ABSTRACT
Reversible or phenotypic tolerance to antibiotics within microbial populations has been implicated in treatment failure of chronic infections and development of persister cells. However, the molecular mechanisms regulating phenotypic drug tolerance are largely unknown. In this study, we identified a four-gene operon in Streptococcus pneumoniae that contributes to phenotypic tolerance to vancomycin (ptv). RNA sequencing, quantiative reverse transcriptase PCR, and transcriptional luciferase reporter experiments revealed that transcription of the ptv operon (consisting of ptvR, ptvA, ptvB, and ptvC) is induced by exposure to vancomycin. Further investigation showed that transcription of the ptv operon is repressed by PtvR, a PadR family repressor. Transcriptional induction of the ptv operon by vancomycin was achieved by transcriptional derepression of this locus, which was mediated by PtvR. Importantly, fully derepressing ptvABC by deleting ptvR or overexpressing the ptv operon with an exogenous promoter significantly enhanced vancomycin tolerance. Gene deletion analysis revealed that PtvA, PtvB, and PtvC are all required for the PtvR-regulated phenotypic tolerance to vancomycin. Finally, the results of an electrophoretic mobility shift assay with recombinant PtvR showed that PtvR represses the transcription of the ptv operon by binding to two palindromic sequences within the ptv promoter. Together, the ptv locus represents an inducible system in S. pneumoniae in response to stressful conditions, including those caused by antibiotics.
IMPORTANCE Reversible or phenotypic tolerance to antibiotics within microbial populations is associated with treatment failure of bacterial diseases, but the underlying mechanisms regulating phenotypic drug tolerance remain obscure. This study reports our finding of a multigene locus that contributes to inducible tolerance to vancomycin in Streptococcus pneumoniae, an important opportunistic human pathogen. The vancomycin tolerance phenotype depends on the PtvR transcriptional repressor and three predicted membrane-associated proteins encoded by the ptv locus. This represents the first example of a gene locus in S. pneumoniae that is responsible for antibiotic tolerance and has important implications for further understanding bacterial responses and phenotypic tolerance to antibiotic treatment in this and other pathogens.
KEYWORDS: Streptococcus pneumoniae, phenotypic tolerance, vancomycin, PadR family regulator, gene regulation, transcriptional derepression
INTRODUCTION
Inherited microbial resistance to antibiotics has posed serious challenges to treatment of infections caused by numerous bacterial pathogens (1). Inherited or genotypic resistance is caused by generation of endogenous resistance mutations or acquisition of exogenous resistance genes (2). Bacteria can also withstand lethal concentrations of bactericidal antibiotics through formation of reversible or phenotypic drug-tolerant cells (3–5), which has been implicated in undesirable outcomes in treating bacterial infections (4, 6, 7). Phenotypically tolerant cells were originally identified in penicillin-treated Staphylococcus aureus infections by Joseph Bigger, and the bacteria were referred to as persister cells (8). Persisters represent small fractions of nongrowing cells in bacterial populations that are refractory to the killing actions of antibiotics (9). In contrast to antibiotic-resistant bacteria, persisters are genetically identical to their siblings that are otherwise susceptible to the antibiotic. Persisters have been described in many other pathogenic bacteria (10).
The current understanding of phenotypic antibiotic tolerance is mainly based on studies of persisters in Escherichia coli (9, 10). The formation of persisters can be a result of a stochastic switch in metabolic status (3, 11–13). A number of additional factors have been shown to induce the formation of persister cells, such as DNA damage (14), oxidative stress (15, 16), indole signaling (17), antibiotics (14, 18, 19), and starvation (13, 20, 21). Many of these factors induce bacterial persistence by inhibiting translation through the activities of toxin-antitoxin (TA) systems (10, 21). Gram-positive bacteria have also been documented to develop phenotypic tolerance to antibiotics (22), but the mechanisms are poorly understood. The GraSR two-component system is required for colistin-induced reversible tolerance of S. aureus to vancomycin (23). It is likely that GraSR can regulate the surface charge of S. aureus via d-alanylation of teichoic acids and lysinylation of phosphatidylglycerol and thus enable bacterial resistance to cationic antimicrobial peptides (24, 25). Also, ATP depletion is associated with persister formation in S. aureus (22). LexA, a DNA-binding protein that represses the SOS response in bacteria, is involved in the formation of Streptococcus mutans persisters induced by the competence-stimulating peptide (CSP) (26). Lastly, Streptococcus pneumoniae (pneumococcus) exhibits phenotypic tolerance to vancomycin and penicillin in a capsule-dependent fashion (27), and this tolerance is epigenetically inherited by daughter cells (28). Along this line, prior exposure of S. pneumoniae to erythromycin induces phenotypic tolerance to vancomycin (29, 30). However, the genetic basis of phenotypic tolerance to antibiotics in S. pneumoniae remains to be defined.
S. pneumoniae naturally resides in the nasopharynx of humans and also causes multiple infections (e.g., pneumonia, otitis media, and meningitis) (31). Pneumococcal disease is estimated to result in up to 1 million deaths globally every year (32, 33). Global emergence of pneumococcal strains resistant to β-lactams and other antibiotics has made it increasingly difficult to treat invasive diseases caused by S. pneumoniae, particularly meningitis (34–36). Vancomycin is regarded as an antimicrobial agent of last resort to treat pneumococcal meningitis, because no vancomycin-resistant pneumococcal strain has been reported to date (34). However, pneumococcal isolates with inherited vancomycin tolerance have been described in multiple countries (37). This phenomenon is associated with the polysaccharide capsule (27, 38), activity of the LytA autolysin (27, 38, 39), and the CiaR/CiaH two-component regulatory system (38). Previous studies in animal models by Harry Eagle showed that curing infections caused by S. pneumoniae and several other pathogens with penicillin depended on the infection dose and duration of the infection (40–42), suggesting a significant impact of bacterial tolerance to the drug on the treatment outcome, a phenomenon also referred to as “drug indifference” (4).
Haas et al. reported that transient exposure of pneumococci to a lethal concentration of vancomycin led to significant alterations in the transcription of 175 genes (43). Among these genes was a cluster of four neighboring genes with unknown functions (SP97 to SP100 in type 4 strain TIGR4 and SPD_93 to SPD_96 in Avery strain D39) which were cotranscribed, as described in this study, and named the ptv operon. We confirmed the previous result that exposure of S. pneumoniae to vancomycin significantly induces the transcription of the ptv operon in strain D39, and we discovered that the induction relies on the transcriptional repressor PtvR, which is encoded by the first gene of the operon. Importantly, experimental overexpression of the ptv operon significantly enhanced phenotypic tolerance to vancomycin.
RESULTS
Vancomycin induces transcription of the ptv gene operon.
Haas et al. reported that exposure to a lethal concentration of vancomycin led to significant alteration in the transcription of many genes in multiple S. pneumoniae strains (43). Among these was a cluster of four genes with unknown functions (SPD_0093, SPD_0094, SPD_0095, and SPD_0096 in type 2 strain D39 and SP0097, SP0098, SP0099, and SP0100 in type 4 strain TIGR4). These genes are highly conserved in all of the fully sequenced S. pneumoniae genomes that were publically available at the time of our BLAST search. In our study, we characterized the impact of these genes on the response to vancomycin in strain D39. In the current D39 genome (accession number NC_008533) (44), SPD_0096 (108 amino acids [aa]) is annotated as a PadR family transcriptional regulator, whereas SPD_0093 (354 aa), SPD_0094 (303 aa), and SPD_0095 (197 aa) are described as hypothetical proteins. Based on our later observation that this locus is involved in phenotypic tolerance to vancomycin (ptv) (see below), we designated SPD_0096, SPD_0095, SPD_0094, and SPD_0093 as ptvR, ptvA, ptvB, and ptvC, respectively (Fig. 1A).
FIG 1.
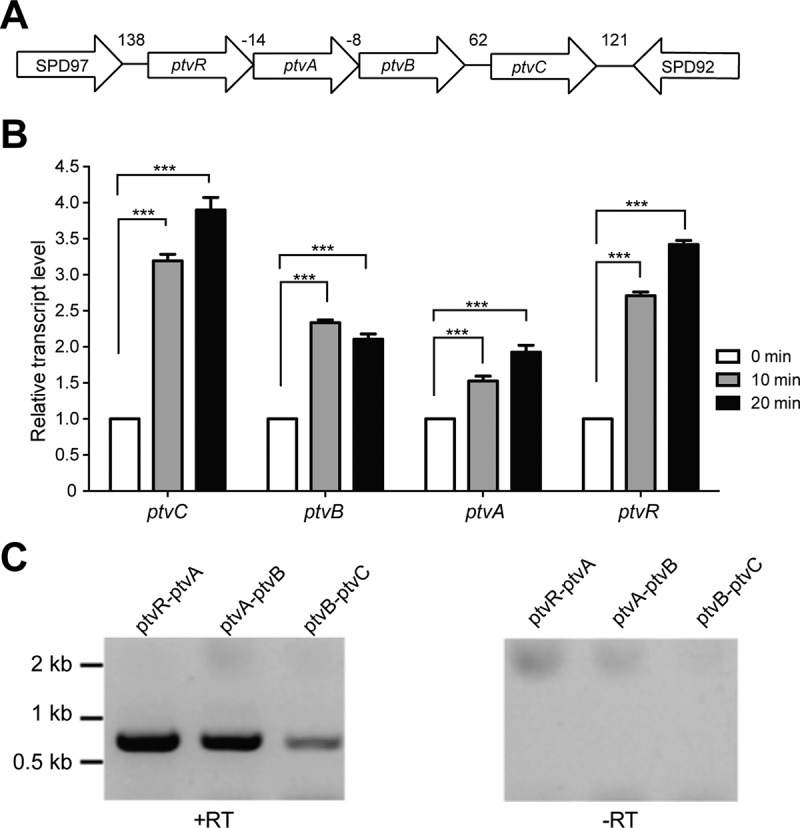
Transcriptional response of the ptv gene cluster to vancomycin. (A). Diagram of the ptv locus in strain D39. The nucleotides between two adjacent genes are marked in base pairs; negative numbers indicate that the nucleotides are shared by the coding regions of two genes. (B) mRNA levels of the ptv genes in S. pneumoniae D39 before and after vancomycin treatment. Mid-log-phase cultures were mixed with vancomycin to a final concentration of 5 μg/ml; portions of the mixtures were removed 10 or 20 min later for extraction of total RNA and measurement of the mRNA level for each of the four genes by qRT-PCR. The results from one of three independent experiments are shown, with mRNA levels of each target gene immediately before (0 min) and after (10 and 20 min) the addition of vancomycin. The results represent the relative gene transcription levels of vancomycin-treated cells (10 or 20 min) after normalization of their mean cycle threshold values to those of untreated cells (0 min), as described in Materials and Methods. ***, P < 0.001. (C) Detection of cotranscription in the ptv locus. The mRNA molecules covering the intergenic sequences of neighboring genes were amplified by RT-PCR in the presence (+RT) or absence (-RT) of reverse transcriptase and separated by DNA electrophoresis. The sizes of the molecular standards are indicated.
We first determined the precise impact of vancomycin on transcription of ptvR, ptvA, ptvB, and ptvC in strain D39 by quantitative reverse transcriptase PCR (qRT-PCR). Pneumococci were treated with a lethal dose of vancomycin (5 μg/ml) before being processed for total RNA extraction. In concordance with the previous report (43), transient treatment of S. pneumoniae D39 with vancomycin for 10 or 20 min led to a significant increase in the mRNA levels of ptvR, ptvA, ptvB, and ptvC by 2.7-, 1.5-, 2.4-, and 3.2-fold at 10 min, respectively, and by 3.4-, 1.9-, 2.1-, and 3.9-fold at 20 min, respectively (Fig. 1B). We also obtained similar levels of induction in transcription of ptvR, ptvA, ptvB, and ptvC in ST556, a type 19F isolate of S. pneumoniae (45) (see Fig. S1 in the supplemental material). This result confirmed the previous study's results, that exposure of pneumococci to lethal concentrations of vancomycin leads to a significant enhancement in transcription of the ptv locus.
As illustrated in Fig. 1A, there are relatively short intergenic sequences in the ptv locus. While the predicted coding sequences of ptvR and ptvA overlap by 14 bp, ptvA and ptvB share an 8-bp coding region. This feature suggested that these four genes are cotranscribed. This possibility was tested by RT-PCR with primers that spanned the intergenic regions of the neighboring genes. The experiment yielded a PCR product with the predicted molecular size for each pair of the adjacent genes in this locus, whereas no PCR products were detected with any of the primer pairs in similar reaction mixtures that did not contain reverse transcriptase (Fig. 1C). This result demonstrated that ptvR, ptvA, ptvB, and ptvC of D39 are transcribed at some level as an operon from the common promoter upstream of ptvR.
PtvR is a transcriptional repressor of the ptv operon.
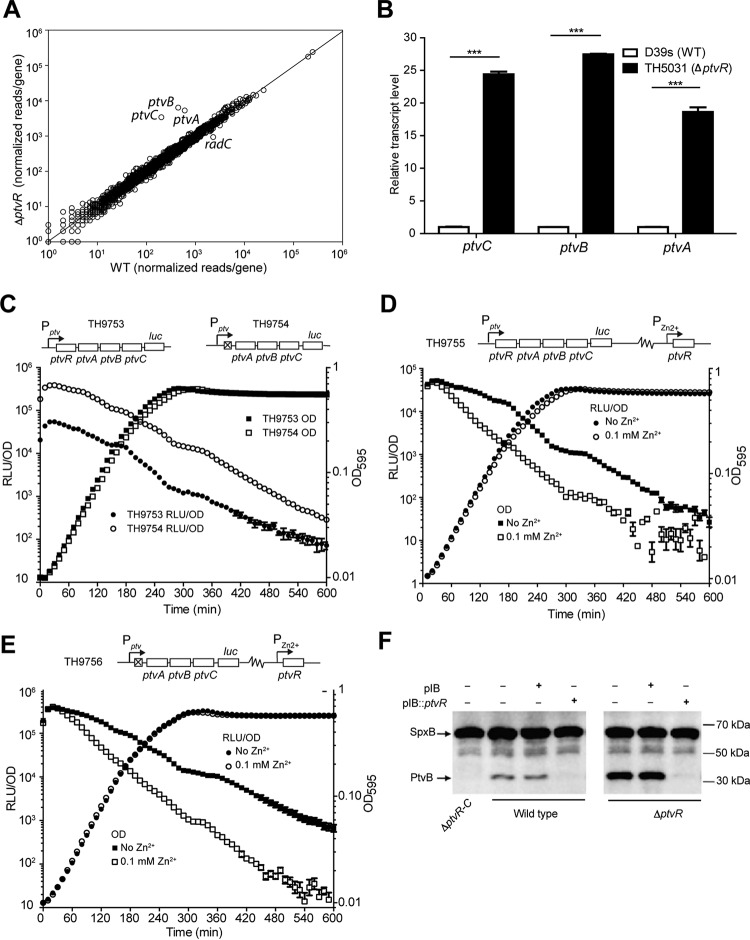
Because the N-terminal protein sequence of ptvR possesses a helix-turn-helix domain for DNA binding, a signature of the PadR-like family proteins (46), we assessed the impact of ptvR on the transcriptome of D39. A ΔptvR mutant strain (TH5031) was generated by unmarked deletion of a 291-bp coding sequence of ptvR (327 bp in total), maintaining the translation signals for the downstream overlapped ptvA gene in D39s (TH4527), a streptomycin-resistant derivative of D39 (referred to as the wild type). This mutant strain was used to determine the transcriptome along with that of wild-type D39s by RNA sequencing (RNA-Seq). Comparative analysis of the RNA-Seq results revealed a total of 4 genes with more than a 2-fold transcriptional increase (3 genes) or decrease (1 gene) in the ΔptvR deletion strain (Fig. 2A) (Table S2). As expected, the transcripts of ptvR were diminished in the ΔptvR mutant. The transcript levels of ptvA, ptvB, and ptvC, the three immediate downstream genes of ptvR (Fig. 1A), were increased 8.7-, 14.3-, and 16.9-fold in this strain, respectively (Fig. 2A). The radC gene (SPD_0975) was reduced in the ptvR mutant strain by 2.5-fold (Fig. 2A). Since the ptvA, ptvB, and ptvC genes showed the most dramatic changes in the ptvR mutant strain, we focused on these genes in the following experiment. This result was subsequently verified by qRT-PCR. In line with the RNA-Seq data, the ΔptvR strain showed significantly higher levels of transcription of ptvA (18.6-fold), ptvB (27.5-fold), and ptvC (24.3-fold) than observed with the parental strain (Fig. 2B).
FIG 2.
Transcriptional repression of the ptv operon by PtvR. (A) Comparison of the transcriptomes of strain D39s and the isogenic ptvR-deficient (ΔptvR) strain by RNA-Seq. Reads per gene were normalized to reads per kilobase per million reads. The line represents y = x. (B) Transcription derepression of the ptv operon in the ΔptvR strain. The transcripts of ptvA, ptvB, and ptvC in the ΔptvR mutant strain were measured by qRT-PCR and normalized to the corresponding value of each gene in D39s (WT), as for Fig. 1B. ***, P < 0.001. (C) Transcriptional repression of the ptv locus by PtvR as assessed by fusing the luc reporter gene to the 3′ end of ptvC in wild-type (TH9753, Pptv_luc) or ΔptvR (TH9754, Pptv_luc) cells. The cell density (OD595) and luciferase activity (RLU/OD) were measured every 10 min for 10 h. (D and E) Additional repression of the ptv locus by trans overexpression of ptvR. The OD595 and RLU/OD values in the presence (D) or absence (E) of the endogenous ptvR were measured with or without Zn2+, as for panel C. (F) Detection of transcription repression by PtvR at the protein level. PtvB was detected by immunoblotting in D39s (wild type), ΔptvR-C, and ΔptvR in the presence (+) or absence (-) of the empty pIB166 (pIB) and the ptvR complementation construct (pIB::ptvR). SpxB was similarly detected as a loading control. The sizes of the protein standards are indicated.
We further tested the transcriptional impact of PtvR on the ptv locus via a luciferase transcription reporter construct (47). The firefly luciferase gene (luc) was fused to the 3′ end of ptvC in the presence (strain TH9753) or absence (strain TH9754) of ptvR. While both the ptvR+ (TH9753) and ptvR-deficient (TH9754) strains had similar growth patterns, the latter displayed approximately 10-fold more luciferase expression from the ptv promoter than the former in all growth phases (Fig. 2C), thus confirming a repressive role of PtvR in regulating transcription of the ptv locus. We further verified this observation by trans expression of ptvR in the luc reporter strains. The second copy of ptvR was inserted at the bgaA locus and driven by a zinc-inducible promoter (PZn2+). In strain TH9755, which possesses the endogenous ptvR gene along with the inducible ptvR gene (Fig. 2D), addition of the inducer Zn2+ led to a substantial reduction of luciferase activity. This result showed not only that the merodiploid copy of ptvR was able to repress the transcription of the ptv locus but also that the genes in this locus are not completely turned off at the transcriptional level under our experimental conditions. Consistently, trans expression of ptvR in strain TH9756, which lacks the endogenous ptvR gene (Fig. 2E), repressed the transcriptional activity of the ptv promoter. It should be noted that, prior to the addition of the inducer (i.e., 0 min), the luciferase activity in TH9755 (with the endogenous ptvR gene) (Fig. 2D) was approximately 10- to 15-fold lower than in strain TH9756 (without the endogenous ptvR gene) (Fig. 2E), which is consistent with the role of PtvR as a transcriptional repressor.
Lastly, we verified transcriptional repression of PtvR in the ptv locus at the protein level. We restored the ΔptvR strain (TH5031) by trans complementation with pTH6862, a pIB166 derivative containing the intact ptvR gene driven by the promoter of the spxB gene (P-spxB, a 206-bp 5′ noncoding sequence of spxB). spxB was among the pneumococcal genes with the highest transcription levels under this growth condition in our earlier RNA-Seq experiment (Fig. 2A; Table S2). The effect of ptvR on protein production of PtvB was assessed by immunoblotting with an antiserum against PtvB. The PtvB protein was completely undetectable in the ptvR-C null strain TH4741 (missing the operon comprising ptvR to ptvC and the genetic material between the two genes, i.e., including ptvA and ptvB) (Fig. 2F, lane 1) but was weakly detected in the parental strain (Fig. 2F, lane 2), indicating weak constitutive expression of PtvB, consistent with the result of the luciferase assay shown in Fig. 2D. While the protein remained visible in the presence of the empty vector (pIB166), it became completely undetectable in the presence of the ptvR complementation construct (Fig. 2F, lanes 3 and 4). This result indicated that overexpression of PtvR from the P-spxB promoter further repressed the expression of the operon, and the low level of the PtvB protein in strain D39 represented basal expression of ptvB caused by incomplete repression by PtvR. Moreover, the ΔptvR mutant strain (TH5031) showed a substantially higher level of PtvB than the parent strain in the absence of episomally encoded ptvR (Fig. 2F, lanes 5 and 6). ptvR complementation from the replicative plasmid completely blocked the expression of the PtvB protein, even in the absence of the endogenous chromosomal copy of ptvR (Fig. 2F, lane 7). Taken together, these results demonstrated that PtvR represses the transcription of the ptv genes.
The ptv promoter is required for PtvR-mediated transcriptional repression.
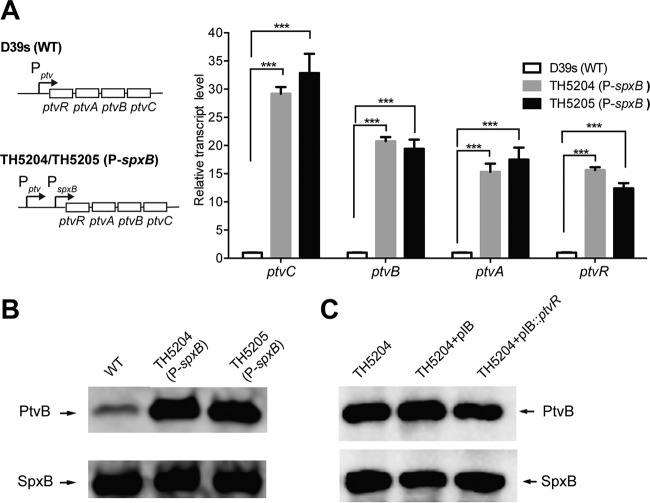
To assess the role of the ptv promoter in PtvR-mediated transcriptional repression, we constructed a promoter replacement derivative in strain D39 in which the ptv promoter was separated from the downstream coding region by inserting the spxB promoter immediately upstream of the ptvR start codon (strains TH5204 and TH5205) (Fig. 3A). We reasoned that insertion of an exogenous promoter immediately upstream of the ptvR start codon would separate the potential cis-acting regulatory/promoter sequence of the operon from the ptv coding region and thereby abolish the repressive impact of PtvR on expression of the ptv operon. The qRT-PCR analysis of TH5204 and TH5205, two clones of the resulting strain, showed a dramatic increase in transcription levels of ptvR (15.7-fold), ptvA (15.3-fold), ptvB (20.8-fold), and ptvC (29.2-fold) in both clonal derivatives of D39s (Fig. 3A). The insertion of the spxB promoter also led to a substantial increase in the amount of the PtvB protein above that in the parental strain D39s (Fig. 3B). In contrast to the repressive effect of the PtvR complementation construct on the expression of PtvB in D39s and its ptvR-deficient mutant, TH5031 (Fig. 2F), the same construct caused no obvious repression on the expression of PtvB in the promoter-bypassing derivative TH5204 of D39s (Fig. 3C). Together with the RNA-Seq data, these results demonstrated that the ptv promoter region is crucial for the PtvR-mediated transcriptional repression in the ptv locus. Because ptvR shares the same promoter with the three downstream genes (ptvA to -C), this finding also revealed that PtvR represses the transcription of its own coding region, an autoregulation mechanism in this locus.
FIG 3.
Overriding of PtvR-dependent transcriptional repression by the spxB promoter. (A) The transcripts of the ptv genes in the spxB promoter insertion strains. The transcripts of ptvR, ptvA, ptvB, and ptvC were quantified by qRT-PCR in strain D39s (wild type [WT]) and two clonal derivatives with an insertion of the spxB promoter sequence immediately upstream of the ptvR start codon (TH5204 and TH5205) as in Fig. 1. ***, P < 0.001. (B) The PtvB protein of the spxB promoter insertion strains was detected by immunoblotting as for Fig. 2F. (C) The PtvB protein of the spxB promoter insertion strain TH5204 in the presence of the empty vector (pIB) or ptvR complementation construct (pIB::ptvR). Proteins were detected as for panel B. Episomal ptvR was unable to repress the expression of PtvB when the ptv promoter was separated from the downstream coding region by the spxB promoter (lane 3).
Transcriptional derepression of the ptvABC genes significantly enhances pneumococcal tolerance to vancomycin.
Based on the transcriptional induction of the ptv genes by vancomycin (Fig. 1) (43), we tested the contribution of this operon to pneumococcal tolerance to vancomycin. The survival rates of mutant strain ΔptvR (TH5031) and promoter-bypassing (TH5204) derivatives of D39s were compared with the parent strain upon exposure to vancomycin (5 μg/ml, 10 times the MIC), a condition that was used in previous studies (30). While both mutant strains with increased ptvABC expression displayed virtually identical growth kinetics as the wild-type D39 in the absence of vancomycin (Fig. 4A, 0 μg/ml), they showed significantly lower levels of vancomycin-induced autolysis, as reflected by the differences in the culture turbidity after addition of vancomycin (Fig. 4A, 5 μg/ml). In addition, the mutant strains displayed significantly higher levels of viable bacteria in the presence of vancomycin for 12 h; no viable pneumococci were detected for any of the three strains after a 16-hour incubation (Fig. 4B, 5 μg/ml). At 6 h, the survival rates of TH5031 and TH5204 were higher than that of D39s by 13- and 20-fold, respectively (Fig. 4B). These data indicated that expression of the ptvABC operon is able to enhance the ability of S. pneumoniae D39 to tolerate a lethal dose of vancomycin.
FIG 4.
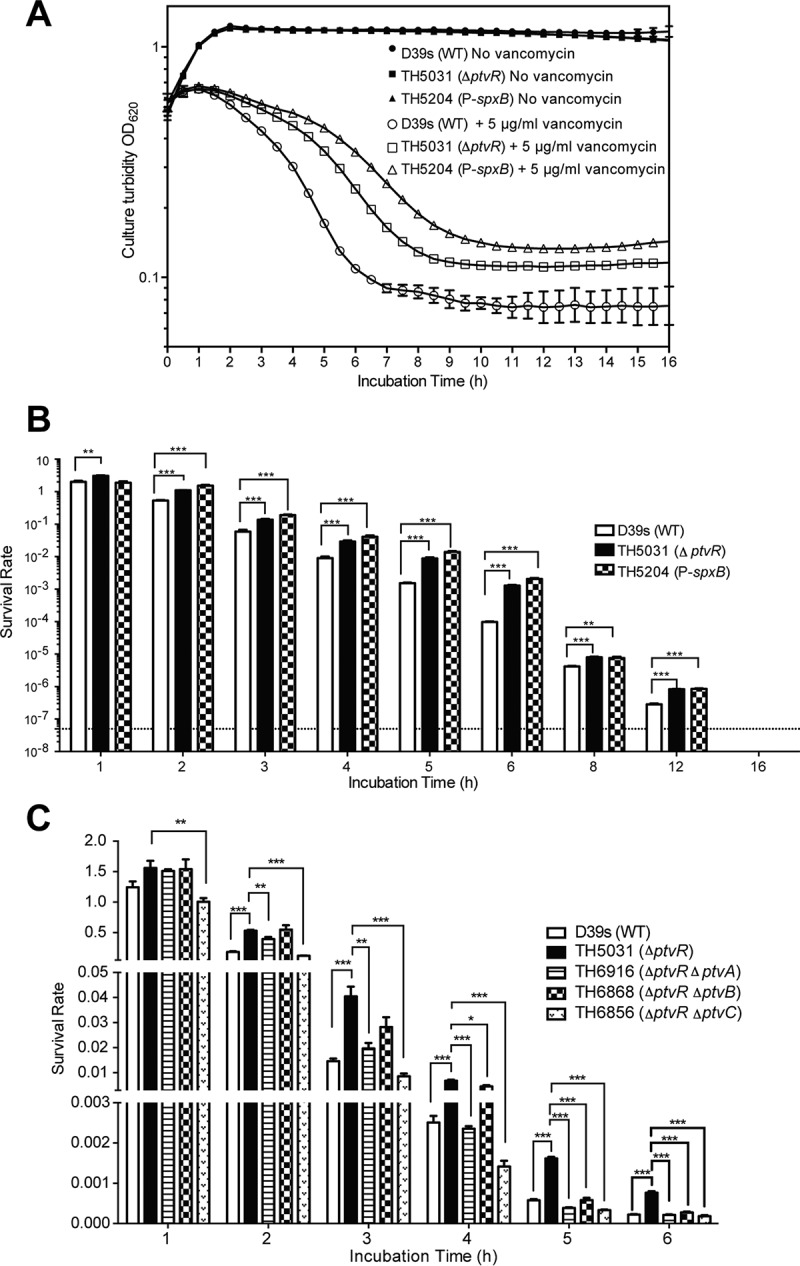
Impact of ptv operon derepression on vancomycin tolerance of S. pneumoniae. (A) Pneumococcal growth and cellular integrity in the presence or absence of vancomycin. Mid-log-phase cultures of strain D39s and its derivatives lacking ptvR (TH5031) or with insertion of the spxB promoter upstream of ptvR (TH5204) in THY broth were mixed with vancomycin to a final concentration of 5 μg/ml. The culture turbidity was monitored by spectrophotometry immediately before (0 h) and after the addition of vancomycin every 0.5 h for 16 h. (B) Survival rates of strain D39s and its derivatives with the derepressed ptv operon in the presence of vancomycin. The experiement was the same as that shown in panel A, except we plated cultures on blood agar dishes at various time points before (0 h) and after (1 to 16 h) the addition of vancomycin. The survival rate represents the CFU ratio of the same cultures before and after addition of vancomycin. The dashed line represents the limit of detection. Asterisks indicate significant increases for strains TH5204 and TH5031 over strain D39s in survival after vancomycin treatment. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Specific contributions of ptvA, ptvB, and ptvC to vancomycin tolerance of S. pneumoniae. D39s and its mutants with an unmarked in-frame deletion(s) of ptvR (TH5031), ptvR and ptvA (TH6916), ptvR and ptvB (TH6868), and ptvR and ptvC (TH6856) were evaluated for their survival rates after treatment with vancomycin for 1 to 6 h, with results displayed as in panel B.
We further determined the impact of different drug concentrations on the ptv-dependent tolerance to vancomycin. Exposure of pneumococci to 0 μg/ml (Fig. S2A) or a low concentration of vancomycin (1 μg/ml, 2 times the MIC) (Fig. S2A) did not yield an obvious impact on culture turbidity for any of the three strains, whereas higher doses of the drug (3, 5, and 10 μg/ml) (Fig. S2A) revealed substantial but similar levels of enhancement in vancomycin tolerance for the ptvR-null (TH5031) and ptvR-bypassing (TH5204) derivatives of D39. We also observed a similar level of increased vancomycin tolerance, based on culture turbidity detection, for the ptv overexpression strains (TH5031and TH5204) with pneumococcal cultures at the exponential phase (optical density at 620 nm [OD620], 0.15) or stationary phase (OD620, 1.0) (data not shown). These results suggested that the ptv locus enhances pneumococcal tolerance to high doses of vancomycin in a drug concentration- and growth phase-independent manner.
To determine the individual impact of ptvA, ptvB, and ptvC on pneumococcal tolerance to vancomycin, we constructed an unmarked in-frame deletion of each gene in the TH5031 (ΔptvR) background and tested the survival capability of each resulting mutant in the presence or absence of vancomycin. Consistent with the aforementioned results (Fig. 4A and B), the ptvR-deficient strain TH5031 was significantly less susceptible to vancomycin (Fig. 4C). However, the enhanced tolerance to vancomycin was abolished in the mutant strain lacking both ptvA and ptvR (TH6916) after 2 h of treatment with a lethal dose of vancomycin. A similar but more pronounced reduction in vancomycin tolerance was observed with the ΔptvC ΔptvR mutant strain (TH6856) throughout the course of the test (Fig. 4C). The ΔptvB ΔptvR mutant strain (TH6868) had the weakest phenotype with regard to vancomycin tolerance among the three mutant strains. The drug tolerance level of TH6868 remained comparable with or marginally different from that of TH5031 (ΔptvR) at the early time points (hours 1 to 4) but became similar to those of the ΔptvA ΔptvR (TH6916) and ΔptvC ΔptvR (TH6856) mutant strains at the last two time points. These results indicated that the ptvA, ptvB, and ptvC genes are all required for pneumococcal tolerance to vancomycin once the operon is transcriptionally derepressed.
Because the expression of the ptvABC genes significantly enhanced the pneumococcal tolerance to vancomycin (Fig. 4), we wondered if this function is applicable to tolerance to other antibiotics. This possibility was tested by comparing the survival rates of D39s and its ΔptvR mutant TH5031 in the presence of lethal doses of penicillin, ampicillin, chloramphenicol, and erythromycin. While no significant difference was observed between TH5031 and the wild-type strain in the presence of penicillin or ampicillin, the ΔptvR mutant showed significantly higher survival rates in the presence of chloramphenicol at the late time points (Fig. S3). Lastly, the experiment with erythromycin yielded a similar difference between TH5031 and the wild-type strain at most of the time points (Fig. S3). These results indicated that expression of the ptvABC genes enhances pneumococcal tolerance to certain nonvancomycin antibiotics (e.g., chloramphenicol and erythromycin) but not others (e.g., penicillin and ampicillin). It is worthwhile to mention that ptvR deletion had a much weaker impact on pneumococcal tolerance to chloramphenicol and erythromycin compared with vancomycin. The differences in survival rates were all within 1.5-fold between the chloramphenicol- or erythromycin-treated ΔptvR mutant strain and the parent strain.
Transcriptional induction of the ptv operon by vancomycin depends on PtvR.
To test whether PtvR is involved in transcriptional induction of the ptv operon by vancomycin, we tested the impact of vancomycin on the transcription of the ptv locus in the presence (Fig. 5A) or absence (Fig. 5B) of ptvR. Vancomycin was mixed with the duplicate cultures of strains with (TH9753) or without (TH9754) ptvR to final concentrations of 0 to 5 μg/ml. While treatment with 0.05 μg/ml vancomycin did not make any obvious difference in the luciferase activity, similar treatment with higher concentrations of the drug (0.5 μg/ml, 2.5 μg/ml, and 5 μg/ml) resulted in a significant increase in the reporter value (Fig. 5A). This result revealed that transcription of the ptv locus is induced by vancomycin when the concentration of the drug is near the MIC value (0.5 μg/ml). In sharp contrast, the isogenic ΔptvR derivative (TH9754) of D39 did not show any increase in the luciferase activity with any of the concentrations of vancomycin tested in this experiment (Fig. 5B). Moreover, the concentrations of vancomycin that induced transcription of the ptv locus also inhibited pneumococcal growth (the OD595) in both reporter strains. This result provides an association between vancomycin-mediated abnormality in cellular structure/metabolism and transcriptional induction of the ptv operon. Together, we conclude from these results that transcriptional induction of the ptv operon by vancomycin requires the transcriptional repressor PtvR of this locus.
FIG 5.
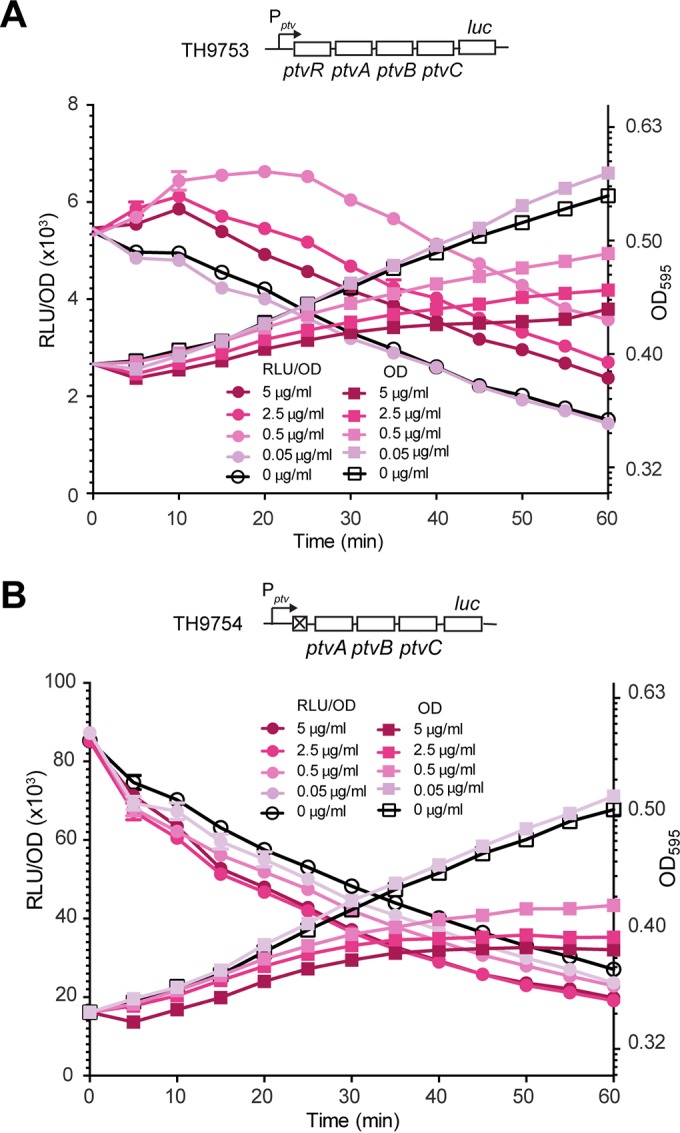
Requirement of ptvR for transcriptional induction of the ptv operon by vancomycin. The transcriptional luc reporter derivatives TH9753 (Pptv_luc) and TH9754 (Δptv mutant strain; Pptv_luc) of D39, constructed in the presence (A) or absence (B) of ptvR, were cultured until the OD595 reached ∼0.4 and subsequently exposed to various concentrations of vancomycin. Cell density and luciferase activity of the cultures are presented as the optical density (OD595) and the relative luminescence units per OD (RLU/OD), respectively.
We tested the impact of vancomycin induction on drug tolerance of the wild-type D39 strain because treatment with certain concentrations of vancomycin (0.5 to 5 μg/ml) significantly induced transcription of the ptv operon (Fig. 5A). Before being exposed to a high concentration of vancomycin (5 μg/ml), D39s and its ptv derivatives were first treated with 0.5 μg/ml vancomycin (the MIC value), a concentration inducing the highest transcription of the ptv operon with a modest inhibition of bacterial growth (Fig. 5A). Consistent with the prior observation (Fig. 4), the ptvR-bypassing strain (TH5204, P-spxB) showed significantly enhanced tolerance to vancomycin after being pretreated with a low dose of vancomycin. However, the pretreated wild-type D39 displayed similar killing kinetics as the ptv-null mutant in terms of both antibiotic-induced autolysis (Fig. S4A) and survival rate (Fig. S4B). This result suggests that vancomycin is not a natural inducer of this operon.
PtvR recognizes the ptv promoter by binding to two palindromic sequences.
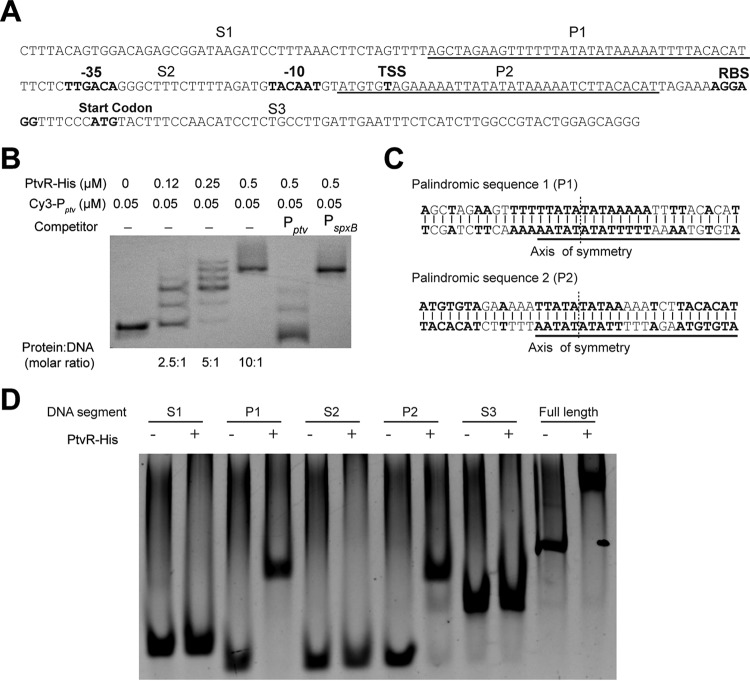
To investigate whether PtvR realizes its repressor function through direct interaction with the ptv promoter, we characterized the promoter and its physical affinity to PtvR. We initially determined the transcriptional start site of the ptv operon by 5′ rapid amplification of cDNA ends (5′ RACE). This sequence was mapped to the 48th T nucleotide upstream of the ptvR start codon (Fig. 6A). The upstream region of the start site contains sequence elements that somewhat resemble canonical −35 (5′-TTGACA-3′) and −10 (5′-TACAAT-3′) promoter motifs. To detect a potential physical interaction of PtvR with the promoter sequence, we performed an electrophoretic mobility shift assay (EMSA) with His-tagged recombinant PtvR and a Cy3-labeled ptv promoter (a 234-bp sequence spanning the translational start codon of ptvR [Fig. 6A]). Addition of His-tagged PtvR (PtvR-His) led to an obvious mobility shift of the ptv promoter sequence in a dose-dependent manner, which was indicative of direct binding of the ptv promoter with the repressor (Fig. 6B, lanes 1 to 4). Furthermore, the mobility shift of the DNA fragment was blocked by adding excess unlabeled ptv promoter sequence (Pptv) but not by the spxB promoter sequence (PspxB). Multiple bands were observed with the reaction mixtures that contained lower PtvR concentrations (e.g., 0.13 and 0.25 μM), which may have been caused by variabilities in the number of PtvR proteins bound to the promoter sequence, as described previously for the promoter-PadR interaction in Bacillus subtilis (48). The EMSA result revealed that PtvR is able to bind to the ptv promoter sequence.
FIG 6.
Specific recognition by PtvR of two palindromic sequences in the ptv promoter. (A) The diagrammatic illustration of the sequence elements (234 bp) spanning the translational start codon of ptvR. In bold are the predicted promoter motifs (−35 and −10), transcriptional start site (TSS), ribosome-binding site (RBS), and start codon of ptvR. The spacing sequences (S1, S2, and S3) are separated by two palindromic sequences (P1 and P2). (B) Biochemical binding of PtvR with the full-length ptv promoter, as determined in an EMSA. A recombinant PtvR protein was used to test binding with the Cy3-labeled ptv promoter; unlabeled promoter sequences of the ptv (Pptv) and spxB (PspxB) genes were used as competitors. (C) Sequences of the P1 and P2 elements in the ptv promoter. The 24-bp repeat sequence in P1 and P2 is underlined. The axis of P1-P2 symmetry is marked with dashed lines. (D) EMSA results with the recombinant PtvR and various segments of the ptv promoter sequence, as depicted in panel A. DNA was detected by staining with GelRed.
Sequence analysis revealed two 36-bp palindromic sequences (P1 and P2) in the ptv promoter (Fig. 6A). P1 and P2 are separated by a 36-bp sequence (Fig. 6A). While P1 is located 5 bp upstream of the predicted −35 promoter motif, P2 is found 2 bp downstream of the −10 promoter motif. Interestingly, P2 overlaps with the transcriptional start site. P1 and P2 also share a 24-bp repeat sequence, with only a 1-nucleotide discrepancy (Fig. 6C, underlined). We further determined the PtvR-binding sequence motifs in the ptv promoter sequence by performing EMSAs with 5 segments of this region: S1 (46 bp), P1 (36 bp), S2 (36 bp), P2 (36 bp), and S3 (80 bp). P1 and P2, along with the full-length ptv promoter sequence (as a positive control), showed a dramatic shift in the presence of PtvR, whereas individually mixing PtvR with each of the three spacing DNA fragments (S1, S2, and S3) did not result in an apparent shift (Fig. 6D). All together, these results showed that PtvR specifically binds to the ptv promoter by recognizing the P1 and P2 palindromic sequences.
DISCUSSION
Previous studies have implicated that phenotypic tolerance of S. pneumoniae and other pathogenic bacteria can significantly affect clinical treatment of the infections caused by these pathogens (4, 37). While phenotypic antibiotic tolerance has been well characterized as a property of persisters in E. coli (4, 10), the genetic basis of phenotypic antibiotic tolerance in S. pneumoniae is currently unknown. This study shows that the ptv operon of S. pneumoniae contributes to phenotypic tolerance to vancomycin in an inducible manner. Exposure of the pneumococci to vancomycin led to a significant increase in the transcription of this operon. RNA-Seq analysis and subsequent experiments revealed that the operon is autoregulated by PtvR, which is encoded by ptvR, the first gene of the operon. Full derepression of the ptv locus enabled pneumococci to become tolerant to vancomycin at a level as high as 10 μg/ml, a concentration exceeding what is clinically achievable in cerebral spinal fluid (5 μg/ml) (49). Vancomycin is frequently used to treat bacterial meningitis caused by β-lactam-resistant S. pneumoniae (31). Interestingly, further analysis showed that the vancomycin-induced upregulation of the ptv operon relies on PtvR. These lines of evidence suggest that the ptv operon could contribute to phenotypic tolerance to vancomycin of S. pneumoniae in an inducible fashion, and the induction of the ptv operon is achieved by derepression of PtvR-mediated autoregulation in response to an unknown environmental cue(s). To the best of our knowledge, the ptv operon represents the first autoregulated locus in S. pneumoniae that contributes to phenotypic antibiotic tolerance.
The transcriptional repression activity of PtvR agrees with its sequence homology, with many DNA-binding transcriptional regulators of the PadR family (Pfam accession PF03551). PadR represses transcription of the phenolic acid detoxification locus in B. subtilis and several other Gram-positive bacteria (50–52). Other members of this protein family are shown to regulate diverse processes, including multidrug resistance (53–55), virulence (56), and cellular metabolism (57, 58). Many PadR family regulators repress the transcription of the target genes by binding to specific sequences of the promoter regions with a conserved N-terminal winged helix-turn-helix domain and thereby blocking the promoter-RNA polymerase interactions in the absence of inducers (e.g., phenolic acid and antibiotic) (46, 59). Transcriptional derepression occurs when the regulator “falls off” the target DNA sequence after binding to the cognate inducers (e.g., phenolic acid and other toxic compounds) (46, 59). The four copies of the B. subtilis PadR repressor are predicted to bind to four different sequence motifs in each promoter sequence of the phenolic acid response gene locus in the absence of phenolic acid (inducer) (48). PtvR showed 36% identity with LmrR, a PadR family protein studied structurally and functionally (55, 59). Further investigation on PtvR in this study showed that PtvR binds to two palindromic sequences (P1 and P2) in the ptv promoter, each of which contains two inverted repeats. Interestingly, P1 is located 5 bp upstream of the predicted −35 promoter motif, and P2 overlaps with the transcriptional start site. P1 and P2 also share a 24-bp repeat sequence with only a 1-nucleotide discrepancy (Fig. 6). PtvR contains a C-terminal alpha-helix motif that can form a dimer, and our gel filtration result confirmed the existence of the PtvR dimer (data not shown), similar to LmrR (59). Taken together, all of the above observations indicate that each ptv promoter may recruit four copies of the apo form of PtvR, which would prevent RNA polymerase from accessing the major promoter motifs (−35 sequence, −10 sequence, and transcriptional start site).
PtvA, PtvB, and PtvC are all predicted to be associated with the cell membrane of S. pneumoniae. PtvA and PtvB are annotated as conserved hypothetical proteins in the genome of D39. While PtvA is homologous to many uncharacterized membrane proteins without any obvious function, the first half of PtvB contains a domain of unknown function (DUF4098) shared by many bacterial proteins. PtvC is annotated as a putative membrane protein with significant similarity with the merozoite surface protein in Plasmodium falciparum (60). Because individual deletion of ptvA, ptvB, or ptvC resulted in reduction of vancomycin tolerance and they all contain predicted transmembrane domains, it is tempting to hypothesize that PtvA, PtvB, and PtvC could cooperatively contribute to antibiotic tolerance by modulating the properties of the pneumococcal cell membrane through formation of a membrane-associated structure(s) or modification of unknown membrane molecules. As an example, changes in the cell surface charge are associated with reversible tolerance of S. aureus to antibiotics (23).
An unknown molecule other than vancomycin may act as the natural inducer(s) that directly interacts with PtvR to derepress the transcription of the operon when pneumococci are under stressful conditions, such as those caused by lysozyme and antimicrobial peptides produced by the host. These enzymes and peptides often damage the cell wall or even form pores in the bacterial cell membrane and might influence the proton motive force that then triggers expression of the ptv operon. Along this line, the transcriptional derepression of the ptv locus upon vancomycin exposure may reflect a response to cell envelope stress caused by the antibiotic. This hypothesis is supported by multiple lines of evidence. First, vancomycin treatment led to partial derepression of the operon compared with full derepression by the deletion of ptvR. This result is consistent with our observation that inducing the ptv operon by pretreatment with vancomycin did not lead to obvious enhancement in vancomycin tolerance of the wild-type strain D39. Second, vancomycin does not effectively penetrate cell membranes, which explains its poor efficacy against Gram-negative bacteria and intracellular S. aureus (61, 62). Therefore, vancomycin is unlikely to interact directly with PtvR, which is presumably located in the cytoplasm of the pneumococci. Lastly, addition of vancomycin in the EMSA did not influence the binding of PtvR to the ptv promoter (data not shown). This result argues for a scenario in which the ptv locus is responsive to multiple stress conditions. This notion is also supported by a previous report that the transcription of the genes in the ptv locus is significantly repressed when S. pneumoniae is exposed to acidic conditions (63).
The phenotypic antibiotic tolerance regulated by PtvR differs from antibiotic tolerance of the persisters, as defined in Gram-negative bacteria (4, 10). The tolerance of persisters to bactericidal antibiotics appears to occur in a drug-independent manner (11). However, among the antibiotics tested in this study, the tolerance mediated by the ptv operon was most effective for vancomycin and, to a much lesser extent, toward chloramphenicol and erythromycin. In an opposite manner, the expression of the ptv operon did not affect pneumococcal susceptibility to penicillin and ampicillin. These results implicate that vancomycin and β-lactams kill pneumococci by different mechanisms, although the LytA autolysin is required for the bactericidal activities of both vancomycin and penicillin (27, 64). Alternatively, persisters in Gram-negative and -positive bacteria may be formed through different mechanisms, and/or behave differently in terms of tolerance efficacy to different bactericidal drugs, as evidenced in a recent study of persister cells in Staphylococcus aureus (22). Taken together, future investigations are warranted to decipher whether (i) induction of the ptv operon by a natural signal(s) confers vancomycin tolerance in wild-type pneumococci, (ii) the increased vancomycin tolerance observed in the ptv derepression/overexpression strains is related to LytA, (iii) induction of this operon is mechanistically related to the drug tolerance of typical persisters.
The four genes in the ptv operon not only are conserved in all S. pneumoniae strains tested thus far but also exist in many other Streptococcus species as partial or complete gene sets. There is a complete set of ptvRABC in each of eight Streptococcus species that are closely related to S. pneumoniae, known as the “great mitis group” (S. pseudopneumoniae, S. mitis, S. oralis, S. infantis, S. dentisani, S. peroris, S. tigurinus, and S. parasanguinis). Many other Streptococcus species possess only ptvR and ptvA (and not ptvB and ptvC), including the human pathogens S. pyogenes and S. suis. The partial presence of ptvR and ptvA in the majority of Streptococcus species indicates that PtvA can function in the absence of PtvB and PtvC, or it has evolved to partner with other proteins in these species. The conservative nature of this locus implies that the ptv genes are important for the fitness of this genus, perhaps in coping with antibiotics and/or other stressful agents. The phylogenetic-dependent distribution of the ptv genes may reflect variable functional contributions of this locus to the dynamic lifestyles of different Streptococcus species.
MATERIALS AND METHODS
Bacteria and chemical reagents.
The strains and plasmids used in this study are listed in Table 1. The pneumococci were grown at 37°C under aerobic conditions with 5% CO2 in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) broth or on plates containing Trypticase soy agar with 5% sheep blood, as described previously (65). For the luciferase assay, C+Y medium (66) was used instead of THY broth. Escherichia coli was cultured in LB medium or on LB agar plates at 37°C under aerobic conditions in the presence or absence of chloramphenicol (20 μg/ml) or ampicillin (100 μg/ml) as described previously (67). All ingredients for bacterial culture media and other chemicals used in this work were obtained from Sigma (Shanghai, China) unless otherwise stated. All DNA enzymes were purchased from New England BioLabs (NEB; Beijing, China). All oligonucleotides were synthesized by Sangon Biotech (Beijing, China) and are listed in Table S1 in the supplemental material.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Descriptiona | Antibiotic resistanceb | Source |
|---|---|---|---|
| S. pneumoniae strains | |||
| D39 | NCTC7466, serotype 2, encapsulated, TH4533 | NCTC | |
| D39s | D39 derivative, rpsL1, TH4527 | Sm | 73 |
| ST556 | S. pneumoniae type 19F | 45 | |
| TH4737 | D39s derivative, ΔptvR-C::JC (replacement of entire ptvR-C coding region with JC) | Kan | This study |
| TH4631 | D39s derivative, ΔptvR::JC (replacement of ptvR coding region with JC) | Kan | This study |
| TH5139 | D39s derivative, JC insertion in front of ptvR start codon | Kan | This study |
| TH6854 | TH5031 derivative, ΔptvR ΔptvC::JC (replacement of ptvC with JC in TH5031) | Kan | This study |
| TH6855 | TH5031 derivative, ΔptvR ΔptvB::JC (replacement of ptvB with JC in TH5031) | Kan | This study |
| TH6910 | TH5031 derivative, ΔptvR ΔptvA::JC (replacement of ptvA with JC in TH5031) | Kan | This study |
| TH4741 | TH4737 derivative, ΔptvR-C (unmarked deletion of the entire ptvR-C coding region) | Sm | This study |
| TH5031 | TH4631 derivative, ΔptvR (unmarked deletion of ptvR) | Sm | This study |
| TH5204 | TH5139 derivative, unmarked insertion of P-spxB immediately before first nucleotide of ptvR start codon | Sm | This study |
| TH5205 | Same as TH5204, clonal sibling | Sm | This study |
| TH6916 | TH5031 derivative, ΔptvR ΔptvA (unmarked deletions of ptvR and ptvA) | Sm | This study |
| TH6868 | TH5031 derivative, ΔptvR ΔptvB (unmarked deletions of ptvR and ptvB) | Sm | This study |
| TH6856 | TH5031 derivative, ΔptvR ΔptvC (unmarked deletions of ptvR and ptvC) | Sm | This study |
| TH6908 | D39s derivative D39s::pTH6862 containing episomal ptvR in pTH6862 | Cm, Sm | This study |
| TH6881 | TH5031 derivative TH5031::pTH6862 containing episomal ptvR in pTH6862 | Cm, Sm | This study |
| TH6876 | TH5204 derivative TH5204::pTH6862 containing episomal ptvR in pTH6862 | Cm, Sm | This study |
| TH6906 | D39s derivative D39s::pIB166 containing empty plasmid pIB166 | Cm, Sm | This study |
| TH6879 | TH5031 derivative TH5031::pIB166 containing empty plasmid pIB166 | Cm, Sm | This study |
| TH6875 | TH5204 derivative TH5204::pIB166 containing empty plasmid pIB166 | Cm, Sm | This study |
| TH9753 | TH4527 derivative, ptv::luc | Kan | This study |
| TH9754 | TH5031 derivative, ptv::luc | Kan | This study |
| TH9755 | TH4527 derivative, ptv::luc bgaA::PZn2+-ptvR | Kan, Tet | This study |
| TH9756 | TH5031 derivative, ptv::luc bgaA::PZn2+-ptvR | Kan, Tet | This study |
| MK134 | D39 PssbB-ssbB_luc Kanr | Kan | 70 |
| E. coli TH5272 | Rosetta containing pET22b::ptvR | Amp | This study |
| Plasmids | |||
| pIB166 | E. coli-S. pneumoniae shuttle vector | Cm | 69 |
| pTH6862 | pIB166 derivative expressing C-terminally His-tagged PtvR driven by spxB promoter, pIB166::ptvR | Cm, Sm | This study |
| pTH2700 | pGEX-2T derivative with a modified multiple-cloning site | Amp | 77 |
| pTH2604 | pTH2700 derivative pTH2700::ptvB expressing N-terminally GST-tagged PtvB | Amp | This study |
| pTH2760 | pTH2700 derivative pTH2700::spxB expressing N-terminally GST-tagged SpxB | Amp | This study |
JC, Janus cassette.
Antibiotic abbreviations: Amp, ampicillin; Cm, chloramphenicol; Kan, kanamycin; Sm, streptomycin; Tet, tetracycline.
Construction of pneumococcal mutants and the ptvR complementation construct.
All of the pneumococcal mutant strains were constructed in D39s, a streptomycin-resistant derivative of strain D39, as described previously (65). The replacement mutant TH4737 of the ptvR-C operon region was generated by amplifying the up- and downstream regions with the primer pairs Pr7187/Pr7188 and Pr7197/Pr7198, digesting the amplicons with XbaI and XhoI, and ligating the resulting PCR products to the XbaI/XhoI-digested amplicon of the Janus cassette (JC; kan-rpsL+) before being used to transform strain TH4527 (D39s) as described previously (68). JC was amplified from S. pneumoniae ST588 (65) with primers Pr1097 and Pr1098. The unmarked ptv operon deletion mutant strain TH4741 was constructed by a counterselection method (68) through transformation of TH4737 with a fusion PCR product of two separate PCR amplicons, which was generated by amplifying the up- and downstream sequences of ptvR-C with the primer pairs of Pr7187/Pr7275 and Pr7276/Pr7198, respectively, and ligating the amplicons by fusion PCR with the most-upstream (Pr7187) and most-downstream (Pr7198) primers as described previously (65). The same procedure was used to generate sequentially the JC replacement and unmarked deletion mutants in other target sequences. The JC replacement (TH4631) and unmarked (TH5031) mutant strains for ptvR were prepared with primer sets Pr7189/Pr7190 with Pr7187/Pr7188 and Pr7187/Pr7207 with Pr7190/Pr7208, respectively. The JC replacement mutants in the downstream genes were generated in the TH5031 background in a similar manner with primer sets of Pr7195/Pr7196 with Pr7197/Pr7198 (TH6854; ΔptvC::JC ΔptvR), Pr8515/Pr8516 with Pr8517/Pr8518 (TH6855; ΔptvB::JC ΔptvR), and Pr8551/Pr8550 with Pr8514/Pr8513 (TH6910; ΔptvA::JC ΔptvR). The counterselections were carried out to remove JC in the replacement strains and create unmarked deletion mutants with the fusion PCR products amplified with the primer sets of Pr7195/Pr7212 and Pr7213/Pr7198 (TH6856; ΔptvC ΔptvR), Pr8515/Pr8522 and Pr8518/Pr8521 (TH6868; ΔptvB ΔptvR), and Pr8551/Pr8548 and Pr8547/Pr8514 (TH6916; ΔptvA ΔptvR). The spxB promoter insertion strains TH5204 and TH5205 were generated as follows. The upstream sequence of the ptvR start codon and the ptvR coding region were amplified with primer pairs Pr8059/Pr8060 and Pr8061/Pr8062, digested with XbaI and XhoI, and ligated to the XbaI/XhoI-digested amplicon of JC as described above. Transformation of the ligation product into D39s resulted in strain TH5139, which was subsequently used to knock-in the spxB promoter (P-spxB) in front of the ptvR start codon. The P-spxB and ptvR coding region were separately amplified from genomic DNA of D39 with primer sets of Pr8065/Pr8139 and Pr8064/Pr8062, respectively, and subsequently linked by overlap PCR using primers Pr8139 and Pr8062. The P-spxB-ptvR amplicon was digested with XhoI and then ligated with the XhoI-digested amplicon of the sequence immediately upstream of the ptvR start codon (amplified with primers Pr8059 and Pr8121). The ligation product was used for transformation of TH5139 to generate strains TH5204 and TH5205 by counterselection.
The spxB promoter-driven complementation construct pTH6862 was prepared in the E. coli-Streptococcus shuttle vector pIB166 (69) by amplifying the hybrid sequence of the spxB promoter and ptvR coding region from genomic DNA of TH5204, using primers Pr10379 and Pr10380 and cloning the amplicon in the XmaI/ApaI site of pIB166. All of the resulting mutant strains and plasmids were confirmed by DNA sequencing.
For construction of luciferase reporter strains, the luc gene insertion fragment was generated as follows: first, a DNA fragment containing luc and kanamycin resistance marker was amplified from genomic DNA of MK134 (70) with primers Pr12576 and Pr12577, followed by BglII and NotI digestion. Second, upstream and downstream fragments of the insertion site were amplified from the genomic DNA of strain D39 with primer pairs Pr12578/Pr12579 and Pr12580/Pr12581, followed by BglII and NotI digestion, respectively. Finally, the three fragments were ligated before the entire fragment was transformed into strains TH4527 and TH5031, generating strains TH9753 and TH9754, respectively. Based on strains TH9753 and TH9754, complementation strains of ptvR, TH9755 and TH9756, were constructed as follows: ptvR was amplified with primers Pr12382 and Pr12583 from genomic DNA of D39, followed by digestion with EcoRI and SpeI. The digested PCR fragment was subcloned into pMK11 (71) and transformed into TH9753 and TH9754, resulting in TH9755 and TH9756, respectively.
RNA extraction and transcriptome analysis.
Total RNA was isolated from mid-log-phase cultures of S. pneumoniae (OD620, ∼0.5) with TRIzol reagent (Invitrogen, Beijing, China) based on the supplier's instructions. The amount, purity, and integrity of all RNA samples isolated were assessed by standard RNA gel electrophoresis (72), detection of the A260/A280 ratio by using a ThermoFisher Scientific Nanodrop 2000 spectrophotometer and Agilent 2100 BioAnalyzer. RNA-Seq was carried out using the Illumina Hiseq 2500 sequencer at the Tsinghua University Genomics Center according to the manufacturer's instructions. Briefly, rRNA was removed from total RNA (∼2 μg) with the RiboZero magnetic kit (EpiCentre, Cleveland, OH). The processed RNA samples were used to prepare double-stranded cDNA libraries with Moloney murine leukemia virus reverse transcriptase and the NEBNext mRNA second-strand synthesis module (27). The cDNA molecules were fragmented by sonication and further processed for end repair, adaptor attachment, and sequence amplification. The quality of the raw reads was checked by using FastQC v.0.11.5 (Babraham Bioinformatics, UK) followed by read trimming with Trimmomatic v.0.32. Trimmed reads were mapped to the reference genome of S. pneumoniae D39 (NC_008533) by using Rockhopper 2.0.3. The complete RNA-Seq result is described in Table S2 in the supplemental material.
Intergenic PCR and qRT-PCR.
The cotranscription of the ptvR-C genes was assessed by intergenic RT-PCR as described previously (73). A cDNA library was prepared using the total RNA of S. pneumoniae D39 (described above) with an iScript cDNA synthesis kit (Bio-Rad, Beijing, China) according to the supplier's instructions and used to amplify the intergenic regions of the ptvR-C genes with the following primer pairs: ptvR-ptvA (Pr7879/Pr7880), ptvA-ptvB (Pr7881/Pr7882), and ptvB-ptvC (Pr7883/Pr7884). A negative control was included for each reaction mixture, with a mixture without reverse transcriptase as a template.
The ptv transcripts were quantified by qRT-PCR using Taq Universal SYBR green supermix (Bio-Rad) as described previously (74) with the following primer pairs: ptvR (Pr7878/Pr7979), ptvA (Pr7881/Pr8202), ptvB (Pr8203/Pr7891), and ptvC (Pr7893/Pr8201). The S. pneumoniae era gene was amplified with primers Pr7932 and Pr7933 as a reference for quantification. The expression level of each target gene was calculated using the average mean cycle threshold value (CT) for the genes of interest for each sample and normalized to the value with the reference gene, according to the following equation: 1.8e(CT era − CT gene of interest). The relative gene transcription levels of each gene after a 10- or 20-min treatment with vancomycin were obtained by dividing data by the expression values for untreated cells. The results of representative experiments are presented as means from three replicates ± standard deviations.
5′-RACE.
The 5′ terminus of mRNA or the transcriptional start site of the ptvR-C operon was identified by 5′-rapid amplification of cDNA ends (5′-RACE), using the SMARTer RACE 5′/3′ kit (Clontech, Mountain View, CA) as instructed by the supplier. A cDNA library was prepared with 1 μg of total RNA of S. pneumoniae D39, with SMARTScribe reverse transcriptase as described previously (75), and used to amplify the 5′ terminus of the ptvR transcript with primer Pr10383 and a universal primer. The PCR product was cloned into pRACE.
Immunoblotting.
Bacterial proteins were detected by immunoblotting as described previously (65). Pneumococcal cultures were grown in THY broth to mid-log phase (OD620 of ∼0.5). The cells were harvested by centrifugation at 4°C and washed with cold phosphate-buffered saline (PBS) before being processed for cell lysis, SDS-PAGE, and immunoblotting. The PtvB and SpxB proteins were detected with polyclonal antisera prepared for this study (1:2,000 dilution of anti-PtvB serum and 1:30,000 dilution of anti-SpxB serum), with a peroxidase-conjugated secondary goat anti-rabbit IgG at a dilution of 1:10,000 (Bio-Rad). Protein bands were visualized by the Clarity Western enhanced chemiluminescence reagent (Bio-Rad) according to the supplier's instructions.
The antisera against PtvB and SpxB were prepared with recombinant forms of the proteins in New Zealand White rabbits, essentially as described previously (76). PtvB and SpxB were expressed as glutathione S-transferase (GST) (67) fusion proteins in E. coli cells by amplifying their coding regions from D39 genomic DNA with the primer sets Pr2773/Pr2774 (PtvB) and Pr3129/Pr3130 (SpxB) and cloning each of the PCR products in the NotI/AscI site of plasmid pTH2700 (Table 1), a modified pGEX-2T plasmid with additional cloning sites. pTH2700 was constructed by insertion of a multiple-cloning site (MCS) sequence into the BamHI/EcoRI site of pGEX-2T (GE Healthcare BioScience, Piscataway, NJ). The MCS fragment was generated by annealing reverse complementary oligonucleotides Pr3110 and Pr3111. The resulting plasmids pTH2604 (PtvB) and pTH2760 (SpxB) were used to produce recombinant proteins by affinity chromatography with glutathione-Sepharose 4 Fast Flow resins (GE Healthcare BioSciences) according to the supplier's instructions. The fusion proteins were visualized by SDS-PAGE, quantified with a bicinchoninic acid (BCA) assay kit (Solarbio, Beijing, China), and used to immunize rabbits for antiserum production. Immunoblotting detection of pneumococcal proteins was carried out as described previously (76).
EMSA.
The C-terminal His-tagged PtvR was expressed in E. coli Rosetta (strain TH5272) cells with recombinant plasmid pET22b::ptvR. To construct pET22b::ptvR, ptvR was amplified from genomic DNA of D39 with primers Pr8171 and Pr8172, digested with NdeI and XhoI, and ligated into similarly digested pET22b. The His-tagged PtvR was purified by affinity chromatography as described previously (77). The 234-bp ptv promoter with and without Cy3 labeling at the 5′ end was amplified with primer pairs Pr10024/Pr10025 and Pr10025/Pr12588, respectively, from genomic DNA of D39 and purified by gel extraction. The binding reaction was performed in a total volume of 20 μl binding buffer essentially as described previously (48, 78). Briefly, His-tagged PtvR and each of the promoter sequences were used at the final concentrations described in Results. For the Cy3-labeled DNA, the promoter sequence of ptv (Pptv) or spxB (PspxB) was added to a final concentration of 0.5 μM as a competitive DNA. PspxB was amplified with primers Pr12584 and Pr12585 from genomic DNA of D39. After 20 min of incubation at 28°C, the samples were processed in 6% polyacrylamide gels to separate the free DNA and that bound to PtvR, and samples were scanned with a Fuji LAS-4000 fluorescence imaging system. Similar procedures were used for an electrophoretic mobility shift assay (EMSA) with the ptv promoter segments, with the following exceptions: (i) higher concentrations of PtvR (50 μM) and target sequences (5 μM), (ii) use of unlabeled DNA fragments, and (iii) detection of DNA by staining with GelRed and scanning with a Bio-Rad molecular imager ChemiDoc XRS system. Target sequences for EMSA were initially synthesized as single-stranded oligonucleotides and subsequently annealed as double-stranded DNA fragments: P1 (Pr10657 and Pr10658), P2 (Pr10661 and Pr10662), S1 (Pr10655 and Pr10656), S2 (Pr10659 and Pr10660), and S3 (Pr10663 and Pr10664), as described previously (67).
Luciferase assay.
Luciferase activity was detected as described previously (47). Briefly, pneumococci were cultured in C+Y broth to an OD600 of 0.1, followed by 1:10 dilution into fresh C+Y medium with or without 0.1 mM ZnCl2–0.01 mM MnCl2. To detect induction of ptv by vancomycin, pneumococci were similarly cultured to an OD600 of 0.4, followed by addition of vancomycin to different final concentrations. Each bacterial culture (250 μl) was mixed with 50 μl of luciferin solution (2.7 mg/ml; d-luciferin sodium salt from Synchem OHG) in 96-well plates in triplicates. The optical density at 595 nm and luminescence were detected by using a Tecan Infinite F200 Pro microtiter plate reader.
Antibiotic tolerance assay.
Antibiotic tolerance of S. pneumoniae was assessed essentially as described previously (30). Pneumococci were grown to an OD620 of 0.2 (early log phase) or 0.5 (late log phase) in THY broth. Aliquots of the cultures were mixed with each of the following antibiotics: vancomycin (5 μg/ml), penicillin (0.12 μg/ml), ampicillin (0.5 μg/ml), chloramphenicol (10 μg/ml), or erythromycin (0.6 μg/ml). Culture turbidity and bacterial viability were monitored every hour by measuring optical density (OD620) and plating for CFU. The level of antibiotic tolerance was calculated by comparing the mean values of four replicates immediately before and at different time points after the addition of each antibiotic.
Statistical analysis.
Statistical significance of the data from the qRT-PCR and antibiotic tolerance experiments was determined via two-tailed unpaired Student's t test. Significant differences were defined by P values of <0.05, <0.01, and <0.001.
Accession number(s).
The raw data from our RNA-Seq analysis are available from the NCBI GEO database under accession number GSE95709.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the staff members in the Tsinghua Genomic Center for their assistance with RNA-Seq.
This work was supported by grants from the National Natural Science Foundation of China (no. 81671972 and no. 31530082), the Ministry of Science and Technology of China (no. 2012CB518702), the Ministry of Education of China (no. 2012Z02293), and the Grand Challenges Exploration of the Bill and Melinda Gates Foundation (no. OPP1021992).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00054-17.
REFERENCES
- 1.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun MN, Levy SB. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 4.Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat Rev Microbiol 4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 5.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 6.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauvart M, De Groote VN, Michiels J. 2011. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- 8.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilization. Lancet ii:497–500. [Google Scholar]
- 9.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 10.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 11.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 12.Germain E, Roghanian M, Gerdes K, Maisonneuve E. 2015. Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci U S A 112:5171–5176. doi: 10.1073/pnas.1423536112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 14.Dorr T, Lewis K, Vulic M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet 5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J Bacteriol 192:1946–1955. doi: 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vega NM, Allison KR, Khalil AS, Collins JJ. 2012. Signaling-mediated bacterial persister formation. Nat Chem Biol 8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. 2004. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 19.Debbia EA, Roveta S, Schito AM, Gualco L, Marchese A. 2001. Antibiotic persistence: the role of spontaneous DNA repair response. Microb Drug Resist 7:335–342. doi: 10.1089/10766290152773347. [DOI] [PubMed] [Google Scholar]
- 20.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versees W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 23.Haaber J, Friberg C, McCreary M, Lin R, Cohen SN, Ingmer H. 2015. Reversible antibiotic tolerance induced in Staphylococcus aureus by concurrent drug exposure. mBio 6:e02268–. doi: 10.1128/mBio.02268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Gotz F. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 3:e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 26.Leung V, Ajdic D, Koyanagi S, Levesque CM. 2015. The formation of Streptococcus mutans persisters induced by the quorum-sensing peptide pheromone is affected by the LexA regulator. J Bacteriol 197:1083–1094. doi: 10.1128/JB.02496-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernebro J, Andersson I, Sublett J, Morfeldt E, Novak R, Tuomanen E, Normark S, Normark BH. 2004. Capsular expression in Streptococcus pneumoniae negatively affects spontaneous and antibiotic-induced lysis and contributes to antibiotic tolerance. J Infect Dis 189:328–338. doi: 10.1086/380564. [DOI] [PubMed] [Google Scholar]
- 28.Sorg RA, Veening JW. 2015. Microscale insights into pneumococcal antibiotic mutant selection windows. Nat Commun 6:8773. doi: 10.1038/ncomms9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas W, Sublett J, Kaushal D, Tuomanen EI. 2004. Revising the role of the pneumococcal vex-vncRS locus in vancomycin tolerance. J Bacteriol 186:8463–8471. doi: 10.1128/JB.186.24.8463-8471.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson GT, Zhao J, Desai BV, Coleman WH, Nicas TI, Gilmour R, Grinius L, Morrison DA, Winkler ME. 2002. Vancomycin tolerance induced by erythromycin but not by loss of vncRS, vex3, or pep27 function in Streptococcus pneumoniae. J Bacteriol 184:6987–7000. doi: 10.1128/JB.184.24.6987-7000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musher DM. 2010. Streptococcus pneumoniae, p 2623–2642. In Mandell GL, Bennett JE, Dolin RD (ed), Principles and practice of infectious diseases, 7th ed, vol 2 Elsevier Churchill Livingstone, New York, NY. [Google Scholar]
- 32.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 33.Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O'Brien KL, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song JH, Dagan R, Klugman KP, Fritzell B. 2012. The relationship between pneumococcal serotypes and antibiotic resistance. Vaccine 30:2728–2737. doi: 10.1016/j.vaccine.2012.01.091. [DOI] [PubMed] [Google Scholar]
- 35.Tan TQ. 2003. Antibiotic resistant infections due to Streptococcus pneumoniae: impact on therapeutic options and clinical outcome. Curr Opin Infect Dis 16:271–277. doi: 10.1097/00001432-200306000-00015. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan SL. 2002. Management of pneumococcal meningitis. Pediatr Infect Dis J 21:589–591. doi: 10.1097/00006454-200206000-00034. [DOI] [PubMed] [Google Scholar]
- 37.Moscoso M, Domenech M, Garcia E. 2011. Vancomycin tolerance in Gram-positive cocci. Environ Microbiol Rep 3:640–650. doi: 10.1111/j.1758-2229.2011.00254.x. [DOI] [PubMed] [Google Scholar]
- 38.Moscoso M, Domenech M, Garcia E. 2010. Vancomycin tolerance in clinical and laboratory Streptococcus pneumoniae isolates depends on reduced enzyme activity of the major LytA autolysin or cooperation between CiaH histidine kinase and capsular polysaccharide. Mol Microbiol 77:1052–1064. doi: 10.1111/j.1365-2958.2010.07271.x. [DOI] [PubMed] [Google Scholar]
- 39.Sung H, Shin HB, Kim MN, Lee K, Kim EC, Song W, Jeong SH, Lee WG, Park YJ, Eliopoulos GM. 2006. Vancomycin-tolerant Streptococcus pneumoniae in Korea. J Clin Microbiol 44:3524–3528. doi: 10.1128/JCM.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eagle H. 1949. The effect of the size of the inoculum and the age of the infection on the curative dose of penicillin in experimental infections with streptococci, pneumococci, and Treponema pallidum. J Exp Med 90:595–607. doi: 10.1084/jem.90.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eagle H, Fleischman R, Musselman AD. 1950. The bactericidal action of penicillin in vivo: the participation of the host, and the slow recovery of the surviving organisms. Ann Intern Med 33:544–571. doi: 10.7326/0003-4819-33-3-544. [DOI] [PubMed] [Google Scholar]
- 42.Eagle H. 1952. Experimental approach to the problem of treatment failure with penicillin. I. Group A streptococcal infection in mice. Am J Med 13:389–399. [DOI] [PubMed] [Google Scholar]
- 43.Haas W, Kaushal D, Sublett J, Obert C, Tuomanen EI. 2005. Vancomycin stress response in a sensitive and a tolerant strain of Streptococcus pneumoniae. J Bacteriol 187:8205–8210. doi: 10.1128/JB.187.23.8205-8210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G, Hu FZ, Yang X, Cui Y, Yang J, Qu F, Gao GF, Zhang JR. 2012. Complete genome sequence of Streptococcus pneumoniae strain ST556, a multidrug-resistant isolate from an otitis media patient. J Bacteriol 194:3294–3295. doi: 10.1128/JB.00363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fibriansah G, Kovacs AT, Pool TJ, Boonstra M, Kuipers OP, Thunnissen AM. 2012. Crystal structures of two transcriptional regulators from Bacillus cereus define the conserved structural features of a PadR subfamily. PLoS One 7:e48015. doi: 10.1371/journal.pone.0048015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen TK, Tran NP, Cavin JF. 2011. Genetic and biochemical analysis of PadR-padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168. J Bacteriol 193:4180–4191. doi: 10.1128/JB.00385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rybak M, Lomaestro B, Rotschafer JC, Moellering R, Craig W, Billeter M, Dalovisio JR, Levine DP. 2009. Therapeutic monitoring of vancomycin in adult patients: a consensus review of the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, and the Society of Infectious Diseases Pharmacists. Am J Health System Pharm 66:82–98. doi: 10.2146/ajhp080434. [DOI] [PubMed] [Google Scholar]
- 50.Barthelmebs L, Lecomte B, Divies C, Cavin JF. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J Bacteriol 182:6724–6731. doi: 10.1128/JB.182.23.6724-6731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gury J, Barthelmebs L, Tran NP, Divies C, Cavin JF. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl Environ Microbiol 70:2146–2153. doi: 10.1128/AEM.70.4.2146-2153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran NP, Gury J, Dartois V, Nguyen TK, Seraut H, Barthelmebs L, Gervais P, Cavin JF. 2008. Phenolic acid-mediated regulation of the padC gene, encoding the phenolic acid decarboxylase of Bacillus subtilis. J Bacteriol 190:3213–3224. doi: 10.1128/JB.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huillet E, Velge P, Vallaeys T, Pardon P. 2006. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol Lett 254:87–94. doi: 10.1111/j.1574-6968.2005.00014.x. [DOI] [PubMed] [Google Scholar]
- 54.Agustiandari H, Lubelski J, van den Berg van Saparoea HB, Kuipers OP, Driessen AJ. 2008. LmrR is a transcriptional repressor of expression of the multidrug ABC transporter LmrCD in Lactococcus lactis. J Bacteriol 190:759–763. doi: 10.1128/JB.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lubelski J, de Jong A, van Merkerk R, Agustiandari H, Kuipers OP, Kok J, Driessen AJM. 2006. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol 61:771–781. doi: 10.1111/j.1365-2958.2006.05267.x. [DOI] [PubMed] [Google Scholar]
- 56.Kovacikova G, Lin W, Skorupski K. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J Bacteriol 185:4825–4836. doi: 10.1128/JB.185.16.4825-4836.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Florez AB, Alvarez S, Zabala D, Brana AF, Salas JA, Mendez C. 2015. Transcriptional regulation of mithramycin biosynthesis in Streptomyces argillaceus: dual role as activator and repressor of the PadR-like regulator MtrY. Microbiology 161:272–284. doi: 10.1099/mic.0.080895-0. [DOI] [PubMed] [Google Scholar]
- 58.Morabbi Heravi K, Lange J, Watzlawick H, Kalinowski J, Altenbuchner J. 2015. Transcriptional regulation of the vanillate utilization genes (vanABK operon) of Corynebacterium glutamicum by VanR, a PadR-like repressor. J Bacteriol 197:959–972. doi: 10.1128/JB.02431-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madoori PK, Agustiandari H, Driessen AJ, Thunnissen AM. 2009. Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition. EMBO J 28:156–166. doi: 10.1038/emboj.2008.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulhern TD, Howlett GJ, Reid GE, Simpson RJ, McColl DJ, Anders RF, Norton RS. 1995. Solution structure of a polypeptide containing four heptad repeat units from a merozoite surface antigen of Plasmodium falciparum. Biochemistry 34:3479–3491. doi: 10.1021/bi00011a001. [DOI] [PubMed] [Google Scholar]
- 61.Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, DePalatis L, Raab H, Hazenbos WL, Hiroshi Morisaki J, Kim J, Park S, Darwish M, Lee BC, Hernandez H, Loyet KM, Lupardus P, Fong R, Yan D, Chalouni C, Luis E, Khalfin Y, Plise E, Cheong J, Lyssikatos JP, Strandh M, Koefoed K, Andersen PS, Flygare JA, Wah Tan M, Brown EJ, Mariathasan S. 2015. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527:323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 62.Barcia-Macay M, Seral C, Mingeot-Leclercq MP, Tulkens PM, Van Bambeke F. 2006. Pharmacodynamic evaluation of the intracellular activities of antibiotics against Staphylococcus aureus in a model of THP-1 macrophages. Antimicrob Agents Chemother 50:841–851. doi: 10.1128/AAC.50.3.841-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin-Galiano AJ, Overweg K, Ferrandiz MJ, Reuter M, Wells JM, de la Campa AG. 2005. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151:3935–3946. doi: 10.1099/mic.0.28238-0. [DOI] [PubMed] [Google Scholar]
- 64.Tomasz A, Albino A, Zanati E. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 65.Lu L, Ma Y, Zhang JR. 2006. Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J Biol Chem 281:15464–15474. doi: 10.1074/jbc.M602404200. [DOI] [PubMed] [Google Scholar]
- 66.Martin B, Garcia P, Castanie MP, Claverys JP. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol Microbiol 15:367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 67.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (ed). 2000. Current protocols in molecular biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 68.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sorg RA, Kuipers OP, Veening JW. 2015. Gene expression platform for synthetic biology in the human pathogen Streptococcus pneumoniae. ACS Synth Biol 4:228–239. [DOI] [PubMed] [Google Scholar]
- 71.Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang J-R, Veening J-W. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13:931. doi: 10.15252/msb.20167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 73.Wen Z, Sertil O, Cheng Y, Zhang S, Liu X, Wang WC, Zhang JR. 2015. Sequence elements upstream of the core promoter are necessary for full transcription of the capsule gene operon in Streptococcus pneumoniae strain D39. Infect Immun 83:1957–1972. doi: 10.1128/IAI.02944-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shainheit MG, Mule M, Camilli A. 2014. The core promoter of the capsule operon of Streptococcus pneumoniae is necessary for colonization and invasive disease. Infect Immun 82:694–705. doi: 10.1128/IAI.01289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30:892–897. [DOI] [PubMed] [Google Scholar]
- 76.Su J, Asare R, Yang J, Nair MK, Mazurkiewicz JE, Abu-Kwaik Y, Zhang JR. 2011. The capBCA locus is required for intracellular growth of Francisella tularensis LVS. Front Microbiol 2:83. doi: 10.3389/fmicb.2011.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He L, Nair MK, Chen Y, Liu X, Zhang M, Hazlett KR, Deng H, Zhang JR. 2016. The protease locus of Francisella tularensis LVS is required for stress tolerance and infection in the mammalian host. Infect Immun 84:1387–1402. doi: 10.1128/IAI.00076-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ringgaard S, Lowe J, Gerdes K. 2007. Centromere pairing by a plasmid-encoded type I ParB protein. J Biol Chem 282:28216–28225. doi: 10.1074/jbc.M703733200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.