Abstract
Adeno-associated viruses (AAVs) such as AAV5 that transduce airway epithelia from the apical surface are attractive vectors for gene transfer in cystic fibrosis (CF). However, their utility in CF has been limited because packaging of the insert becomes inefficient when its length exceeds ≈4,900–5,000 bp. To partially circumvent this size constraint, we previously developed a CF transmembrane conductance regulator (CFTR) transgene that deleted a portion of the R domain (CFTRΔR). In this study, we focused on shortening the other elements in the AAV expression cassette. We found that portions of the CMV immediate/early (CMVie) enhancer/promoter could be deleted without abolishing activity. We also tested various intervening sequences, poly(A) signals, and an intron to develop an expression cassette that meets the size restrictions imposed by AAV. We then packaged these shortened elements with the CFTRΔR transgene into AAV5 and applied them to the apical surface of differentiated CF airway epithelia. Two to 4 weeks later, the AAV5 vectors partially corrected the CF Cl– transport defect. These results demonstrate that a single AAV vector can complement the CF defect in differentiated airway epithelia and thereby further the development of effective CF gene transfer.
Keywords: CMV, promoter, gene therapy
Cystic fibrosis (CF) is an autosomal recessive disease resulting from mutations in the gene encoding the CF transmembrane conductance regulator (CFTR) Cl– channel (1). Airway disease causes most of the morbidity and mortality in CF. Transfer of the CFTR cDNA to airway epithelia could potentially offer a new prevention and treatment for the people who suffer from this disease (for review see refs. 2–5). Previous studies established the feasibility of gene transfer to CF airway epithelial cells, both in vitro and in people with CF. However, such studies also revealed numerous barriers, which have so far prevented gene transfer from becoming an effective treatment.
Adeno-associated viruses (AAVs) overcome some of these barriers and have advantages that make them attractive vectors for CF gene transfer (5–10). AAVs cause no known human disease, and AAV vectors have a good safety record. AAV vectors retain no viral genes, thereby minimizing cell-mediated immune responses. They transduce nondividing cells, such as airway epithelia. Transgene expression from AAV vectors can be prolonged. In addition, recent studies have identified AAV types that can efficiently transduce well differentiated human airway epithelia, including AAV5, which has a receptor on the apical surface of airway epithelia (11–13).
A limitation of AAV vectors is the relatively small AAV genome. Studies testing the insert size suggest that 4,100–4,900 bp is the optimal genome size for packaging (14). Other studies and our own unpublished data also suggest that packaging becomes very inefficient whenever insert sizes exceed 4,900–5,000 bp (15, 16). This poses a problem for genes with large coding sequences, such as CFTR, which has a cDNA of 4,450 bp (17). With the addition of the two AAV inverted terminal repeats (ITRs), 3′ and 5′ untranslated regions, and enhancer/promoter elements, the expression cassette can easily surpass the packaging limits. A number of strategies have been explored to circumvent this limitation. Some studies have used short endogenous AAV sequences as a promoter (with or without proteosomal inhibitors to increase expression) with variable results in differentiated airway epithelia (15, 18, 19). Other studies have produced AAV vectors with a CFTR cDNA that deletes the N terminus (15), and other deletions have been proposed for AAV vectors (16, 18). However, their function in airway epithelia requires additional study. A different approach has involved coadministration of two independent vectors followed by intermolecular joining of the DNA or RNA to generate expression (20–23). Although promising, such strategies require that each cell be infected with two different vectors.
In an earlier study, we developed a transgene encoding a CFTR Cl– channel with a shortened R domain (deleting residues 708–759) (24). For simplicity, we call this CFTRΔR. The CFTRΔR variant showed normal biosynthesis, targeting to the apical membrane, and Cl– channel activity. Like the wild type, CFTRΔR complemented the Cl– transport defect in differentiated CF airway epithelia in vitro and in nasal mucosa of CF mice in vivo. Although shortening the CFTR cDNA takes us closer to a workable expression cassette, by itself it is not sufficient to meet the size constraints imposed by AAV. In this study we endeavored to minimize the length of the other components of an AAV expression cassette. After the CFTR cDNA, the next largest element is the enhancer/promoter. Therefore, we attempted to design a small but effective promoter. In addition, we minimized the size of other required elements in the expression vector. Then we tested the hypothesis that the expression cassette would function in well differentiated human airway epithelia and correct the CF Cl– transport defect.
Materials and Methods
Plasmids. We began with pCMV5, which contains a full-length CMV immediate/early (CMVie) enhancer/promoter with a multiple cloning site and a human growth hormone (hGH) poly(A) signal (25). We inserted the coding sequence for β-gal from pCMVβ (Clontech) to make pCMV5–β-gal and used this for making all of the other plasmids.
The truncated promoter constructs 348CMV and 222CMV were made by excising the enhancer region from –587 to –348 or –222, respectively. We produced construct Δ470–173 by deleting nucleotides 470–173 from pCMV5–β-gal. We generated construct 173CMV by deleting nucleotides from –587 to –173 and construct 113CMV by deleting nucleotides from –587 to –113. All truncated promoter constructs in plasmids retained nucleotides –600 to –587, which contain no known transcription factor binding sites. These nucleotides were removed when we generated the AAV5 vector.
We varied the length (44, 51, and 69 bp) of the intervening sequence (IVS) between the transcription start site and the translation start site of 173CMV and 113CMV. The 19S/16S intron (96-bp) from pCMVβ (Clontech) was excised with intact splice sites and ligated into 173CMV and 113CMV (with a 44-bp IVS). The synthetic poly(A) signal (SPA) (26) was introduced into pCMV5–β-gal, 173CMV, and 113CMV by using PCR mutagenesis (QuikChange, Stratagene). All constructs were sequenced from promoter to poly(A) signal for verification.
AAV Vectors. To make AAV5-173CMV-CFTRΔR and AAV5-113CMV-CFTRΔR, we excised CFTRΔR from pAd-CFTRΔR (24) and ligated it into 173CMV and 113CMV in place of the β-gal coding sequence. The NotI restriction sites were engineered upstream of the 173CMV and 113CMV promoters and downstream of the SPA in 173CMV-CFTRΔR and 113CMV-CFTRΔR. We also engineered a NotI site into the multiple cloning site of the AAV5 plasmid (27, 28) to insert 173CMV-CFTRΔR and 113CMV-CFTRΔR. AAV5-173CMV-CFTRΔR and AAV5-113CMV-CFTRΔR were produced as previously described (12) with the following modification: vectors were produced by transfection of 293T cells and harvested 48–60 h posttransfection. DNase-resistant particle titers were determined by quantitative PCR (29) with the following primers: forward, CTGCTTATATAGACCTCCCACCGTA, and reverse, AATAACCCCGCCCCGTT. Titers ranged from 5 × 1011 to 3 × 1012 particles per ml. In light of previous studies with AAV5 vectors expressing β-gal and GFP in COS cells, we estimated the ratio of transducing units to DNase-resistant particles as 1:104 (ref. 27 and unpublished observations). We used this value to calculate multiplicity of infection (moi) as transducing units per cell.
Cell Culture. A549 cells [CCL-185, American Type Culture Collection (ATCC)] were cultured in DMEM/10% Ham's F-12 medium with 10% FCS, 1% l-glutamine, and 1% penicillin/streptomycin. H441 cells (HTB-174, ATCC) were cultured in RPMI medium 1640 with 10% FCS and 1% penicillin/streptomycin. Primary cultures of non-CF and CF airway epithelia were cultured as previously described (30).
Transfection and Assays. A549 and H441 cell lines were transfected with 3 μg of DNA per 18 μl of Lipofectamine (Invitrogen). Airway cells were transfected with 3 μg of DNA per 18 μl of Lipofectamine with 200 moi of Ad-GFP for 1.5–2 h (31). β-gal activity was measured 48 h later by using the Galactolite assay (Applied Biosystems). Proteins were quantitated from soluble cell lysate using the Bio-Rad protein assay. For some studies, TNF-α (10 ng/ml) and IL-1β (50 ng/ml) were added together to the apical surface of A549 cells 12 h after transfection. Cells were lysed, and luminescence and protein were measured 12, 36, and 60 h later.
Differentiated CF airway epithelia were infected with 1 or 10 mois of AAV5 vectors for 4 h from the apical surface. After 14 or 28 days, epithelia were studied in Ussing chambers, and transepithelial current was measured as described (24, 30). CFTR Cl– current is shown as cAMP-stimulated bumetanide-sensitive short-circuit current.
Results and Discussion
An AAV5 Expression Cassette. Fig. 1 graphically depicts the 4,642-bp AAV5 genome (27), including the left and right flanking ITRs and the sequences encoding Rep and Cap genes. Below that we show a potential starting point for a CFTR expression cassette, which includes the viral ITRs, a CMVie enhancer/promoter, the IVS between the promoter and the transgene, the CFTR cDNA, and a poly(A) signal. The 6,065-bp total length exceeds the packaging capacity of AAV (refs. 14–16 and unpublished observations). Substituting the recently developed shortened CFTR transgene, CFTRΔR (4,287 bp) (24), reduced the cassette length to 5,902 bp. However, this still exceeds the packing limits. To achieve additional reduction, we first focused on the next largest element in the expression cassette, the enhancer/promoter.
Fig. 1.
AAV5 genome and expression cassette. The schematic shows the relative length of the left and right ITRs and the coding sequence for the Rep and Cap genes. Also shown is the relative size of the elements of a CMVie CFTR expression cassette.
We began these studies by using the CMVie enhancer/promoter because it has been successfully used in both viral and nonviral vectors, and it drives transgene expression in many different types of cells and tissues, including airway epithelia (24, 32–35). We also chose the CMVie enhancer/promoter because it is a very strong promoter (18, 32, 33, 35). Compared to another promoter used for CFTR gene transfer, the adenovirus E1a promoter, the CMVie promoter produced ≈100-fold more β-gal activity and ≈20-fold more CFTR mRNA (35). However, this very high level of expression may be more than is needed in CF because in airway epithelia the rate of transepithelial Cl– flow can be limited by basolateral membrane transporters rather than the amount of CFTR in the apical membrane. For example, the adenovirus E1a promoter was sufficient to correct Cl– current in CF airway epithelia while the much stronger CMVie promoter generated only ≈3-fold more transepithelial Cl– current (35). Thus, if activity fell as we shortened the CMVie promoter, we anticipated that it would still be more than sufficient for CFTR expression.
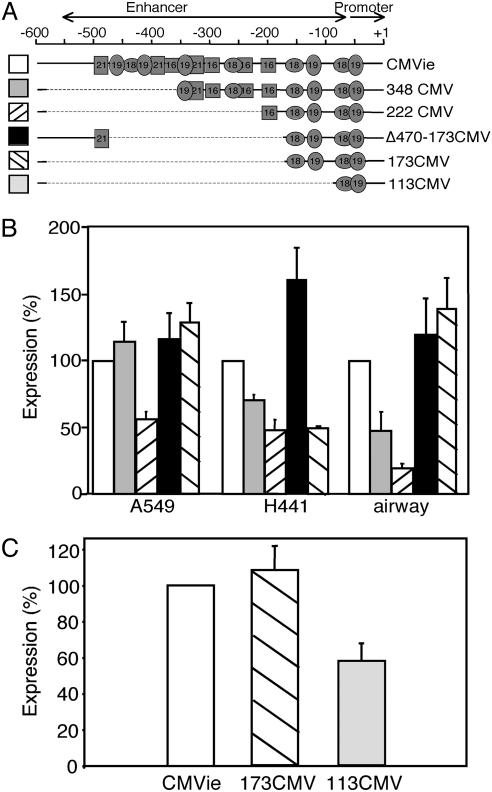
A schematic of the CMVie enhancer/promoter is shown in Fig. 2A. It includes the promoter itself (–50 to 1) and the enhancer region (–550 to –51) with motifs containing transcription factor binding sites: 16-, 18-, 19-, and 21-bp repeats. These binding sites are influenced by numerous cell factors and are major modulators of CMVie promoter activity. We focused on the 18-bp repeat bound by NF-κB/Rel and the 19-bp repeat bound by CREB/ATF (32).
Fig. 2.
Truncated CMVie enhancer/promoter constructs express β-gal. (A) A schematic of the CMVie enhancer/promoter with location of transcription factor binding sites. A diagram of some promoter constructs is shown below. (B) β-gal activity produced by the indicated constructs in A549, H441, and primary cultures of airway epithelial cells. Data are expressed as the percentage relative to that produced by the full-length CMVie promoter (n = 6–14 experiments). (C) β-gal activity produced by 173CMV and 113CMV in A549 cells, relative to that produced by the full-length CMVie promoter (n = 10 experiments).
Shortening the CMVie Enhancer/Promoter. To test shortened CMVie promoters, we made promoter constructs driving the reporter β-gal and measured transgene expression in two airway epithelial cell lines derived from human lung carcinomas, A549 and H441 cells. We also studied primary cultures of human airway epithelial cells. Earlier work had shown that truncating the CMVie promoter at two unique restriction sites, NdeI(–348) and NcoI (–222), yielded active promoters driving chloramphenicol acetyltransferase (CAT) activity in HeLa cells (25). We found that both of these truncated promoters retained substantial activity in airway epithelial cells, although the longer 348CMV promoter was more active than the 222CMV promoter (Fig. 2B). Because the CMVie promoter is such a strong promoter (25, 32–35), we were encouraged that the truncated constructs retained so much activity. Therefore, we tested whether the promoter could be shortened further.
The 222CMV promoter contained two 18- and two 19-bp repeats and a single 16-bp repeat. We asked whether retaining just the two 18- and 19-bp repeats would be sufficient for expression. We produced two constructs: 173CMV, which included the enhancer/promoter region up to –173, and Δ470–173CMV, which deleted the sequence between –470 and –173 and thus included the 21-bp repeat at –470 plus upstream sequences. Both truncated promoters generated β-gal activity equivalent to that generated by the full-length CMVie enhancer/promoter in A549 cells and primary cultures of airway epithelia (Fig. 2B). The 173CMV promoter produced less activity in H441 cells. These results suggested that two 18- and 19-bp repeats were sufficient for substantial promoter activity and led us to test a promoter with a single set of 18- and 19-bp repeats. A promoter truncated at –113 (113CMV) still generated substantial activity (Fig. 2C). Other constructs containing more 18- and 19-bp repeats did not produce greater β-gal activity (data not shown). When coupled with the CFTRΔR transgene, both the 173CMV and 113CMV truncated promoters were potentially short enough to fit in an AAV expression cassette. In addition, the strength of the full-length CMVie promoter suggested that a modest reduction in its activity would still be more than sufficient to effectively drive CFTR expression (34, 35). Therefore, we proceeded to optimize the other elements in an expression cassette containing these promoters.
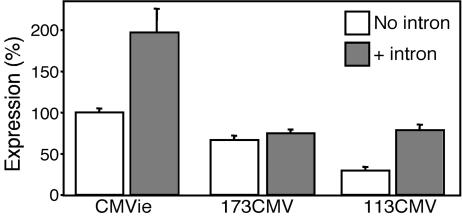
Evaluation of an Intron to Enhance Expression. Introns have been included in gene transfer expression cassettes to enhance transcriptional activity (33, 36). Consistent with this, we found that a full-length CMVie enhancer/promoter expression cassette that contained a simian virus 40 (SV40) 19S/16S intron generated more β-gal activity than did a construct that lacked the intron. When we added the SV40 19S/16S intron (96 bp) (37) to the 173CMV promoter, expression did not change, although expression from the 113CMV promoter increased (Fig. 3). Because of the relatively small effects of adding an intron and the additional length that it would introduce, we did not include it in the AAV expression cassette.
Fig. 3.
The 19S/16S intron from simian virus 40 (SV40) does not enhance activity in the 173CMV or 113CMV promoters. β-gal activity in A549 cells from full-length CMVie, 173CMV, or 113CMV promoters alone or with the 19S/16S intron from SV40 is shown. Data are expressed as the percentage relative to that produced by the full-length CMVie promoter with no intron (n = 8–12 experiments).
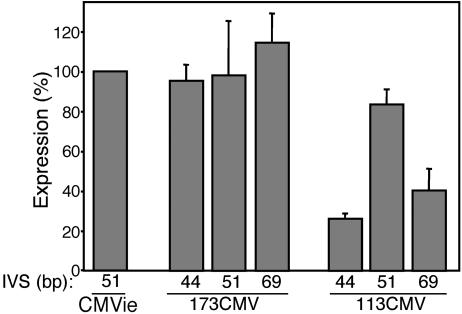
Evaluation of the Length of an IVS. The length of the IVS between a transcription and a translation start site can vary greatly. Although there is no consensus sequence for an IVS, minimal secondary structure seems to be important (38). We varied the length of the IVS from 44 to 69 bp, inserted these variations in the 173CMV and 113CMV promoter constructs, and tested expression in A549 cells (Fig. 4). Length of the IVS had little effect on expression from the 173CMV promoter, but the 51-bp IVS generated maximal expression from the 113CMV promoter. Therefore, we used the 51-bp IVS for the expression cassette.
Fig. 4.
A 51-bp IVS allows function of the 173CMV and 113CMV promoters. Expression from 173CMV and 113CMV promoters with IVSs of 44, 51, and 69 bp is shown as a percentage of β-gal activity produced by the full-length CMVie promoter with a 51-bp IVS in A549 cells (n = 4–17 experiments).
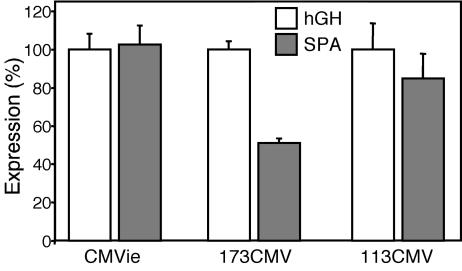
Assessment of a SPA. Poly(A) signals range from 600 to >1,000 bp. However, previous studies have shown that the required consensus sequence, AATAAAX22GTx/Tx, in a 49-bp sequence produces protein expression equivalent to that of a full-length poly(A) signal in HeLa cells (26). We compared this 49-bp SPA with the 624-bp hGH poly(A) signal present in pCMV5–β-gal. When coupled with the full-length and 113CMV promoters, the SPA sequence produced β-gal expression equivalent to that from the hGH sequence (Fig. 5). Although the SPA sequence showed reduced expression when combined with the 173CMV promoter, expression remained high. We therefore included the minimal 49-bp SPA in the expression cassette.
Fig. 5.
A SPA retains β-gal expression in A549 cells. Expression of β-gal driven by full-length CMVie, 173CMV, and 113CMV promoters with either the hGH poly(A) signal or the 49-bp SPA in A549 cells. Data are expressed as percentage of each promoter with the hGH poly(A) signal (n = 9 experiments).
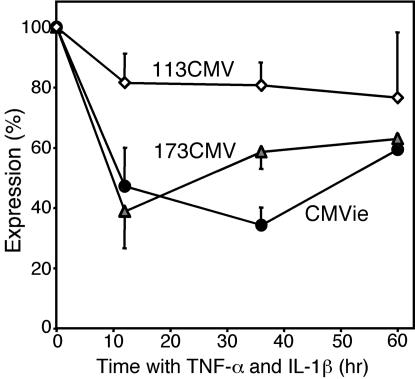
Effect of Cytokines on Expression from a Shortened Expression Cassette. Airways of patients with CF contain elevated levels of proinflammatory cytokines such as TNF-α and IL-1β (1, 39). Previous studies suggested that TNF-α can alter activity of the CMVie promoter and other widely used viral promoters (40, 41). We found that adding TNF-α and IL-1β reduced reporter expression from the full-length CMVie and 173CMV promoters but had minimal effects on the 113CMV promoter (Fig. 6). Thus, the shortened promoters show no greater susceptibility to cytokine effects, and the 113CMV promoter may be less prone to cytokine-dependent attenuation than the CMVie enhancer/promoter.
Fig. 6.
Short promoters are not more susceptible to the cytokines TNF-α and IL-1β than is the full-length CMVie promoter. A549 cells were transfected with pCMV5-β-gal, 173CMV, or 113CMV. Cells were treated 12 h later with TNF-α (10 ng/ml) and IL-1β (50 ng/ml) or medium alone and incubated for an additional 12, 36, or 60 h. β-gal activity is expressed as percentage of β-gal activity in the absence of cytokines for each promoter (n = 6 experiments).
Construction of an AAV5 Expression Cassette for CFTR. Because of these studies, we constructed two expression cassettes by combining the optimized elements. The AAV5-173CMV-CFTRΔR vector contained the following elements: left ITR (167 bp), retained plasmid sequence with NotI site (34 bp), promoter (173 bp), IVS (51 bp), CFTRΔR (4,287 bp), SPA (49 bp) retained plasmid sequence with NotI site (12 bp), and right ITR (167 bp) for a total length of 4,940 bp. The AAV5-113CMV-CFTRΔR vector was identical except that the promoter was 113 bp, for a total length of 4,880 bp. Both expression cassettes were predicted to fall in the range for optimal AAV packaging (14). No difference in packaging efficiency or vector production was observed with either vector. Therefore, we used these expression cassettes to generate recombinant AAV5 vectors.
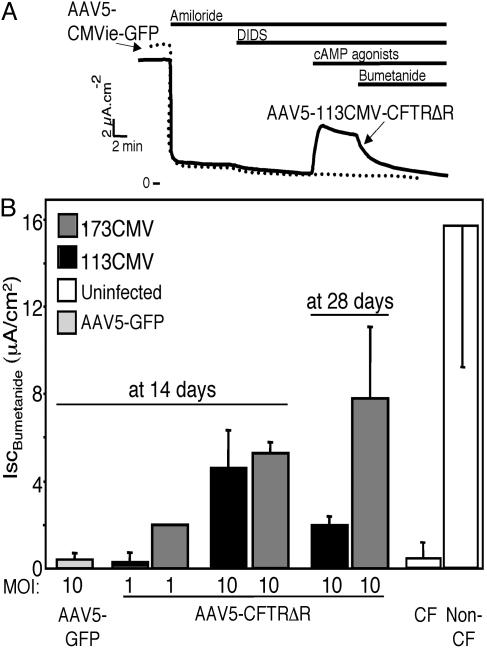
Correction of the Transepithelial Cl– Transport Defect in CF Epithelia. To learn whether these vectors could correct the CF Cl– transport defect, we applied them for 4 h to the apical surface of well differentiated primary cultures of CF airway epithelia. Fourteen to 28 days later, we measured transepithelial Cl– current. Both AAV5-173CMV-CFTRΔR and AAV5-113CMV-CFTRΔR generated cAMP-stimulated bumetanide-sensitive current (Fig. 7). Correction of Cl– transport increased as the moi increased and persisted for at least 28 days after infection. Consistent with the β-gal expression data, AAV5-173CMV-CFTRΔR tended to produce more current than AAV5-113CMV-CFTRΔR. Importantly, levels of Cl– current correction approached those obtained in non-CF airway epithelia.
Fig. 7.
AAV5–173CMV-CFTRΔR and AAV5-113CMV-CFTRΔR vectors drive expression of regulated CFTR Cl– currents in well differentiated CF airway epithelia. Epithelia were infected with indicated moi of vector, and cAMP-stimulated bumetanide-sensitive current was measured 14 or 28 days later. (A) Representative current traces. DIDS, 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid. (B) Data are average values from vector-treated and untreated CF and non-CF epithelia. At 14 days, GFP, 113CMV, and 173CMV at both mois (n = 3 epithelia). At 28 days, 113CMV (n = 12 epithelia) and 173CMV (n = 6 epithelia). ISC, short-circuit current.
Conclusion and Future Studies. These data provide, to our knowledge, the first evidence that a single AAV vector administered by itself can correct the CF Cl– transport defect in differentiated CF airway epithelia. Thus, they set the stage for additional studies designed to develop a CF gene transfer vector. Nevertheless, additional questions remain to be answered.
Will AAV5 vectors target the apical surface of differentiated airway epithelia with enough efficiency to correct CF electrolyte transport defects and prevent disease? We chose AAV5 because it binds receptors in the apical membrane: 2,3-linked sialic acid and the platelet-derived growth factor (PDGF) receptor (11–13). However, it remains uncertain whether another AAV type might be more efficient (42–44). If so, the expression cassette we have developed may be of value for such vectors. The minimal elements in the expression cassette we describe may also be of value in transferring other cDNAs that exceed the AAV packaging limit.
Will the expression cassette we have engineered produce prolonged CFTR expression? This question may need to be answered in differentiated human airway epithelia and in the context of AAV5 delivery. It is also possible that results could vary between in vitro and in vivo studies, and in CF and non-CF airways.
What level of expression will be sufficient to correct the CF Cl– transport defect and prevent or treat the disease? Previous studies showed that the endogenous CFTR promoter is very weak, producing perhaps one to two transcripts per cell (45). Even as little as 8% of normal transcripts seem to preserve normal lung function (46). However, in those cases the endogenous CFTR promoter drove expression in most, if not all, of the airway epithelial cells. In contrast, a gene transfer vector will express CFTR in a smaller fraction of cells. In this regard, we were encouraged that the shortened expression cassettes we developed retained such a large fraction of CMVie enhancer/promoter activity. This level of promoter activity will likely be more than sufficient for CF gene transfer because the CMVie promoter is very strong (18, 32, 33) and because promoters with 1–10% of its activity correct the Cl– transport nearly as well as the CMVie promoter does (34, 35).
Development of an AAV5 vector that can correct the Cl– transport defect in differentiated CF airway epithelia will allow the pursuit of these and other questions that are key for CF gene transfer.
Acknowledgments
We thank Tamara Nesselhauf, Daniel Vermeer, Autumn Ruiz, Beverly Handelman, and Theresa Mayhew for assistance; the University of Iowa In Vitro Models and Cell Culture Core [supported by National Heart, Lung, and Blood Institute (NHLBI) Grant HL51670, Cystic Fibrosis Foundation (CFF) Grants R458-CR02 and ENGLH9850, and National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant DK54759] and Vector Core (supported by the Roy J. Carver Charitable Trust, NHLBI Grant HL51670, CFF Grant R458-CR02, and NIDDK Grant DK54759) for valuable assistance; and Amgen Biologicals for the gift of recombinant human keratinocyte growth factor for cell culture. This work was supported by NHLBI Grant HL51670 (to M.J.W.), CFF Grant R458-CR02 (to M.J.W.), and National Institute of Allergy and Infectious Diseases Grant AI-13562 (to M.F.S.). J.A.C. is supported by an intramural grant from the National Institute of Dental and Craniofacial Research. M.J.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: CF, cystic fibrosis; CFTR, CF transmembrane regulator; AAV, adeno-associated virus; ITR, inverted terminal repeat; CMVie, CMV immediate/early; hGH, human growth hormone; IVS, intervening sequence; SPA, synthetic poly(A) signal; moi, multiplicity of infection.
References
- 1.Welsh, M. J., Ramsey, B. W., Accurso, F. & Cutting, G. R. (2001) in The Metabolic and Molecular Basis of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B. & Vogelstein, B. (McGraw–Hill, New York), pp. 5121–5189.
- 2.Flotte, T. R. (1999) Curr. Opin. Mol. Ther. 1, 510–516. [PubMed] [Google Scholar]
- 3.Welsh, M. J. (1999) J. Clin. Invest. 104, 1165–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, J. C., Geddes, D. M. & Alton, E. W. (2001) J. Gene Med. 3, 409–417. [DOI] [PubMed] [Google Scholar]
- 5.Driskell, R. A. & Engelhardt, J. F. (2003) Annu. Rev. Physiol. 65, 585–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.High, K. A. (2001) Ann. N.Y. Acad. Sci. 953, 64–74. [DOI] [PubMed] [Google Scholar]
- 7.Grimm, D. & Kay, M. A. (2003) Curr. Gene Ther. 3, 281–304. [DOI] [PubMed] [Google Scholar]
- 8.Conlon, T. J. & Flotte, T. R. (2004) Expert Opin. Biol. Ther. 4, 1093–1101. [DOI] [PubMed] [Google Scholar]
- 9.Halbert, C. L. & Miller, A. D. (2004) Methods Mol. Biol. 246, 201–212. [DOI] [PubMed] [Google Scholar]
- 10.McCarty, D. M., Young, S. M. & Samulski, R. J. (2004) Annu. Rev. Genet. 38, 819–845. [DOI] [PubMed] [Google Scholar]
- 11.Zabner, J., Seiler, M., Walters, R., Kotin, R. M., Fulgeras, W., Davidson, B. L. & Chiorini, J. A. (2000) J. Virol. 74, 3852–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters, R. W., Yi, S. M., Keshavjee, S., Brown, K. E., Welsh, M. J., Chiorini, J. A. & Zabner, J. (2001) J. Biol. Chem. 276, 20610–20616. [DOI] [PubMed] [Google Scholar]
- 13.Di Pasquale, G., Davidson, B. L., Stein, C. S., Martins, I., Scudiero, D., Monks, A. & Chiorini, J. A. (2003) Nat. Med. 9, 1306–1312. [DOI] [PubMed] [Google Scholar]
- 14.Dong, J. Y., Fan, P. D. & Frizzell, R. A. (1996) Hum. Gene Ther. 7, 2101–2112. [DOI] [PubMed] [Google Scholar]
- 15.Flotte, T. R., Afione, S. A., Solow, R., Drumm, M. L., Markakis, D., Guggino, W. B., Zeitlin, P. L. & Carter, B. J. (1993) J. Biol. Chem. 268, 3781–3790. [PubMed] [Google Scholar]
- 16.Zhang, L., Wang, D., Fischer, H., Fan, P. D., Widdicombe, J. H., Kan, Y. W. & Dong, J. Y. (1998) Proc. Natl. Acad. Sci. USA 95, 10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riordan, J. R., Rommens, J. M., Kerem, B. S., Alon, N., Rozmahel, R., Grzelczak, Z., Zielenski, J., Lok, S., Plavsic, N., Chou, J. L., et al. (1989) Science 245, 1066–1073. [DOI] [PubMed] [Google Scholar]
- 18.Wang, D., Fischer, H., Zhang, L., Fan, P., Ding, R. X. & Dong, J. (1999) Gene Ther. 6, 667–675. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, L. N., Karp, P., Gerard, C. J., Pastor, E., Laux, D., Munson, K., Yan, Z., Liu, X., Godwin, S., Thomas, C. P., et al. (2004) Mol. Ther. 10, 990–1002. [DOI] [PubMed] [Google Scholar]
- 20.Nakai, H., Storm, T. A. & Kay, M. A. (2000) Nat. Biotechnol. 18, 527–532. [DOI] [PubMed] [Google Scholar]
- 21.Yan, Z., Zhang, Y., Duan, D. & Engelhardt, J. F. (2000) Proc. Natl. Acad. Sci. USA 97, 6716–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halbert, C. L., Allen, J. M. & Miller, A. D. (2002) Nat. Biotechnol. 20, 697–701. [DOI] [PubMed] [Google Scholar]
- 23.Pergolizzi, R. G., Ropper, A. E., Dragos, R., Reid, A. C., Nakayama, K., Tan, Y., Ehteshami, J. R., Coleman, S. H., Silver, R. B., Hackett, N. R., et al. (2003) Mol. Ther. 8, 999–1008. [DOI] [PubMed] [Google Scholar]
- 24.Ostedgaard, L. S., Zabner, J., Vermeer, D. W., Rokhlina, T., Karp, P. H., Stecenko, A. A., Randak, C. & Welsh, M. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3093–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stinski, M. F. & Roehr, T. J. (1985) J. Virol. 55, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitt, N., Briggs, D., Gil, A. & Proudfoot, N. J. (1989) Genes Dev. 3, 1019–1025. [DOI] [PubMed] [Google Scholar]
- 27.Chiorini, J. A., Kim, F., Yang, L. & Kotin, R. M. (1999) J. Virol. 73, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiorini, J. A., Yang, L., Liu, Y., Safer, B. & Kotin, R. M. (1997) J. Virol. 71, 6823–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaludov, N., Brown, K. E., Walters, R. W., Zabner, J. & Chiorini, J. A. (2001) J. Virol. 75, 6884–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karp, P. H., Moninger, T., Weber, S. P., Nesselhauf, T. S., Launspach, J., Zabner, J. & Welsh, M. J. (2002) in Epithelial Cell Culture Protocols, ed. Wise, C. (Humana, Totowa, NJ), Vol. 188, pp. 115–137. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. H. & Welsh, M. J. (1999) Gene Ther. 6, 676–682. [DOI] [PubMed] [Google Scholar]
- 32.Stinski, M. F. (1999) in Gene Expression Systems: Using Nature for the Art of Expression, eds. Fernandez, J. M. & Holffler, J. P. (Academic, San Diego), pp. 211–233.
- 33.Yew, N. S., Wysokenski, D. M., Wang, K. X., Ziegler, R. J., Marshall, J., McNeilly, D., Cherry, M., Osburn, W. & Cheng, S. H. (1997) Hum. Gene Ther. 8, 575–584. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, C., O'Connor, S. P., Armentano, D., Berthelette, P. B., Schiavi, S. C., Jefferson, D. M., Smith, A. E., Wadsworth, S. C. & Cheng, S. H. (1996) Am. J. Physiol. 271, L527–L537. [DOI] [PubMed] [Google Scholar]
- 35.Zabner, J., Wadsworth, S. C., Smith, A. E. & Welsh, M. J. (1996) Gene Ther. 3, 458–465. [PubMed] [Google Scholar]
- 36.Nott, A., Meislin, S. H. & Moore, M. J. (2003) RNA 9, 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okayama, H. & Berg, P. (1983) Mol. Cell. Biol. 3, 280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak, M. (1986) Proc. Natl. Acad. Sci. USA 83, 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konstan, M. W. & Davis, P. B. (2002) Adv. Drug Deliv. Rev. 54, 1409–1423. [DOI] [PubMed] [Google Scholar]
- 40.Ritter, T., Brandt, C., Prösch, S., Vergopoulos, A., Vogt, K., Kolls, J. & Volk, H. D. (2000) Cytokine 12, 1163–1170. [DOI] [PubMed] [Google Scholar]
- 41.Stein, J., Volk, H. D., Liebenthal, C., Kruger, D. H. & Prosch, S. (1993) J. Gen. Virol. 74, 2333–2338. [DOI] [PubMed] [Google Scholar]
- 42.Halbert, C. L., Allen, J. M. & Miller, A. D. (2001) J. Virol. 75, 6615–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao, G., Alvira, M. R., Somanathan, S., Lu, Y., Vandenberghe, L. H., Rux, J. J., Caldcedo, R., Sanmiguel, J., Abbas, Z. & Wilson, J. M. (2003) Proc. Natl. Acad. Sci. USA 100, 6081–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, M., Katano, H., Bossis, I. & Chiorini, J. A. (2004) J. Virol. 78, 6509–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell, B. C., Chu, C. S., Paakko, P. K., Banks, T. C., Yoshimura, K., Ferrans, V. J., Chernick, M. S. & Crystal, R. G. (1991) Proc. Natl. Acad. Sci. USA 88, 6565–6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu, C. S., Trapnell, B. C., Curristin, S. M., Cutting, G. R. & Crystal, R. G. (1992) J. Clin. Invest. 90, 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]