ABSTRACT
The canonical translation initiation mechanism involves base pairing between the mRNA and 16S rRNA. However, a variety of identified mechanisms deviate from this conventional route. Beck and Janssen (J Bacteriol 199:e00091-17, 2017, https://doi.org/10.1128/JB.00091-17) have recently described another noncanonical mode of translation initiation. Here, we describe how this process differs from previously reported mechanisms, with the hope that it will foster increased awareness of the diversity of regulatory mechanisms that await discovery.
TEXT
The standard model for initiation of translation in bacteria involves base pairing between a Shine-Dalgarno (SD) sequence upstream of the start codon and the complementary anti-SD (aSD) sequence at the 3′ end of 16S rRNA in a 30S ribosomal subunit. This mode of initiation, where the SD is a major component of the ribosome binding site (RBS), is widespread in bacteria and archaea and has until recently been inferred to be the dominant initiation pathway. The model posits that the SD-aSD interaction increases the local concentration of 30S ribosomal subunits in the vicinity of the initiation codon, facilitating subsequent events in initiation complex formation (1). Systematic approaches in Escherichia coli have demonstrated the influence of SD length and its distance from the initiation codon (2). In general, longer SD elements support increased expression; however, the relationship is nonlinear, and binding affinity alone does not fully explain the observed protein expression levels. Weaker SD sequences are particularly susceptible to the effects of mRNA decay and transcription termination (3), while extended (8- to 10-nucleotide [nt]) SD sequences inhibit translation, presumably since such sequences trap the ribosome on the RBS (4). Among E. coli genes, the average SD length is 6.3 nt (5), and the average spacing between the SD and initiation codon is 4.4 nt (4). The SD sequences from Bacillus subtilis, in general, are longer than their E. coli counterparts (6), a property that may be linked to the absence of a large ribosomal protein bS1 in many Gram-positive organisms (7).
30S subunit binding and effective initiation is heavily influenced by the mRNA structure surrounding the RBS. Many of the gene regulatory mechanisms that target initiation operate by altering the local RNA structure encompassing SD sequences. These mechanisms include translational coupling, where translation of an upstream open reading frame (uORF) disrupts the RNA structure surrounding the downstream RBS. The binding of small regulatory RNAs or RNA binding proteins, metabolites (in riboswitches), or increases in temperature (in thermosensors) can all influence the local RNA structure, with corresponding effects on initiation. An additional model is required to explain how certain mRNAs with RBSs sequestered in stable structures can be translated effectively. The “standby binding” model posits that 30S subunits bind first to single-stranded RNA flanking the structured, RBS-containing element. The bound standby 30S subunit can then compete effectively for RBS capture upon transient opening of the adjacent RNA structure (8). Additional examples of standby binding suggest that this initiation mode may be widespread (9).
Alternatives to the SD-aSD mode of initiation.
Some leadered mRNAs lack recognizable SD-like sequences in their untranslated regions (UTRs). Moreover, analyses of sequenced genomes indicate that the SD-aSD interaction is completely absent in some archaea and bacteria (10, 11). Among the alternative initiation mechanisms, proposals involving mRNA-rRNA complementarity outside the aSD region of 16S rRNA have, in general, not withstood scrutiny (12, 13). In some instances, mRNA interactions with ribosomal protein bS1 provide an alternative, SD-independent route to initiation on leadered mRNAs. This large protein has both RNA binding and unwinding activities and aids initiation on both SD-containing and SD-free templates. The protein bS1 has a broad affinity for G-poor RNAs, and at least some of the SD-lacking mRNAs contain A/U-rich leader regions that are required for bS1 binding (4). However, there is no S1 protein in archaea, and in some bacteria bS1 is either absent or nonfunctional in translation, and so additional, undefined initiation mechanisms must exist in these organisms.
Leaderless mRNAs lack a 5′ untranslated region (5′-UTR) entirely, and while rare in E. coli, they are abundant in archaea and in the Actinobacteria and Deinococcus-Thermus groups of bacteria, in which as many as 30% of the genes are leaderless (14). An essential difference between initiation on leadered versus leaderless mRNAs is that the latter begins with the binding of a 70S ribosome to the 5′ end. While the 30S mode of initiation has long been considered dominant, a recent proposal suggested that initiation by scanning 70S ribosomes is the frequent mode of translation of polycistronic mRNAs (15).
Regulation of translation initiation by a 5′-uAUG.
While a variety of noncanonical translation initiation mechanisms have been described, Beck and Janssen have recently described yet another variation of noncanonical translation initiation in E. coli (16). Previous work from this group identified several examples of uORFs positioned at the 5′ terminus that influenced expression of the corresponding downstream cistron (17). The uORF within ptrB mRNA was shown to greatly increase expression of the ptrB coding sequence. Although increased expression depended on the 5′-terminal AUG codon (5′-uAUG), the uORF was not efficiently translated (17). In their most recent study, Beck and Janssen explored this fascinating translation initiation mechanism (16). Notably, instead of relying on translation of the 5′-uORF, the 5′-uAUG functioned as a 70S ribosome recognition signal to attract ribosomes to ptrB mRNA, which facilitated efficient translation of the ptrB coding sequence further downstream. Since strengthening the ptrB SD sequence alleviated the requirement for the 5′-uAUG, it is apparent that recruitment of a 70S ribosome by the 5′-uAUG compensates for a poor ptrB SD sequence. Strikingly, replacing the 5′-UTR of other mRNAs with that from ptrB conferred a similar dependence on the ptrB 5′-uAUG, indicating that the features within the ptrB 5′-UTR are sufficient to control downstream gene expression (16).
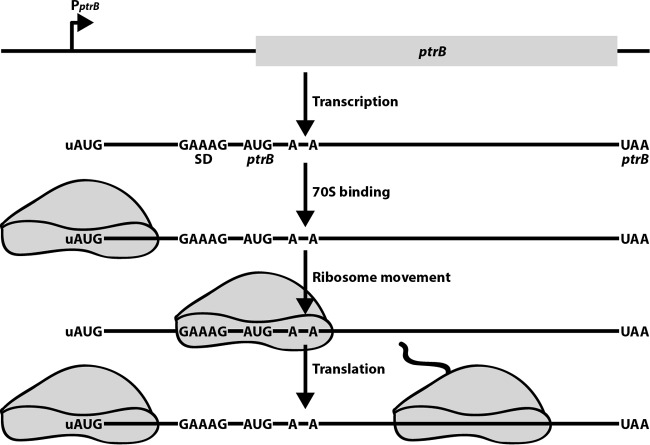
A model was proposed to explain the mechanism of ptrB regulation (16). In this model, the 5′-uAUG acts as a recognition signal and binding platform to attract 70S ribosomes to the transcript, thereby increasing the localized concentration of ribosomes on the ptrB transcript. Following this initial interaction, sequence features within the 5′-UTR stabilize the bound 70S ribosome until it transitions down the mRNA. Additional interactions with two A residues in the early ptrB coding sequences help compensate for the poor ptrB SD sequence, thereby properly positioning the ribosome at the ptrB initiation codon. Together, these sequences function cooperatively, using established recognition signals in a unique combination (Fig. 1).
FIG 1.
Model for 5′-uAUG-dependent activation of ptrB translation. Following transcription, a 70S ribosome binds to the 5′-uAUG and subsequently moves to the ptrB ribosome binding site with the assistance from additional interactions within the 5′-UTR and early ptrB coding sequence. Once ptrB translation initiates, another 70S ribosome can bind to the 5′-uAUG and the process is repeated. The 70S ribosome bound to the 5′-uAUG would be capable of protecting the mRNA from 5′-end-dependent cleavage by RNase E (not shown).
While it is clear that the 5′-uAUG plays a direct role in increasing translation of ptrB by functioning as a recognition signal to attract 70S ribosomes to the transcript, it is likely that a 70S ribosome stably associated at the 5′ end of a transcript would lead to stabilization of the mRNA. E. coli RNase E is an endoribonuclease that cleaves RNA in single-stranded A/U-rich regions (18). This essential enzyme plays a key role in the degradation of mRNA and in a variety of RNA processing reactions (19). In addition to cleaving transcripts by an internal entry pathway (20), RNase E cleavage can occur by a 5′-end-dependent mechanism in which 5′ monophosphorylated mRNAs interact with a 5′ binding pocket on the enzyme (21, 22). Since a small RNA binding protein is capable of stabilizing mRNA by blocking RNase E access to the 5′ end of an mRNA (23), it is reasonable to assume that binding of a 70S ribosome to a 5′-uAUG would protect the mRNA from 5′-end-dependent cleavage. Such a mechanism would be in addition to the general mRNA stabilization caused by translating ribosomes (24).
Once viewed as a peculiarity, it is now evident that noncanonical translation initiation mechanisms are commonplace. It has been estimated that approximately half of bacterial genes lack an SD sequence (25). Furthermore, the observation that mRNAs with 5′-uAUG sequences are abundant in E. coli (17) suggests that the mechanism described by Beck and Janssen (16) could be widespread. Considering the frequency and diversity of these atypical mechanisms, it seems likely that additional noncanonical translation initiation mechanisms await discovery.
ACKNOWLEDGMENTS
The Babitzke laboratory is supported by National Institutes of Health grants GM059969 and GM098399 and by grant KA2016-85222 from the Charles E. Kaufman Foundation of The Pittsburgh Foundation. The O'Connor laboratory is supported by funds from the School of Biological Sciences, University of Missouri—Kansas City.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
For the article discussed, see https://doi.org/10.1128/JB.00091-17.
REFERENCES
- 1.Gualerzi CO, Pon CL. 2015. Initiation of mRNA translation in bacteria: structural and dynamic aspects. Cell Mol Life Sci 72:4341–4367. doi: 10.1007/s00018-015-2010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L. 1992. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol 6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 3.Eriksen M, Sneppen K, Pedersen S, Mitarai N. 2017. Occlusion of the ribosome binding site connects the translational initiation frequency, mRNA stability and premature transcription termination. Front Microbiol 8:362. doi: 10.3389/fmicb.2017.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komarova AV, Tchufistova LS, Supina EV, Boni IV. 2002. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 8:1137–1147. doi: 10.1017/S1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schurr T, Nadir E, Margalit H. 1993. Identification and characterization of E.coli ribosomal binding sites by free energy computation. Nucleic Acids Res 21:4019–4023. doi: 10.1093/nar/21.17.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hockenberry AJ, Pah AR, Jewett MC, Amaral LA. 2017. Leveraging genome-wide datasets to quantify the functional role of the anti-Shine-Dalgarno sequence in regulating translation efficiency. Open Biol 7:pii160239. doi: 10.1098/rsob.160239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farwell MA, Roberts MW, Rabinowitz JC. 1992. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol Microbiol 6:3375–3383. doi: 10.1111/j.1365-2958.1992.tb02205.x. [DOI] [PubMed] [Google Scholar]
- 8.de Smit MH, van Duin J. 2003. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J Mol Biol 331:737–743. doi: 10.1016/S0022-2836(03)00809-X. [DOI] [PubMed] [Google Scholar]
- 9.Unoson C, Wagner EG. 2007. Dealing with stable structures at ribosome binding sites: bacterial translation and ribosome standby. RNA Biol 4:113–117. doi: 10.4161/rna.4.3.5350. [DOI] [PubMed] [Google Scholar]
- 10.Accetto T, Avgustin G. 2011. Inability of Prevotella bryantii to form a functional Shine-Dalgarno interaction reflects unique evolution of ribosome binding sites in Bacteroidetes. PLoS One 6:e22914. doi: 10.1371/journal.pone.0022914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa S, Niimura Y, Miura K, Gojobori T. 2010. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci U S A 107:6382–6387. doi: 10.1073/pnas.1002036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connor M, Asai T, Squires CL, Dahlberg AE. 1999. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc Natl Acad Sci U S A 96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor M, Dahlberg AE. 2001. Enhancement of translation by the epsilon element is independent of the sequence of the 460 region of 16S rRNA. Nucleic Acids Res 29:1420–1425. doi: 10.1093/nar/29.7.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng X, Hu GQ, She ZS, Zhu H. 2011. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics 12:361. doi: 10.1186/1471-2164-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto H, Wittek D, Gupta R, Qin B, Ueda T, Krause R, Yamamoto K, Albrecht R, Pech M, Nierhaus KH. 2016. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc Natl Acad Sci U S A 113:E1180–E1189. doi: 10.1073/pnas.1524554113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck HJ, Janssen GR. 2017. Novel translation initiation regulation mechanism in Escherichia coli ptrB mediated by a 5′-terminal AUG. J Bacteriol 199:e00091-17. doi: 10.1128/JB.00091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck HJ, Fleming IM, Janssen GR. 2016. 5′-terminal AUGs in Escherichia coli mRNAs with Shine-Dalgarno sequences: identification and analysis of their roles in non-canonical translation initiation. PLoS One 11:e0160144. doi: 10.1371/journal.pone.0160144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDowall KJ, Lin-Chao S, Cohen SN. 1994. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem 269:10790–10796. [PubMed] [Google Scholar]
- 19.Stead MB, Marshburn S, Mohanty BK, Mitra J, Castillo LP, Ray D, van Bakel H, Hughes TR, Kushner SR. 2011. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res 39:3188–3203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kime L, Jourdan SS, Stead JA, Hidalgo-Sastre A, McDowall KJ. 2010. Rapid cleavage of RNA by RNase E in the absence of 5′ monophosphate stimulation. Mol Microbiol 76:590–604. doi: 10.1111/j.1365-2958.2009.06935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackie GA. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 22.Richards J, Luciano DJ, Belasco JG. 2012. Influence of translation on RppH-dependent mRNA degradation in Escherichia coli. Mol Microbiol 86:1063–1072. doi: 10.1111/mmi.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P. 2013. CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87:851–866. doi: 10.1111/mmi.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 25.Chang B, Halgamuge S, Tang SL. 2006. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene 373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]