Abstract
Preoperative chemoradiotherapy (CRT) followed by mesorectal excision is the standard treatment for patients with locally advanced rectal cancer (LARC). The balance between treatment efficacy and toxicity is a major issue in the clinical management of these patients. There is a requirement for the identification of predictive molecular biomarkers for the response of patients to CRT. The present study aimed to analyze the association between microRNA (miRNA/miR) expression and treatment efficacy in patients with LARC who were treated with preoperative CRT. From previous clinical trials, 55 patients for the test cohort and 130 patients for the validation cohort met the criteria for the present investigation. Through reverse transcription-quantitative polymerase chain reaction analysis, the expression of miR-21, −31, −125b, −145 and −630 in the diagnostic biopsies was analyzed. The primary endpoint of tumor regression was evaluated according to Mandard's Tumor Regression Grade (TRG) system. In the test cohort, a significant association was identified between low miRNA-145 expression and TRG1+2 (P=0.0003). Similarly, this association was identified in the validation cohort, although it did not reach statistical significance. Furthermore, a significant association between high miRNA-21 expression and TRG1+2 (P=0.035) was observed in the validation cohort. The remaining miRNAs analyzed were not associated with TRG. The results of the present study highlight the clinical importance of miRNAs in LARC and underline the necessity for validation studies in this setting.
Keywords: biological marker, microRNA, preoperative chemoradiotherapy, rectal cancer, validation
Introduction
Preoperative 5-fluorouracil-based chemoradiotherapy (CRT) followed by mesorectal excision is the standard treatment for patients with locally advanced rectal cancer (LARC) (1–3). Several previous studies have demonstrated that CRT significantly downstages the disease and reduces the risk of local tumor recurrence (4–6). The response rate to CRT in this group varies, with 9–30% of patients having a pathological complete response (pCR) and 46–60% achieving some degree of tumor downstaging (7,8). An improved long-term outcome has been demonstrated for patients with pCR compared with patients who are non-responsive to CRT (9–12); however, this treatment has several long-term side effects. Therefore, the appropriate selection of patients who will respond to CRT is important. At present, only certain clinical parameters and radiological investigations are available for use in the selection of patients for CRT, which is insufficient. There is a requirement for biomarkers that are able to predict the effect of preoperative CRT.
MicroRNAs (miRNAs/miRs) are short (18–25 nucleotides) non-coding RNA molecules that act as negative gene regulators at the post-transcriptional level. miRNAs serve an important role in the regulation of biological processes, such as cell differentiation, proliferation and apoptosis. Numerous miRNAs interfere with the expression of oncogenes and tumor suppressor genes with a direct involvement in carcinogenesis (13,14). Due to their association with cancer, miRNAs are being investigated as potential diagnostic and prognostic biomarkers, and predictors of treatment response (15,16).
In the present study, 5 miRNAs were chosen for study based on the literature and our group's previous methodological work. In rectal cancer tissue, reference genes (miR-193a-5p, miR −27a and let-7g) for the relative quantification of miRNAs have previously been identified (17) and the intratumoral heterogeneity of the present panel of miRNAs (miR-21, −31, −125b, −145 and −630) has been assessed (18).
miRNA-21 is overexpressed in rectal cancer and its downregulation following neoadjuvant CRT has been suggested (19–21). Furthermore, overexpression of miRNA-21 has been associated with complete tumor regression following neoadjuvant CRT (22). Increased expression of miRNA-31 has been detected in colorectal cancer (CRC), with a positive correlation between its expression and the stage of the disease (23,24). In addition, the upregulation of miRNA-125b and miRNA-145 in rectal cancer tissue following neoadjuvant CRT has been reported (19,25). Della Vittoria Scarpati et al (26) demonstrated that miRNA-630 had 100% sensitivity and specificity in selecting patients with complete tumor regression following CRT. However, only one of the studies described above included a validation of their results (22). Consequently, the majority of them serve as hypothesis-generating investigations. The present study aimed to analyze the association between miRNA expression and the treatment efficacy of preoperative CRT in a test cohort of LARC patients, and to subsequently validate the results in an independent cohort.
Materials and methods
Patient populations
Reporting of all data in the present study is in accordance with the Reporting recommendations for tumor marker prognostic studies criteria (27). The present study consisted of a test and a validation cohort of patients with LARC. The patients were enrolled in previously conducted clinical trials, and the details of patients, treatment and follow-up interval times are available from the pertaining publications (28–30). Briefly, the test cohort began with 85 patients treated in one of two following trials: i) 50 patients (enrollment, November 2002-June 2004) with resectable T3 rectal cancer treated with preoperative CRT (60 Gy/30 fractions, and concurrent tegafur-uracil (300 mg/m2) and L-leucovorin (22.5 mg/day) combined with an endorectal boost (5 Gy/1 fraction) (28); ii) 35 patients (enrollment, June 2004-January 2005) with resectable T3-4 rectal cancer receiving similar treatment combined with the cyclooxygenase-2-inhibitor celecoxib (400 mg twice a day) (30). The validation cohort (enrollment, March 2005-November 2009) was based on a previous randomized phase III trial (29) of 224 patients with resectable T3-4 rectal cancer receiving neoadjuvant CRT (50.4 Gy/28 fractions, and concomitant tegafur-uracil (300 mg/m2) and L-leucovorin (22.5 mg/day) with (arm B) or without (arm A) an endorectal boost (10 Gy/2 fractions)).
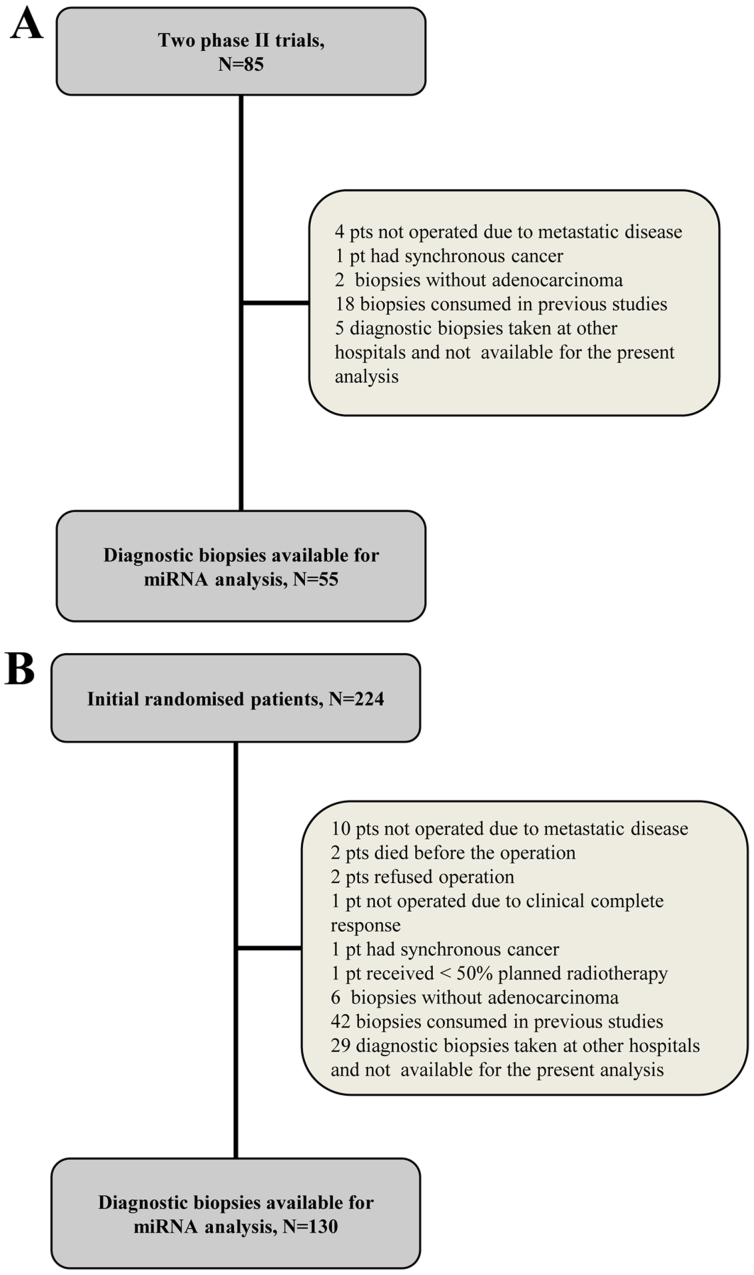
The present study was approved by the Regional Committee on Health Research Ethics of Southern Denmark and the requirement for written informed consent was waived (protocol ID no. S-20140083). The study was registered with The Danish Data Protection Agency and The Danish Registry of Human Tissue Utilization was consulted prior to any tissue samples being used. The inclusion criteria were the availability of formalin-fixed paraffin-embedded (FFPE) diagnostic biopsies containing adenocarcinoma tissue and tumor specimens for the pathological evaluation of tumor regression grade (TRG). The exclusion criteria were the presence of synchronous malignant diseases (with the exception of non-melanoma skin cancer and carcinoma in situ of the cervical uterus). A total of 55 and 130 patients from the test and validation cohorts, respectively, met the criteria for the present investigation. Flowcharts of the study populations are presented in Fig. 1 and a summary of their clinicopathological characteristics are presented in Table I.
Figure 1.
CONSORT flow diagram of the (A) test and (B) validation populations. miRNA, microRNA.
Table I.
Clinicopathological characteristics and response to chemoradiotherapy of patients in the test and validation study cohorts.
| Clinicopathological characteristic | Test cohort (N=55), no. of patients (%) | Validation cohort (N=130), no. of patients (%) |
|---|---|---|
| Gender | ||
| Female | 19 (34.5) | 50 (38) |
| Male | 36 (65.5) | 80 (62) |
| T-stage | ||
| T3 | 48 (87) | 110 (85) |
| T4 | 7 (13) | 20 (15) |
| N-stage | ||
| N0 | 16 (29) | 14 (11) |
| N+ | 39 (71) | 116 (89) |
| TRG | ||
| TRG1 | 13 (23.5) | 26 (20) |
| TRG2 | 13 (23.5) | 17 (13) |
| TRG3 | 28 (51) | 75 (58) |
| TRG4 | 1 (2) | 12 (9) |
In the test cohort the median age was 62 (range, 40–78), in the validation cohort the median age was 64 (range, 35–79). No patients had TRG5. TRG, tumor regression grade according to Mandard et al (31); T, tumor; N, node, according to the tumor-node-metastasis (TNM) classification of malignant tumours (UICC, international union against cancer).
Samples
Diagnostic biopsies from the rectal tumors followed routine preservation (FFPE), and were transported and stored at room temperature. The median storage duration from archiving to analysis was 11.5 years in the test cohort and 9 years in the validation cohort. Histological sections stained with hematoxylin and eosin (H&E) were examined by a pathologist in order to ensure the presence of tumor cells in the analyzed sections. From the corresponding tissue blocks, 8-µm-thick sections were cut for use in subsequent reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. In the test population, areas of tumor cells were identified by a pathologist and encircled as regions of interest (ROI) on an image of the H&E-stained section. The ROI were isolated through membrane-based laser microdissection (LMD) using a Leica LMD6500 Microsystems (Leica Microsystems GmbH, Wetzlar, Germany) and collected in the caps of 0.5 ml RNase-free PCR tubes with a drop of ethanol (99%). In the validation population, LMD was performed as described above. However, in a subset of cases in the validation cohort LMD was not required, since the whole biopsy consisted of tumor tissue without marked inflammation. In these cases (N=26) the tissue was removed from the slides using a scalpel and collected into 1.5 ml RNase-free PCR tubes containing a drop of ethanol (99%).
Expression analysis via RT-qpcr
Normalizer miRNAs
Recently, our group performed a study (17) on miRNA expression profiling to identify and validate reference genes for the relative quantification of miRNAs in rectal cancer tissue. miR-193a-5p, miR-27a and let-7g were identified as the most stably expressed miRNAs in rectal cancer tissue, and the mean expression value of these three miRNAs were subsequently used for normalization in the present study.
RNA extraction
Total RNA was isolated from the FFPE tissue using the miRNeasy FFPE kit for the test study and the AllPrep DNA/RNA FFPE kit (both Qiagen GmbH, Hilden, Germany) for the validation study according to the manufacturer's protocol. Total RNA was eluted into 14 µl RNase-free water.
RT-qPCR
RT and preamplification were performed as previously described (18). Custom TaqMan® MicroRNA Single Assays (Thermo Fisher Scientific, Inc., Waltham, MA, USA) for hsa-let-7g (cat. no. 002282), hsa-miR-193a-5p (cat. no. 002281), hsa-miR-27a (cat. no. 000408), hsa-miR-21 (cat. no. 000397), hsa-miR-31 (cat. no. 002279), hsa-miR-125b (cat. no. 000449), hsa-miR-145 (cat. no. 002278) and hsa-miR-630 (cat. no. 001563) were used, according to the manufacturer's protocol. The qPCR analyses were carried out using a QuantStudio™ 12K Flex Real-Time PCR system (Thermo Fisher Scientific, Inc.) with 1 µl diluted preamplification product, TaqMan MicroRNA Assays and TaqMan® Universal Master Mix II NoAmpErase® UNG in a total reaction volume of 20 µl. All reactions were performed in triplicate. Data analysis was performed using Quantstudio 12K Flex software (version 1.2.2; Thermo Fisher Scientific, Inc.) and relative quantification was performed using the 2−ΔΔCq method as described by Eriksen et al (18). The mean expression value of miR-193a-5p, miR-27a and let-7g was used as the normalization factor. Water was used as the negative control. A no-template control was included in the entire process and analyzed together with samples. The analyses were performed by staff blinded to the patient outcome.
Endpoints
The primary endpoint of tumor regression was determined by assessing the operative specimens according to Mandard's Tumor Regression Grade (TRG) system (31,32), as follows: TRG1, no residual tumor; TRG2, microscopic residual tumor; TRG3, moderate response; TRG4, minor response; and TRG5, no response. Overall survival (OS) and disease-free survival (DFS) were the secondary endpoints. OS was defined as the time from inclusion in the primary study until mortality from any cause. DFS was calculated as the time from inclusion in the primary study until the first documented tumor progression or mortality from any cause.
Statistical analysis
Wilcoxon rank-sum test was used for comparison of medians between groups. The prognostic value of variables was analyzed using the log rank-test and survival curves were produced using the Kaplan-Meier estimator. Patients with additional malignancies were excluded from the DFS analysis (test study, N=9; validation study, N=16). Clinical outcome data were last updated in April 2016. Possible correlations between continuous data were analyzed and visualized using the parametric linear regression analysis. Univariate Cox's regression analysis was used to estimate the prognostic value of individual variables and those with P<0.1 were included in a multivariate Cox's regression analysis for independent prognostic value. All tests were two-tailed. All statistical analyses were performed using NCSS Statistical Software 2007 (version 07.1.20; NSCC, LLC, Kaysville, UT, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Test population
The test cohort analyses consisted of 55 patients and the treatment compliance was high (28). The median follow-up time was 11.6 years. All patients were responsive to preoperative CRT. Patient characteristics and treatment responses are presented in Table I. An expression of miR-145 below the median expression level was significantly associated with major response (TRG1+2) to treatment (P<0.001; Table II). For the other investigated miRNAs no significant association between their expression and TRG was identified (Table II). For survival analysis, patients were grouped according to their median expression of the miRNA of interest [above (high) or below (low) the median]. No significant difference was identified between the high and low expression groups for the investigated miRNAs following OS and DFS analyses (data not shown).
Table II.
Association between TRG and miR expression in the test cohort (N=55).
| miR expression, median (95% CI) | |||
|---|---|---|---|
| miR | TRG1+2 | TRG3+4 | P-value |
| 125b | 1.030 (0.558–1.447) | 1.708 (0.951–2.026) | 0.174 |
| 145 | 9.854 (5.501–13.846) | 17.370 (13.515–25.070) | <0.001 |
| 21 | 14.206 (11.529–18.195) | 17.463 (16.057–19.450) | 0.062 |
| 3 | 0.128 (0.051–0.235) | 0.136 (0.066–0.262) | 0.463 |
| 630 | 0.004 (0.003–0.005) | 0.003 (0.002–0.004) | 0.104 |
TRG, tumor regression grade according to Mandard et al (31); CI, confidence interval; miR, microRNA.
Validation population
A total of 130 patients were included in the validation population. Patient characteristics and treatment responses are presented in Table I. All patients, with the exception of two, received >80% of the planned radiotherapy with a curative intent. Eight patients had developed distant metastases in the liver and/or lungs at the time of surgery. In the validation population, an expression of miR-21 above the median expression level was significantly associated (P=0.035) with major response (TRG1+2) to treatment (Table III). The association between miR-145 expression and TRG was similar to the one demonstrated in the test cohort, although it did not reach statistical significance (P=0.085). For miR-125b, −31 and −630, no significant association between expression and TRG was detected (Table III).
Table III.
Association between TRG and miR expression in the validation cohort (N=130).
| miR expression, median (95% CI) | |||
|---|---|---|---|
| miR | TRG1+2 | TRG3+4 | P-valuea |
| 125b | 1.338 (1.009–1.729) | 1.566 (1.252–1.922) | 0.337 |
| 145 | 18.735 (13.923–22.943) | 22.015 (18.164–27.612) | 0.085 |
| 21 | 26.861 (22.906–29.768) | 21.107 (19.615–22.829) | 0.035 |
| 3 | 0.109 (0.072–0.391) | 0.135 (0.090–0.210) | 0.761 |
| 630 | 0.001 (0.001–0.002) | 0.001 (0.001–0.001) | 0.333 |
TRG1+2 vs. TRG3+4. TRG, tumor regression grade according to Mandard et al (31); CI, confidence interval; miR, microRNA.
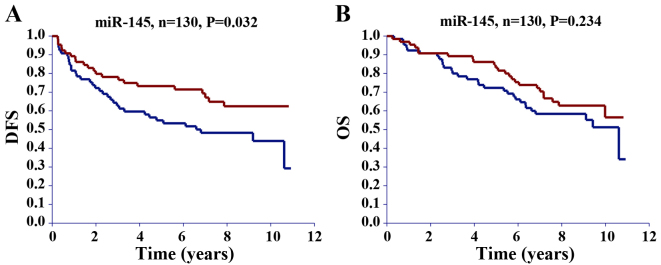
Grouping patients in the validation cohort according to their median expression of the miRNA of interest revealed that patients with miR-125b and miR-145 expression below the median had a significantly better DFS compared with those with expression above the median (Table IV). Survival curves for miR-145 are illustrated in Fig. 2. No significant difference in DFS was detected for miR-21, −31 and −630 (Table IV). No significant difference was demonstrated between the high and low expression of any of the investigated miRNAs and OS (Table IV).
Table IV.
Univariate Cox's regression analysis of the association between clinicopathological characteristics and DFS and OS in the validation cohort (N=130).
| DFS | OS | |||||
|---|---|---|---|---|---|---|
| Clinicopathological characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Gender | 0.50–1.42 | 0.517 | 0.64–1.93 | 0.711 | ||
| Female | 1 | 1 | ||||
| Male | 0.84 | 1.11 | ||||
| Age | 0.84–2.41 | 0.187 | 1.22–3.74 | 0.008 | ||
| <64 | 1 | 1 | ||||
| ≥64 | 1.42 | 2.14 | ||||
| T-stage | 0.78–2.92 | 0.220 | 0.47–2.09 | 0.973 | ||
| T3 | 1 | 1 | ||||
| T4 | 1.51 | 0.98 | ||||
| N-stage | 0.44–2.16 | 0.954 | 0.42–2.04 | 0.837 | ||
| N0 | 1 | 1 | ||||
| N+ | 0.98 | 0.92 | ||||
| Distance from anal verge (cm) | 0.42–2.65 | 0.913 | 0.54–3.50 | 0.497 | ||
| ≤5 | 1 | 1 | ||||
| >5 | 1.05 | 1.38 | ||||
| TRG | 0.28–0.94 | 0.032 | 0.46–1.44 | 0.473 | ||
| TRG1-2 | 0.52 | 0.81 | ||||
| TRG3-4 | 1 | 1 | ||||
| Resection status | 1.81–7.19 | <0.001 | 1.78–7.59 | <0.001 | ||
| R0 | 1 | 1 | ||||
| Not R0 | 3.61 | 3.68 | ||||
| miR-125b expression | 1.14–3.34 | 0.015 | 0.74–2.15 | 0.404 | ||
| Above median | 1 | 1 | ||||
| Below median | 1.95 | 1.26 | ||||
| miR-145 expression | 1.04–3.03 | 0.035 | 0.81–2.37 | 0.237 | ||
| Above median | 1 | 1 | ||||
| Below median | 1.78 | 1.38 | ||||
| miR-21 expression | 0.48–1.36 | 0.418 | 0.43–1.25 | 0.255 | ||
| Above median | 1 | 1 | ||||
| Below median | 0.81 | 0.73 | ||||
| miR-31 expression | 0.75–2.12 | 0.389 | 0.74–2.15 | 0.402 | ||
| Above median | 1 | 1 | ||||
| Below median | 1.26 | 1.26 | ||||
| miR-630 expression | 0.68–1.96 | 0.602 | 0.79–2.32 | 0.269 | ||
| Above median | 1 | 1 | ||||
| Below median | 1.15 | 1.35 | ||||
DFS, disease-free survival; OS, overall survival; HR, hazard ratio; TRG, tumor regression grade according to Mandard et al (31); CI, confidence interval; miR, microRNA; T, tumor; N, node.
Figure 2.
Kaplan-Meier survival curves for (A) DFS and (B) OS based on miR-145 expression. The red curves represent patients with miR expression below the median and the blue curves patients with miR expression above the median. miR, microRNA; DFS, disease-free survival; OS, overall survival.
For a parameter to be included in the multivariate Cox's regression analysis, a cut-off significance level of P<0.1 in the univariate analysis was pre-specified. In the linear regression analysis, a significant positive correlation was demonstrated between miR-125b and miR-145 expression (r=0.73; P<0.0001; Fig. 3), and therefore they were included in a separate multivariate analysis (Table V). Resection status (P=0.002, Table V) and miR-125b expression (P=0.026, Table V) remained significant in predicting DFS, and a borderline significance was demonstrated for miR-145 expression (P=0.053). Multivariate analysis was not performed for OS, as none of the investigated miRNAs were significantly associated with OS in the univariate analysis.
Figure 3.
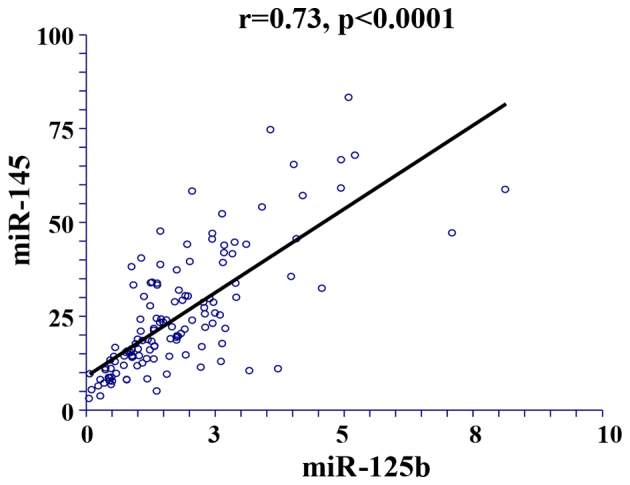
Linear regression analysis of the correlation between miRNA-125b and −145. miR, microRNA.
Table V.
Multivariate Cox's regression analysis of the association between clinicopathological characteristics and disease-free survival in the validation cohort (N=130).
| Multivariate analysis including miR-125b | Multivariate analysis including miR-145 | |||||
|---|---|---|---|---|---|---|
| Clinicopathological characteristic | HR | 95% CI | P-value | HR | 95% CI | P-value |
| TRG | 0.36–1.28 | 0.232 | 0.34–1.20 | 0.168 | ||
| TRG1-2 | 0.68 | 0.64 | ||||
| TRG3-4 | 1 | 1 | ||||
| Resection status | 1.54–6.47 | 0.002 | 1.49–6.22 | 0.002 | ||
| R0 | 1 | 1 | ||||
| Not R0 | 3.15 | 3.04 | ||||
| miR-125b expression | 1.08–3.20 | 0.026 | ||||
| Above median | 1 | |||||
| Below median | 1.86 | |||||
| miR-145 expression | 0.99–2.91 | 0.053 | ||||
| Above median | 1 | |||||
| Below median | 1.70 | |||||
HR, hazard ratio; TRG, tumor regression grade according to Mandard et al (31); CI, confidence interval; miR, microRNA.
Discussion
The balance between treatment efficacy and toxicity is a major issue in the clinical management of patients with LARC. Identifying molecular biomarkers capable of predicting the response of patients to preoperative CRT is therefore important. The ‘watchful waiting’ strategy is emerging as an alternative the typical preoperative CRT followed by surgery; thus far, a number of studies have revealed encouraging outcomes for this strategy (33–37). If tumor response to neoadjuvant CRT could be predicted it would facilitate in providing more individualized treatment planning, whereby patients with pCR could avoid the standard resection procedure, which is frequently followed by a high risk of morbidity.
It has previously been demonstrated by Ryan et al (38) that patients with TRG1 and TRG2 tumors can be regarded as having pCR to preoperative CRT. In addition, Lindebjerg et al (39) demonstrated that 28% of tumors originally classified as TRG1 were reclassified as TRG2 tumors following step sectioning (39). Based on these results, patients with TRG1 and TRG2 were pooled as complete responders in the present study and compared with patients with a poorer response (TRG3+4).
In the present study, the expression of selected miRNAs in diagnostic biopsies from patients with locally advanced T3-4 rectal cancer was analyzed. The results demonstrated a significant association between miR-145 expression and the overall response of patients with rectal cancer to preoperative CRT. Furthermore, the results suggest a possible association between miR-21 and TRG, and between miR-125b and DFS.
In the test cohort, a significant association between low miR-145 expression and major treatment response (TRG1+2) was detected. Similarly, this association was identified in the validation cohort; however, it did not reach statistical significance (P=0.085). Drebber et al (19) reported a significant correlation between a major response to neoadjuvant CRT and a high level of miR-145 expression. However, the results of this study are difficult to compare with those of the present study, since they used a different tumor regression grading system (40) and performed macrodissection, whereas the majority of the samples in the current study underwent laser microdissection.
The significant association between a high expression of miR-21 and TRG1+2 in the validation population is in accordance with a previous study by Lopes-Ramos et al (22), which revealed that the overexpression of miR-21-5p is predictive of complete tumor regression following neoadjuvant CRT in patients with rectal cancer with a sensitivity and specificity of 78 and 86%, respectively. Furthermore, Bandres et al (41) revealed that an upregulation of miR-21 was associated with TRG1 or TRG2 tumors. The results of the test study population in the present study did not identify the same association between miR-21 expression and treatment response. Results from the test cohort suggested an association between a low expression of miR-21 and a major treatment response, but this did not reach statistical significance. However, in the validation population there was a significant association between a high miR-21 expression and a major response to preoperative CRT, in agreement with Lopes-Ramos et al (22) and Bandres et al (41).
miR-125b and miR-31 were included in the present study based on previous literature on miRNAs associated with rectal cancer (23–25). In the current study, no association between their expression and response to preoperative CRT was detected. However, the significant association between a low expression of miR-125b and increased DFS in the validation cohort requires further investigation.
The results from a study by Della Vittoria Scarpati et al (26), which demonstrated that miR-630 had 100% sensitivity and specificity in selecting patients having pCR after undergoing CRT (N=38), were not confirmed in the current study. The expression of miR-630 was analyzed in diagnostic biopsies from a total of 185 patients in the present study. In general, miR-630 was sparsely expressed, and it was undetected in 24 samples. Previously, high intratumoral heterogeneity has been reported regarding miR-630 expression in rectal cancer (18).
In conclusion, the partially diverging results between the test and the validation cohorts in the present study underline the importance of the validation of biomarker studies. This issue has not been adequately addressed in the previous literature. The majority of studies include <50 patients with no validation. The risk of positive results being considered false positives in investigations with small cohorts is high. This approach has hampered or delayed the clinical application of diagnostic and prognostic biomarkers. The results of the present study underline the necessity for large prospective trials of miRNA biomarkers in the future.
Acknowledgements
The authors would like to thank Laboratory Technician Birgit Roed Sørensen (Department of Clinical Pathology, Vejle Hospital, Denmark) and Laboratory Technician Pia Nielsen (Department of Clinical Biochemistry, Vejle Hospital) for their technical assistance, Research Secretary, PA Karin Larsen (Department of Oncology, Vejle Hospital) for the linguistic editing of the manuscript and University Academic Fellow in Radiotherapy Research, Ane Lindegaard Appelt (Faculty of Medicine and Health, University of Leeds, & St James Institute of Oncology, St James's Hospital, Leeds, UK), for the initial data collection.
References
- 1.Cellini F, Valentini V. Current perspectives on preoperative integrated treatments for locally advanced rectal cancer: A review of agreement and controversies. Oncology (Williston Park) 2012;26:730–735. [PubMed] [Google Scholar]
- 2.Valentini V, Glimelius B, Haustermans K, Marijnen CA, Rödel C, Gambacorta MA, Boelens PG, Aristei C, van de Velde CJ. EURECCA consensus conference highlights about rectal cancer clinical management: The radiation oncologist's expert review. Radiother Oncol. 2014;110:195–198. doi: 10.1016/j.radonc.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 3.Julien LA, Thorson AG. Current neoadjuvant strategies in rectal cancer. J Surg Oncol. 2010;101:321–326. doi: 10.1002/jso.21480. [DOI] [PubMed] [Google Scholar]
- 4.Sauer R, Becker H, Hohenberger W, Rödel C, Wittekind C, Fietkau R, Martus P, Tschmelitsch J, Hager E, Hess CF, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351:1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 5.Kapiteijn E, Marijnen CA, Nagtegaal ID, Putter H, Steup WH, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van Krieken JH, et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N Engl J Med. 2001;345:638–646. doi: 10.1056/NEJMoa010580. [DOI] [PubMed] [Google Scholar]
- 6.Kim JC, Kim TW, Kim JH, Yu CS, Kim HC, Chang HM, Ryu MH, Park JH, Ahn SD, Lee SW, et al. Preoperative concurrent radiotherapy with capecitabine before total mesorectal excision in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2005;63:346–353. doi: 10.1016/j.ijrobp.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 7.Lim SH, Chua W, Henderson C, Ng W, Shin JS, Chantrill L, Asghari R, Lee CS, Spring KJ, de Souza P. Predictive and prognostic biomarkers for neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Crit Rev Oncol Hematol. 2015;96:67–80. doi: 10.1016/j.critrevonc.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Kuremsky JG, Tepper JE, McLeod HL. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys. 2009;74:673–688. doi: 10.1016/j.ijrobp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Maas M, Nelemans PJ, Valentini V, Das P, Rödel C, Kuo LJ, Calvo FA, García-Aguilar J, Glynne-Jones R, Haustermans K, et al. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol. 2010;11:835–844. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 10.Fokas E, Liersch T, Fietkau R, Hohenberger W, Beissbarth T, Hess C, Becker H, Ghadimi M, Mrak K, Merkel S, et al. Tumor regression grading after preoperative chemoradiotherapy for locally advanced rectal carcinoma revisited: Updated results of the CAO/ARO/AIO-94 Trial. J Clin Oncol. 2014;32:1554–1562. doi: 10.1200/JCO.2013.54.3769. [DOI] [PubMed] [Google Scholar]
- 11.Rödel C, Martus P, Papadoupolos T, Füzesi L, Klimpfinger M, Fietkau R, Liersch T, Hohenberger W, Raab R, Sauer R, Wittekind C. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23:8688–8696. doi: 10.1200/JCO.2005.02.1329. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Aguilar J, de Anda E Hernandez, Sirivongs P, Lee SH, Madoff RD, Rothenberger DA. A pathologic complete response to preoperative chemoradiation is associated with lower local recurrence and improved survival in rectal cancer patients treated by mesorectal excision. Dis Colon Rectum. 2003;46:298–304. doi: 10.1007/s10350-004-6545-x. [DOI] [PubMed] [Google Scholar]
- 13.Slaby O, Svoboda M, Michalek J, Vyzula R. MicroRNAs in colorectal cancer: Translation of molecular biology into clinical application. Mol Cancer. 2009;8:102. doi: 10.1186/1476-4598-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riordan AM, Thomas MK, Ronnekleiv-Kelly S, Warner T, Geiger PG, Kennedy GD. Utility of micro-ribonucleic acid profile for predicting recurrence of rectal cancer. J Surg Res. 2012;177:87–92. doi: 10.1016/j.jss.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mestdagh P, van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, Vandesompele J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nugent M, Miller N, Kerin MJ. MicroRNAs in colorectal cancer: Function, dysregulation and potential as novel biomarkers. Eur J Surg Oncol. 2011;37:649–654. doi: 10.1016/j.ejso.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Eriksen AH, Andersen RF, Pallisgaard N, Sørensen FB, Jakobsen A, Hansen TF. MicroRNA expression profiling to identify and validate reference genes for the relative quantification of microRNA in rectal cancer. PLoS One. 2016;11:e0150593. doi: 10.1371/journal.pone.0150593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eriksen AH, Andersen RF, Nielsen BS, Sorensen FB, Appelt AL, Jakobsen A, Hansen TF. Intratumoral heterogeneity of MicroRNA expression in rectal cancer. PloS One. 2016;11:e0156919. doi: 10.1371/journal.pone.0156919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drebber U, Lay M, Wedemeyer I, Vallböhmer D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP, Odenthal M. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol. 2011;39:409–415. doi: 10.3892/ijo.2011.1036. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Leong MH, Harms B, Kennedy G, Chen L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J Gastroenterol. 2013;19:5615–5621. doi: 10.3748/wjg.v19.i34.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 22.Lopes-Ramos CM, Habr-Gama A, Bde S Quevedo, Felicio NM, Bettoni F, Koyama FC, Asprino PF, Galante PA, Gama-Rodrigues J, Camargo AA, et al. Overexpression of miR-21-5p as a predictive marker for complete tumor regression to neoadjuvant chemoradiotherapy in rectal cancer patients. BMC Med Genomics. 2014;7:68. doi: 10.1186/s12920-014-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, Chen HY, Sun XF. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27–34. doi: 10.1155/2009/921907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurila EM, Kallioniemi A. The diverse role of miR-31 in regulating cancer associated phenotypes. Genes Chromosomes Cancer. 2013;52:1103–1113. doi: 10.1002/gcc.22107. [DOI] [PubMed] [Google Scholar]
- 25.Svoboda M, Holla L Izakovicova, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541–547. [PubMed] [Google Scholar]
- 26.Della Vittoria, Scarpati G, Falcetta F, Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK, D'Incalci M, De Placido S, Pepe S. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83:1113–1119. doi: 10.1016/j.ijrobp.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 27.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics: Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji446. [DOI] [PubMed] [Google Scholar]
- 28.Jakobsen A, Mortensen JP, Bisgaard C, Lindebjerg J, Hansen JW, Rafaelsen SR. Preoperative chemoradiation of locally advanced T3 rectal cancer combined with an endorectal boost. Int J Radiat Oncol Biol Phys. 2006;64:461–465. doi: 10.1016/j.ijrobp.2005.07.969. [DOI] [PubMed] [Google Scholar]
- 29.Jakobsen A, Ploen J, Vuong T, Appelt A, Lindebjerg J, Rafaelsen SR. Dose-effect relationship in chemoradiotherapy for locally advanced rectal cancer: A randomized trial comparing two radiation doses. Int J Radiat Oncol Biol Phys. 2012;84:949–954. doi: 10.1016/j.ijrobp.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Jakobsen A, Mortensen JP, Bisgaard C, Lindebjerg J, Rafaelsen SR, Bendtsen VO. A COX-2 inhibitor combined with chemoradiation of locally advanced rectal cancer: A phase II trial. Int J Colorectal Dis. 2008;23:251–255. doi: 10.1007/s00384-007-0407-7. [DOI] [PubMed] [Google Scholar]
- 31.Mandard AM, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF, Roussel A, Jacob JH, Segol P, Samama G. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680–2686. doi: 10.1002/1097-0142(19940601)73:11<2680::AID-CNCR2820731105>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 32.Bouzourene H, Bosman FT, Seelentag W, Matter M, Coucke P. Importance of tumor regression assessment in predicting the outcome in patients with locally advanced rectal carcinoma who are treated with preoperative radiotherapy. Cancer. 2002;94:1121–1130. doi: 10.1002/cncr.10327.abs. [DOI] [PubMed] [Google Scholar]
- 33.Habr-Gama A, Perez RO, Nadalin W, Sabbaga J, Ribeiro U, Jr, e Sousa AH Silva, Jr, Campos FG, Kiss DR, Gama-Rodrigues J. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: Long-term results. Ann Surg. 2004;240:711–718. doi: 10.1097/01.sla.0000141194.27992.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habr-Gama A, Gama-Rodrigues J, São Julião GP, Proscurshim I, Sabbagh C, Lynn PB, Perez RO. Local recurrence after complete clinical response and watch and wait in rectal cancer after neoadjuvant chemoradiation: Impact of salvage therapy on local disease control. Int J Radiat Oncol Biol Phys. 2014;88:822–828. doi: 10.1016/j.ijrobp.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Maas M, Beets-Tan RG, Lambregts DM, Lammering G, Nelemans PJ, Engelen SM, van Dam RM, Jansen RL, Sosef M, Leijtens JW, et al. Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer. J Clin Oncol. 2011;29:4633–4640. doi: 10.1200/JCO.2011.37.7176. [DOI] [PubMed] [Google Scholar]
- 36.Appelt AL, Pløen J, Harling H, Jensen FS, Jensen LH, Jørgensen JC, Lindebjerg J, Rafaelsen SR, Jakobsen A. High-dose chemoradiotherapy and watchful waiting for distal rectal cancer: A prospective observational study. Lancet Oncol. 2015;16:919–927. doi: 10.1016/S1470-2045(15)00120-5. [DOI] [PubMed] [Google Scholar]
- 37.Habr-Gama A, De Souza PM, Ribeiro U, Jr, Nadalin W, Gansl R, Sousa AH, Jr, Campos FG, Gama-Rodrigues J. Low rectal cancer: Impact of radiation and chemotherapy on surgical treatment. Dis Colon Rectum. 1998;41:1087–1096. doi: 10.1007/BF02239429. [DOI] [PubMed] [Google Scholar]
- 38.Ryan R, Gibbons D, Hyland JM, Treanor D, White A, Mulcahy HE, O'Donoghue DP, Moriarty M, Fennelly D, Sheahan K. Pathological response following long-course neoadjuvant chemoradiotherapy for locally advanced rectal cancer. Histopathology. 2005;47:141–146. doi: 10.1111/j.1365-2559.2005.02176.x. [DOI] [PubMed] [Google Scholar]
- 39.Lindebjerg J, Rafaelsen S, Pløen J, Jakobsen A. Step-sectioning of paraffin block from ‘completely regressed’ rectal carcinoma after preoperative chemorediation. Ann Oncol. 2009 [Google Scholar]
- 40.Schneider PM, Baldus SE, Metzger R, Kocher M, Bongartz R, Bollschweiler E, Schaefer H, Thiele J, Dienes HP, Mueller RP, Hoelscher AH. Histomorphologic tumor regression and lymph node metastases determine prognosis following neoadjuvant radiochemotherapy for esophageal cancer: Implications for response classification. Ann Surg. 2005;242:684–692. doi: 10.1097/01.sla.0000186170.38348.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandres E, Arias F, Guerrero D, Lopez I, Gonzalez-Huarriz M, Dorronsoro ML Gomez, Montes M, Monzon F, Torrea N, Armendariz P, Balen E. Association between a specific miRNA signature and pathological response to neoadjuvant chemoradiotherapy (CRT) in locally advanced rectal cancer (LARC) patients. ASCO. 2012:e14057–e14057. [Google Scholar]