Abstract
Plasma membrane receptors can be endocytosed through clathrin-dependent and clathrin-independent pathways. Here, we show that the epidermal growth factor (EGF) receptor (EGFR), when stimulated with low doses of EGF, is internalized almost exclusively through the clathrin pathway, and it is not ubiquitinated. At higher concentrations of ligand, however, a substantial fraction of the receptor is endocytosed through a clathrin-independent, lipid raft-dependent route, as the receptor becomes ubiquitinated. An ubiquitination-impaired EGFR mutant was internalized through the clathrin pathway, whereas an EGFR/ubiquitin chimera, that can signal solely through its ubiquitin (Ub) moiety, was internalized exclusively by the non-clathrin pathway. Non-clathrin internalization of ubiquitinated EGFR depends on its interaction with proteins harboring the Ub-interacting motif, as shown through the ablation of three Ub-interacting motif-containing proteins, eps15, eps15R, and epsin. Thus, eps15s and epsin perform an important function in coupling ubiquitinated cargo to clathrin-independent internalization.
Keywords: internalization, rafts, caveolae, ubiquitination, ubiquitin-interacting motif
Ubiquitination is a posttranslational modification whereby substrate proteins are conjugated to a short highly conserved peptide, ubiquitin (Ub), through the action of Ub ligases (E3 enzymes). Polyubiquitination, in which a chain of Ub is appended, targets proteins to proteasomal degradation (1). However, when a single Ub moiety is appended (monoubiquitination), the modification functions as a signaling device through interactions with intracellular proteins harboring Ub-binding domains, such as the Ub-interacting motif (UIM) (2). In yeast, monoubiquitination has been known to act as an internalization signal for quite some time (3). In mammals, however, this connection has remained more elusive.
We are interested in the mechanisms of internalization of receptor tyrosine kinases and, in particular, the epidermal growth factor receptor (EGFR). The EGFR is monoubiquitinated at multiple sites (4) through the action of the E3 enzyme Cbl. Although there is consensus on the function of Cbl and receptor ubiquitination in intracellular sorting of the EGFR, their role in the internalization step of endocytosis is less clear (5, 6). To gain insight into this issue, we generated a chimera in which the extracellular and transmembrane domains of the EGFR are fused to a mutant Ub (Ubmut), unable to form polyUb chains (EGFR/Ubmut). With this chimera, we showed that ubiquitination is sufficient for internalization (4). The present studies were undertaken to elucidate the molecular mechanisms through which receptor ubiquitination directs internalization.
Materials and Methods
Transfection and Biochemical Studies. Transfections were performed by using Lipofectamine or Oligofectamine (Invitrogen). For biochemical experiments, cells were serum-starved and then stimulated with EGF (100 ng/ml, unless otherwise indicated) at 37°C. Lysis, immunoprecipitation, and immunoblotting were performed as described in ref. 7. Plasmids, pharmacological agents, and antibodies are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Gene Silencing. Gene silencing of eps15, eps15R, or epsin was obtained by pSUPER vectors encoding specific targeting sequences. Two distinct small interfering RNAs/targets were tested, with comparable results. Stable clones were obtained by cotransfection with a plasmid carrying puromycin resistance. Three independent clones were analyzed with similar results. Silencing of clathrin heavy chain was by transient transfection of two different short interfering RNA oligos (which yielded comparable results) directed against the two previously described sequences (8, 9). Further details are in Supporting Materials and Methods.
Immunofluorescence and Internalization Studies. Immunofluorescence, internalization of rhodaminated ligands, 125I-EGF or 13A9 antibody were performed as described in ref. 4. At the single cell level, endocytosis was monitored both by rhodamine-EGF and by anti-EGFR 13A9 antibody (which does not interfere with EGFR internalization) with comparable results (only one of the two procedures is shown in the pictures). Rhodaminated EGF (1 μg/ml) or the 13A9 antibody (20 μg/ml in the presence of EGF) were added for 1 h at 0°C, followed by wash and shift at 37°C for 20–30 min to allow internalization. After fixation, 13A9 was detected with Cy3-conjugated secondary antibody. In assays with anti-hemagglutinin (anti-HA) (see Fig. 6B), 20 μg/ml anti-HA was substituted for 13A9. Quantitation was performed on at least two experiments, in duplicate (>100 cells per condition). Plasma membrane staining was compared with intracellular vesicle-associated staining. Internalizing cells displayed preponderant intracellular staining but little, if any, plasma membrane staining. Noninternalizing cells displayed the opposite phenotype.
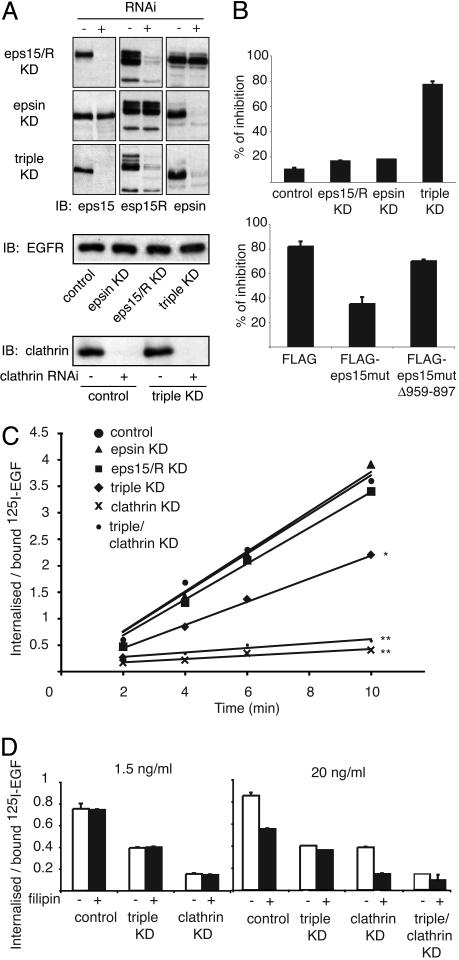
Fig. 6.
Effects of multiple KDs on internalization. (A)(Top) Lysates from HeLa KD stable clones (+) for eps15/eps15R, epsin, or eps15/eps15R/epsin (triple KD) or from control clones obtained with mismatched oligos (–) were immunoblotted as indicated. (Middle) EGFR levels were comparable in all clones. (Bottom) The clathrin levels in the clathrin KDs. (B)(Upper) The indicated HeLa KD clones were transfected with a HA-EGFR/Ubmut chimera (see Supporting Materials and Methods) to circumvent the presence of endogenous EGFR. Internalization was assessed with an anti-HA antibody. Data are expressed as the percentage of cells in which internalization was inhibited. (Lower) A HeLa triple KD clone expressing HA-EGFR/Ubmut was transfected with the indicated eps15mut constructs (see Supporting Materials and Methods). Internalization assays and bar graph are as in Upper. Actual images of these experiments are in Fig. 11. (C) HeLa clones harboring the indicated KDs were incubated with 1.5 ng/ml 125I-EGF. The rate of internalization is expressed as internalized/surface-bound radioactivity. *, P = 0.0164; **, P = 0.003; linear regression analysis. (D) The indicated HeLa KD clones were treated with filipin (+) or mock treated (–), followed by incubation with 1.5 or 20 ng/ml 125I-EGF for 6 min at 37°C. The rate of internalization is expressed as internalized/surface-bound radioactivity.
Immunoelectron Microscopy. For preembedding immunolabeling (Figs. 1A and 2B Left and Center and Table 1), starved prechilled cells were incubated on ice for 30 min with 20 μg/ml anti-EGFR 13A9, then with rabbit anti-mouse (code no. ZO412, DAKO), and finally with 10-nm protein A-gold. Cells were then incubated at 37°C for 2 min with EGF (at the indicated doses), fixed for 15 min at room temperature (2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2), and processed as described in ref. 10. ImmunoGold labeling on ultrathin cryosections (Figs. 2B Right and 4B) was performed as reported in ref. 10. Further details are in Supporting Materials and Methods.
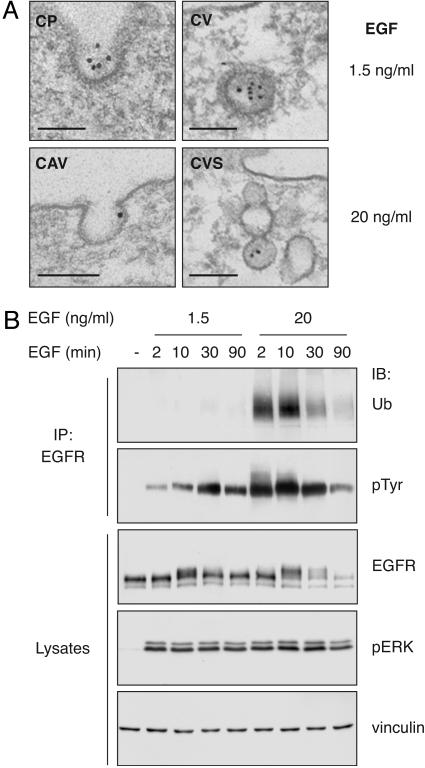
Fig. 1.
Internalization of EGFR at different doses of EGF. (A) Immunoelectron microscopy of EGFRwt in HeLa cells in coated pits (CP), coated vesicles (CV), caveolae (CAV), or caveosome-like structures (CVS). (Bar, 111 nm.) (B) Lysates (1 mg) from HeLa cells were immunoprecipitated (IP) or directly loaded (50 μg) and immunoblotted (IB) with the indicated antibodies.
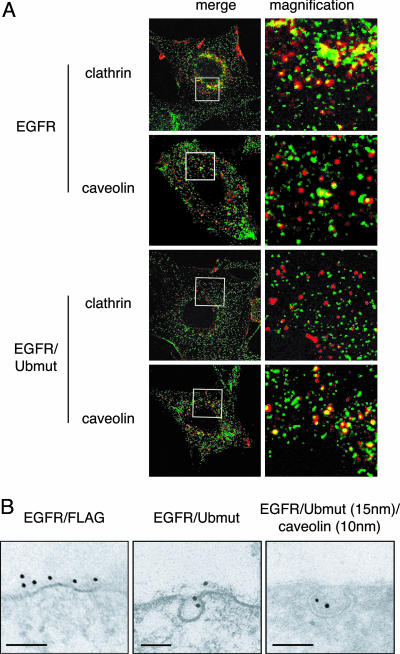
Fig. 2.
Internalization of EGFR/Ubmut. (A) NR6 cells expressing EGFRwt or EGFR/Ubmut were treated (15 min) with rhodamine-EGF (red) and stained for either clathrin or caveolin (green). (Left) Boxes enclose areas magnified in Right (magnification, ×4). Similar results were obtained when the internalization assay was performed with the 13A9 anti-EGFR antibodies in the presence or absence of EGF (data not shown). (B) Immunoelectron microscopy on NR6 expressing EGFR/Ubmut or EGFR/FLAG [a Ub-less control chimera (4)]. EGFR/Ubmut is exclusively detected in caveolae (20 cell profiles analyzed). (Bar, 83 nm.)
Table 1. Morphometry from 10 cell profiles.
| Organelles | EGF at 1.5 ng/ml, % (n) | EGF at 20 ng/ml, % (n) |
|---|---|---|
| Labeled caveolae | 4 (72) | 22 (106) |
| Labeled coated pits | 40 (35) | 30 (46) |
| Gold in caveolae | 1 (296) | 12 (383) |
| Gold in coated pits | 16 (296) | 15 (383) |
Shown is the percentage of labeled organelles, with values in parentheses representing the number of caveolae or coated pits counted, or the percentage of gold particles in organelles, with the values in parentheses representing the total gold particles detected on the plasma membrane.
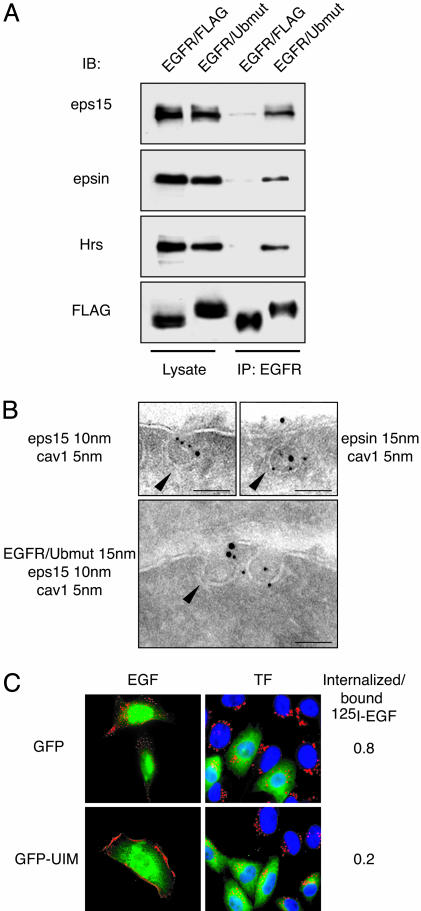
Fig. 4.
UIM proteins and EGFR/Ubmut. (A) φ-NX cells were transfected as indicated above the gels. Lysates (5 mg) were immunoprecipitated (IP) and immunoblotted (IB) as indicated. (B) Immunoelectron microscopy of NR6 expressing EGFR/Ubmut. (Bar, 83 nm.) Arrowheads point to caveolar membranes. Note that in cells expressing the EGFR/FLAG control chimera, eps15 and epsin were never detected in caveolae (data not shown). (C) CHO cells were cotransfected with EGFR/Ubmut and either GFP or GFP-UIM (amino acids 842–897 of eps15) and incubated with rhodamine-EGF (red, Left) or rhodamine-transferrin (red, Center). Merged images are shown. (Right) A quantitative assessment of EGF internalization (3 ng/ml 125I-EGF for 15 min at 37°C).
Results
In HeLa cells, expressing endogenous EGFR, we found that EGFR follows different internalization routes, depending on the dose of EGF. We used two concentrations of EGF (low EGF, 1.5 ng/ml; high EGF, 20 ng/ml), both within the range of physiological EGF levels (1–2 ng/ml in serum; 10–100 ng/ml in several other biological fluids, see Supporting Materials and Methods). At low EGF, EGFR localized predominantly into clathrin-coated pits, whereas, at high EGF, the receptor was roughly equally partitioned between coated pits and caveolae (Fig. 1 A and Table 1). At low EGF, the receptor was significantly tyrosine phosphorylated and fully competent for downstream signaling (as demonstrated by activation of ERK), whereas significant ubiquitination was only detectable at high EGF (Fig. 1B). Thus, there is correlation between the state of EGFR ubiquitination and its partition into caveolae.
We have shown that a chimera encompassing the extracellular and transmembrane domain of the EGFR fused to Ub (EGFR/Ubmut) is constitutively internalized (4). By exploiting this chimera, whose internalization exclusively depends on the signaling abilities of the Ub moiety, we endeavored to determine how ubiquitination directs internalization. In EGFR-negative NR6 (Fig. 2 A) or Chinese hamster ovary (CHO) (data not shown) cells, the internalized EGFR/Ubmut colocalized extensively with caveolin-1, a marker of the non-clathrin lipid raft/caveolar pathway, but poorly with clathrin, whereas a transfected EGFR (EGFRwt) showed the opposite behavior (Fig. 2 A). By immunoelectron microscopy, we detected EGFR/Ubmut exclusively in caveolae (Fig. 2B), whereas EGFRwt partitioned between coated pits and caveolae, with preference for coated pits (data not shown). Consistently, by lipid raft fractionization experiments, the EGFR/Ubmut partitioned 10-fold more than the EGFRwt in the raft fractions (Fig. 7, which is published as supporting information on the PNAS web site).
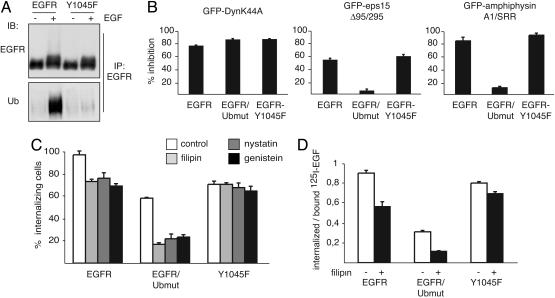
Next, we used pharmacological and dominant negative inhibitors of endocytosis and comparatively tested their effects on EGFRwt, EGFR/Ubmut, and a mutant EGFR (Y1045F), that cannot efficiently recruit Cbl (11), thus showing considerably reduced ubiquitination (Fig. 3A). All these constructs were expressed in EGFR-negative CHO or NR6 cells.
Fig. 3.
Ubiquitination and internalization. (A) Lysates (5 mg) from CHO cells expressing EGFRwt or Y1045F were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies. (B) CHO cells were cotransfected with the indicated GFP-tagged and various EGFR-based constructs and incubated with rhodamine-EGF. The bar graphs represent the percentage of cells in which EGF internalization was inhibited (normalized to control GFP-transfected cells). (C) NR6 cells expressing the indicated constructs were treated with the indicated drugs. Internalization was monitored with the anti-EGFR 13A9 antibody in the presence of high EGF. (D) NR6 cells expressing the indicated constructs were treated with filipin and then incubated with 20 ng/ml 125I-EGF for 15 min at 37°C. The rate of internalization is expressed as internalized/surface-bound radioactivity. Actual images of the experiments described for B and C are in Fig. 8.
In an initial set of experiments, we monitored internalization of rhodamine-conjugated EGF (Fig. 3 B and C; see Fig. 8, which is published as supporting information on the PNAS web site). DynaminII-K44A, a general endocytic inhibitor (12, 13), abrogated internalization of EGFR/Ubmut, EGFRwt, and Y1045F (Fig. 3B). Conversely, a mutant eps15 (Δ95/295), which interferes predominantly with AP2 (14), or an amphiphysin fragment (A1/SRR) that selectively interferes with clathrin (15) was ineffective on endocytosis of EGFR/Ubmut, while reducing internalization of EGFRwt and Y1045F (Fig. 3B). Pharmacological tools (filipin, nystatin, and low-dose genistein) that allow preferential interference with non-clathrin internalization (16, 17) effectively inhibited the internalization of EGFR/Ubmut (Fig. 3C). Of note, a different chimeric receptor harboring the extracellular domain of the transferrin receptor and intracellular Ub, was also internalized through a nystatin and filipin-sensitive pathway, whereas the wild-type transferrin receptor, which is internalized exclusively through coated pits, was insensitive to these drugs (Fig. 9, which is published as supporting information on the PNAS web site). EGFRwt was inhibited by the drugs to a lesser, albeit significant, extent (Fig. 3C). Conversely, the internalization of the Y1045F mutant was insensitive to these inhibitors (Fig. 3C).
A quantitative assessment was obtained by measuring the internalization of 125I-EGF (Fig. 3D). At high EGF, although the internalization of EGFRwt was partially sensitive to filipin, the drug had no significant effect on endocytosis of the Y1045 mutant, but it reduced the internalization of EGFR/Ubmut to background levels (Fig. 3D).
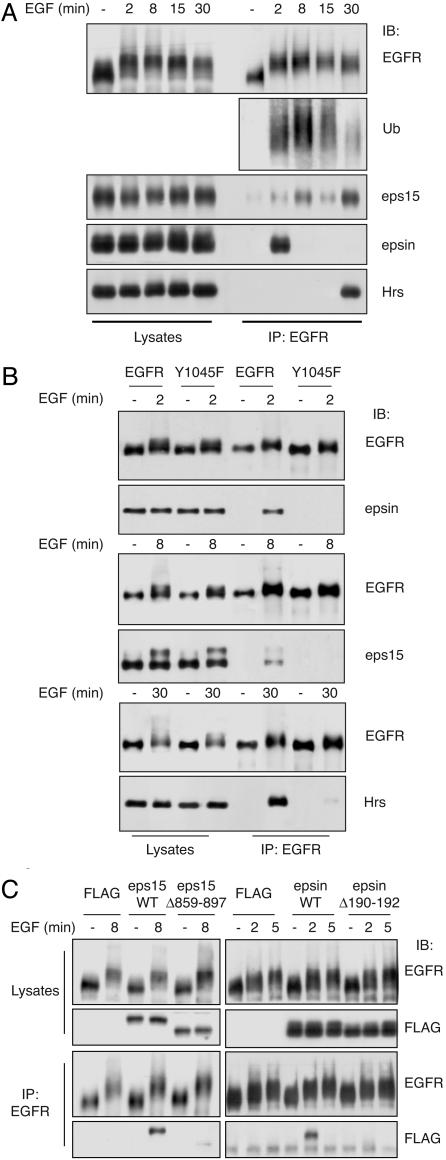
How does Ub couple with the intracellular machinery of non-clathrin endocytosis? A family of UIM-containing proteins has been implicated in endocytosis, with eps15 and epsin functioning at the plasma membrane (10, 18, 19) and hepatocyte growth factor receptor substrate (HRS) in endosomes (20). These proteins coimmunoprecipitated with EGFR/Ubmut (Fig. 4A) and were also found in caveolae containing the chimera and caveolin-1 (Fig. 4B). In addition, the overexpression of an isolated UIM inhibited the internalization of EGFR/Ubmut but not of the transferrin receptor (Fig. 4C). Finally, when EGFR/Ubmut was modified, such as to harbor a Ub moiety incapable of interacting with UIMs (21), it could not be internalized any longer (Fig. 10, which is published as supporting information on the PNAS web site). UIM proteins also coimmunoprecipitated with endogenous EGFR in a ligand-dependent fashion and with a time kinetic compatible with function(s) at different stations of the endocytic route (Fig. 5A). Of interest, eps15 showed a second peak of association at late time points, suggesting an additional role(s) at the endosomal level, as also recently suggested (22). The association depended on the presence of Ub and functional UIMs, as shown by the lack of coimmunoprecipitation of Y1045F with the three UIM proteins (Fig. 5B) and the lack of endogenous EGFR with mutant eps15 and epsin, which were carrying deletions in their UIMs (Fig. 5C). Thus, Ub, either biosynthetically added to EGFR/Ubmut or posttranslationally appended to EGFR, might direct the receptors to non-clathrin internalization through an interaction(s) with UIM proteins.
Fig. 5.
UIM proteins bind to ubiquitinated EGFR. (A) Lysates (5 mg) from HeLa cells were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies. (B) CHO cells were transiently cotransfected with either EGFRwt or Y1045F and individually transfected with epsin, eps15, and hepatocyte growth factor receptor substrate (Hrs). Lysates (2 mg) were immunoprecipitated/immunoblotted as indicated. (C) HeLa cells were transfected with the indicated FLAG-tagged constructs encoding wild-type eps15 or an eps15-UIM-deleted mutant [eps15Δ859–897 (29)], or wild-type epsin, or an epsin-UIM-deleted mutant [epsinΔ190–192 (29)]. FLAG is an empty control vector. Lysates (2 mg) were immunoprecipitated/immunoblotted as indicated.
To address this point, we knocked down (KD) eps15, eps15R (a related protein), and epsin in HeLa cells (Fig. 6A). The endocytosis of EGFR/Ubmut was abrogated in triple eps15/eps15R/epsin KD cells but not in individual KD cells (Fig. 6B; see Fig. 11 A, which is published as supporting information on the PNAS web site). The reexpression of eps15 but not of an eps15-UIM-deleted mutant in the triple KD cells restored the internalization of EGFR/Ubmut (Fig. 6B, see Fig. 11B). Thus, when a membrane protein is exclusively internalized as a function of its ubiquitination in a nystatin/filipin-sensitive pathway, the interaction with the UIM proteins eps15/eps15R/epsin is essential. In addition, the functions of eps15/eps15R and epsin are redundant in this pathway.
We then investigated the behavior of the endogenous EGFR. Initially, we investigated the role of clathrin because discordant results have been reported (8, 9, 23). We reasoned that the scarce effect of clathrin KD in one of these studies could be explained by the use of high EGF in the internalization assay (8). According to our present data, under those conditions the contribution of a clathrin-independent pathway (defined on the basis of its nystatin/filipin sensitivity) would be substantial. As shown in Fig. 6C, at low EGF, the KD of clathrin (levels of clathrin in KD cells are in Fig. 6A) almost completely abrogated internalization, demonstrating that, under this condition, clathrin is absolutely required. Next, we tested whether UIM-containing proteins are required in this pathway. At low EGF, although the individual KDs of eps15/eps15R or epsin had no effect, the triple eps15/eps15R/epsin KD exhibited a significant decrease in EGFR internalization (Fig. 6C). Treatment with filipin had no effect in any of the KDs at low EGF (Fig. 6D). These results are in agreement with those reported by Huang et al. (23) and demonstrate that eps15/eps15R and epsin participate (redundantly) also in the clathrin pathway. However, they are not strictly essential, as clathrin endocytosis proceeds also in their absence.
At high EGF, clathrin KD reduced internalization by ≈50%. However, filipin treatment of clathrin KD cells reduced the residual internalization to background levels (Fig. 6D). These results show the existence of two modalities of internalization, one relying on clathrin (at low EGF) and one relying both on clathrin-dependent and clathrin-independent pathways (at high EGF). The triple eps15/eps15R/epsin KD also reduced EGFR endocytosis at high EGF, albeit in a filipin-insensitive fashion, demonstrating that, in the triple KD, the non-clathrin nystatin/filipin-sensitive component of EGFR endocytosis is completely abolished (Fig. 6D). Individual KDs of eps15/eps15R or epsin had no effect, reinforcing the notion of redundancy among these proteins (data not shown). Finally, the quadruple eps15/eps15R/epsin/clathrin KD reduced EGFR internalization at high and low doses to background levels (Fig. 6 C and D). Together with data obtained with EGFR/Ubmut, these results prove that eps15/eps15R and epsin are (redundantly) indispensable to the non-clathrin internalization of the EGFR.
Discussion
Our studies unveil important differences in the mechanisms through which the EGFR couples with the machinery of internalization as a function of EGF dose. At low doses of ligand, the receptor is almost exclusively internalized through a clathrin-dependent mechanism, whereas, at higher doses, a non-clathrin pathway defined on the basis of its nystatin/filipin-sensitivity emerges and becomes as relevant as the clathrin pathway. Of note, low and high EGF concentrations are physiologically relevant, as discussed in Supporting Materials and Methods. The emergence of the non-clathrin pathway correlates with the appearance of detectable ubiquitinated EGFR. A molecular genetics approach provided a mechanistic basis for this correlation, because a mutant EGFR that can no longer be ubiquitinated (Y1045F) was internalized through the clathrin pathway, even at high doses of EGF.
The dependence of the non-clathrin pathway on ubiquitination was further confirmed by a reductionistic approach, which exploited chimeric molecules that can signal solely through an Ub moiety. It should also be pointed out that such molecules are not “true” representations of receptors. In the case of the EGFR-based chimera, we have previously demonstrated that the chimera is constitutively internalized and routed to the degradative compartment, even in the absence of ligand stimulation (4). Once, however, the relevance of ubiquitination in non-clathrin endocytosis was established on the holo-EGFR, as discussed above, the chimera proved very useful in simplifying the plethora of signals present on an activated receptor and in allowing studies of internalization solely as a function of cargo ubiquitination.
Our results are consistent with data showing that EGFR internalization through clathrin-coated pits requires Cbl but not receptor ubiquitination (5, 24). These latter findings also indirectly stress the importance of Cbl-executed ubiquitination of other downstream endocytic proteins for proper clathrin endocytosis. A recent report by Stang et al. (25), which shows that Cbl-dependent ubiquitination is required for progression of the EGFR into coated pits, further reinforces this notion. Taken together with these results, our findings suggest a mechanism through which ubiquitination of endocytic machinery, but not cargo ubiquitination, is required for internalization into coated pits. At higher doses of EGF, however, when cargo ubiquitination becomes substantial, endocytosis is partially switched to a non-clathrin pathway.
How is this “switch” achieved? Our results directly implicate the recruitment of eps15/eps15R and epsin to the ubiquitinated cargo, as also shown by findings that expression of the EGFR/Ub chimera was able to induce relocalization of these proteins into caveolae. In addition, we showed that these proteins are redundantly essential in non-clathrin endocytosis of the EGFR. How the signal is further propagated downstream of these molecules in this pathway is not known and warrants further investigation. Furthermore, other Ub-binding proteins have been implicated in endocytosis. Thus, their potential role in the internalization of ubiquitinated cargo, possibly in a cell-specific fashion, also appears worth investigating.
Our results also indicate a role (albeit nonessential) for eps15/eps15R and epsin in clathrin internalization. Indeed, these proteins have been implicated in the formation of coated pits, and they have been proposed to function as adaptors between ubiquitinated cargo and components of the coat, such as AP2 and clathrin (2, 26). This latter possibility seems unlikely based on our present data, at least under a condition in which the clathrin route is paramount at low EGF concentrations, simply because there is not sufficient cargo ubiquitination at these doses. However, we have previously shown that clathrin-dependent synaptic vesicle recycling in Caenorhabditis elegans is negatively affected by the removal of eps15 through a mechanism that involves a genetic interaction with dynamin (27). It is possible, therefore, that, at least in the case of eps15, participation to clathrin endocytosis involves mechanisms different from interactions with ubiquitinated cargo.
Finally, we note that the TGF-β receptor internalizes through coated pits and caveolae (28). The two pathways were associated with increased receptor signaling and rapid receptor degradation, respectively (28). In the case of EGFR, at low EGF (when internalization proceeds exclusively through coated pits), the receptor is already fully competent for effector signaling (Fig. 1B). Conversely, at high EGF (when non-clathrin endocytosis becomes significant), there is no increase in signaling abilities but readily detectable increase in EGFR down-regulation (Fig. 1B). Thus, at minimum, caveolar/raft internalization does not apparently contribute to EGFR signaling, and possibly it is preferentially associated with receptor degradation.
Supplementary Material
Acknowledgments
We thank A. Benmerah (Université Paris 5, Paris), P. De Camilli (Yale University School of Medicine, New Haven, CT), G. Gill (University of California at San Diego, La Jolla), Genentech (South San Francisco, CA), and N. Sidenius and A. Andolfo (Instituto FIRC di Oncologia Molecolare) for reagents; P. Rubini and E. Cavallaro for technical assistance; and P. De Camilli, A. Musacchio, S. Confalonieri, and G. Scita for discussions. This work is supported by grants from the Italian Association for Cancer Research (to S.P., P.P.D.F., and C.T.), the Human Science Frontier Program, the European Community (VI Framework), and the Italian Ministry of Health (to P.P.D.F.).
Abbreviations: KD, knockdown; Ub, ubiquitin; UBmut, mutant Ub; UIM, Ub-interacting motif; HA, hemagglutinin.
See Commentary on page 2679.
References
- 1.Pickart, C. M. (2000) Trends Biochem. Sci. 25, 544–548. [DOI] [PubMed] [Google Scholar]
- 2.Di Fiore, P. P., Polo, S. & Hofmann, K. (2003) Nat. Rev. Mol. Cell Biol. 4, 491–497. [DOI] [PubMed] [Google Scholar]
- 3.Hicke, L. (1999) Trends Cell Biol. 9, 107–112. [DOI] [PubMed] [Google Scholar]
- 4.Haglund, K., Sigismund, S., Polo, S., Szymkiewicz, I., Di Fiore, P. P. & Dikic, I. (2003) Nat. Cell Biol. 5, 461–466. [DOI] [PubMed] [Google Scholar]
- 5.Jiang, X. & Sorkin, A. (2003) Traffic 4, 529–543. [DOI] [PubMed] [Google Scholar]
- 6.Duan, L., Miura, Y., Dimri, M., Majumder, B., Dodge, I. L., Reddi, A. L., Ghosh, A., Fernandes, N., Zhou, P., Mullane-Robinson, K., et al. (2003) J. Biol. Chem. 278, 28950–28960. [DOI] [PubMed] [Google Scholar]
- 7.Fazioli, F., Bottaro, D. P., Minichiello, L., Auricchio, A., Wong, W. T., Segatto, O. & Di Fiore, P. P. (1992) J. Biol. Chem. 267, 5155–5161. [PubMed] [Google Scholar]
- 8.Hinrichsen, L., Harborth, J., Andrees, L., Weber, K. & Ungewickell, E. J. (2003) J. Biol. Chem. [DOI] [PubMed]
- 9.Motley, A., Bright, N. A., Seaman, M. N. & Robinson, M. S. (2003) J. Cell Biol. 162, 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Confalonieri, S., Salcini, A. E., Puri, C., Tacchetti, C. & Di Fiore, P. P. (2000) J. Cell Biol. 150, 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levkowitz, G., Waterman, H., Ettenberg, S. A., Katz, M., Tsygankov, A. Y., Alroy, I., Lavi, S., Iwai, K., Reiss, Y., Ciechanover, A., et al. (1999) Mol. Cell 4, 1029–1040. [DOI] [PubMed] [Google Scholar]
- 12.Damke, H., Baba, T., Warnock, D. E. & Schmid, S. L. (1994) J. Cell Biol. 127, 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henley, J. R., Krueger, E. W., Oswald, B. J. & McNiven, M. A. (1998) J. Cell Biol. 141, 85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benmerah, A., Bayrou, M., Cerf-Bensussan, N. & Dautry-Varsat, A. (1999) J. Cell Sci. 112, 1303–1311. [DOI] [PubMed] [Google Scholar]
- 15.Slepnev, V. I., Ochoa, G. C., Butler, M. H. & De Camilli, P. (2000) J. Biol. Chem. 275, 17583–17589. [DOI] [PubMed] [Google Scholar]
- 16.Simons, K. & Toomre, D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 17.Pelkmans, L., Puntener, D. & Helenius, A. (2002) Science 296, 535–539. [DOI] [PubMed] [Google Scholar]
- 18.Chen, H., Fre, S., Slepnev, V. I., Capua, M. R., Takei, K., Butler, M. H., Di Fiore, P. P. & De Camilli, P. (1998) Nature 394, 793–797. [DOI] [PubMed] [Google Scholar]
- 19.Ford, M. G., Mills, I. G., Peter, B. J., Vallis, Y., Praefcke, G. J., Evans, P. R. & McMahon, H. T. (2002) Nature 419, 361–366. [DOI] [PubMed] [Google Scholar]
- 20.Raiborg, C., Bache, K. G., Gillooly, D. J., Madshus, I. H., Stang, E. & Stenmark, H. (2002) Nat. Cell Biol. 4, 394–398. [DOI] [PubMed] [Google Scholar]
- 21.Nakatsu, F., Sakuma, M., Matsuo, Y., Arase, H., Yamasaki, S., Nakamura, N., Saito, T. & Ohno, H. (2000) J. Biol. Chem. 275, 26213–26219. [DOI] [PubMed] [Google Scholar]
- 22.Bache, K. G., Raiborg, C., Mehlum, A. & Stenmark, H. (2003) J. Biol. Chem. 278, 12513–12521. [DOI] [PubMed] [Google Scholar]
- 23.Huang, F., Khvorova, A., Marshall, W. & Sorkin, A. (2004) J. Biol. Chem. 279, 16657–16661. [DOI] [PubMed] [Google Scholar]
- 24.Waterman, H., Katz, M., Rubin, C., Shtiegman, K., Lavi, S., Elson, A., Jovin, T. & Yarden, Y. (2002) EMBO J. 21, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stang, E., Blystad, F. D., Kazazic, M., Bertelsen, V., Brodahl, T., Raiborg, C., Stenmark, H. & Madshus, I. H. (2004) Mol. Biol. Cell 15, 3591–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wendland, B. (2002) Nat. Rev. Mol. Cell Biol. 3, 971–977. [DOI] [PubMed] [Google Scholar]
- 27.Salcini, A. E., Hilliard, M. A., Croce, A., Arbucci, S., Luzzi, P., Tacchetti, C., Daniell, L., De Camilli, P., Pelicci, P. G., Di Fiore, P. P. & Bazzicalupo, P. (2001) Nat. Cell Biol. 3, 755–760. [DOI] [PubMed] [Google Scholar]
- 28.Di Guglielmo, G. M., Le Roy, C., Goodfellow, A. F. & Wrana, J. L. (2003) Nat. Cell. Biol. 5, 410–421. [DOI] [PubMed] [Google Scholar]
- 29.Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P. & Di Fiore, P. P. (2002) Nature 416, 451–455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.