Abstract
The ligand-binding domains of steroid hormone receptors possess a conserved structure with 12 α-helices surrounding a central hydrophobic core. On agonist binding, a repositioned helix 12 forms a pocket with helix 3 (H3) and helix 5 (H5), where transcriptional coactivators bind. The precise molecular interactions responsible for activation of these receptors remain to be elucidated. We previously identified a H3–H5 interaction that permits progesterone-mediated activation of a mutant mineralocorticoid receptor. We were intrigued to note that the potential for such interaction is widely conserved in the nuclear receptor family, indicating a possible functional significance. Here, we demonstrate via transcriptional activation studies in cell culture that alteration of residues involved in H3–H5 interaction consistently produces a gain of function in steroid hormone receptors. These data suggest that H3–H5 interaction may function as a molecular switch regulating the activity of nuclear receptors and suggest this site as a general target for pharmacologic intervention. Furthermore, they reveal a general mechanism for the creation of nuclear receptors bearing increased activity, providing a potentially powerful tool for the study of physiologic pathways in vivo.
Keywords: gain of function, interhelix interaction
Steroid hormone receptors, a subgroup of the larger family of nuclear receptors (NRs), are ligand-activated transcription factors that play a critical regulatory role in a wide variety of physiologic processes, including development, cellular differentiation, and maintenance of cellular homeostasis and blood pressure. NRs have a conserved domain structure, consisting of an N-terminal domain, a central DNA binding domain, a linker domain, and a C-terminal ligand-binding domain (LBD). The LBDs of NRs share a common architecture, with 12 α-helices and one β-turn arranged around a central hydrophobic core (1). Comparisons of the crystal structures of ligand-free and ligand-bound NRs have shown that, upon ligand binding, NRs undergo a conformational change, including movement of helix 12 and a bending of helix 3 (H3) toward helix 5 (H5). The repositioned helix 12 combines with H3 and H5 to form a pocket where transcriptional coactivators can bind (1).
The precise molecular interactions that mediate these molecular events are beginning to be understood. Key residues within the LBDs of nuclear and steroid hormone receptors that interact with specific steroid functional groups have been identified, providing a structural basis for the steroid specificity of these receptors (2–8). For example, in the mineralocorticoid receptor (MR), it has been proposed that conserved residues in H3 and H5, Gln-776 and Arg-817, form hydrogen bonds with the 3-ketone group of steroid hormones to anchor aldosterone's A-ring, whereas conserved residues in H3 and helix 12, Asn-770 and Thr-945, mediate binding of the D-ring (2). Mineralocorticoid specificity of MR is thought to be provided, at least in part, by a hydrogen bond between Asn-770 on H3 and the C21-OH group of mineralocorticoids (2). Such ligand–receptor interactions have now been described for a number of different steroid/nuclear hormone receptors (9). However, the relationship between ligand binding and receptor activation remains poorly understood, and events required beyond ligand binding for receptor activation have not been well defined.
We previously demonstrated an interhelix interaction in MR via human genetic studies that markedly alters receptor activity (10). A single amino acid substitution in MR of leucine for serine at residue 810 in H5 (MRL810) causes severe early onset hypertension in humans with exacerbation during pregnancy. Transcriptional activation studies of the mutant MRL810 receptor indicated that antagonists of wild-type MR such as progesterone and spironolactone function as agonists of MRL810, thus providing a pathophysiologic explanation for the observed worsening of hypertension during pregnancy (10). To better understand how a single amino acid change could so dramatically alter the activity of the receptor, we created a structural model of the MR-LBD based on the known crystal structure of the highly homologous progesterone receptor (PR) LBD (11). Our model suggested that the mutant leucine, but not the wild-type serine, at residue 810 on H5 is in van der Waals (vdW) contact with alanine 773 on H3. We proposed that the interaction between Leu-810 and Ala-773 led to progesterone-mediated activation of MRL810. This model was tested by assaying a series of mutant MRs bearing site-specific mutations at these positions. We found that progressive shortening of side-chain length at residue 810 on H5 resulted in a progressive loss of progesterone, but not aldosterone, mediated activation of these mutant receptors. Furthermore, we found evidence for second-site complementation; the loss of activity observed with the shortening of the H5 side chain could be reversed via lengthening of the H3 side chain (10). These findings strongly suggested that progesterone-mediated activation of MRL810 is due at least in part to an H3–H5 interaction between leucine 810 and alanine 773, which renders unnecessary the Asn770-C21-OH interaction otherwise required for MR activity.
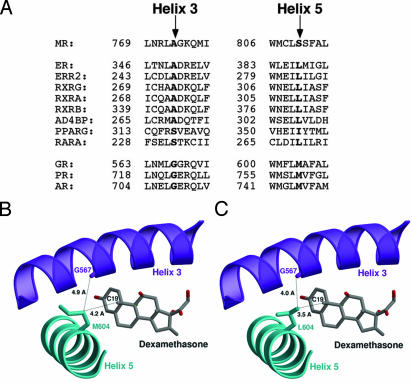
Our interest in this H3–H5 interaction was heightened by the finding that the potential for similar H3–H5 interactions exist in a number of different NRs. These two residues are coconserved within the NR family. Human receptors bearing leucine (or isoleucine) on H5 frequently carry alanine on H3 (Fig. 1A); methionine and glycine are capable of the same type of interhelix interaction as leucine and alanine, and receptors bearing methionine on H5 bear glycine on H3 (Fig. 1 A). This relationship is not unique to humans, either. These two residues are coconserved in MR, PR, and glucocorticoid receptor (GR) in virtually every vertebrate species thus far examined, the lone exception being a glycine to cysteine H3 substitution in chicken and hamster PR that renders the receptor resistant to RU486 antagonism (12).
Fig. 1.
Structural model for H3–H5 interaction in GRL604. (A) Coconservation of H3–H5 residues in NRs. The H3 and H5 amino acid sequences of selected NRs were aligned by using clustal w (26), and the residues corresponding to serine 810 on H5 and alanine 773 on H3 in MR are highlighted. Receptors bearing leucine at the H5 position frequently have alanine at the H3 position, whereas receptors bearing methionine at the H5 position have glycine at the H3 position. ER, estrogen receptor; ERR2, estrogen-related receptor type 2; RXRG, rexinoid receptor γ, α, and β; AD4BP, adrenal 4 binding protein (also known as SF1, steroidogenic factor 1); PPARG, peroxisome proliferator-activated receptor γ; RARA, mouse retinoic acid receptor α; AR, androgen receptor. (B) Ribbon drawing of the wild-type GR-LBD (19). H3 is colored in purple, H5 in cyan, and dexamethasone in gray. The steroid, the side chain of Met-604, and the carbonyl oxygen of the Gly-567 main chain are shown as ball-and-stick models and are labeled accordingly. The figure shows that the Met-604 side chain points away from the ligand and is too far from Gly-567 to be in vdW contact. (C) Ribbon drawing of the GRL604 mutant. Unlike Met-604, the side chain of Leu-604 is in vdW contact with the carbonyl oxygen of the Gly-567 main chain. The structure of the GRL604 mutant was generated in silico by replacing Met-604 against leucine. Figures were prepared with the programs molscript, bobscript, and raster3d.
Crystal structure data support the relevance of this interaction as well. The crystal structures of the estrogen receptor and PR LBD (11, 13) show that the corresponding residues (Leu-387, Ala-350; Met-759, Gly-722, respectively) are in vdW contact. Moreover, the potential for H3–H5 interaction is not limited to the steroid hormone receptor family, as the crystal structures of RXRα (14) and PPARγ (15) demonstrate that the corresponding H5 and H3 residues (Leu-309, Ala-272; Ile-354, Ser-317, respectively) are also in vdW contact. The wide conservation of this interaction suggests a possible functional role of this interaction as a general determinant of NR activity and suggested to us a mechanism by which we could generate steroid hormone receptors with increased function. To better understand the functional significance of this H3–H5 interaction, we studied these interactions in related steroid receptors.
Methods
Plasmids. The plasmid pRShGRNX (16) encodes human GR expressed from the Rous sarcoma virus long-terminal repeat promoter. The human PR expression plasmid for full-length PR isoform B has been described previously (17). Site-specific mutations were generated by using the QuikChange procedure (Stratagene) with the desired mutagenic primers. Each resulting mutant receptor gene was sequenced in its entirety to ensure that no undesired mutations were introduced. The plasmid pMTV-Luc (16) encodes luciferase under control of the GR-sensitive mouse mammary tumor virus promoter and was used for assays of GR activity. The progesterone response element (PRE)-luciferase reporter used in this work has been described previously; it contains two copies of the consensus PRE linked to the TATA element from E1b (18). pSV2 (Promega) encodes β-galactosidase from the SV40 early promoter.
Transcriptional Activation Studies. Cos-7 cells (American Type Culture Collection) were cultured in DMEM (GIBCO/BRL) supplemented with 10% heat-inactivated FCS. On the night before transfection, cells were plated at 2 × 105 cells per well. For GR transfections, cells in each well were transfected with 1 μg of pMTV-Luc, 1 μg of receptor plasmid, and 5 ng of pSV2. For PR transfections, cells were transfected with 500 ng of receptor plasmid, 1 μg of PRE-Luc, and 10 ng of pSV2. Transfection was performed by using cationic liposomes (Lipofectamine 2000, Life Technologies, Grand Island, NY), after which cells were incubated in DMEM supplemented with 10% FCS overnight. Cells were washed with PBS, and then serum-free DMEM containing the test steroid was added. The cells were incubated for an additional 16–24 h before assay. Luciferase and β-galactosidase activities were measured as described previously (10). Luciferase activity was normalized to β-galactosidase activity to correct for transfection efficiency and is expressed as a percentage of the wild-type receptor activity at 10 nM dexamethasone (GR) or 10 nM progesterone (PR). All results represent the mean ± SEM of at least nine independent transfections. Two-way ANOVA was used to compare the significance of differences between groups (in all cases, similar significance was obtained by using the two-tailed Student t test).
Receptor Binding Studies. Cos-7 cells (1 × 106) were transfected in 100-mm plates with 5 μg of receptor plasmid by using Lipofectamine 2000 (Life Technologies). On the day after transfection, serum-free media were substituted, and the cells were grown for an additional 24 h. Cells were harvested in 40 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA and lysed by freeze-thaw treatment in hypotonic buffer containing 10 mM Tris·HCl (pH 7.8), 10 mM NaCl, 1 mM EDTA, 10 mM Na2MoO4, 5 mM DTT, antipain (5 μg/ml), leupeptin (5 μg/ml), chymostatin (5 μg/ml), pepstatin A (5 μg/ml), and 500 μM PMSF. After centrifugation at 15,000 × g for 15 min, extracts were adjusted to 100 mM NaCl and 5% glycerol (binding buffer). Extracts were incubated overnight with [3H]dexamethasone (New England Nuclear) and competitor steroid (Sigma) at 0°C in a total volume of 200 μl, and then incubated with 100 μl of a 50% slurry of hydroxyapatite in binding buffer. Samples were spun, washed twice in binding buffer, then resuspended in ethanol and prepared for scintillation counting. The value for 100% binding was determined by subtracting the number of cpm bound in the presence of 500-fold excess of unlabeled dexamethasone from the counts bound in the absence of competitor. Nonspecific binding was determined with a 500-fold excess of unlabeled dexamethasone. No specific binding was seen in mock-transfected cells.
Results
H5 Alteration Induces a Gain of Function in the GR. Like PR, GR bears methionine (Met-604) and glycine (Gly-567) at the relevant H5 and H3 positions (Fig. 1 A). We noted in the crystal structure of the GR LBD in complex with dexamethasone, however, that Met-604 is not in vdW contact with Gly-567, and it forms only a weak interaction with the C19 methyl group of dexamethasone (ref. 19 and Fig. 1B). To better understand the role of H3–H5 interaction in NRs, we created a structural model of GR based on this structure. When we substituted leucine for methionine at residue 604 in silico, our model predicted that one of the methyl groups of the Leu-604 side chain would be within vdW distance of both the C19 methyl group of dexamethasone (≈3.5 Å) and the carbonyl oxygen of the Gly-567 main chain (≈4.0 Å) (Fig. 1C). Based on our experience with the MRL810 mutant, we speculated that such a substitution could increase the activity of the receptor. Conversely, when we substituted alanine for glycine on H3 (amino acid 567), our model predicted that the alanine methyl side chain would sterically interfere with the steroid ring and thus decrease receptor activity (data not shown).
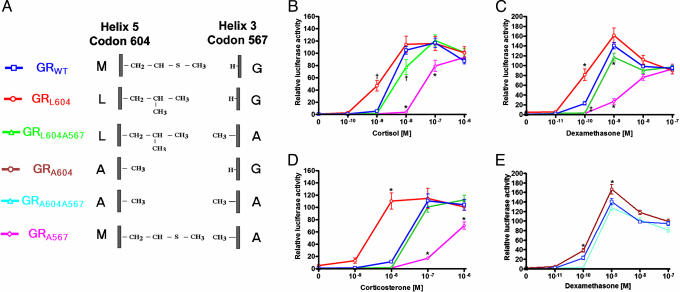
To determine whether our model correctly predicted molecular activity, we created a series of GR mutants designed to alter H3–H5 interaction, altering the H5 methionine (amino acid 604) to leucine, isoleucine, valine, or alanine or the H3 glycine to alanine. We studied the transcriptional activity of these mutant receptors in Cos7 cells, assessing the ability of the mutant receptors to drive luciferase expression from the mouse mammary tumor virus promoter. The activity of the wild-type receptor in the presence of 10 nM dexamethasone was considered as 100% activity.
We first studied the activity of GR bearing leucine at the H5 position in place of the wild-type methionine (Fig. 2A). GR bearing the Leu-604 substitution (GRL604) had increased activity, being activated by 10-fold lower steroid concentrations than GRWT. For example, whereas GRWT had no significant transcriptional activation in the presence of 1 nM cortisol, the GRL604 receptor was 47% active (P < 0.001, Fig. 2B). A similar effect was observed with dexamethasone as well; GRL604, but not GRWT, showed significant transcriptional activity at 0.1 nM (Fig. 2C). This proved to be true for all steroids tested; GRL604 demonstrated maximal activity at 10-fold lower corticosterone concentrations than did GRWT (Fig. 2D) and even in the presence of nonglucocorticoids, such as 11-deoxycorticosterone and 17-hydroxyprogesterone (data not shown), indicating that this gain of activity is not dependent on a particular steroid substituent.
Fig. 2.
GRL604 is active at 10-fold lower steroid concentrations than GRWT. The ability of wild-type and mutant GRs to induce luciferase expressed under control of the mouse mammary tumor virus promoter was assessed in Cos7 cells in the absence or presence of the indicated steroids. Luciferase activity is expressed as percent of maximal induction of GRWT by dexamethasone (10 nM). All data points represent the mean of at least nine independent transfections. (A) Schematic of the GR mutants studied. (B–D) Dose–response curves for induction of luciferase by GRWT, GRL604, GRL604A567, and GRA567 in response to cortisol (B), dexamethasone (C), and corticosterone (D). (E) Dose–response curves for induction of luciferase by GRWT, GRA604, and GRA604A567 by dexamethasone. *, P < 0.001 vs. GRWT...., P < 0.01 vs. GRWT.
To further investigate the role of residues involved in H3–H5 interaction, we examined the activity of GR mutants bearing further hydrophobic substitutions at the H5 position. Our model predicts steric hindrance between the C19 methyl group of glucocorticoids and the branched side chain of isoleucine (or valine), and consistent with this, receptors bearing these residues at amino acid 604 had minimal activity (data not shown). Interestingly, however, the GRA604 mutant (Fig. 2E) proved to be slightly more active than GRWT showing a minor, but statistically significant, increase in transcriptional activity at 0.1 and 1 nM dexamethasone (Fig. 2E). Again, this increase in transcriptional activity is not dependent on a particular steroid moiety, as it is observed in the presence of all glucocorticoids tested (data not shown).
To determine the contribution of H3 alteration to receptor activity, we created a second set of receptor mutations in which Gly-567 was altered to alanine. Our structural model predicts that the single carbon alanine side chain will interfere with ligand binding, but not with the H3–H5 interaction observed between Leu-604 and the carbonyl group of residue 567, and our transcriptional activation studies supported this notion. Substitution of alanine for glycine at residue 567 reduces the sensitivity of the receptor by ≈10-fold, regardless of the H5 residue. For example, GRWT is 100% active at 10 nM cortisol, whereas GRA567 is ≈80% active at 100 nM cortisol. Similarly, GRL604A567 and GRA604A567 each required ≈10-fold higher glucocorticoid concentrations as GRL604 and GRA604 for equivalent activity, respectively (Fig. 2 B–E), indicating that the loss of activity observed with substitution of alanine for glycine at residue 567 is independent of the H5 residue.
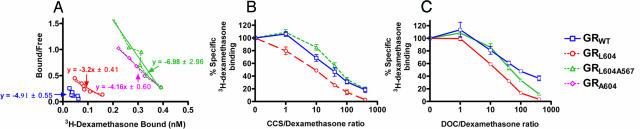
GRL604 Has Increased Affinity for Glucocorticoids. If our model that GRL604 has increased activity because of the creation of an H3–H5 interaction is correct, it suggests that GRL604 should bind ligand more tightly than does GRWT. Therefore, we assessed the binding of these receptors for glucocorticoids. Scatchard binding showed GRWT had a Kd of 4.91 ± 0.55 nM for dexamethasone (Fig. 3A), in good agreement with the published literature (20). GRL604 demonstrated increased binding affinity for dexamethasone (Kd = 3.2 ± 0.41 nM), whereas the affinity of GRL604A567 for dexamethasone was decreased (Kd = 6.98 ± 2.96 nM). GRA604 binding affinity for dexamethasone (Kd = 4.16 ± 0.60 nM) was slightly higher than that of GRWT, which is also consistent with our transcriptional activation results. These data suggest that the increased steroid sensitivity of GRL604 and GRA604 are because of increased affinity for the steroids.
Fig. 3.
GRL604 has increased affinity for glucocorticoids. (A) Scatchard analysis of the binding of [3H]dexamethasone in extracts expressing GRWT, GRL604, GRL604A567, or GRA604. Each data point was assayed at least four times; one representative set of data for binding to GRWT, GRL604, GRL604A567, and GRA604 is shown, and the mean Kd values are indicated. (B) Competition of varying concentrations of corticosterone and 5 nM [3H]dexamethasone for binding to GRWT, GRL604, and GRL604A567 in Cos-7 cell extracts. Each data point represents the mean ± SEM of at least four independent experiments. (C) Competition of varying concentrations of 11-deoxycorticosterone and 5 nM [3H]dexamethasone for binding to GRWT, GRL604, and GRL604A567 in Cos-7 cell extracts. Each data point represents the mean ± SEM of at least four independent experiments.
To determine whether this increased affinity of GRL604 for glucocorticoids is specific for dexamethasone or more general in nature, we used an in vitro competition assay to study the affinity of GRL604 for corticosterone and 11-deoxycorticosterone. In this assay, the relative affinity of a receptor for the ligand is compared to its affinity for dexamethasone. We found that 4-fold lower concentrations of corticosterone and 11-deoxycorticosterone effectively competed with dexamethasone for binding to GRL604 compared to GRWT (Fig. 3 B and C). For example, a 40:1 ratio of corticosterone to dexamethasone was required to compete for 50% dexamethasone binding to GRWT, but a 10:1 ratio was sufficient for similar competition with GRL604. This indicates an increased affinity of GRL604 for corticosterone relative to dexamethasone. As GRL604 itself binds dexamethasone more tightly than does GRWT (Fig. 3A), this suggests an even more marked affinity of GRL604 for corticosterone compared to GRWT. Again, this is consistent with our finding that the added increased activity of GRL604 compared to GRWT is even more pronounced in the presence of corticosterone than in the presence of dexamethasone (Fig. 2D).
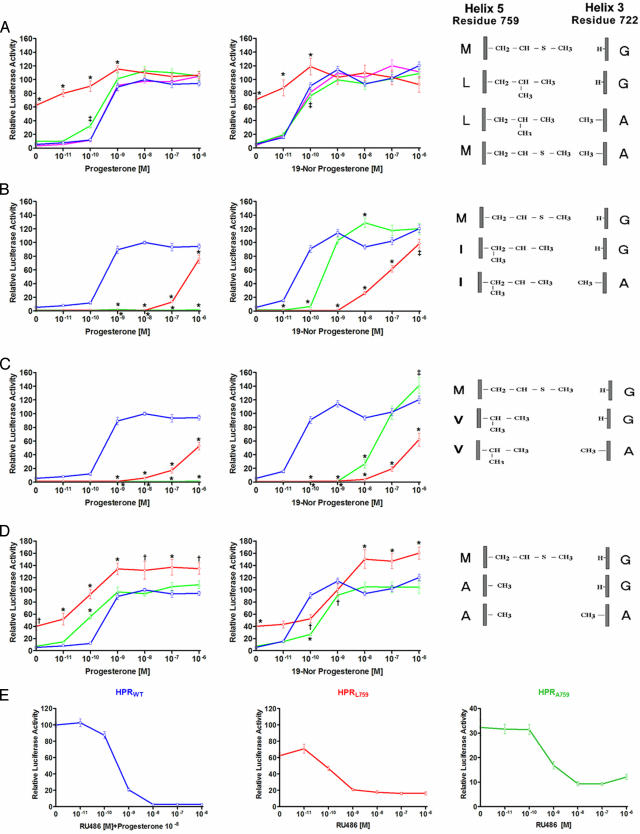
H5 Alteration Induces a Gain of Function in the PR. Having identified a critical role of the H3–H5 interaction in regulating receptor activity in GR, we sought to investigate the role of this interaction in another member of the steroid hormone receptor family, the PR. Like GR, PR bears glycine (Gly-722) and methionine (Met-759), respectively, at the critical H3 and H5 positions; however, analysis of the crystal structure of the PR-LBD (11) indicates that these residues are in vdW contact. To understand the role of this interaction, we created a similar substitution series as we did for GR, and we determined transcriptional activation activity of the mutant receptors. The activity of the wild-type receptor at 10 nM progesterone was considered 100% active.
Interestingly, PR bearing a single leucine substitution for methionine at the H5 position (amino acid 759) also proved to be a gain-of-function receptor. In contrast to what we observed with GRL604, however, the gain in activity of PRL759 was evidenced by constitutive activity, the receptor exhibiting 62% of maximal activity in the absence of progesterone (P < 0.001 vs. PRWT, Fig. 4A). The constitutive activity of PRL759 was not specific to Cos-7 cells, as we observed similar activity in HEK293 cells (data not shown). Addition of 10 nM progesterone causes a further increase in receptor activity up to 110% of PRWT activity (P = NS vs. PRWT); the progesterone concentration required to achieve maximal activation is identical for PRWT and PRL759. Reminiscent of what we observed with GRL604, second-site substitution of alanine for glycine at the H3 position (amino acid 722) causes PRL759 to lose its constitutive activity; the activity of PRL759A722 is virtually indistinguishable from that of PRWT. However, in contrast to our observations in GR, substitution of alanine for glycine at position 722 had no significant effect on the activity of wild-type PR.
Fig. 4.
PRL759 and PRA759 are constitutively active. The ability of PR mutants to induce luciferase expressed under control of the PRE promoter was assessed in Cos7 cells in the presence of progesterone and 19-norprogesterone. Luciferase activity is expressed as percent of maximal induction of PRWT by progesterone. (A) Transcriptional activity of PRWT, PRL759, PRL759A722, and PRA722 in the presence of progesterone and 19-norprogesterone. (B) Transcriptional activity of PRWT, PRI759, and PRI759A722 in the presence of progesterone and 19-norprogesterone. (C) Transcriptional activity of PRWT, PRV759, and PRV759A722 in the presence of progesterone and 19-norprogesterone. (D) Transcriptional activity of PRWT, PRA759, and PRA759A722 in the presence of progesterone and 19-norprogesterone. (E) RU486 inhibits constitutive activity of PRL759 and PRA759. The ability of RU486 to inhibit PRWT, PRL759, or PRA759 induced luciferase expression from the PRE promoter was assessed in Cos-7 cells in the presence (PRWT) or absence (PRL759 or PRA759) of 10 nM progesterone. Luciferase activity is expressed as percent of maximal induction of PRWT by 10 nM progesterone. All data points represent the mean ± SEM of at least nine independent transfections. *, P < 0.001 vs. GRWT...., P < 0.01 vs. GRWT; ‡, P < 0.05 vs. GRWT.
To further investigate the importance of residues 759 and 722 to receptor function, we created additional mutants bearing nonpolar amino acids at these positions. PRs bearing isoleucine or valine at the H5 amino acid 759 have minimal activity in the presence of progesterone (Fig. 4 B and C). This lack of activity is likely due to steric hindrance of the branched side chains of these amino acids with the 19-methyl group of progesterone, as marked increases in receptor activity are observed in the presence of 19-norprogesterone, which lacks the 19-methyl group (Fig. 4 B and C). Importantly, PRI759A722 and PRV759A722 are substantially more active than PRI759 and PRV759, respectively, in the presence of 19-norprogesterone, suggesting that the substitution of alanine for glycine on H3 in these mutants may have recreated an important H3–H5 interaction necessary for receptor activity.
Because the side chains of valine and isoleucine likely clash with the C19 methyl group of steroids, we created an Ala-759 mutant to determine the effect of complete disruption of H3–H5 interaction. Interestingly, PRA759 shares many of the transcriptional properties of PRL759. It possesses significant constitutive activity in Cos-7 cells (and HEK293 cells), ≈40% of PRWT maximal activity, and it also can be further activated by progesterone to reach maximal activity (Fig. 4D). As was seen with PRL759, the constitutive and increased maximal activities of PRA759 are lost when we add the H3 Ala-722 mutation.
To better understand the mechanism underlying the observed constitutive activity of PRL759 and PRA759, we tested the effect of the PR antagonist RU486 on receptor activity. As seen in Fig. 4E, concentrations of RU486 sufficient to inhibit PRWT also inhibit much of the constitutive activity of both PRL759 and PRA759, although these receptors retain 10–20% of maximal activity in the presence of high concentrations of RU486.
Discussion
The molecular contacts necessary for steroid hormone receptor activity are beginning to be understood. Such work has typically focused on the contacts between functional groups of individual steroids and amino acid residues within the receptor ligand-binding pocket. Here, we describe a mechanism of receptor activation, an interhelix interaction between receptor residues, that serves as a general regulator of NR activity and specificity. We demonstrate here that alterations in H3–H5 residues leads to profound alterations in receptor sensitivity in all steroid hormone receptors thus far tested.
Our GR data are consistent with the notion that a vdW interaction between one of the methyl groups of the Leu-604 side chain on H5 and the carbonyl oxygen of Gly-567 on H3 is responsible for the increased activity of GRL604 (Fig. 1C). As such, the increased activity of this receptor closely correlates with what we previously observed with MRL810, that a mutation enabling a new H3–H5 interaction increases receptor activity. Whereas second-site substitution of alanine for glycine causes a loss of the observed gain of function, Ala-567 caused a loss of activity in all GR mutants we tested, suggesting that the effect of Ala-567 is independent of the H5 residue, and consistent with the model that Ala-567 sterically interferes with steroid binding as opposed to inhibiting an H3–H5 interaction.
The underlying mechanism for the constitutive activity of PRL759 and PRA759 is distinct. Mutant receptors exhibiting constitutive activity have been observed for a number of other members of the NR family (21, 22), but the mechanism of this constitutive activity has not always been established. The mutation may allow contacts within the receptor that allow it to fold into an active conformation independent of ligand or alter receptor interactions with coactivators in a ligand-independent fashion. Alternatively, mutations in the LBD may allow normally inactive cellular sterols to function as agonists by forming critical contacts with the receptor. For example, cellular fatty acids have been shown to activate the mutant receptor RXRF318A, thus giving this receptor the appearance of constitutive activity (22). Our finding that RU486 inhibits the constitutive activity of PRL759 is consistent with this model, but at this point, other models cannot be excluded.
We have now examined H3–H5 interactions in three distinct steroid hormone receptors: MR, GR, and PR. In all three instances, we have demonstrated that mutations at relevant H3 and H5 positions markedly alter receptor sensitivity and/or specificity, allowing us to create gain-of-function mutations for each of these receptors. Furthermore, an estrogen receptor α bearing a substitution of phenylalanine for alanine at this precise H3 residue is constitutively active (23). Whereas loss-of-function mutations in NRs are relatively easy to devise, mutations which increase receptor activity are rare. To our knowledge, the MRL810 mutation (10) and the PRL759 (and PRA759) mutations are the only known gain-of-function mutations in these receptors. Furthermore, we are aware of only three previously reported gain-of-function mutations in GR (20, 24, 25), two of which lie on H3 in close proximity to GRG567. These findings, coupled with the wide coconservation of H3–H5 residues in the NR family, suggest that the H3–H5 interaction plays a critical role by functioning as a molecular switch, which regulates the specificity and sensitivity of steroid hormone receptors and perhaps of other NRs as well.
What might be the role of H3–H5 interaction in receptor activity? In the case of MRL810, we have noted that the H3–H5 interaction renders the interaction between H3 and the steroid C21 group unnecessary, and we speculated that bending of H3 toward the steroid ligand is a necessary step for MR activation (10). Similarly, our structural model predicts that the increased activity of GRL604 is due to a vdW contact between H3 and H5. Given the widespread and almost universal involvement of H3 in the gain-of-function mutations described above, it is tempting to speculate that interactions influencing the bending of H3 represent a principal mode of steroid hormone receptor activation and that alteration of H3–H5 interaction may represent a general mechanism by which gain-of-function mutations in steroid hormone receptors and perhaps other NRs can be created. With the ongoing advances in our ability to manipulate genomes of model organisms, the availability of gain-of-function NRs may provide a potentially powerful new tool for the study of physiologic roles of these receptors in vivo. For example, tissue-specific expression of overactive steroid receptors would provide a mechanism to enable study of steroid effects in individual tissues, a mechanism which would be operative in utero as well.
In summary, our work has demonstrated a critical role of H3–H5 interaction in regulating the sensitivity and/or specificity of all steroid hormone receptors tested to date. Pharmacologic agents that modify or interfere with this interaction, perhaps via the addition of C19 or C11 substitutions on the steroid ring, would be expected to have major effect on the activities of these receptors. Determining the precise structural contacts necessary for steroid hormone receptor ligand binding will improve our ability to design compounds that regulate steroid hormone receptors and other NRs.
Acknowledgments
We thank S. Lee for help in figure preparation, H. E. Xu and GlaxoSmith Kline for making the coordinates of the GR-LBD available to us, and R. P. Lifton for numerous scientific discussions and helpful critique of this manuscript. F.T.F.T. is supported by American Heart Association Grant AHA-0130124N. D.S.G. is supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant K08-DK02765 and National Heart, Lung, and Blood Institute Grant P50-HL55007.
Abbreviations: NR, nuclear receptor; LBD, ligand-binding domain; MR, mineralocorticoid receptor; PR, progesterone receptor; vdW, van der Waals; GR, glucocorticoid receptor; H3, helix 3; H5, helix 5; PRE, progesterone response element.
References
- 1.Moras, D. & Gronemeyer, H. (1998) Curr. Opin. Cell Biol. 10, 384–391. [DOI] [PubMed] [Google Scholar]
- 2.Fagart, J., Wurtz, J. M., Souque, A., Hellal-Levy, C., Moras, D. & Rafestin-Oblin, M. E. (1998) EMBO J. 17, 3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivat, V., Gofflo, D., Garcia, T., Wurtz, J. M., Bourguet, W., Philibert, D. & Gronemeyer, H. (1997) J. Mol. Endocrinol. 18, 147–160. [DOI] [PubMed] [Google Scholar]
- 4.Poujol, N., Wurtz, J. M., Tahiri, B., Lumbroso, S., Nicolas, J. C., Moras, D. & Sultan, C. (2000) J. Biol. Chem. 275, 24022–24031. [DOI] [PubMed] [Google Scholar]
- 5.Dey, R., Roychowdhury, P. & Mukherjee, C. (2001) Protein Eng. 14, 565–571. [DOI] [PubMed] [Google Scholar]
- 6.Lind, U., Greenidge, P., Gillner, M., Koehler, K. F., Wright, A. & Carlstedt-Duke, J. (2000) J. Biol. Chem. 275, 19041–19049. [DOI] [PubMed] [Google Scholar]
- 7.Marhefka, C. A., Moore, B. M., II, Bishop, T. C., Kirkovsky, L., Mukherjee, A., Dalton, J. T. & Miller, D. D. (2001) J. Med. Chem. 44, 1729–1740. [DOI] [PubMed] [Google Scholar]
- 8.Lind, U., Greenidge, P., Gustafsson, J. A., Wright, A. P. & Carlstedt-Duke, J. (1999) J. Biol. Chem. 274, 18515–18523. [DOI] [PubMed] [Google Scholar]
- 9.Steinmetz, A. C., Renaud, J. P. & Moras, D. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 329–359. [DOI] [PubMed] [Google Scholar]
- 10.Geller, D. S., Farhi, A., Pinkerton, N., Fradley, M., Moritz, M., Spitzer, A., Meinke, G., Tsai, F. T., Sigler, P. B. & Lifton, R. P. (2000) Science 289, 119–123. [DOI] [PubMed] [Google Scholar]
- 11.Williams, S. P. & Sigler, P. B. (1998) Nature 393, 392–396. [DOI] [PubMed] [Google Scholar]
- 12.Benhamou, B., Garcia, T., Lerouge, T., Vergezac, A., Gofflo, D., Bigogne, C., Chambon, P. & Gronemeyer, H. (1992) Science 255, 206–209. [DOI] [PubMed] [Google Scholar]
- 13.Brzozowski, A. M., Pike, A. C., Dauter, Z., Hubbard, R. E., Bonn, T., Engstrom, O., Ohman, L., Greene, G. L., Gustafsson, J. A. & Carlquist, M. (1997) Nature 389, 753–758. [DOI] [PubMed] [Google Scholar]
- 14.Egea, P. F., Mitschler, A., Rochel, N., Ruff, M., Chambon, P. & Moras, D. (2000) EMBO J. 19, 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gampe, R. T., Jr., Montana, V. G., Lambert, M. H., Miller, A. B., Bledsoe, R. K., Milburn, M. V., Kliewer, S. A., Willson, T. M. & Xu, H. E. (2000) Mol. Cell 5, 545–555. [DOI] [PubMed] [Google Scholar]
- 16.Rupprecht, R., Reul, J. M., van Steensel, B., Spengler, D., Soder, M., Berning, B., Holsboer, F. & Damm, K. (1993) Eur. J. Pharmacol. 247, 145–154. [DOI] [PubMed] [Google Scholar]
- 17.Vegeto, E., Allan, G. F., Schrader, W. T., Tsai, M. J., McDonnell, D. P. & O'Malley, B. W. (1992) Cell 69, 703–713. [DOI] [PubMed] [Google Scholar]
- 18.Nawaz, Z., Stancel, G. M. & Hyder, S. M. (1999) Cancer Res. 59, 372–376. [PubMed] [Google Scholar]
- 19.Bledsoe, R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., et al. (2002) Cell 110, 93–105. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborti, P. K., Garabedian, M. J., Yamamoto, K. R. & Simons, S. S., Jr. (1991) J. Biol. Chem. 266, 22075–22078. [PubMed] [Google Scholar]
- 21.Weis, K. E., Ekena, K., Thomas, J. A., Lazennec, G. & Katzenellenbogen, B. S. (1996) Mol. Endocrinol. 10, 1388–1398. [DOI] [PubMed] [Google Scholar]
- 22.Bourguet, W., Vivat, V., Wurtz, J. M., Chambon, P., Gronemeyer, H. & Moras, D. (2000) Mol. Cell 5, 289–298. [DOI] [PubMed] [Google Scholar]
- 23.Chen, S., Zhou, D., Yang, C. & Sherman, M. (2001) J. Biol. Chem. 276, 28465–28470. [DOI] [PubMed] [Google Scholar]
- 24.Yu, C., Warriar, N. & Govindan, M. V. (1995) Biochemistry 34, 14163–14173. [DOI] [PubMed] [Google Scholar]
- 25.Warriar, N., Yu, C. & Govindan, M. V. (1994) J. Biol. Chem. 269, 29010–29015. [PubMed] [Google Scholar]
- 26.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]