ABSTRACT
Under unfavorable growth conditions, bacteria enter stationary phase and can maintain cell viability over prolonged periods with no increase in cell number. To obtain insights into the regulatory mechanisms that allow bacteria to resume growth when conditions become favorable again (outgrowth), we performed global transcriptome analyses at different stages of growth for the alphaproteobacterium Rhodobacter sphaeroides. The majority of genes were not differentially expressed across growth phases. After a short stationary phase (about 20 h after growth starts to slow down), only 7% of the genes showed altered expression (fold change of >1.6 or less than −1.6, corresponding to a log2 fold change of >0.65 or less than −0.65, respectively) compared to expression at exponential phase. Outgrowth induced a distinct response in gene expression which was strongly influenced by the length of the preceding stationary phase. After a long stationary phase (about 64 h after growth starts to slow down), a much larger number of genes (15.1%) was induced in outgrowth than after a short stationary phase (1.7%). Many of those genes are known members of the RpoHI/RpoHII regulons and have established functions in stress responses. A main effect of RpoHI on the transcriptome in outgrowth after a long stationary phase was confirmed. Growth experiments with mutant strains further support an important function in outgrowth after prolonged stationary phase for the RpoHI and RpoHII sigma factors.
IMPORTANCE In natural environments, the growth of bacteria is limited mostly by lack of nutrients or other unfavorable conditions. It is important for bacterial populations to efficiently resume growth after being in stationary phase, which may last for long periods. Most previous studies on growth-phase-dependent gene expression did not address outgrowth after stationary phase. This study on growth-phase-dependent gene regulation in a model alphaproteobacterium reveals, for the first time, that the length of the stationary phase strongly impacts the transcriptome during outgrowth. The alternative sigma factors RpoHI and RpoHII, which are important regulators of stress responses in alphaproteobacteria, play a major role during outgrowth following prolonged stationary phase. These findings provide the first insight into the regulatory mechanisms enabling efficient outgrowth.
KEYWORDS: Alphaproteobacteria, alternative sigma factors, gene regulation, growth adaptation, growth phases, Rhodobacter sphaeroides
INTRODUCTION
Distinct phases of bacterial growth can be observed not only under laboratory conditions in batch cultures but also in natural environments. Under favorable conditions, cells grow exponentially and replicate at a maximal rate. When conditions become unfavorable due to, e.g., limitation of substrates or oxygen, the majority of bacterial cells stop replicating, enter stationary phase, and switch from a state of active growth promoting the production of cell mass and daughter cells to a maintenance metabolism. Cell viability can be maintained over prolonged periods with no net increase in cell number. With entry into stationary phase, bacteria may become more resistant to various environmental stresses, including oxidative stress, acidic pH, and nutrient deprivation (1). The stationary-phase characteristics can also help bacteria survive in host cells. Conditions that sustain constant growth are rarely found in nature. As bacteria often face nutrient limitation and unfavorable conditions, they may be forced to remain in stationary phase for long time periods. When nutrients become available, bacteria resume growth (outgrowth) until nutrients are exhausted, and again they enter stationary phase. The accumulation of metabolic waste products after prolonged stationary phase leads to cell death (2).
To date, regulatory factors controlling growth-phase-dependent gene expression are known only for a limited number of species and conditions. For example, the entrance into stationary phase is a very well regulated process involving diverse transcriptional regulators in the systems studied so far (3). In many Gram-negative bacteria, starvation triggers the alternative sigma factor RpoS, which may control up to 10% of the genes. Initially considered a stationary-phase sigma factor, RpoS is now recognized as a general stress response sigma factor. Furthermore, nucleoid-associated and nucleoid-structuring proteins, like Lrp, Fis, and integration host factor (IHF), have a reported role in stationary-phase regulation in enteric bacteria (3–6). Stationary phase often goes along with amino acid starvation and initiates the stringent response, which downregulates DNA replication and the syntheses of rRNAs and ribosomal proteins, but it induces levels of RpoS and stress proteins, as well as amino acid biosynthesis (3, 7).
Despite the importance of alphaproteobacteria as plant and animal symbionts and pathogens and their impact on global metabolic cycles through photosynthesis, nitrogen fixation, and carbon dioxide fixation, our knowledge of growth phase regulation in members of the alphaproteobacteria is very limited. Alphaproteobacteria do not contain RpoS homologs with a function similar to that of RpoS of enteric bacteria, and the general stress response is instead mediated by extracytoplasmic function (ECF) sigma factors (8). For many alphaproteobacteria, an important role in stress responses, including oxidative stress, photooxidative stress, and stationary phase, was also ascribed to RpoH proteins belonging to the sigma-32 family (9–13). In Rhodobacter sphaeroides, the RpoHI and RpoHII sigma factors are able to complement the temperature-sensitive phenotype of an Escherichia coli rpoH mutant (14). The alternative sigma factor RpoE activates the rpoHII gene in response to singlet oxygen, and RpoHII consequently activates many genes with functions in the detoxification of peroxides or methylglyoxal, singlet oxygen scavenging, and iron and redox homeostasis (12). RpoHI also has an important role in the singlet oxygen response, and there is a big overlap of the RpoHI and RpoHII regulons (15, 16). More recently, it was demonstrated that several stress factors besides heat and singlet oxygen, like organic peroxides, hydrogen peroxide, superoxide, and CdCl2, can activate RpoHI/RpoHII-dependent promoters (17).
To learn more about the regulatory mechanisms involved in stationary-phase survival in a member of the alphaproteobacteria, we monitored the growth-phase-dependent transcriptome response in R. sphaeroides 2.4.1. This facultative anoxygenic phototrophic bacterium has been intensely studied in the past regarding the formation of photosynthetic complexes, carbon dioxide fixation, nitrogen fixation, other metabolic processes, and underlying regulatory mechanisms. It also serves as a model organism to study the oxidative stress and photooxidative stress responses in bacteria. For this reason, several mutants lacking certain regulatory factors are available. R. sphaeroides is a free-living aquatic bacterium which does not undergo a special life cycle. It is highly likely that factors involved in growth phase regulation of Rhodobacter have a similar function in other alphaproteobacteria.
Our data assign a major role in outgrowth after extended stationary phase to the alternative sigma factor RpoHI.
RESULTS AND DISCUSSION
Growth and sampling of R. sphaeroides.
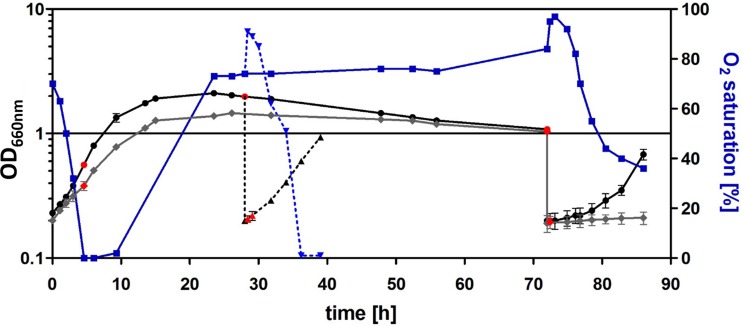
R. sphaeroides 2.4.1 was inoculated from overnight cultures in late exponential phase to an optical density at 660 nm (OD660) of 0.2 into 400 ml of medium in 500-ml Erlenmeyer flasks and constantly shaken (140 rpm). We chose these conditions since anoxygenic phototrophic bacteria in nature are rarely found in environments with continuous high aeration. Furthermore, the control experiments revealed little differences in gene expression in high- or low-aeration cultures, with the exception of photosynthesis genes, whose oxygen-dependent gene expression was intensively studied in Rhodobacter species in the past (18, 19) (data not shown). Oxygen levels were monitored throughout growth, and the results are displayed in Fig. 1. The wild-type culture showed maximal growth up to an OD of 0.8, when transition into stationary phase starts. The maximal OD was reached about 16 h after inoculation. Twenty-eight hours after inoculation, wild-type cells were diluted into fresh medium and quickly resumed growth.
FIG 1.
Growth curves and oxygen levels of R. sphaeroides wild-type 2.4.1 and 2.4.1 ΔrpoHI. Strains were grown under microaerobic conditions, and the optical density at 660 nm (OD660) was determined over time; growth is indicated as a black line (wild type) or gray line (2.4.1 ΔrpoHI). The cultures were diluted 28 h (dashed line) after inoculation (early stationary phase) or 72 h after inoculation (late stationary phase) into fresh medium to an OD of 0.2. Oxygen saturation in the wild-type culture (blue) was similar to that in the mutant (not shown). RNA samples were taken at different time points (red dots). Both data sets represent the means of results from at least three independent experiments, and the error bars indicate the standard deviation.
Samples for RNA isolation were taken in mid-exponential phase (OD, 0.5 to 0.6), 28 h after inoculation (OD, 1.8 to 2.0; herein referred to as early stationary phase), 72 h after inoculation (OD, 1.0 to 1.3; herein referred to as late stationary phase), and 20 min and 90 min after inoculation into fresh medium (OD, 0.2 to 0.3) after both early and late stationary phase. RNA samples from six independent experiments were used for RNA sequencing (RNA-seq) and microarray analyses, as described in Materials and Methods. Classification of genes according to the expression in different growth phases is based on the microarray data sets, while the differential RNA-seq (dRNA-seq) analysis provided information on transcriptional start sites (TSS). The reproducibility of microarray replicates was very high, as reflected by the Pearson correlation coefficient r ranging between 0.96 and 0.98 (see Fig. S1 in the supplemental material). Relative changes in RNA levels from the different growth phases were compared to the levels at exponential phase and considered to be regulated in the case of a fold change of >1.6 or less than −1.6 (Table S1).
Duration of stationary phase has a strong impact on changes in gene expression in outgrowth.
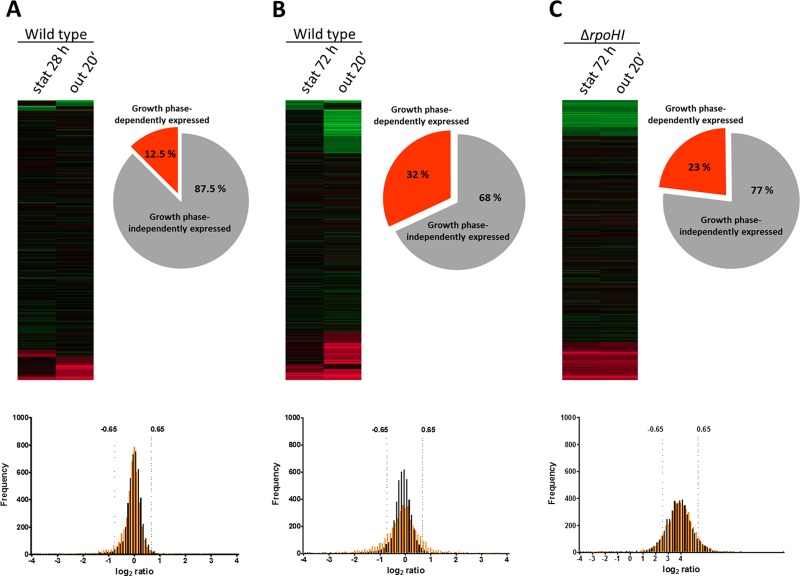
When gene expression in early stationary phase (28 h after inoculation) or the following outgrowth was compared to that at exponential phase, 4,214 of a total of 4,303 protein-encoding genes passed our average signal intensity for a gene across all arrays (A-value) cutoff. Of the genes, 87.5% showed changes in expression levels compared to those at exponential phase of more than −1.6-fold and less than 1.6-fold, which we consider to indicate expression independent of growth phase (Fig. 2A). However, when cultures were kept in stationary phase for an extended period (72 h after inoculation, followed by outgrowth), only 66.5% of 4,116 protein-encoding genes with reliable A-values showed growth-phase-independent expression (Fig. 2B). These numbers reflect the strongly altered expression during outgrowth after prolonged stationary phase. Overall, 2,628 genes belonging to many different clusters of orthologous groups (COGs) were growth-phase-independently expressed irrespective of the duration of stationary phase.
FIG 2.
Distribution and expression kinetics of the whole transcriptome. Wild-type cells were grown for 28 h (A) or 72 h (B) and ΔrpoHI mutant cells were grown for 72 h (C) after inoculation, and cells were then diluted into fresh medium (outgrowth [out]). Relative changes of RNA levels in stationary phase (stat) directly before dilution and 20 min after dilution were monitored by microarray analysis of total RNA and normalized to levels in exponential phase. Changes are illustrated as heat maps, with a color code ranging from red (a log2 ratio of −2) to green (a log2 ratio of 2). Pie chart diagrams show the distribution of growth-phase-independently and -dependently regulated genes. Frequency distribution analysis was performed on bins of all genes in the data set corresponding to the log2 ratio. In the frequency graphs, data for stationary phase are in black and data for the following outgrowth are in orange.
Changes in gene expression in stationary phase.
In early stationary phase, 71 genes showed at least 1.6-fold-higher expression than in mid-exponential phase. Several genes with high expression in early stationary phase have a predicted function in transport (of peptides, sugar, C4-dicarboxylate, or spermidine), including iron/metal transport (znuA and RSP_3571 for Zn transport, sitA for Mn transport, and RSP_0904 or RSP_1438 for Fe-hydroxamate transport). This may reflect an increasing demand for nutrients, including iron; however, the exact metabolic functions for many predicted transporters are not known.
One hundred sixty genes showed at least 1.6-fold-lower expression in early stationary phase than in exponential phase. Of the 97 protein-encoding genes with annotated functions downregulated in stationary phase, 67 genes (69%) encode proteins required for translation (e.g., ribosomal proteins and elongation factor TufA) or energy metabolism (e.g., Atp proteins for ATP synthase, FbcC cytochrome c1, and CycA cytochrome c2), including photosynthesis (e.g., all Puf and Puc pigment binding proteins and BchJ, BchN, BchX, BchZ, and BchM for bacteriochlorophyll synthesis) (Fig. S2).
In late stationary phase, 123 genes showed higher expression than that in exponential phase, demonstrating that the duration of stationary phase influences gene expression. When we compared the 100 genes with the highest increases in expression compared to that at exponential phase, 57 genes were commonly induced in early and late stationary phase (Fig. S3). No specific function could be assigned to the genes which are induced exclusively in late stationary phase. Among these genes were several genes coding for hypothetical proteins and four small RNAs (sRNAs) with unknown function. A role in stationary-phase-dependent gene regulation was described for several sRNAs in other species (20–24).
In early stationary phase, 160 genes showed at least 1.6-fold-lower expression than in exponential phase; in late stationary phase, 232 genes showed this decrease in expression. Of the 100 genes with the strongest decrease in expression in each phase, 78 genes were identical (Fig. S3). Among these were mostly photosynthesis genes and genes for ribosomal proteins, cytochromes, and ATP synthase. Surprisingly, the transcript levels of both nuo operons (RSP_ 0110 to RSP_0112 and RSP_2512 to RSP_2530) were decreased, although increased respiratory activity during outgrowth is expected. During outgrowth following early stationary phase, transcripts for one of the nuo operons (RSP_2512 to RSP_2530) showed increased transcript levels, in agreement with this expectation.
Changes in gene expression in outgrowth phase.
Twenty minutes after the addition of new growth medium following early stationary phase, 73 genes showed >1.6-fold upregulation compared to exponential phase (Table S1). Only a very weak correlation of global gene expression in early stationary phase and the following outgrowth phase was observed (Fig. 2A), while there was a good correlation between expression changes 20 min and 90 min after the addition of new medium (Fig. S4). We conclude that outgrowth indeed leads to quick reprogramming of gene expression. Among the genes induced in outgrowth following early stationary phase were several genes for siderophores and TRAP-T family transporters and nuo genes for subunits of NADH dehydrogenase (operon RSP_2512 to RSP_2530) (Table S1). This most likely reflects the need for the import of iron and other molecules and increased respiratory activity.
Following early stationary phase, 345 genes showed at least 1.6-fold-lower expression after 20 min of outgrowth than at exponential phase. Among the 100 genes with the strongest downregulation in stationary phase or the following 20 min of outgrowth, 47 genes were identical (Fig. 2A and Table S1), indicating that for those genes, no specific response was initiated directly after dilution. Most of those were photosynthesis genes.
The prolonged stationary growth phase (72 h after inoculation) resulted in a much stronger change in gene expression and a much larger number of genes with changed expression during the following outgrowth (Fig. 2B and Table S1): 624 protein-encoding genes showed at least 1.6-fold-higher expression than in exponential phase, and 539 protein-encoding genes showed at least 1.6-fold-lower expression. There was a very good correlation of the expression patterns between 20 min and 90 min of outgrowth (Fig. S4). The correlation of gene expression was, however, weak between late stationary phase and the following outgrowth (Fig. 2B). Of the 100 genes with the strongest induction after 20 min of outgrowth, only 13 genes encoding proteins with diverse functions were also induced in late stationary phase compared to exponential phase. Likewise, there is only a small correlation between gene expression in outgrowth in early stationary phase and that in late stationary phase (Fig. S3). We conclude that outgrowth after prolonged stationary phase induces a very strong and distinct response at the transcriptome level.
Fifteen percent (624 genes) of all genes with a reliable A-value showed >1.6-fold-increased expression after 20 min of outgrowth following prolonged stationary phase. Two hundred eighty-seven genes were induced by a factor of more than two compared to exponential phase (Table S1). The products of many of those genes have functions in protein stabilization or turnover (19 genes) and in glutathione-dependent defense systems (10 genes), DNA repair (4 genes), and iron metabolism (15 genes), and several of them are induced by photooxidative stress, generated by the presence of methylene blue in the light (Table S1) (12, 16, 25–27).
Many of the genes induced during outgrowth following late stationary phase are preceded by promoters which were proven or predicted to be recognized by the alternative sigma factor RpoHI (Fig. 3) or by an RpoHII- or RpoHI/RpoHII-dependent promoter (Table S1) (15, 16). Since a role of the alternative RpoHI/RpoHII sigma factors in the response to singlet oxygen was observed in previous studies (12, 16, 25, 28), we also tested for a correlation of genes induced during outgrowth (following prolonged stationary phase of 72 h after inoculation) (this study) and genes induced by singlet oxygen, as determined in the study by Berghoff et al. (25) (Fig. S5). We observe a good correlation between genes induced in outgrowth following long stationary phase and genes induced after 7, 45, and 90 min of singlet oxygen stress (Pearson correlation coefficient r = 0.50, 0.53, and 0.52, respectively) (Fig. S5). This indicates that outgrowth after prolonged stationary phase and the presence of singlet oxygen stimulate similar responses.
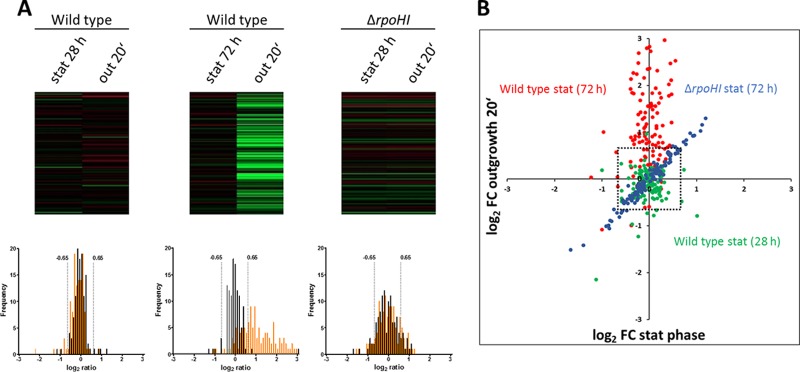
FIG 3.
Distribution and expression kinetics of the RpoHI regulon. Wild-type cells were grown for 28 h or 72 h and ΔrpoHI cells were grown for 72 h after inoculation, and cells were incubated for another 20 min of outgrowth, as indicated. Relative changes of RNA levels in different growth phases compared to levels in exponential phase were monitored by microarray analysis of total RNA. (A) Changes are illustrated as heat maps, with a color code ranging from red (a log2 ratio of −2) to green (a log2 ratio of 2). Frequency distribution analysis was performed on bins corresponding to the log2 ratio. In the frequency graphs, data for stationary phase are in black and data for the following outgrowth are in orange. stat, stationary phase. (B) Correlation between stationary phase and outgrowth is shown as a scatter plot. The dotted square indicates the cutoff log2 fold change of >0.65 or less than −0.65.
Major role for RpoHI/RpoHII in survival in outgrowth after long stationary phase.
Among the 265 genes which showed at least 2-fold higher expression after 20 min of outgrowth than in exponential phase (following late stationary phase), 105 genes were known or predicted to be transcribed by the alternative sigma factor RpoHI, RpoHII, or both (Table S1) (12, 15, 16). Members of the RpoH family of sigma factors often control genes during a heat shock response. In R. sphaeroides, both RpoH sigma factors are also implicated in the response to singlet oxygen (12, 16) and other oxidative stresses (17, 29). Since dilution of the cells from low-aeration stationary-phase cultures resulted in a sudden rise in oxygen levels (Fig. 1), it is tempting to assume that these genes indeed respond to oxidative stress during outgrowth. However, many genes, including RpoHI/RpoHII-dependent genes, showed strongly increased expression during outgrowth only following late stationary phase, although the sudden rise of oxygen upon dilution was identical to that observed after short stationary phase (Fig. 1). Of the 105 RpoHI/RpoHII-dependent genes with more than 2-fold-increased expression after 20 min of outgrowth (following late stationary phase), only 8 genes (sRNAs 0826 and 0827, RSP_1548, and RSP_6006 with a role in iron metabolism, pqqA and expE coding for a hemolysin-type calcium binding protein, RSP_3095 coding for an alternative sigma factor, and a tRNA gene) also showed more than 2-fold-increased expression in late stationary phase before dilution. This demonstrates that for most genes high levels of expression are not a remnant from prolonged stationary phase but that a distinct response is initiated upon subsequent outgrowth (Fig. 3).
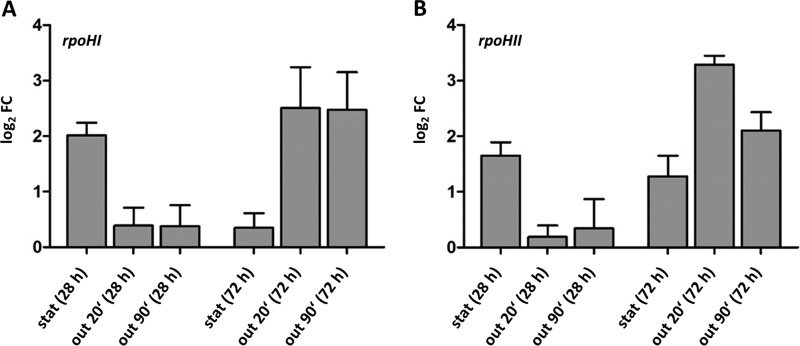
Our microarray data complemented by real-time reverse transcription-PCRs (RT-PCRs) revealed that the expression of the rpoHI (RSP_2410) and rpoHII (RSP_0601) genes is increased in early stationary phase but drops upon subsequent outgrowth or during late stationary phase (Fig. 4). Their expression levels increase quickly during outgrowth following late stationary phase. This further supports an important role of RpoHI and RpoHII in outgrowth after prolonged stationary phase.
FIG 4.
Log2 fold changes in rpoHI and rpoHII mRNA levels in stationary phase (stat) or outgrowth (out; 20 and 90 min) compared to the levels in exponential phase as determined by real-time RT-PCR. Values were normalized to the input RNA amount. The data represent the means of the results from at least three independent experiments, and error bars indicate standard deviation.
However, not all genes known or predicted to be transcribed by RpoHI, RpoHII, or both showed increased expression during outgrowth after prolonged stationary phase. For example, the genes of one of the nuo operons (RSP_0100 to RSP_0112) are regulated by RpoHI and RpoHII, and binding of the sigma factors upstream of the first gene of the operon was demonstrated (15), but these nuo genes mostly showed decreased expression in outgrowth phase, which was also strongly dependent on RpoHI in our transcriptome analysis. The facts that not all promoters induced in outgrowth following late stationary phase in R. sphaeroides have RpoH recognition motifs and, further, that genes preceded by RpoH binding sites show different growth-phase-dependent expression suggest that additional factors are responsible for the differential expression patterns of some genes.
In order to test our hypothesis that RpoHI and RpoHII have an important role in adaptation to outgrowth, we monitored the growth of rpoHI, rpoHII, and rpoHI rpoHII mutants under the same conditions as those described for the wild type. Figure 1 compares the growth curve of the rpoHI mutant to that of the wild type. While the wild type reached an OD of about 1.2 at 8 h after initial inoculation (start OD, 0.2; doubling time, 2 h 45 min in high- or low-aeration cultures), the rpoHI mutant reached ODs of only 0.7 to 0.8. The initial doubling time of the rpoHI mutant was 3 h 45 min, but growth slowed down earlier than that of the wild type. The rpoHII mutant had an identical doubling time and reached an OD similar to that of the rpoHI mutant, and the outgrowth phase after inoculation was longer than that for the rpoHI mutant (Fig. S6A). The rpoHI rpoHII double mutant showed the same growth kinetics as the single mutants (initial doubling time, 3 h 45 min) and reached a similar OD 28 h after inoculation (Fig. S6A). The impaired growth of the mutants demonstrates that RpoHI and RpoHII are required for optimal growth in all growth stages. Following late stationary phase, wild-type cultures resumed growth quickly after dilution into fresh medium. None of the mutants resumed growth within 4 h after dilution following late stationary phase and then showed a very slow increase in OD, reflecting an important function of the RpoH sigma factors in outgrowth. Plating of the cultures indicated very low survival rates of the mutants (data not shown). In outgrowth following short stationary phase, the RpoHI and RpoHII mutants showed the same growth kinetics as the wild type (Fig. S7A). This is in agreement with the observation that no major induction of RpoHI/RpoHII-dependent genes was observed in this growth phase. When the rpoHI mutant was complemented by the plasmid-carried rpoHI gene [in strain 2.4.1 ΔrpoHI(pRK2.4.1rpoHI)] (16), it resumed growth after prolonged stationary phase as quickly as the wild type (Fig. S7B).
Since RpoHI and RpoHII are known to regulate genes in R. sphaeroides in response to oxidative stress, we also determined reactive oxygen species (ROS) levels at different growth stages for wild-type and mutant strains in low-aeration cultures. Fig. S6B shows the ROS levels normalized to the OD of the cultures. ROS levels increased directly after inoculation in the wild type, dropped when mid-exponential phase was reached, and increased again slowly when the cells reached stationary phase. Directly after dilution of the culture, ROS levels increased further, as well upon dilution following early stationary phase (not shown) as after dilution following late stationary phase (Fig. S6B). This excludes the possibility that the strong response during outgrowth after prolonged stationary phase is due to increased ROS levels. In all mutant strains, ROS levels were higher than in the wild type throughout growth. The ROS levels were highest in the double mutant.
RpoHI has a main role in induction of genes during outgrowth following late-stationary phase.
Since the growth experiments proved an important role of RpoHI and RpoHII in outgrowth after prolonged stationary phase, and the RpoHI regulon is mostly activated in outgrowth after prolonged stationary phase (Fig. 3), we also tested the effect of RpoHI on gene expression under these conditions. Microarray analysis was performed with RNA isolated from the rpoHI mutant following late stationary phase and 20 min of outgrowth in comparison to exponential phase. Indeed, many genes upregulated during outgrowth after prolonged stationary phase in the wild type depend on RpoHI. Of the 100 genes with the strongest induction in outgrowth after 60 h of stationary phase in the wild type, only 16 genes were also induced in the rpoHI mutant. For those genes, the factor of induction was clearly lower than in the wild type. Only 11% of all genes with a fold increase of >1.6 in outgrowth after prolonged stationary phase were also induced in the rpoHI mutant. A clear dependence on RpoHI was also observed for several genes which were assigned to the overlapping RpoHI and RpoHII regulons. Despite the overlap of the two regulons, previous reports indicated main functions of RpoHI in the heat shock response and of RpoHII in the singlet oxygen stress response, respectively (16). It is therefore conceivable that RpoHI has a dominant function in activating genes during outgrowth. The growth experiments imply, however, that RpoHII is equally important for survival after prolonged stationary phase.
For one-third of the genes we identified as RpoHI dependent, an RpoHI promoter was neither predicted nor identified prior to this study. Thus, RpoHI affects the expression of more genes than anticipated before, including genes involved in iron metabolism (e.g., sufBCD, hemP, and exbB-exbD-tonB) or chemotaxis (fli and flg) and genes for several transcriptional regulators or sigma factors (RSP_0415, RSP_3094 to RSP_3095, and fecI).
Despite the lack of RpoHI, the mutant shows a strong transcriptional response to late stationary phase and the following outgrowth (Fig. 2C). This also excludes the possibility that the lack of induction of RpoHI-dependent genes is due to the growth defects of the mutant that impair the transcriptional machinery. However, the induced genes strongly differ from those genes induced in the wild type (Fig. S4) and do not belong to the RpoHI regulon (Fig. 3). For genes with lower expression levels than in exponential phase, the overlap of the expression pattern of the wild type is bigger but still limited (among the 100 most repressed genes, 41 genes are for stationary phase and 29 genes are for outgrowth).
In contrast to the response in the wild type, there is a very big overlap of genes induced in prolonged stationary phase (422 genes induced >1.6-fold) and genes induced during the following outgrowth (450 genes induced >1.6-fold) in the rpoHI mutant (Fig. 2C). The fold changes under the two conditions are also very similar, indicating that gene expression stays high during outgrowth following the prolonged stationary phase in the mutant, while the wild type induces a distinct response. The genes induced in the rpoHI mutant belong to many different COGs, and no groups of genes with similar functions emerge.
Four hundred sixty-four genes showed ≤1.6-fold expression in the mutant in prolonged stationary phase compared to exponential phase, as did 492 genes in the following outgrowth. Again, the overlap of those genes is large (Fig. 2C). In summary, the data support a central role of RpoHI in mounting a distinct response to outgrowth after prolonged stationary phase in R. sphaeroides.
In enteric bacteria, the alternative sigma factor RpoS has an important function in stationary phase and other stress responses (30). Our results demonstrate that in R. sphaeroides, RpoHI and RpoHII contribute to survival in outgrowth after long stationary phase and thus imply an RpoS-like function. There is, however, no significant overlap of the R. sphaeroides RpoHI/RpoHII regulons and the E. coli RpoS regulon, as described by Weber et al. (31). Some of the R. sphaeroides genes with homology to genes of the E. coli RpoS regulon were regulated in stationary phase in R. sphaeroides but were not identified as members of the RpoHI/RpoHII regulons. For many genes of the E. coli RpoS regulon, no homologs exist in R. sphaeroides.
Our study reveals that levels of rpoHI and rpoHII mRNAs vary with growth phase and are strongly induced in outgrowth after prolonged stationary phase (Fig. 4). The level of rpoH mRNA also varies with growth phase and cell cycle in E. coli (32), and RpoH, together with RpoE, contributes to survival under starvation in Salmonella (33). Like R. sphaeroides, the plant symbiont Sinorhizobium meliloti harbors two RpoH sigma factors. Both sigma factors play a role in stationary-phase survival (13), and the rpoHI and rpoHII mRNA levels increase during stationary phase (34).
In alphaproteobacteria, a major function in the general stress response was assigned to the PhyR-NepR-σEcfG cascade (35). None of the genes encoding homologs of PhyR, NepR, or RpoE2 (RSP_1274 to RSP_1272) showed growth-phase-dependent expression in our study (Table S1), and a PhyR mutant did not differ significantly in growth behavior from the wild type (data not shown). Thus, we exclude a major role of the PhyR-NepR-σEcfG cascade in growth phase adaptation in R. sphaeroides.
Search for promoter motifs of genes with growth-phase-dependent expression.
In order to find additional promoter motifs which may be responsible for growth-phase-dependent gene expression, besides those for RpoHI/RpoHII, the RNA-seq and dRNA-seq data (36) were used to map the TSS on the R. sphaeroides 2.4.1 chromosomes. The MEME program (http://meme-suite.org/) was applied to search for common motifs in the regions upstream of the TSS of genes which were induced or repressed under certain growth conditions, according to the data in Table S1 (see also Fig. 5).
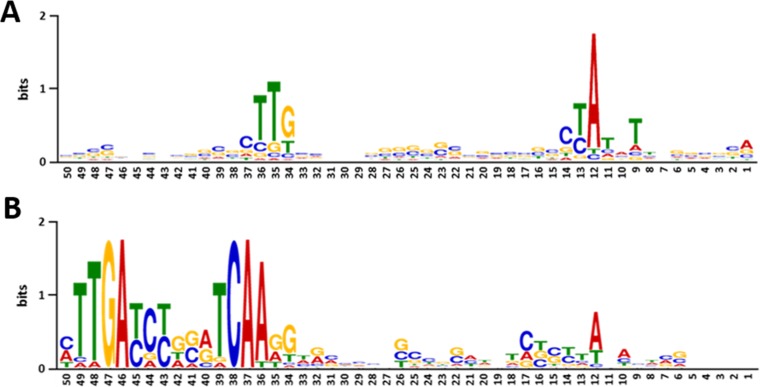
FIG 5.
Sequence logos for promoter regions of strongly repressed genes. Shown are the consensus motifs for 238 promoters (among 294) that were induced in late stationary phase (A) and for 16 promoters (among 97) that were repressed during outgrowth following early stationary phase (B).
The analysis revealed a conserved TTG at position −35 of the TSS for most promoters under most of the tested conditions. Likewise, an A residue was conserved at position −11/−12 in almost all promoter regions (data not shown). In total, 294 promoters were induced during outgrowth following late stationary phase. A consensus motif was detected for 238 promoters out of these 294 (Fig. 5A). This consensus sequence perfectly matches the known R. sphaeroides RpoHI promoter consensus sequence [(T/G)TG(N18/N19)(C/A)(T/C)AT(A/C/G)T] and is very similar to the RpoHII consensus sequence [(T/G)(C/T)(C/T)N17–19CTAG(A/C/G)T] (15, 16).
An unique common motif was found for 16 promoters which were repressed during outgrowth after short stationary phase (Fig. 5B). A TTGA motif is present at positions −49 to −46, while at positions −39 to −36, a TCAA is conserved. At position −12, the A residue is less conserved than in other promoters. This motif is known as a consensus binding site for FnrL [(T/C/A)TGAN6TCAA], a transcriptional regulator that activates photosynthesis genes under low oxygen tension in R. sphaeroides (37). The decreased expression of the FnrL-dependent genes may reflect the lack of activation under conditions with transiently increased oxygen levels. Since many genes without a recognizable promoter consensus sequence in addition to the −35 TTG and the −11/−12 A show growth-phase-dependent expression, it is likely that not only promoter recognition by specific transcription factors is responsible for their expression pattern. It is highly conceivable that similar to those in enteric bacteria, nucleoid-associated and nucleoid-structuring proteins influence growth-phase-dependent gene expression (3, 4, 6), a possibility that needs to be elucidated in the future.
Concluding remarks.
Our study demonstrated a distinct response of R. sphaeroides to outgrowth after prolonged stationary phase at the transcriptome level and a strong impact of RpoH sigma factors on this response. Nevertheless, not only rpoH genes but also genes for RpoE and sigma factors of unknown function (RSP_0415 and RSP_3095), genes for putative transcriptional regulators, and genes for sRNAs of unknown function were induced during outgrowth after prolonged stationary phase, suggesting a complex regulatory network in this response. Many genes induced in outgrowth after prolonged stationary phase have established functions in stress defense, indicating that efficient stress defense is a prerequisite for resuming growth. We excluded ROS as the trigger for the RpoH-dependent stress response, and therefore, further investigation needs to be done to identify the factors triggering this response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The R. sphaeroides 2.4.1 wild-type strain (DSM 158) and the mutants 2.4.1 ΔrpoHI, 2.4.1 ΔrpoHII, 2.4.1 ΔrpoHI rpoHII, and 2.4.1 ΔrpoHI(pRKrpoHI) (see Table S2 in the supplemental material) were grown at 32°C in minimal salt medium containing malate (0.022 M) as the carbon source (38). Low-aeration growth conditions were established by continuous shaking of Erlenmeyer flasks at 140 rpm with a culture volume of 80%, leading to a dissolved oxygen concentration of approximately 25 to 30 μM in exponential phase with strong changes throughout growth, as measured using the GMH 3610 (Greisinger) digital oxygen measuring device (Fig. 1). When necessary, kanamycin (25 μg · ml−1), spectinomycin (10 μg · ml−1), or tetracycline (1.5 μg · ml−1) was added to growth media in precultures of the mutant strains but was omitted during the 87 h of the experiment.
Fluorescence measurements.
The fluorescence measurements were performed as described elsewhere (38).
RNA isolation and quantification.
Cells were rapidly cooled on ice and harvested by cooled centrifugation. Total RNA for RNA-seq or microarray analysis was isolated using the hot phenol method, followed by two chloroform-isoamyl alcohol treatments and precipitation with sodium acetate and ethanol (39). For quantitative real-time RT-PCR, RNA was isolated using the peqGOLD TriFast kit (Peqlab), as described by the manufacturer. After DNA digestion, RNA was purified using a mixture of phenol-chloroform–isoamyl alcohol and chloroform-isoamyl alcohol (for RT-PCR) or RNeasy MinElute spin columns (Qiagen) (for microarray and RNA-seq). RNA was resolved in RNase-free water (Roth), and concentrations were determined using a NanoDrop 1000 spectrophotometer (Peqlab).
Quantitative real-time RT-PCR.
The one-step Brilliant III quantitative RT-PCR (qRT-PCR) master mix kit (Agilent) was used for reverse transcription, followed by PCR, as described in the manufacturer's manual. RT-PCR samples containing 4 ng of total RNA/μl were run in a Rotor-Gene 3000 real-time PCR cycler (Corbett Research) for relative quantification of mRNAs in each of three independent experiments using primers rpoHI_RT and rpoHII_RT (Table S2). Crossing points (Cp) with a fluorescence threshold of 0.002 were visualized with the Rotor-Gene software 6.0 (Corbett Research). The relative mRNA levels were normalized to the input amount of RNA and calculated according to Pfaffl (40).
Microarray analysis.
Microarray analysis was performed as described elsewhere (41). In brief, RNA from three cultures harvested in exponential phase was pooled with RNA from cultures harvested either in stationary phase or during outgrowth and applied to high-density oligonucleotide R. sphaeroides microarrays (Agilent gene chips corresponding to the whole 4.6-Mb genome). Two arrays with probes against 4,304 protein-encoding genes, 79 rRNA and tRNA genes, and 144 intergenic regions were performed for each condition; construction and performance analysis were carried out according to the instructions of Agilent. The ULS fluorescent labeling kit for Agilent arrays (Kreatech) was used for RNA labeling and fragmentation. Multiarray analysis and normalization according to locally weighted smoothing (LOESS) were accomplished with the Bioconductor package Limma for R and performed as described elsewhere (42, 43). On the basis of calculated microarray analysis (MA) plots, genes were considered reliable if the average signal intensity [A-value = 1/2 log2(Cy3 × Cy5)] was above the average background signal. The data shown in this study represent the results from two individual microarrays (biological replicates), each containing a pool of three independent experiments for each sample. For expression cluster analysis, log2 ratios were imported to MeV (Multi Experiment Viewer version 4.7.4) from the TM4 microarray software suite (44, 45) and visualized as heat maps. Clustering was based on a k-means clustering (KMC) method according to Euclidean distance, with a maximum of 50 iterations.
TEX treatment.
For the depletion of processed transcripts, equal amounts of RNA were incubated with Terminator 5′-phosphate-dependent exonuclease (TEX) (catalog no. TER51020; Epicentre), as previously described (36).
Library construction and sequencing.
The transcripts were not fragmented in order to obtain mainly sequencing reads of the 5′ end of the transcripts. Libraries for Illumina sequencing of cDNA were constructed by Vertis Biotechnology AG, Germany, as described previously (38). The resulting cDNA libraries were sequenced on an Illumina HiSeq 2000 machine in single-read mode running 100 cycles.
Read mapping and coverage plot construction.
In order to ensure high sequence quality, the Illumina reads in FASTQ format were trimmed with a cutoff phred score of 20 by the program fastq_quality_trimmer from FASTX toolkit version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/). The following steps were performed using the subcommands “create,” “align,” and “coverage” of the tool READemption version 0.3.4 (46), with default parameters. The poly(A) tail sequences were removed, and a size-filtering step was applied in which sequences shorter than 12 nucleotides (nt) were eliminated. The collections of remaining reads were mapped to the reference genome sequences comprising two circular chromosomes, CI (3.19 Mb) and CII (0.94 Mb), and five endogenous plasmids, A (0.11 Mb), B (0.11 Mb), C (0.11 Mb), D (0.10 Mb), and E (0.04 Mb) (accession numbers NC_007488.2, NC_007489.1, NC_007490.2. NC_007493.2, NC_007494.2, NC_009007.1, and NC_009008.1, respectively, downloaded from the NCBI ftp server), using segemehl version 0.1.7 (47). Mapping statistics (input, aligned, uniquely aligned reads, etc.) can be found in Table S3. Coverage plots in wiggle format representing the number of aligned reads per nucleotide were generated based on the aligned reads and visualized in the Integrated Genome Browser (48). The raw coverage values of the graphs were normalized to the total number of reads that could be aligned for the respective library and multiplied by the minimum number of mapped reads of all libraries (i.e., 4,528,755). This restores the original data range and prevents over- or underestimation due to the normalized values. Relative changes in gene expression as determined by RNA-seq are displayed in Table S1.
Transcription start site prediction.
Transcriptional start sites (TSS) were predicted with TSSpredator (49) based on the normalized coverage files. Parameter optimization for TSSpredator was done with ANNOgesic (S.-H. Yu, J. Vogel, and K. U. Förstner, unpublished data) guided by a manually curated TSS set.
Promoter motif detection.
Sequence motif detection was performed with MEME 4.10.1 (PMID 19458158) based on the 50 nt upstream of the TSS and the TSS position itself using a motif width of 50 nt.
Reproducibility of the data analysis.
A shell script that covers the main RNA-seq data processing steps is deposited at https://zenodo.org/record/34192.
Accession number(s).
The microarray data and the RNA-seq data are available at the NCBI Gene Expression Omnibus database (50) under accession numbers GSE75345 and GSE71844, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Olga Tsoy and Mikhail Gelfand for initial data analysis and Sabrina Brückmann and Andrea Weisert for technical support.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00249-17.
REFERENCES
- 1.Roop RM Jr, Gee JM, Robertson GT, Richardson JM, Ng WL, Winkler ME. 2003. Brucella stationary-phase gene expression and virulence. Annu Rev Microbiol 57:57–76. doi: 10.1146/annurev.micro.57.030502.090803. [DOI] [PubMed] [Google Scholar]
- 2.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 3.Navarro Llorens JM, Tormo A, Martinez-Garcia E. 2010. Stationary phase in Gram-negative bacteria. FEMS Microbiol Rev 34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 4.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. 1999. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol 181:6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landgraf JR, Boxer JA, Calvo JM. 1999. Escherichia coli Lrp (leucine-responsive regulatory protein) does not directly regulate expression of the leu operon promoter. J Bacteriol 181:6547–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin DJ, Cagliero C, Zhou YN. 2012. Growth rate regulation in Escherichia coli. FEMS Microbiol Rev 36:269–287. doi: 10.1111/j.1574-6976.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staron A, Mascher T. 2010. General stress response in alpha-proteobacteria: PhyR and beyond. Mol Microbiol 78:271–277. doi: 10.1111/j.1365-2958.2010.07336.x. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Salazar JM, Sandoval-Calderon M, Guo X, Castillo-Ramirez S, Reyes A, Loza MG, Rivera J, Alvarado-Affantranger X, Sanchez F, Gonzalez V, Davila G, Ramirez-Romero MA. 2009. The Rhizobium etli RpoH1 and RpoH2 sigma factors are involved in different stress responses. Microbiology 155:386–397. doi: 10.1099/mic.0.021428-0. [DOI] [PubMed] [Google Scholar]
- 10.Delory M, Hallez R, Letesson JJ, De Bolle X. 2006. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J Bacteriol 188:7707–7710. doi: 10.1128/JB.00644-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lucena DK, Puhler A, Weidner S. 2010. The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol 10:265. doi: 10.1186/1471-2180-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuss AM, Glaeser J, Klug G. 2009. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J Bacteriol 191:220–230. doi: 10.1128/JB.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnett MJ, Bittner AN, Toman CJ, Oke V, Long SR. 2012. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J Bacteriol 194:4983–4994. doi: 10.1128/JB.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green HA, Donohue TJ. 2006. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J Bacteriol 188:5712–5721. doi: 10.1128/JB.00405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour YS, Imam S, Koo BM, Green HA, Donohue TJ. 2012. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet 8:e1002929. doi: 10.1371/journal.pgen.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuss AM, Glaeser J, Berghoff BA, Klug G. 2010. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J Bacteriol 192:2613–2623. doi: 10.1128/JB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billenkamp F, Peng T, Berghoff BA, Klug G. 2015. A cluster of four homologous small RNAs modulates C1 metabolism and the pyruvate dehydrogenase complex in Rhodobacter sphaeroides under various stress conditions. J Bacteriol 197:1839–1852. doi: 10.1128/JB.02475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregor J, Klug G. 1999. Regulation of bacterial photosynthesis genes by oxygen and light. FEMS Microbiol Lett 179:1–9. doi: 10.1111/j.1574-6968.1999.tb08700.x. [DOI] [PubMed] [Google Scholar]
- 19.Zeilstra-Ryalls JH, Kaplan S. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell Mol Life Sci 61:417–436. doi: 10.1007/s00018-003-3242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Ke Y, Xu J, Wang L, Wang T, Liang H, Zhang W, Gong C, Yuan J, Zhuang Y, An C, Lei S, Du X, Wang Z, Li W, Yuan X, Huang L, Yang X, Chen Z. 2015. Identification of a novel small non-coding RNA modulating the intracellular survival of Brucella melitensis. Front Microbiol 6:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppee JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabharwal D, Song T, Papenfort K, Wai SN. 2015. The VrrA sRNA controls a stationary phase survival factor Vrp of Vibrio cholerae. RNA Biol 12:186–196. doi: 10.1080/15476286.2015.1017211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagamitsu H, Murata M, Kosaka T, Kawaguchi J, Mori H, Yamada M. 2013. Crucial roles of MicA and RybB as vital factors for sigma-dependent cell lysis in Escherichia coli long-term stationary phase. J Mol Microbiol Biotechnol 23:227–232. doi: 10.1159/000350370. [DOI] [PubMed] [Google Scholar]
- 24.Lu P, Wang Y, Zhang Y, Hu Y, Thompson KM, Chen S. 2016. RpoS-dependent sRNA RgsA regulates Fis and AcpP in Pseudomonas aeruginosa. Mol Microbiol 102:244–259. doi: 10.1111/mmi.13458. [DOI] [PubMed] [Google Scholar]
- 25.Berghoff BA, Konzer A, Mank NN, Looso M, Rische T, Forstner KU, Kruger M, Klug G. 2013. Integrative “omics”-approach discovers dynamic and regulatory features of bacterial stress responses. PLoS Genet 9:e1003576. doi: 10.1371/journal.pgen.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anthony JR, Warczak KL, Donohue TJ. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A 102:6502–6507. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glaeser J, Zobawa M, Lottspeich F, Klug G. 2007. Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter. J Proteome Res 6:2460–2471. doi: 10.1021/pr060624p. [DOI] [PubMed] [Google Scholar]
- 28.Braatsch S, Moskvin OV, Klug G, Gomelsky M. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J Bacteriol 186:7726–7735. doi: 10.1128/JB.186.22.7726-7735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adnan F, Weber L, Klug G. 2015. The sRNA SorY confers resistance during photooxidative stress by affecting a metabolite transporter in Rhodobacter sphaeroides. RNA Biol 12:569–577. doi: 10.1080/15476286.2015.1031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengge R. 2008. The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv Exp Med Biol 631:40–53. doi: 10.1007/978-0-387-78885-2_4. [DOI] [PubMed] [Google Scholar]
- 31.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner MA, Zahrl D, Rieser G, Koraimann G. 2009. Growth phase- and cell division-dependent activation and inactivation of the {sigma}32 regulon in Escherichia coli. J Bacteriol 191:1695–1702. doi: 10.1128/JB.01536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. 2002. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol 43:771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- 34.Oke V, Rushing BG, Fisher EJ, Moghadam-Tabrizi M, Long SR. 2001. Identification of the heat-shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti. Microbiology 147:2399–2408. doi: 10.1099/00221287-147-9-2399. [DOI] [PubMed] [Google Scholar]
- 35.Francez-Charlot A, Kaczmarczyk A, Fischer HM, Vorholt JA. 2015. The general stress response in Alphaproteobacteria. Trends Microbiol 23:164–171. doi: 10.1016/j.tim.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 37.Kumka JE, Bauer CE. 2015. Analysis of the FnrL regulon in Rhodobacter capsulatus reveals limited regulon overlap with orthologues from Rhodobacter sphaeroides and Escherichia coli. BMC Genomics 16:895. doi: 10.1186/s12864-015-2162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Remes B, Berghoff BA, Forstner KU, Klug G. 2014. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genomics 15:794. doi: 10.1186/1471-2164-15-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janzon L, Lofdahl S, Arvidson S. 1986. Evidence for a coordinate transcriptional control of alpha-toxin and protein a synthesis in Staphylococcus aureus. FEMS Microbiol Lett 33:193–198. doi: 10.1111/j.1574-6968.1986.tb01270.x. [DOI] [Google Scholar]
- 40.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peuser V, Metz S, Klug G. 2011. Response of the photosynthetic bacterium Rhodobacter sphaeroides to iron limitation and the role of a Fur orthologue in this response. Environ Microbiol Rep 3:397–404. doi: 10.1111/j.1758-2229.2011.00245.x. [DOI] [PubMed] [Google Scholar]
- 42.Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 43.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 44.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. 2006. TM4 microarray software suite. Methods Enzymol 411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 45.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 46.Förstner KU, Vogel J, Sharma CM. 2014. READemption–a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30:3421–3423. doi: 10.1093/bioinformatics/btu533. [DOI] [PubMed] [Google Scholar]
- 47.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dugar G, Herbig A, Forstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM. 2013. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet 9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.