of cells that spontaneously enter a dormant, nondividing state” (7, 8), a definition based on survival of bacterial cell populations after antibiotic treatment (9, 10). A complementary definition for VBNC is “living cells that have lost the ability to grow on routine media” (1). This VBNC definition is based on evidence that these cells are metabolically active, respiration competent, and capable of incorporating amino acids into proteins (11, 12). However, these definitions have caveats for defining actual dormancy states. Differentiating and isolating VBNC and persister cells is difficult at best because both states exhibit enhanced survival under antibiotic treatment. It is also likely that these states coexist and stochastically arise in any dormant cell population (13, 14). There is general agreement that VBNC and persistence states are interrelated, have reduced metabolic activity, and are important for antibiotic tolerance, latent infection, and reemergence of active infections after resolution of stress (13). From these considerations and for the purposes of this article, we propose to describe persister cells as proliferation-competent cells that can spontaneously resuscitate after resolution of stress. VBNC cells are induced into a nonproliferative state by external stress and will not resuscitate spontaneously until an appropriate external signal is provided. These definitions were chosen to specifically address the experimental system described in this article, but they are consistent with observations in the field and provide a viable starting point for experiments and discussion (1, 7).
Dormancy has been implicated in persistence and VBNC stress responses in chronic infection, latency, and resuscitation of active infections (15, 16). Targeting resuscitation has been a primary focus by the World Health Organization (WHO) as an avenue for treatment of widespread tuberculosis infection (17). A link between latency and dormancy was suggested by Wayne from results showing initiation of dormancy in oxygen-poor (hypoxic) tissue environments (18), an observation leading to an in vitro hypoxia M. tuberculosis model of dormancy (19–23). The actual state and underlying mechanisms of tuberculosis latency are not well understood, and diverse definitions exist for latent and dormant phenotypic states (24). However, other bacteria, including Mycobacterium bovis, Mycobacterium smegmatis, Escherichia coli, Listeria monocytogenes, and Micrococcus luteus, can exhibit VBNC, persistence, and other dormancy stress responses under nutrient deprivation and long-term antibiotic treatment (9, 25–29). These observations are not unique, and several recent reviews compiling data from many laboratories using different bacterial strains and conditions suggest that different stresses may not generate equivalent dormancy states (30, 31, 32). There is even diversity of specific genetically encoded pathways that are critical for the regulation of persistence, including quorum sensing (33, 34), (p)ppGpp (35–37), toxin-antitoxin systems (10, 38–40), and even the age of the inoculation (41). These data suggest diverse stress response states that are dependent on diverse sets of proteins. We note that drawing general conclusions from combining unrelated studies using different bacteria and experimental conditions should be approached with caution. However, this level of complexity presents experimental challenges, and our definitions may require amendment in different bacterial systems.
Rather than focusing on diversity, we ask the question, is there a common core of stress response proteins across different dormancy states and conditions? A conserved dormancy-related set of proteins has remarkable implications. These protein data would be complementary to and enhance genetic and transcriptomic profiles, because the relationship between transcripts and expressed proteins may not be directly comparable in a metabolically reduced state. These proteins would be a signature for the dormant state and facilitate diagnostics for latent bacterial infections. The biological functions of these proteins could define baseline mechanisms that drive the dormancy stress response. Extension to structural studies and protein-protein interactions can provide viable targets for drug development against bacteria exhibiting reduced metabolic activity, which is a limitation of current antibiotics. There is even the potential, outlined in a recent review, of identifying the “Achilles heel” for persister cells, i.e., a protein that is essential for maintaining the dormant state (7). These considerations suggested the importance of identifying conserved dormancy-related proteins.
Here, we report using Micrococcus luteus NCTC 2665 (MI-2665) as a nonpathogenic actinobacterial model to identify VBNC stress response proteins. MI-2665 has unique advantages for these studies, including rapid growth (doubling time of ∼40 min compared to over 24 h for M. tuberculosis), a small, 2.5-Mb genome comprising a “minimal set” of gene products and proteins necessary for dormancy, and homology with pathogenic M. tuberculosis, M. bovis, and Mycobacterium leprae. Importantly, MI-2665 exhibits a well-defined and reproducible VBNC state under nutrient deprivation (42). Finally, the resuscitation-promoting factor (Rpf) protein from MI-2665 can induce resuscitation from the VBNC state in M. tuberculosis (43). These reports suggest that mechanisms driving VBNC and, potentially, other dormancy states may be conserved among actinobacteria and that MI-2665 has a uniquely tractable VBNC state for study. An additional consideration was that MI-2665 VBNC bacteria do not resuscitate spontaneously, which circumvents the potential complication presented by persister cells combined with VBNC cells in the biological sample, based on our definition. We note that many other bacterial systems can resuscitate spontaneously after the removal of external stress (1), but it is unclear to us whether the restored viability is from VBNC or persistence cell states.
We used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantitatively identify proteins in MI-2665 grown in logarithmic-growth and dormant phases, using nutrient deprivation to generate a physiologically relevant and long-term dormancy state (44). We have identified 18 proteins that are differentially increased in dormant MI-2665. Given the caveats for generalization of our data identified above, we noted that four tuberculosis latency-related proteins, including universal stress protein A (UspA) and proteins involved in the anaplerotic glyoxylate pathway, were quantitatively increased in the MI-2665 VBNC state (45, 46, 47). The other 14 proteins we observed to be quantitatively increased in the MI-2665 VBNC state have not been previously reported but are conserved across many, if not a majority, of bacterial species and correlate to known important amino acid metabolism and ribosomal regulation pathways. These proteins present a proteomic signature for the VBNC state in MI-2665 that complements other transcriptomic studies (47–50). Our data support the idea that conserved proteins and pathways could regulate dormancy across actinobacteria and, potentially, across diverse bacterial species. While it is not possible at this time to conclusively generalize our results, the conservation of the proteins identified in these data warrants further exploration. The unique properties of MI-2665 present a relevant and tractable bacterial system for these future studies.
RESULTS AND DISCUSSION
Reduced global protein diversity in VBNC MI-2665.
MI-2665 bacteria were grown on acetate minimal medium and harvested in logarithmic, stationary, and VBNC phases (confirmed by CFU counts). Bacterial samples were analyzed by SDS-PAGE, followed by LC-MS/MS. Proteins were identified using OMSSA by searching MS/MS spectra against the NCBI M. luteus protein FASTA database appended with a randomized decoy database (51, 52). To evaluate any bias in our protein-sampling procedures, we subjected all protein identifications to gene ontology analysis to identify functional classifications that were statistically represented at levels above the background level. We could not reliably identify overrepresentation of any functional classifications in our data, indicating that our experimental approach did not enrich for particular protein classes.
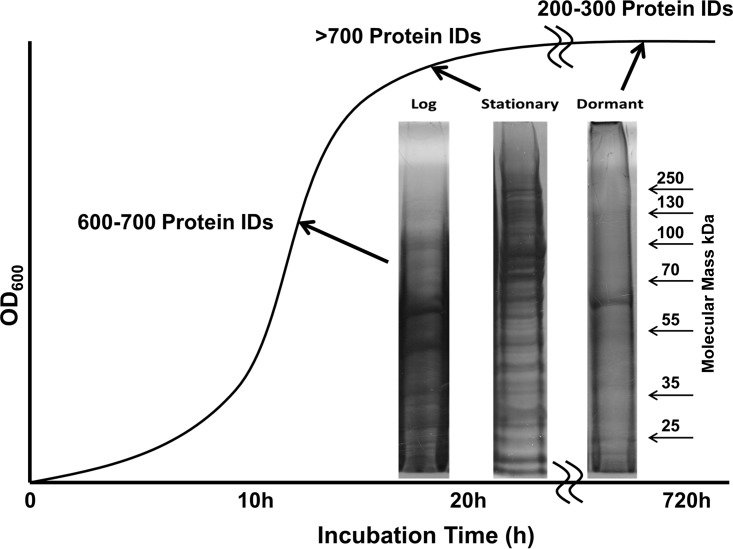
An obvious decrease of protein diversity and protein levels in the MI-2665 VBNC dormancy state was observed from Coomassie staining (Fig. 1). This observation is consistent with VBNC states in other bacterial species, such as Vibrio vulnificus (13). LC-MS/MS identified approximately 700 proteins in both logarithmic- and stationary-phase samples but only 200 to 300 proteins in the dormant sample. Using larger quantities (4 times the total number of cells compared to the number in the logarithmic-phase sample) of dormant MI-2665 cells did not increase protein identifications. The majority of proteins observed in the dormant phase were also found in the logarithmic phase, with only 24 proteins uniquely observed in dormancy compared to the protein expression in the logarithmic state (see Fig. S1 in the supplemental material). A cursory evaluation indicates that the majority of proteins observed in the VBNC state (315) were also observed in logarithmic-phase cells. Preliminary stationary-phase data show an overlap of 603 proteins with the logarithmic state, and only 173 were observed uniquely in the logarithmic-phase and 79 in the stationary-phase cells. A similar protein overlap between VBNC and either stationary or logarithmic phases was noted. We caution that this approach only considers the protein identifications. Quantitative proteomics data provided a much more nuanced picture. The quantity of protein in the VBNC cells was drastically reduced, as is evident from the results in Fig. 1. Our quantitative proteomics data confirmed this gross observation based on both fewer total protein identifications (339 in VBNC, 776 in logarithmic phase, and 682 in stationary phase) and a lower total number of peptide spectra annotated to proteins.
FIG 1.
Gel electrophoresis comparison of the protein complements of MI-2665 in log, stationary, and dormant phases. Note the significant reduction in total protein observed in dormant-phase gel electrophoresis. Total protein identifications by mass spectrometry were consistent with ∼700 proteins for logarithmic- and stationary-phase MI-2665 and 200 to 300 for dormant-phase MI-2665. The 60-kDa band in the dormant sample is highly abundant catalase, but this protein does not exhibit a reproducibly increased abundance in dormancy.
The drastic reduction in both the diversity and quantity of proteins accompanying transition into VBNC suggests a global shutdown of metabolic and protein machinery. This is not unexpected. The overlap between proteins identified in the logarithmic and VBNC states indicates that 315 proteins were identified in both states, but 461 protein identifications were lost in the VBNC state. There was much more observed overlap in protein identifications between the logarithmic and stationary phases. This overlap may be related to the time proximity of the samples, but we have noted that stationary-phase MI-2665 has limited growth potential, even when transferred to a highly growth-supportive medium. Obviously there is a shift in metabolism predisposing stationary MI-2665 cells toward the VBNC state, but the mechanisms constituting this shift are unknown. We must caution that considering these protein identifications in exclusion of other data may be misleading because of the sensitivity of LC-MS/MS. The quantitative data do support the idea that many housekeeping proteins are present in the VBNC state and may be important for reconstitution of cellular respiration and proliferation. However, more detailed quantitative experiments are required to differentiate proteins under transcriptional, translational, or degradative regulation and to discern when this regulation occurs. Such experiments will be necessary to identify proteins that are synthesized or retained specifically in the VBNC state rather than being identified because of the sensitivity of the experimental technology.
The most abundant band observed in the dormant gel lane was identified as a bifunctional catalase, KatG, which is highly abundant in all growth phases based on mass spectrometry. The data shown in Fig. 1 and our mass spectrometry data indicate that this protein is retained at high levels in the VBNC state compared to the loss of other proteins. KatG is a primary peroxidase, a defense against reactive oxygen species (ROS), and apparently activates the first-line antituberculosis antibiotic isoniazid (28, 53). We anticipate that KatG's retention in VBNC cells may be related to anti-ROS activity in stress survival or VBNC induction, as observed in V. vulnificus (54, 55).
Multiple proteins are differentially expressed in MI-2665 in VBNC and logarithmic-growth phases.
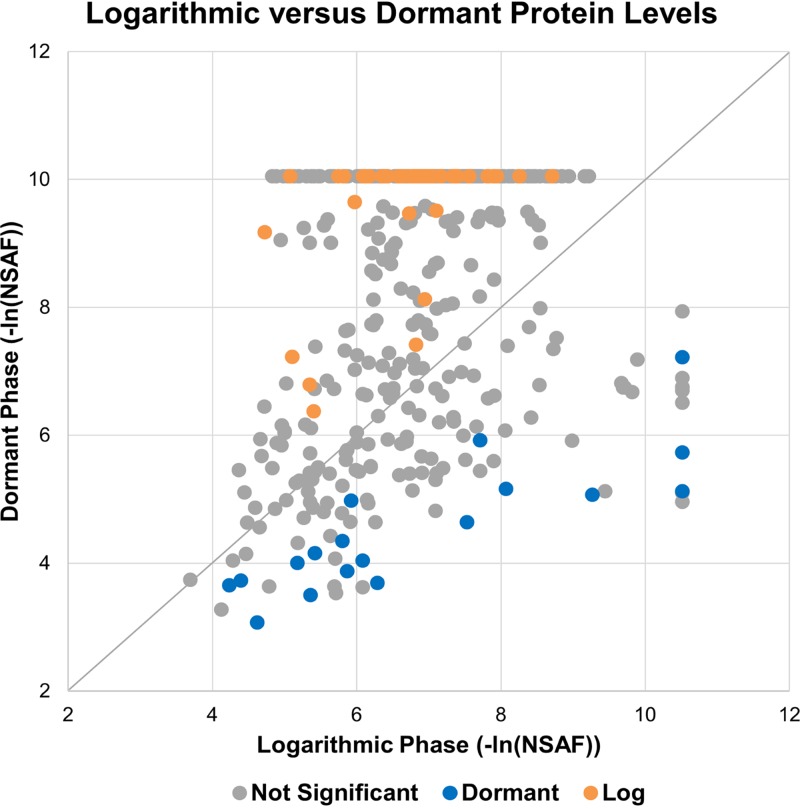
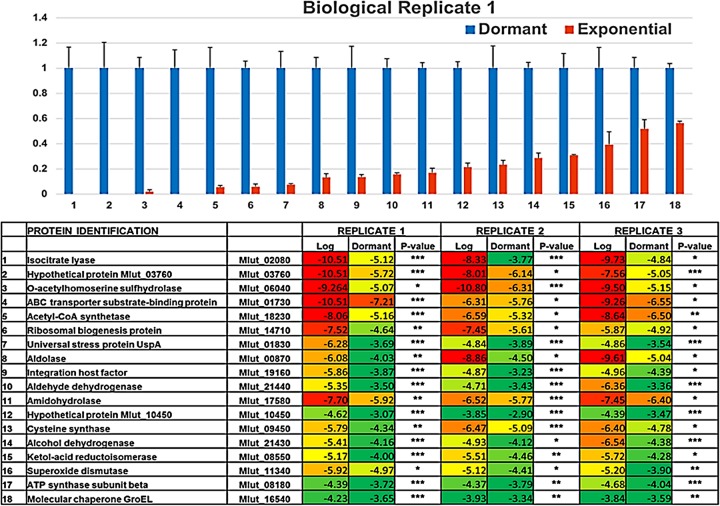
A more definitive measure of the differential quantities of proteins in MI-2665 during the VBNC state was obtained by quantitative proteomics analysis. The proteins identified were quantified using natural log-transformed normalized spectral abundance factors (NSAF) for statistical comparison of protein levels between the bacterial growth phases (56). The ln(NSAF) data were analyzed for normality by the Shapiro-Wilks test. The scatter plot analysis comparing negative ln(NSAF) values in the logarithmic and dormant phases showed a wide distribution of protein quantities, as expected from the large differences in observed proteins (Fig. 2). We observed 18 proteins that were statistically more abundant in dormant growth than in logarithmic growth (P < 0.05) in all biological replicates. Any of these proteins could potentially serve as part of a molecular signature for the dormant stress response, and the implications of such a signature are described in the introduction. The small number of identifications precluded functional organization by gene ontology, but quantitative data identified important differences in protein levels that suggest that both metabolic and ribosomal regulatory processes are important for the VBNC state (Fig. 3). Several of the proteins we identified are homologous to proteins important in M. tuberculosis hypoxia and latency stress responses, i.e., universal stress protein A, isocitrate lyase, O-acetylhomoserine sulfhydrylase, and O-acetylserine sulfhydrylase.
FIG 2.
Scatter plot of –ln(NSAF) values for proteins identified in dormant and logarithmic growth phases. Proteins identified and quantified in dormant and logarithmic phases were natural log transformed and converted to positive values for comparison by correlation scatter plot. Proteins statistically overrepresented are colored blue (dormant) and yellow (log) and exhibit P values of <0.05 for all biological replicate experiments. These data indicate that quantitation statistics are required to identify proteins overrepresented in different growth phases. Distance from the diagonal correlation line (i.e., ≥2-fold change) is not a sufficient criterion. Note that negative natural log transformation generates positive x and y axes, but larger numbers correspond to smaller quantitative abundances.
FIG 3.
Bar graph comparison of the 18 proteins increased in dormant versus logarithmic phase (top) and heat map of quantitative data comparison by ln(NSAF) for dormant and logarithmic phases (bottom). (Top) NSAF protein quantities in dormant and logarithmic phases were plotted as a bar graph after normalizing to the level of each protein in the dormant phase, set as 1. (Bottom) Heat map comparing natural log-transformed data between logarithmic (Log) and dormant growth phases for each biological replicate experiment (replicates 1, 2, and 3). Statistical quality was evaluated by P value and is represented by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.005). Numbering in the bar graph corresponds to the same number listing in the heat map. Error bars show standard deviations.
Universal stress protein A.
The results for VBNC MI-2665 demonstrate that only one of the three universal stress protein A (UspA) variants is increased in the VBNC state, WP_010079616.1 (Mlut_01830) (Fig. 3). This protein is conserved across many mycobacterial species (see Fig. S2 in the supplemental material). In M. tuberculosis, the UspA homolog Rv2623 is an ATP-binding protein that facilitates tuberculosis infection in animal models. Included in an animal study were structural studies of the role of Rv2623 in establishing chronic and persistent infection (discussed below) (45). Usp-type proteins are observed to be differentially regulated under different external stress conditions, including hypoxia, nutrient deprivation, heat, oxidants, and antibiotics (57, 58). In E. coli, UspA has been associated with oxidative stress and modulation of carbon metabolism, acetyl coenzyme A (acetyl-CoA) synthetase, and the glyoxylate shunt proteins isocitrate lyase (Icl1) and malate synthase (58, 59). We note that acetyl-CoA synthetase and Icl1 were identified in our data and malate synthase was statistically significantly increased in two of the biological replicate data sets (Fig. S9) Additionally, E. coli UspA induction occurs through sigma 70, which is also activated during carbon starvation (60). Stress regulation of UspA and other Usp proteins is also subject to the stringent (ppGpp) response system, with the uspA gene being upregulated most rapidly of the usp genes (61, 62). UspA appears to be regulated by proteins regulating the bacterial cold shock stress response and even negatively regulated by the lipid biosynthesis regulator FadR (63, 64). These reports provide further evidence that UspA may be an important component of and even mechanistically linked to many different stress responses. The tuberculosis genome contains 10 different Usp-type proteins with apparent functional redundancy, but data suggest that there may be incomplete redundancy of particular Usp proteins for different stresses (65, 66). The data linking Rv2623 to chronic infection indicate this protein may have a specific importance in the infection process that cannot be replicated by other Usp proteins. Therefore, the combination of our observations and data in the literature indicate that UspA has a role in modulating the VBNC state in response to nutrient deprivation and that the homologous Rv2623 may be associated with the same type of response in Mycobacterium tuberculosis.
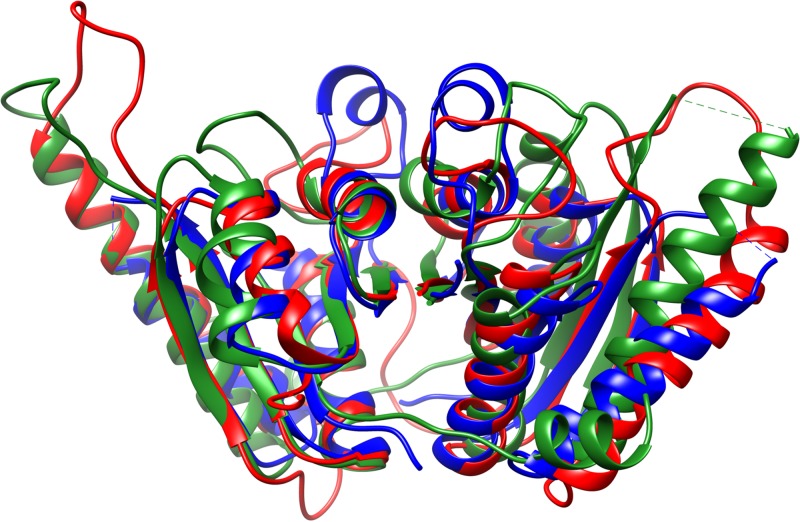
To define the relationships between UspA proteins, sequences from Rv2623, the MI-2665 UspA, and homologous sequences from M. tuberculosis, M. smegmatis, and Lactobacillus plantarum were analyzed. The results indicate sequence conservation for regions important in the Rossman fold, including the magnesium- and nucleotide-binding-site residues (see Fig. S2 in the supplemental material). We and others have observed that Usp proteins exist as either single- or double-domain structures, with the majority of double-domain structures in actinobacteria (45). The M. smegmatis and L. plantarum Usp proteins are single-domain variants but are crystallized as a dimer of dimers. The UspA proteins from M. tuberculosis and MI-2665 are double-domain proteins. Therefore, we combined two single-domain sequences for the alignment of M. smegmatis and L. plantarum Usp proteins to M. tuberculosis and MI-2665 UspA proteins. While the MI-2665 UspA structure is not known, the structures of these other Usp-type proteins, including Rv2623, have been solved by crystallography. We used Chimera to visualize and overlay multiple UspA structures from M. tuberculosis, M. smegmatis, and L. plantarum (67). These structures exhibit the same Rossman fold, important in signaling and nucleotide-binding proteins (Fig. 4) (45). Confirming the alignment for these structures, the nucleotide substrate overlay is displayed in Fig. 5. These data suggest UspA and Rv2623 have similar structures with two connected ATP-binding domains. While the biochemical function of Usp proteins is currently unknown, these data and the homology between Rv2623 and UspA enable us to propose that MI-2665 UspA contains two Rossman fold ATP-binding domains and that Usp proteins may act as dimers to regulate the VBNC transition through interactions and signaling with other stress response proteins. Additional support for this hypothesis is the observation of phosphorylation of other UspA-type proteins in E. coli and the correlation that UspA knockout affects the levels of acetyl-CoA synthase and the glyoxylate shunt protein isocitrate lyase, both of which were identified as robustly increased in our VBNC data (59, 60).
FIG 4.
Overlap of Usp structures. Usp protein structures from M. tuberculosis (PDB no. 3cis; red), M. smegmatis (PDB no. 5ahw; blue), and L. plantarum (PDB no. 3fg9; green) were overlaid in Chimera using a 3-Å cutoff with an RMSD of 1.139 Å between 101 atom pairs. The overlap demonstrates that Usp proteins from diverse organisms exhibit almost identical core structures, with sequence homology at ∼35% identity and ∼50% similarity. The sequence homology between these UspA proteins is similar to that observed for M. luteus MI-2665. These images were created in Chimera.
FIG 5.
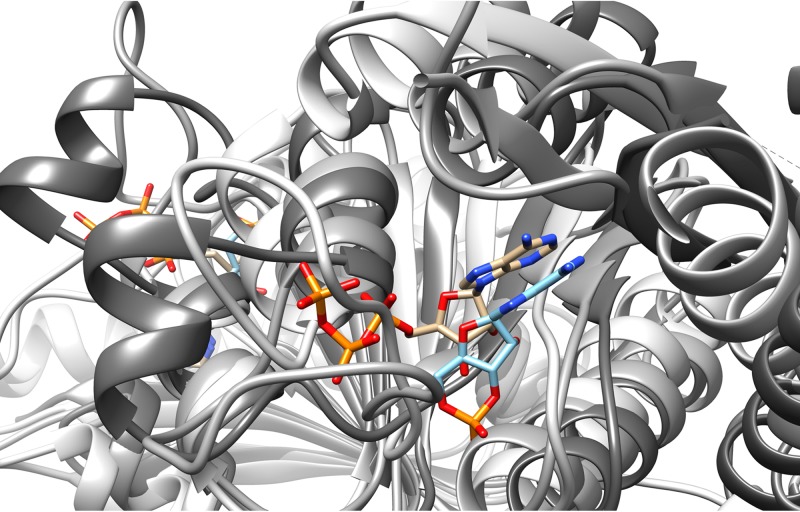
Overlay of nucleotide substrates for Usp homologues from M. tuberculosis (PDB no. 3cis) and M. smegmatis (PDB no. 5ahw). The ATP nucleotide substrate (tan) from M. tuberculosis Rv2623 is similarly oriented and overlays the cyclic AMP (cAMP) substrate (light blue) in M. smegmatis Usp protein. The Usp protein from L. plantarum (PDB no. 3fg9) did not have a nucleotide substrate in the crystal structure but is included to emphasize the structural similarity in this protein family. These images were created in Chimera.
Isocitrate lyase.
We observed a remarkable increase in isocitrate lyase (Icl1), an enzyme in the glyoxylate pathway. This enzyme is also induced in M. tuberculosis hypoxia (46). Hypoxia would downregulate the citric acid cycle because oxidative phosphorylation requires oxygen. Icl1 catalyzes the conversion of isocitrate to malate, bypassing two oxidative decarboxylation steps in the citric acid cycle, reducing NADH formation, and preserving a limited carbon pool. The MI-2665 Icl1 structure has not been determined, but the core structures of Icl1 from M. tuberculosis and Aspergillus nidulans show remarkable structural conservation (see Fig. S3 in the supplemental material). We compared the sequences for Icl1 from M. luteus to those of M. tuberculosis and the fungus A. nidulans and found sequence homologies at 79% identity and 87% similarity for M. tuberculosis and 39% identity and 54% similarity for A. nidulans (Fig. S4). We hypothesize that these proteins perform the same function across actinobacteria and that significant mutation of the Icl1 core structure would result in a compromised enzyme that would not support survival under stress conditions. This hypothesis is consistent with the knockout of the icl1 gene in M. tuberculosis, which severely impairs survival in both acute and persistent infection processes (68, 69). It is also of note that FadR, a protein involved in lipid biosynthesis regulation and a negative regulator of UspA (see “Universal stress protein A” above), also affects the regulation of the glyoxylate shunt proteins, including Icl1 (70).
Cysteine and methionine metabolism.
Two related proteins involved in amino acid metabolism were observed to be increased in VBNC MI-2665. Cysteine synthase (CysK1, or O-acetylserine sulfhydrylase) is a critical enzyme in cysteine metabolism and catalyzes the synthesis of cysteine from O-acetylserine and hydrogen sulfide. An alignment between the homologous proteins in MI-2665 and M. tuberculosis demonstrates 70% sequence identity and 85% sequence similarity (Fig. S5). Similar levels of sequence homology are observed from other mycobacterial species, and the overlay of several available protein structures demonstrates structural conservation for CysK1 across different mycobacteria (Fig. S6). A related protein involved in methionine synthesis, O-acetylhomoserine sulfhydrylase, was also observed to be greatly increased in VBNC MI-2665. This enzyme catalyzes a reaction similar to the one catalyzed by CysK1 but uses O-acetylhomoserine condensation with methylthiol to generate methionine. An alignment between the MI-2665 and M. tuberculosis O-acetylhomoserine sulfhydrylase proteins demonstrates 65% sequence identity and 77% similarity (Fig. S7). This is approximately the level of sequence identity and similarity observed between the same proteins from M. tuberculosis, Mycobacterium ulcerans, and Mycobacterium marinum. Because the structure of O-acetylhomoserine sulfhydrylase was not found for M. tuberculosis, we used the structure of the homologous protein O-succinylhomoserine sulfhydrylase from M. tuberculosis, which catalyzes a similar sulfur transfer reaction in methionine synthesis (Fig. S8). The alignment between MI-2665 and M. tuberculosis O-acetylhomoserine sulfhydrylase proteins demonstrates 65% sequence identity and 77% similarity. This is approximately the level of sequence identity and similarity observed between the same proteins from M. tuberculosis, M. ulcerans, and M. marinum, whose structures are compared (Fig. S5 and S6). As we describe below (next section), these proteins are amino acid metabolism enzymes and would signal a pivot away from complete catabolic breakdown of carbon sources to conservation of carbon species under nutrient deprivation. However, these enzymes synthesize cysteine and methionine, which would also be important to stabilize the oxidation-reduction environment within bacterial cells under starvation stress.
Other proteins increased in dormancy.
Our initial focus on UspA, isocitrate lyase and sulfhydrylases derives from the reported associations of these proteins in latency, antibiotic resistance, hypoxia, and nutrient stress in M. tuberculosis (45–50, 71, 72). These proteins may represent components of a reproducible protein signature for the VBNC state and mediate general responses to different external stresses, making them important biomarkers for stress and viable drug targets. Currently, Icl1 and O-acetylserine sulfhydrylase are targets for antituberculosis drug therapeutics (73, 74).
We also observed 14 other proteins not previously identified as stress responsive but robustly increased in the nutrient deprivation-induced VBNC state, with P values of ≤0.05 in all biological replicate experiments (Fig. 3). What could these proteins be doing in the VBNC state? Superoxide dismutase and the molecular chaperone GroEL have obvious implications for stress response. Superoxide dismutase is another enzyme important for detoxification of ROS and would complement the retention of KatG. GroEL would facilitate refolding of proteins under stress conditions. Others of the observed proteins are linked to metabolic pathways and generally signal a pivot away from catabolic metabolism toward anabolic construction of metabolites important for stress survival. In evaluating protein homology with other bacteria, we identified likely biological functions for many proteins without informative names in the M. luteus FASTA database. MI-2665 aldolase has homology with citrate lyase, which generates oxaloacetate and acetyl-CoA from citrate. This metabolic process would complement the glyoxylate shunt by further metabolizing citrate to acetyl-CoA, which can be used for anabolic synthesis of proteins and metabolites. Aldehyde and alcohol dehydrogenases affect glyceraldehyde-3-phosphate oxidation and lactate fermentation. These enzymes signify a shift away from aerobic respiration and complete combustion of sugars and toward the utilization of carbon sources for anabolic metabolism. Amidohydrolase has homology with acyl-amino acid degradation enzymes, linking this enzyme with ketol-acid reductoisomerase and the O-acetylserine and O-acetylhomoserine sulfhydrylases (described above) for amino acid metabolism.
A highly significant observation in our data was the identification of the ribosomal regulatory protein ribosomal biogenesis protein and DNA-binding integration host factor. Sequence and structural analysis for ribosomal biogenesis protein indicates homology with ribosomal hibernation-promoting factors (HPFs) (75). HPFs are important for stationary-phase ribosome stabilization through dimerization of 70S ribosomes into 100S particles (76, 77). This process shuts off translation and stabilizes ribosomes against proteolytic degradation. This protein also has homology to a light-repression transcript found in Synechococcus sp. strain PCC7002, indicating that this mechanism of translational regulation under stress may be general across bacteria (78). Our observation of an HPF homolog upregulated in the VBNC state of MI-2665 suggests this protein may have functional relevance to dormancy. Integration host factor (IHF) is a DNA-binding protein which helps in maintaining bacterial chromosomal architecture and regulates transcription (79, 80, 81). Unlike E. coli IHF, mycobacterial IHF is essential for survival and binds DNA unspecifically (82, 83). Indeed, mycobacterial IHF is one of the most abundant proteins in mycobacteria (84). The crystal structure of IHF bound to DNA is also known (85). A previous study has described the role of IHF as a transcriptional regulator of genes involved in the transition from exponential to stationary phase (79). However, the role of actinobacterial IHF in VBNC is yet to be studied.
We observed two hypothetical proteins that increased in dormancy compared to their levels under logarithmic-growth conditions, Mlut_10450 and Mlut_03760 (Fig. 3) Using the NCBI Conserved Domain Search System (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (86), we were able to identify a domain correlation between Mlut_10450 and an ancestral histone fold in a hyperthermophilic bacterium, Aquifex aeloicus (87). If the correlation is correct, Mlut_10450 could protect the bacterial genome against oxidative and stress damage, as described for another M. tuberculosis protein, Lsr2 (88). The association for Mlut_03760 is more tenuous and identifies a relationship with a predicted IG fold in a putative copper oxidoreductase, FixG. The reproducible observation of these proteins in the VBNC state and their correlation to equivalent proteins in other actinobacteria, including M. tuberculosis, suggests further experimental studies are warranted.
Many more proteins were identified as being increased in VBNC MI-2665 in two of the three replicate measurements, exhibited significant P values when observed, and are important stress response proteins. For example, malate synthase was observed to be upregulated in two of the three biological replicate experiments but is critical for the glyoxylate pathway. As would be expected, this protein was only identified in correlation with isocitrate lyase. Other examples are present in our data and will be the subject of future investigations (Fig. S9).
Conclusions.
We have identified 18 proteins that are upregulated in a VBNC state of Micrococcus luteus strain MI-2665, an actinobacterial relative of pathogenic mycobacteria, including M. tuberculosis, M. bovis, and M. leprae. Four proteins identified in our data have known importance in M. tuberculosis latency and hypoxic states, including universal stress protein A, isocitrate lyase, O-acetylserine sulfhydrylase, and O-acetylhomoserine sulfhydrylase. The other 14 proteins identified are novel and have not been previously identified in any actinobacterial stress response states, but some are correlated to stress responses in other bacteria. The sequence and structural homologies between MI-2665 and other bacterial proteins suggest that VBNC mechanisms may be conserved across actinobacteria and even to more distant bacterial relatives.
An intriguing idea derived from these data is the upregulation of anaplerotic metabolism as a stress response to nutrient deprivation, exemplified by the glyoxylate metabolic pathway. We refer to the definition provided by H. L. Kornberg in his original Colworth Medal Lecture in 1965, namely, that an anaplerotic sequence comprises ancillary pathways that extend from, link to, and replenish a metabolic pathway (89). This lecture provided an overview of the glyoxylate metabolic bypass in the tricarboxylic acid (TCA) cycle and, by logical extension, the foundation of why anaplerotic metabolic pathways could be important components of a stress response. In Fig. 1 of this remarkable lecture, Kornberg identifies the growth delay for E. coli linked to changes in specific activities of isocitrate lyase and malate synthase when transferred to acetate medium. Transition to a new carbon source is an obvious stress that requires the activation of alternative metabolic pathways to restore growth. When we impose limited acetate as a sole alternative carbon source to force MI-2665 to transition into the VBNC state, the upregulated glyoxylate cycle enables survival through two different mechanisms. Glyoxylate metabolism bypasses both oxidative decarboxylation CO2 emission steps in the TCA cycle, thus conserving a limited carbon source. Why should this be important? The anaplerotic nature of this and associated metabolic pathways can shuttle these carbons away from degradation and toward synthesis of other physiologically critical metabolites in accordance with the definition of an anaplerotic pathway. Another logical extension of these data is that interconnected metabolic systems must be regulated by other sensor proteins and metabolites that translate the diminishing acetate (or other limited carbon source) levels into the cellular VBNC response. An anaplerotic metabolic pathway is ideal for such regulation, and Kornberg identifies phosphoenolpyruvate as just such a regulatory molecule for glyoxylate metabolism. It is not surprising that the other metabolic proteins we observe to be upregulated in the VBNC state are components in other important metabolic pathways linked to the TCA cycle in some manner. However, the identities of regulatory sensor proteins and metabolites in the VBNC state remain to be elucidated. This last point is of considerable interest for us, because we expected to find ppGpp stringent response and toxin-antitoxin proteins that could serve as sensor protein systems. These were not observed to be increased in the MI-2665 VBNC state. Another expected class of proteins are the two-component signaling kinase system proteins associated with LuxR, the equivalent system to the DosR/DevR system observed in M. tuberculosis. Most studies identifying DosR and DevR system components in M. tuberculosis used hypoxic stress, so it is possible that the LuxR system is not upregulated in MI-2665 under nutrient deprivation. Alternatively, these proteins may be lower in abundance under our experimental stress conditions and not observed. As we reference below, more sensitive directed mass spectrometry methods are available and may be applicable to identify these potentially important but lower abundance proteins.
Our data at the protein level have importance when correlated to several published studies on gene regulation in M. tuberculosis under different stress conditions, including hypoxia (47, 48, 49) and macrophage engulfment (50). The initial comparisons with these genetic studies were responsible for identifying UspA, isocitrate lyase, and both sulfhydrylase enzymes as potential targets for protein regulation in the MI-2665 VBNC dormancy state. Many of the genes differentially regulated in these genetic studies were not observed in our protein-based experiments. Among the reasons for this may be that stress conditions for these genetic studies were not equivalent to our conditions or that the dynamic range available for genomic studies is higher than that of the protein analysis technologies we employed in this study. Additionally, there may be limitations in the sample preparations used for these experiments that would limit our observations of particular classes of proteins. For example, the use of molecular-weight-cutoff filters and limiting detergents for mass spectrometry analysis may preclude our observation of peptides or smaller proteins and reduce the number of membrane proteins in our experiments. We note that our sample preparations are similar to published procedures that would exhibit many of these same limitations (90). More comprehensive mass spectrometry technologies are available once a complete proteomic analysis of a bacterial sample is completed, and these may be useful for future experiments in MI-2665, especially with the incorporation of multiple different sample preparations (84). Despite these limitations, our data provide many insights for future experiments based on the 18 proteins we rigorously identified as quantitatively increased in the VBNC state. We propose the following experiments: (i) knockout of proteins to identify causative agents and regulators for the VBNC state, (ii) proteomic analysis of alternative stress systems like hypoxia, acidity, and antibiotics to identify core proteins involved in stress responses, (iii) identification of protein interactions and signaling processes that initiate and maintain the VBNC state, (iv) proteomic and functional analysis of the proteins lost in VBNC transition and how loss of these functions affects dormancy, (v) identification of the protein signature and functional mechanisms of resuscitation, and (vi) development of novel strategies to therapeutically target proteins that drive the VBNC and persistent bacterial states and facilitate resuscitation from dormancy. These experiments will answer critically important questions about the proteins enforcing dormancy and resuscitation states and how these proteins affect bacterial survival and antibiotic tolerance. The studies presented in this article are the foundation for building a molecular picture of dormancy; they establish a baseline protein signature for a uniquely well-defined and reproducible VBNC state and mark MI-2665 as a relevant reference organism for understanding the molecular basis of this critical bacterial survival mechanism.
MATERIALS AND METHODS
Buffers, reagents, and chemicals were purchased from Sigma-Aldrich, Fisher Scientific, or VWR and were ACS-certified reagent grade or better. Solvents were purchased from Fisher Scientific and were high-performance liquid chromatography (HPLC) grade, except for solvents used in mass spectrometry, which were LC-MS or Fisher Optima grade. Halt protease inhibitors were purchased from Thermo Fisher Scientific. Trypsin was purchased from Promega or Worthington Biochemicals. Micrococcus luteus NCTC 2665 was purchased from ATCC (www.atcc.org) and stored as glycerol stocks at −80°C until plating and culture experiments.
Exponential-phase and dormant cultures of M. luteus MI-2665 on acetate minimal medium.
Micrococcus luteus NCTC 2665 was plated from a glycerol stock onto a rich medium plate. A single colony was used to inoculate 5 ml of rich medium broth starter culture. After incubating the culture at 30°C for 5 h (optical density at 660 nm [OD660] of between 0.3 and 0.6) with shaking (265 rpm), 2 ml of starter culture was inoculated into 500 ml of acetate minimal medium (AMM) and incubated at 30°C with shaking. The bacterial growth was monitored by OD660 from time point zero until the growth curve entered late stationary phase. The samples were collected every 2 h for OD measurements. When the bacterial growth was in logarithmic (log) phase (i.e., OD660 of 0.5 to 0.9), 40 ml of culture was harvested and collected by centrifugation at 10,000 × g for 10 min at 4°C. The cell pellet was resuspended in 1 ml of Tris-EDTA (TE) buffer (100 mM Tris-HCl and 10 mM EDTA, pH 8) and used immediately or stored at −80°C. To collect dormant M. luteus samples, the culture were incubated at 30°C with agitation for 2 months and then kept at room temperature without agitation for up to 1 month. The viable but nonculturable (VBNC) state of dormant bacteria was confirmed by plating experiments on rich medium agar plates. The dormant culture was harvested similarly to the logarithmic-phase sample.
Rich medium.
In a total volume of 500 ml, 8.5 g tryptone, 1.5 g yeast extract, 1.25 g dextrose, 2.5 g NaCl, and 1.25 g potassium phosphate dibasic (K2HPO4) were added, adjusted to pH 7.4, and autoclaved. Rich medium agar plates contained 1.5% agar.
Acetate minimal medium.
A 500-ml volume of acetate minimal medium (AMM) containing 4 g/liter NH4Cl, 1.4 g/liter K2HPO4 at pH 7.4, 0.1 M sodium acetate trihydrate, 0.5 g inosine, 0.5 g yeast extract (0.1%), and 5 ml 100× trace metal stock solution (14.3 g/liter MgSO4 7H2O, 0.00375 g/liter CuSO4 5H2O, 0.079 g/liter MnCl2 4H2O, 0.183 g/liter FeSO4 7H2O, 0.025 g/liter Na2MoO4, and 0.005 g/liter ZnSO4 7H2O) was autoclaved, and 0.5 ml filter-sterilized 1,000× vitamin supplement consisting of 0.2 g/10 ml methionine, 0.4 g/10 ml thiamine, and 0.05 g/10 ml biotin dissolved in 0.1 M NaPO4 at pH 7.4 was added.
Cellular lysate preparation.
Sample preparation was modified from the procedures in a proteomic study in Streptococcus pyogenes (90). M. luteus MI-2665 cells were harvested from exponentially growing (50 mg) and dormant (200 mg) cultures by centrifugation at 10,000 × g for 5 min, followed by 3 washes with 50 mM Tris HCl, pH 7.2. The cells were lysed in 1 ml of lysis buffer (6 M urea, 50 mM Tris HCl with 1× Halt protease inhibitors [Thermo Scientific]) using a Mini-BeadBeater (BioSpec Products) at 4,200 rpm, with five cycles of 3 min of bead beating and 3 min of incubation on ice. Cellular debris was pelleted by centrifugation (16,000 × g for 10 min), and the protein supernatant isolated and reduced using 5 mM 2-triscarboxyethyl phosphine hydrochloride (TCEP) at 37°C for 30 min. The sample was alkylated by adding iodoacetamide (IAA) to 5 mM and incubated at room temperature for 30 min in the dark. The reduced/alkylated sample was concentrated to 25 μl using a 10-kDa molecular-mass-cutoff filter (GE Healthcare), and protein concentrations were determined by absorbance at 280 nm. The concentrated protein sample (700 μg) was mixed with Laemmli buffer, boiled at 95°C for 10 min, and separated on a 10-well, 4 to 12% Bis-Tris NuPAGE gel (Invitrogen) at 200 V for 40 min using 1× MOPS (morpholinepropanesulfonic acid) running buffer. The gel was stained with Coomassie brilliant blue for an hour and destained overnight.
In-gel digestion.
Gel lanes were cut into 16 molecular-weight-range fractions and placed in separate LoBind Eppendorf tubes. The gel piece was minced into small fragments and washed in 100 mM ammonium bicarbonate-acetonitrile in a 3:1 (vol/vol) ratio at 37°C for 30 min. The cycle was repeated 3 times or until the gel became colorless. The gel was dehydrated with acetonitrile, rehydrated with 100 μl of trypsin (200 ng) in 100 mM ammonium bicarbonate buffer at pH 7.4, and incubated overnight at 37°C. The supernatant was transferred to a new LoBind Eppendorf tube and extracted with 100 μl of 1% formic acid-acetonitrile in a 3:1 (vol/vol) ratio. The extraction was repeated two more times, and the pooled supernatant was lyophilized and stored at −80°C or directly resuspended in 28 μl of 1% formic acid and used for mass spectrometry analysis. For each biological replicate, 48 total liquid chromatography-tandem mass spectrometry (LC-MS/MS) runs were performed for three independent technical replicates.
Liquid chromatography-tandem mass spectrometry.
LC-MS/MS was performed on a Thermo Finnigan LTQ (linear trap quadrupole) linear ion trap mass spectrometer coupled to an Agilent 1290 Infinity ultraperformance liquid chromatography (UPLC) system using solvent A (water plus 0.1% formic acid) and solvent B (methanol plus 0.1% formic acid). The gradient was 20% solvent B to 90% solvent B over 60 min at a flow rate of 40 μl/min on a 500-μm by 6-cm C18 reversed-phase column packed in-house. The electrospray voltage was set to 3.78 kV, with sweep, auxiliary, and sheath gas set to 0 on a standard IonMax electrospray ionization (ESI) source. The capillary temperature was 250°C, and data were acquired using data-dependent dynamic exclusion MS/MS on the three highest intensity masses observed in the MS scan. The RAW data files were transformed to MGF format using the MSConvert utility program from ProteoWizard (http://proteowizard.sourceforge.net/tools.shtml).
Data analysis.
MGF data files were searched using OMSSA against a database of 2,196 M. luteus proteins (NCBI RefSeq GCF_000023205.1 ASM2320v1) appended with a randomized decoy database (51, 52). MGF files for all 16 fractions for each technical replicate were combined into a single merged file prior to search. Default settings were employed for the ion trap, as follows: trypsin digestion with a maximum of 1 missed cleavage, precursor m/z tolerance of 2 Da, product ion tolerance of 0.8 Da, and carbamidomethyl C as a variable modification. The E value threshold was adjusted to a false discovery rate of ≤1%.
Quantitative-analysis-normalized spectral abundance factor (NSAF) (56) values were natural log transformed, and the Shapiro-Wilk test performed to confirm normal distribution of data (http://scistatcalc.blogspot.com/2013/10/shapiro-wilk-test-calculator.html). Proteins with no spectral counts in either the logarithmic or dormant states were replaced by 0.16 spectral counts as previously described to enable natural log transformation (56). Statistical analysis was performed using Microsoft Excel to identify proteins that were significantly upregulated (P value < 0.05) in all three biological replicates in either dormant or log phase. For the data shown in Fig. 2, negative natural log transformation was used to establish positive x and y axes to simplify visualization of the data.
Visualization and comparison of protein structures in Chimera.
Protein structures were visualized and comparisons performed in Chimera. Structure files from the Protein Data Bank (PDB) were imported into Chimera based on their PDB identification numbers and manipulated with the core toolset available in the program. Overlays were used for comparison of protein structures from different organisms (Fig. 4; see also Fig. S3 and S6 in the supplemental material) and were generated using MatchMaker with the following parameters: best-aligning pair of chains between reference and match structure, Needleman-Wunsch alignment algorithm, BLOSUM-62 matrix, and iterate by pruning long atom pairs until no pair exceeds 3.0 Å. All other parameters were the defaults. The root mean square deviations (RMSD) for overlays were calculated at 1.139 Å for 101 atom pairs (UspA structures) (Fig. 4), 1.029 Å for 356 atom pairs (Icl1 structures) (Fig. S3), 1.311 Å for 235 atom pairs (CysK1 structures) (Fig. S6), and 1.205 Å for 216 atom pairs (homoserine sulfhydrylase structures) (Fig. S8).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Amy Cheng Vollmer and Steven Finkel for informative discussions and critical review of the manuscript and all of the experiments and data presented. We also gratefully acknowledge Jody Massey and John Glenn for editorial assistance.
This work was supported by initial laboratory funding from the Division of Research at the University of Houston (S.J.B.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00206-17.
REFERENCES
- 1.Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. 2014. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol 5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 4.Carvalhais V, Cerca N, Vilanova M, Vitorino R. 2015. Proteomic profile of dormancy within Staphylococcus epidermidis biofilms using iTRAQ and label-free strategies. Appl Microbiol Biotechnol 99:2751–2762. doi: 10.1007/s00253-015-6434-3. [DOI] [PubMed] [Google Scholar]
- 5.Ayrapetyan M, Williams TC, Oliver JD. 2014. Interspecific quorum sensing mediates the resuscitation of viable but nonculturable vibrios. Appl Environ Microbiol 80:2478–2483. doi: 10.1128/AEM.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayrapetyan M, Williams TC, Oliver JD. 2015. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol 23:7–13. doi: 10.1016/j.tim.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 8.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 9.Kaprelyants AS, Mukamolova GV, Davey HM, Kell DB. 1996. Quantitative analysis of the physiological heterogeneity within starved cultures of Micrococcus luteus by flow cytometry and cell sorting. Appl Environ Microbiol 62:1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lleò MM, Pierobon S, Tafi MC, Signoretto C, Canepari P. 2000. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl Environ Microbiol 66:4564–4567. doi: 10.1128/AEM.66.10.4564-4567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lleó MM, Tafi MC, Canepari P. 1998. Nonculturable Enterococcus faecalis cells are metabolically active and capable of resuming active growth. Syst Appl Microbiol 21:333–339. doi: 10.1016/S0723-2020(98)80041-6. [DOI] [PubMed] [Google Scholar]
- 13.Ayrapetyan M, Williams TC, Baxter R, Oliver JD. 2015. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect Immun 83:4194–4203. doi: 10.1128/IAI.00404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 15.Barry CE, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7:845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gengenbacher M, Kaufmann SHE. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruggiero A, Marchant J, Squeglia F, Makarov V, De Simone A, Berisio R. 2013. Molecular determinants of inactivation of the resuscitation promoting factor B from Mycobacterium tuberculosis. J Biomol Struct Dyn 31:195–205. doi: 10.1080/07391102.2012.698243. [DOI] [PubMed] [Google Scholar]
- 18.Wayne LG. 1994. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis 13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- 19.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64:2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert OT, Ludwig C, Kogadeeva M, Zimmermann M, Rosenberger G, Gengenbacher M, Gillet LC, Collins BC, Röst HL, Kaufmann SHE, Sauer U, Aebersold R. 2015. Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18:96–108. doi: 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Boon C, Dick T. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J Bacteriol 184:6760–6767. doi: 10.1128/JB.184.24.6760-6767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boon C, Dick T. 2012. How Mycobacterium tuberculosis goes to sleep: the dormancy survival regulator DosR a decade later. Future Microbiol 7:513–518. doi: 10.2217/fmb.12.14. [DOI] [PubMed] [Google Scholar]
- 23.Rosenkrands I, Slayden RA, Crawford J, Aagaard C, Barry CE, Andersen P. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J Bacteriol 184:3485–3491. doi: 10.1128/JB.184.13.3485-3491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipworth S, Hammond RJH, Baron VO, Hu Y, Coates A, Gillespie SH. 2016. Defining dormancy in mycobacterial disease. Tuberculosis (Edinb) 99:131–142. doi: 10.1016/j.tube.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Besnard V, Federighi M, Declerq E, Jugiau F, Cappelier J-M. 2002. Environmental and physico-chemical factors induce VBNC state in Listeria monocytogenes. Vet Res 33:359–370. doi: 10.1051/vetres:2002022. [DOI] [PubMed] [Google Scholar]
- 26.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppée J-Y, Ghigo J-M, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X, Li X, Huang L, Chan J, Chen Y, Deng H, Mi K. 2015. Quantitative proteomics reveals novel insights into isoniazid susceptibility in mycobacteria mediated by a universal stress protein. J Proteome Res 14:1445–1454. doi: 10.1021/pr5011058. [DOI] [PubMed] [Google Scholar]
- 29.Shleeva M, Mukamolova GV, Young M, Williams HD, Kaprelyants AS. 2004. Formation of “non-culturable” cells of Mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology 150:1687–1697. doi: 10.1099/mic.0.26893-0. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y. 2004. Persistent and dormant tubercle bacilli and latent tuberculosis. Front Biosci 9:1136–1156. doi: 10.2741/1291. [DOI] [PubMed] [Google Scholar]
- 31.Chao MC, Rubin EJ. 2010. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol 64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 32.Dutta NK, Karakousis PC. 2014. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol Mol Biol Rev 78:343–371. doi: 10.1128/MMBR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Möker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J Bacteriol 192:1946–1955. doi: 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung V, Lévesque CM. 2012. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol 194:2265–2274. doi: 10.1128/JB.06707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerdes K, Maisonneuve E. 2015. Remarkable functional convergence: alarmone pp;gpp mediates persistence by activating type I and II toxin-antitoxins. Mol Cell 59:1–3. doi: 10.1016/j.molcel.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 36.Gaca AO, Kudrin P, Colomer-Winter C, Beljantseva J, Liu K, Anderson B, Wang JD, Rejman D, Potrykus K, Cashel M, Hauryliuk V, Lemos JA. 2015. From (p)ppGpp to (pp)pGpp: characterization of regulatory effects of pGpp synthesized by the small alarmone synthetase of Enterococcus faecalis. J Bacteriol 197:2908–2919. doi: 10.1128/JB.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corrigan RM, Bellows LE, Wood A, Gründling A. 2016. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci U S A 113:E1710–E1719. doi: 10.1073/pnas.1522179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J Bacteriol 188:8360–8367. doi: 10.1128/JB.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Page R, Peti W. 2016. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat Chem Biol 12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 40.Ramage HR, Connolly LE, Cox JS. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet 5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luidalepp H, Jõers A, Kaldalu N, Tenson T. 2011. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J Bacteriol 193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaprelyants AS, Kell DB. 1993. Dormancy in stationary-phase cultures of Micrococcus luteus: flow cytometric analysis of starvation and resuscitation. Appl Environ Microbiol 59:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu W, Plikaytis BB, Shinnick TM. 2003. Resuscitation factors from mycobacteria: homologs of Micrococcus luteus proteins. Tuberculosis (Edinb) 83:261–269. doi: 10.1016/S1472-9792(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 44.Greenblatt CL, Baum J, Klein BY, Nachshon S, Koltunov V, Cano RJ. 2004. Micrococcus luteus—survival in amber. Microb Ecol 48:120–127. doi: 10.1007/s00248-003-2016-5. [DOI] [PubMed] [Google Scholar]
- 45.Drumm JE, Mi K, Bilder P, Sun M, Lim J, Bielefeldt-Ohmann H, Basaraba R, So M, Zhu G, Tufariello JM, Izzo AA, Orme IM, Almo SC, Leyh TS, Chan J. 2009. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-binding: requirement for establishing chronic persistent infection. PLoS Pathog 5:e1000460. doi: 10.1371/journal.ppat.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wayne LG, Lin KY. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun 37:1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muttucumaru DGN, Roberts G, Hinds J, Stabler RA, Parish T. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb) 84:239–246. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Voskuil MI, Visconti KC, Schoolnik GK. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Bacon J, James BW, Wernisch L, Williams A, Morley KA, Hatch GJ, Mangan JA, Hinds J, Stoker NG, Butcher PD, Marsh PD. 2004. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis (Edinb) 84:205–217. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med 198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. 2004. Open mass spectrometry search algorithm. J Proteome Res 3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 52.Elias JE, Haas W, Faherty BK, Gygi SP. 2005. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods 2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 53.Smaoui S, Siala M, Hadj Fredj S, Kammoun S, Marouane C, Hachicha S, Ghorbel A, Gdoura R, Slim L, Ben Messaoud T, Messadi F. 2016. Molecular characterization of Mycobacterium tuberculosis strains resistant to isoniazid. Int J Mycobacteriol 5(Suppl 1):S151. doi: 10.1016/j.ijmyco.2016.09.070. [DOI] [PubMed] [Google Scholar]
- 54.Kong I-S, Bates TC, Hülsmann A, Hassan H, Smith BE, Oliver JD. 2004. Role of catalase and oxyR in the viable but nonculturable state of Vibrio vulnificus. FEMS Microbiol Ecol 50:133–142. doi: 10.1016/j.femsec.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 55.Rao NV, Shashidhar R, Bandekar JR. 2014. Induction, resuscitation and quantitative real-time polymerase chain reaction analyses of viable but nonculturable Vibrio vulnificus in artificial sea water. World J Microbiol Biotechnol 30:2205–2212. doi: 10.1007/s11274-014-1640-1. [DOI] [PubMed] [Google Scholar]
- 56.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 57.Kvint K, Nachin L, Diez A, Nyström T. 2003. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol 6:140–145. doi: 10.1016/S1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 58.Nyström T, Neidhardt FC. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol 11:537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 59.Nachin L, Nannmark U, Nyström T. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J Bacteriol 187:6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nyström T, Neidhardt FC. 1993. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J Bacteriol 175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gustavsson N, Diez A, Nyström T. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol Microbiol 43:107–117. doi: 10.1046/j.1365-2958.2002.02720.x. [DOI] [PubMed] [Google Scholar]
- 63.Phadtare S. 2004. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol 6:125–136. [PubMed] [Google Scholar]
- 64.Farewell A, Diez AA, DiRusso CC, Nyström T. 1996. Role of the Escherichia coli FadR regulator in stasis survival and growth phase-dependent expression of the uspA, fad, and fab genes. J Bacteriol 178:6443–6450. doi: 10.1128/jb.178.22.6443-6450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hingley-Wilson SM, Lougheed KEA, Ferguson K, Leiva S, Williams HD. 2010. Individual Mycobacterium tuberculosis universal stress protein homologues are dispensable in vitro. Tuberculosis (Edinb) 90:236–244. doi: 10.1016/j.tube.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Toole R, Williams HD. 2003. Universal stress proteins and Mycobacterium tuberculosis. Res Microbiol 154:387–392. doi: 10.1016/S0923-2508(03)00081-0. [DOI] [PubMed] [Google Scholar]
- 67.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Blumenthal A, Trujillo C, Ehrt S, Schnappinger D. 2010. Simultaneous analysis of multiple Mycobacterium tuberculosis knockdown mutants in vitro and in vivo. PLoS One 5:e15667. doi: 10.1371/journal.pone.0015667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muñoz-Elías EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med 11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raman N, Black PN, DiRusso CC. 1997. Characterization of the fatty acid-responsive transcription factor FadR. Biochemical and genetic analyses of the native conformation and functional domains. J Biol Chem 272:30645–30650. [DOI] [PubMed] [Google Scholar]
- 71.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 72.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y, Zhou S, Deng Q, Li X, Meng J, Guan Y, Li C, Xiao C. 2016. Identification of a novel inhibitor of isocitrate lyase as a potent antitubercular agent against both active and non-replicating Mycobacterium tuberculosis. Tuberculosis (Edinb) 97:38–46. doi: 10.1016/j.tube.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Mazumder M, Gourinath S. 2016. Structure-based design of inhibitors of the crucial cysteine biosynthetic pathway enzyme O-acetyl serine sulfhydrylase. Curr Top Med Chem 16:948–959. doi: 10.2174/1568026615666150825142422. [DOI] [PubMed] [Google Scholar]
- 75.Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A. 2008. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem 143:425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 76.Polikanov YS, Blaha GM, Steitz TA. 2012. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science 336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ueta M, Yoshida H, Wada C, Baba T, Mori H, Wada A. 2005. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells 10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 78.Tan X, Varughese M, Widger WR. 1994. A light-repressed transcript found in Synechococcus PCC 7002 is similar to a chloroplast-specific small subunit ribosomal protein and to a transcription modulator protein associated with sigma 54. J Biol Chem 269:20905–20912. [PubMed] [Google Scholar]
- 79.Mangan MW, Lucchini S, Danino V, Cróinín TO, Hinton JCD, Dorman CJ. 2006. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol 59:1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- 80.Lee SY, Lim CJ, Dröge P, Yan J. 2015. Regulation of bacterial DNA packaging in early stationary phase by competitive DNA binding of Dps and IHF. Sci Rep 5:18146. doi: 10.1038/srep18146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chaparian RR, Olney SG, Hustmyer CM, Rowe-Magnus DA, van Kessel JC. 2016. Integration host factor and LuxR synergistically bind DNA to coactivate quorum-sensing genes in Vibrio harveyi. Mol Microbiol 101:823–840. doi: 10.1111/mmi.13425. [DOI] [PubMed] [Google Scholar]
- 82.Pedulla ML, Hatfull GF. 1998. Characterization of the mIHF gene of Mycobacterium smegmatis. J Bacteriol 180:5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mishra A, Vij M, Kumar D, Taneja V, Mondal AK, Bothra A, Rao V, Ganguli M, Taneja B. 2013. Integration host factor of Mycobacterium tuberculosis, mIHF, compacts DNA by a bending mechanism. PLoS One 8:e69985. doi: 10.1371/journal.pone.0069985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schubert OT, Mouritsen J, Ludwig C, Röst HL, Rosenberger G, Arthur PK, Claassen M, Campbell DS, Sun Z, Farrah T, Gengenbacher M, Maiolica A, Kaufmann SHE, Moritz RL, Aebersold R. 2013. The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13:602–612. doi: 10.1016/j.chom.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rice PA, Yang S, Mizuuchi K, Nash HA. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87:1295–1306. doi: 10.1016/S0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 86.Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiu Y, Tereshko V, Kim Y, Zhang R, Collart F, Yousef M, Kossiakoff A, Joachimiak A. 2006. The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold. Proteins 62:8–16. doi: 10.1002/prot.20590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colangeli R, Haq A, Arcus VL, Summers E, Magliozzo RS, McBride A, Mitra AK, Radjainia M, Khajo A, Jacobs WR, Salgame P, Alland D. 2009. The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc Natl Acad Sci U S A 106:4414–4418. doi: 10.1073/pnas.0810126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kornberg HL. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malmström J, Karlsson C, Nordenfelt P, Ossola R, Weisser H, Quandt A, Hansson K, Aebersold R, Malmström L, Björck L. 2012. Streptococcus pyogenes in human plasma: adaptive mechanisms analyzed by mass spectrometry-based proteomics. J Biol Chem 287:1415–1425. doi: 10.1074/jbc.M111.267674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.