Abstract
Background:
Chronic intermittent hypoxia is the most remarkable feature of obstructive sleep apnea/hypopnea syndrome and it can induce the change of hypoxia-inducible factor-1α (HIF-1α) expression and contractile properties in the genioglossus. To clarify the role of HIF-1α in contractile properties of the genioglossus, this study generated and compared high-throughput RNA-sequencing data from genioglossus between HIF-1α conditional knockout (KO) mice and littermate wild-type (WT) mice.
Methods:
KO mice were generated with cre-loxP strategy. Gene expression profile analysis was performed using gene enrichment analysis. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of differently expressed messenger RNAs were performed to identify the related pathways and biological functions. Six differentially expressed genes (DEGs) were validated by qualitative reverse transcription polymerase chain reaction.
Results:
A total of 142 (77 upregulated and 65 downregulated) transcripts were found to exhibit statistically significant difference between the HIF-1α-KO and WT mice. GO and KEGG analyses indicated that DEGs included genes involved in “skeletal muscle cell differentiation,” “muscle organ development,” “glucose metabolic process,” “glycogen biosynthetic and metabolic process,” etc.
Conclusion:
This study might provide evidence that HIF-1α affects the expression of multiple genes involved in the myogenesis, muscle development, and carbohydrate metabolism through transcriptome analysis in conditional HIF-1α-KO mice.
Keywords: Carbohydrate Metabolism, Genioglossus, Hypoxia-inducible Factor-1α, Myogenesis, Transcriptomic Analysis
Introduction
Obstructive sleep apnea/hypopnea syndrome (OSAHS) is a serious breathing disorder characterized by intermittent and recurrent upper airway (UA) collapse during sleep, which has neurobehavioral and cardiovascular consequences and is associated with an increased risk of hypertension, cardiovascular disease, stroke, daytime sleepiness, and diminished quality of life.[1,2]
Chronic intermittent hypoxia (CIH), a result of repetitive narrowing or collapse of UA, is one of the most remarkable features of OSAHS. Several studies have suggested that intermittent hypoxia promoted the expression of hypoxia-inducible factor-1 (HIF-1) and activated the serial reactions to hypoxia.[3,4,5,6] HIF-1 is a heterodimeric transcription factor composed of a hypoxia-inducible HIF-1α subunit, which determines the activation of HIF-1, and a consistently expressed HIF-1β subunit.[7] HIF-1α has been testified with higher expression in skeletal muscles than other tissues even in normoxic conditions, indicating that it plays an important role in skeletal muscles. In our previous study, we have found that CIH induced the expression of HIF-1α in the genioglossus of mice and altered the physical properties toward a more fatigable phenotype, inhibiting the overexpression of HIF-1α that might improve the fatigue resistance.[8] However, it remains unclear about the molecular mechanism of HIF-1α in the influence of genioglossus physical properties.
High-throughput next-generation sequencing (NGS) technologies make it possible, nowadays, to carry out detailed transcriptomic profiling. In this study, we established a HIF-1α knockout (KO) mice model and applied the NGS-based RAN-sequencing (RNA-seq) method to carry out a transcriptional analysis in genioglossus of both HIF-1α-KO mice and littermate wild-type (WT) mice. Gene ontology (GO) and Kyoto Encyclopedia of Genes pathway analyses were then applied to identify the messenger RNA (mRNA) expression discrepancy and related pathways which caused different biological responses to HIF-1α-KO. The study could provide a comprehensive understanding of HIF-1α-related gene expression and help to elucidate the molecular mechanism of HIF-1α in the influence of genioglossus physical properties.
Methods
Ethical approval
The study was approved by the Animal Care Committee of Tongji University, and all the animal care and experimental procedures were conducted in accordance with “The Guide for Care and Use of Laboratory Animals.”
Gene targeting and generation of hypoxia-inducible factor-1α floxed mice
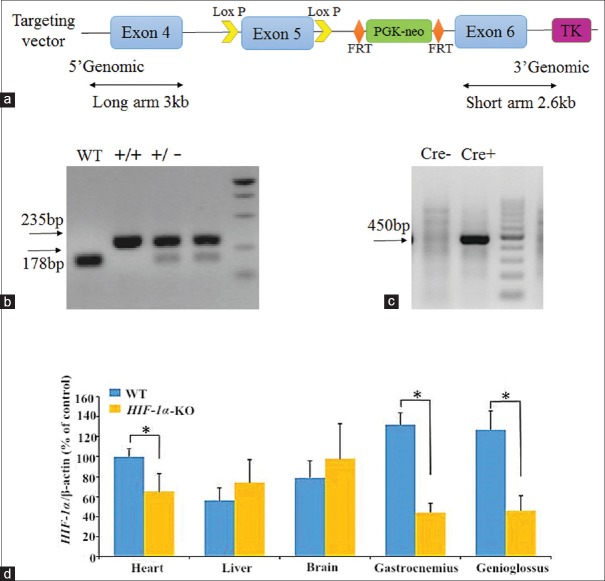
The C57bl/6 mice were provided by Shanghai Biomodel Organism Science and Technology Development Co. Ltd., China. Gene sequence of HIF-1α was obtained from Ensembl database (http://www.ensembl.org/index.html) and used to design and construct the targeting vector. The RecE/RecT-based DNA homologous recombination technology was applied to build a conditional KO targeting vector at the site of exon 5. The bacterial artificial chromosome plasmid was subcloned into two homologous recombination arms. The long arm (LA) was 3 kb while the short arm was 2.6 kb. The region was designed such that the short homologous arm extends 2.6 kb to the 3’end. The LA extends 3 kb to the 5’end of the loxP site. This single loxP site is inserted upstream of exon 5, and the loxP/Flp recognition target (FRT)-flanked neomycin (Neo) cassette is inserted downstream of exon 5 [Figure 1a]. The targeting vector is confirmed by sequencing. The 35 μg targeting vectors were linearized by NotI and then transfected to embryonic stem (ES) cells by electroporation. Then, ES cells were cultured with G418 (300 mg/L) and ganciclovir (2 μmol/L), and macroscopic clones were chosen and expanded for polymerase chain reaction (PCR) analysis to identify recombination ES clones. The targeted ES cells were microinjected into blastocysts. Then, the blastocysts were transplanted into pseudopregnant C57BL/6 mice to generate chimeras (completed by Shanghai Biomodel Organism Science and Technology Development Co. Ltd., China). A high-percentage black coat color was chosen to mate with WT mice to generate heterozygous offspring which was identified with PCR using tail DNA. As Neo cassette was designed outside loxP and FRT-flanked fragment, the heterozygous mice were mated with Flp+ mice to delete Neo cassette. Resulting heterozygous mice without Neo (HIFflox/-) were obtained.
Figure 1.
Targeting of HIF-1α gene. (a) Targeting strategy to create HIF-1α allele. (b) PCR analysis of DNA isolated from tails of HIF-1α-KO mice showing WT, heterozygous (HIF-1αflox/-), and homozygous genotypes (HIF-1αflox/flox) for the conditional targeted allele. (c) PCR analysis of DNA isolated from tails of HIF-1α-KO mice showing Cre- and Cre+ genotypes. (d) HIF-1α messenger RNA expressions in various tissues of WT and HIF-1α-KO mice. HIF-1α: Hypoxia-inducible factor-1α; WT: Wild-type; PCR: Polymerase chain reaction; KO: Knockout. *P < 0.05.
Conditional knockout mice and polymerase chain reaction screening strategy
HIFflox/- mice were bred with muscle creatine kinase (MCK)-Cre+ hemizygous mice to create HIFflox/- Cre+ mice. HIFflox/- Cre+ mice were screened out with PCR. The primers were as follows: HIF-1α: 5’-ATGAATAATTAATCCTGTGAAGTGTGG-3’ (forward), 5’-TTCTGAGGTAAGGCTTGTGTTCC-3’ (reverse); Cre+: 5’-GTGAAACAGCATTGCTGTCACTT-3’ (forward), 5’-TAAGTCTGAACCCGGTCTGC-3’ (reverse). Then, HIFflox/- Cre+ mice were bred with HIFflox/- Cre+ or HIFflox/- mice to generate HIFflox/flox Cre+ mice (HIF-1α-KO mice) as experimental group and littermate HIF-/- mice (WT) as control group. HIF-1α-KO mice and WT mice were screened out with PCR as mentioned above.
RNA isolation
Total RNA was isolated from genioglossus of 2-month-old HIF-1α-KO mice (n = 3) and littermate WT mice (n = 3) using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The quality and concentration of RNA were tested by absorbance ratios of A260/A280 with the NanoDrop ND-1000 spectrophotometer (Nano Drop Technologies, Wilmington, DE, USA). A 2 μl RNA of each sample was used to assess the integrity in denaturing gel electrophoresis. High-quality RNA samples were prepared for RNA-seq analysis and quantitative reverse transcription PCR (qRT-PCR).
RNA-sequencing library preparation and sequencing
RNA-seq applied in this research was to analyze the mRNA expressions in genioglossus from both HIF-1α-KO and WT mice. Sample labeling and array hybridization were performed according to the Agilent One-color Microarray-based Gene Expression Analysis protocol (Agilent Technologies Inc., Santa Clara, CA, USA) with minor modifications. Briefly, mRNA was purified from total RNA with Dynabeads mRNA DIRECT kit (Invitrogen, Carlsbad, CA, USA), then amplified and transcribed into complementary DNA (cDNA) utilizing a random priming method and synthesize double-strand cDNA. After having repaired the ends of cDNA and added A’ bases to 3’end, each sample was ligated, purified, amplified using PCR, and subjected to library preparation. The quality, quantity, and size distribution of the libraries were determined using an Agilent Bioanalyzer 2100 (Agilent Technologies, USA). The libraries were then sequenced with Illumina HiSeq 2000 (Illumina Technology, USA).
The array images were imported into Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies Inc., USA) for further analysis. Quantile normalization and background correction were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies Inc.). The effect of normalization was examined on box plots. Differentially expressed genes (DEGs) were identified by fold change analysis and P value cutoff of 0.05.
Gene ontology and pathway analysis
GO derived from GO (www.geneontology.org) and pathway analysis were applied to check the roles of these DEGs played in these GO terms or biological pathways. Three main associated aspects, biological processes, cellular components, and molecular functions, were shown from GO, according to their annotations.[9] Pathway analysis was based on the pathways available at the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The P value denotes the significance of GO terms’ enrichment and the pathway correlation.
Quantitative reverse transcription-polymerase chain reaction
Total RNA mentioned above was reversely transcribed to cDNA using PrimeScript RT reagent kit (TaKaRa Bio Inc., Japan) in a GeneAmp PCR system 9700 (Foster, CA, USA). Real-time PCR was performed using SYBR Premix Ex Taq™ (TaKaRa Bio Inc.) on a 7500 real-time PCR system (ABI, 7500, USA). Gene-specific primers were described in Table 1. The first step of the PCR protocol was 95°C for 30 s, followed by 45 cycles of 95°C for 5 s, and 60°C for 34 s as the second step. A melting curve analysis was performed to ensure specificity of the PCR products. Each datum presented was expressed relative to β-actin mRNA using the comparative 2−ΔΔCt method.
Table 1.
Nucleotide sequences of primers used for PCR amplification
| Genes | Forward primer | Reverse primer |
|---|---|---|
| JUNB | 5’-AACAGCCCTTCTACCACGAC-3’ | 5’-GCTCGGTTTCAGGAGTTTGTA-3’ |
| MT1 | 5’-ATGGACCCCAACTGCTCCTGCTCCACC-3’ | 5’-GGCTGCAACTGTATAGGAAGACGCTCG-3’ |
| NR4A2 | 5’-GGTGCAGCATAGCCCGATGT-3’ | 5’-GATATCCTGTGGGCTCTTCGGC-3’ |
| AP4S1 | 5’-CTCACCCTCAAGGGGACCCA-3’ | 5’-CGTAGCTGGAACCTGAGCCG-3’ |
| KCNJ2 | 5’-TGCAGGAGCCGCTTTGTGAA-3’ | 5’-AAGGCAGCCGTGAAGCTGTT-3’ |
| TIRAP | 5’-CAGCAGGTGCAAACCCA-3’ | 5’-GGCTTTCCTGGGAGATCGGC-3’ |
| β-actin | 5’-CCTCATGAAGATCCTGACCG-3’ | 5’-TGCCAATAGTGATGACCTGG-3’ |
PCR: Polymerase chain reaction; JUNB: Jun B proto-oncogene; MT1: Metallothionein 1; NR4A2: Nuclear receptor subfamily 4, Group A, member 2; AP4S1: Adaptor-related protein complex AP-4, sigma 1; KCNJ2: Potassium inwardly-rectifying channel, subfamily J, member 2; TIRAP: Toll-interleukin 1 receptor domain-containing adaptor protein.
Statistical analysis
All quantitative results were obtained from triplicate samples. Data were expressed as mean ± standard deviation (SD). Statistical analysis was assessed by the independent samples t-test and performed with Statistical Product and Service Solutions version 17.0 (SPSS Inc., Chicago, IL, USA). A P < 0.05 was considered statistically significant.
Results
Targeted exclusion of hypoxia-inducible factor-1α
LoxP sites were inserted into both upstream and downstream of HIF-1α exon 5, and a FRT-flanked Neo cassette was inserted into upstream of exon 6 to generate a conditional mutant allele, allowing for excision of HIF-1α in a tissue-specific manner [Figure 1a]. Homozygous HIF-1αflox/flox Cre+ mice (HIF-1α-KO, n = 3) and their littermate HIF-/- mice (WT, n = 3) were screened out by PCR using tail DNA [Figure 1b and 1c]. Each pair of HIF-1α-KO mice and WT mice was in the same gender. The KO efficiency of HIF-1α was confirmed by qRT-PCR [Figure 1d]. Mean frequencies of deletion of HIF-1α in genioglossus and gastrocnemius muscles were 63.7 ± 3.7% and 66.4 ± 6.3%, respectively. The mRNA expression of HIF-1α decreased by 34.6 ± 5.2% in cardiac tissue of HIF-1α-KO mice.
RNA quantity and quality
The quantity and quality of the RNA samples of genioglossus in WT and HIF-1α-KO mice were assessed by gel electrophoresis [Figure 2] and the optical density ratio OD260/OD280 [Table 2]. These analyses confirmed that the isolated total RNA was of good quality.
Figure 2.
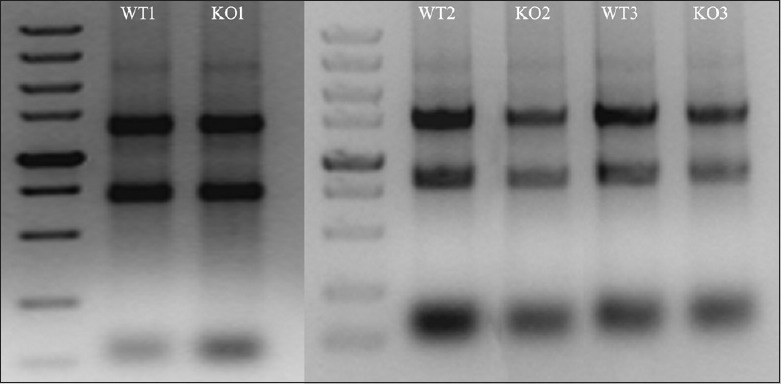
Examination of RNA integrity by denaturing agarose gel electrophoresis. WT: Wild-type; KO: Knockout.
Table 2.
OD of total RNA of genioglossus in WT and HIF-1α-KO mice
| Samples | A260/280 | RIN* | RNA concentration (ng/µl) |
|---|---|---|---|
| WT1 | 1.98 | 8.9 | 1188.0 |
| KO1 | 2.01 | 8.8 | 1212.1 |
| WT2 | 1.97 | 9.2 | 892.3 |
| KO2 | 1.96 | 9.0 | 1024.8 |
| WT3 | 1.99 | 8.9 | 922.3 |
| KO3 | 2.02 | 9.1 | 767.4 |
*High RIN scores (7–10) and a narrow distribution of scores (1–1.5) from an Agilent Bioanalyzer indicated high RNA sample quality. RIN: RNA integrity number; OD: Optical densities; WT: Wild-type; HIF-1α: Hypoxia-inducible factor-1α; KO: Knockout.
Gene expression profile by microarray analysis
After quantile normalization of the raw data, the expression profiles of 22619 mRNAs were obtained from genioglossus muscle of HIF-1α-KO and WT mice. Compared with WT mice, 77 mRNAs were found to be upregulated and 65 to be downregulated in HIF-1α-KO mice based on fold-change differences in expression (all P < 0.05). The top ten upregulated and downregulated mRNAs were listed in Tables 3 and 4, respectively.
Table 3.
Top ten upregulated mRNAs in WT and HIF-1α-KO mice
| Genes | RPKM in WT mice | RPKM in KO mice | Fold changes | P |
|---|---|---|---|---|
| JUNB | 165 | 765 | 2.24 | <0.00001 |
| BTG2 | 242 | 617 | 1.82 | <0.00001 |
| SIK1 | 491 | 1105 | 1.77 | <0.00001 |
| MT1 | 361 | 753 | 1.59 | <0.00001 |
| NR4A2 | 51 | 145 | 1.56 | <0.00001 |
| UT2R | 37 | 91 | 1.55 | <0.00001 |
| KLF10 | 559 | 943 | 1.50 | <0.00001 |
| MT2 | 231 | 609 | 1.48 | <0.00001 |
| FOSL2 | 357 | 698 | 1.48 | <0.00001 |
| MIDN | 371 | 575 | 1.42 | 0.00014 |
RPKM: Reads Per Kilobase of exon model per million mapped reads; JUNB: Jun B proto-oncogene; BTG2: B-cell translocation gene 2; SIK1: Salt-inducible kinase 1; MT1: Metallothionein 1; NR4A2: Nuclear receptor subfamily 4, Group A, member 2; UT2R: Urotensin 2 receptor; KLF10: Kruppel-like factor 10; MT2: Metallothionein 2; FOSL2: Fos-like antigen 2; MIDN: Midnolin; WT: Wild type; HIF-1α: Hypoxia-inducible factor-1α; KO: Knockout; mRNA: Messenger RNA.
Table 4.
Top ten downregulated long noncoding RNAs in WT and HIF-1α-KO mice
| Genes | RPKM in WT mice | RPKM in KO mice | Fold change | P |
|---|---|---|---|---|
| NARF | 1667 | 861 | 0.68 | <0.00001 |
| AP4S1 | 304 | 149 | 0.70 | 0.00028 |
| NT5C1A | 235 | 133 | 0.72 | 0.00024 |
| KCNJ2 | 971 | 553 | 0.75 | 0.00260 |
| MUSK | 413 | 236 | 0.76 | 0.00240 |
| SNAPC1 | 244 | 96 | 0.76 | 0.00370 |
| LRRC2 | 63 | 27 | 0.77 | 0.00290 |
| TIRAP | 786 | 503 | 0.77 | 0.00380 |
| AMYL | 364 | 173 | 0.78 | 0.00820 |
| FZD7 | 940 | 641 | 0.78 | 0.00075 |
NARF: Nuclear prelamin A recognition factor; AP4S1: Adaptor-related protein complex AP-4, sigma 1; NT5C1A: 5’-nucleotidase, cytosolic IA; KCNJ2: Potassium inwardly-rectifying channel, subfamily J, member 2; MUSK: Muscle, skeletal, receptor tyrosine kinase; SNAPC1: Small nuclear RNA-activating complex, polypeptide 1; LRRC2: Leucine-rich repeat containing 2; TIRAP: Toll-interleukin 1 receptor domain-containing adaptor protein; AMYL: Amylase 1, salivary; FZD7: Frizzled class receptor 7; RPKM: Reads Per Kilobase of exon model per million mapped reads; WT: Wild-type; HIF-1α: Hypoxia-inducible factor-1α; KO: Knockout.
Gene ontology analysis and Kyoto Encyclopedia of Genes and Genomes pathway analysis of aberrantly expressed messenger RNAs
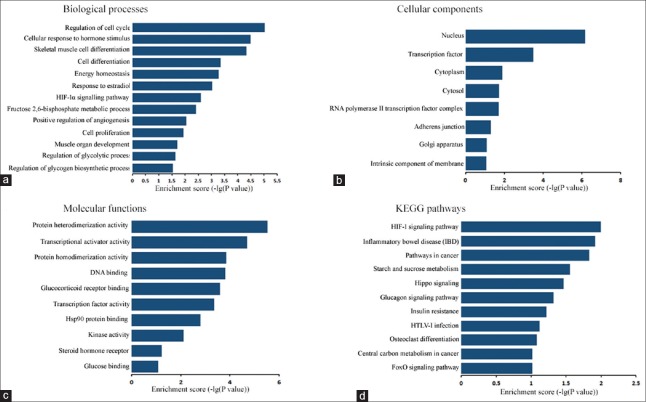
GO analysis was applied to investigate the potential functions of the aberrantly expressed mRNAs in the two groups. In this study, there were 119 aberrantly expressed mRNAs assigned to biological process, 127 assigned to cellular component, and 123 assigned to molecular function. The enrichment of each GO term was assessed. Then, GO terms were filtered by the enrichment scores in aberrantly expressed mRNAs. This study demonstrated that enriched GO terms targeted by different expressed mRNAs involved in “skeletal muscle cell differentiation,” “muscle organ development,” “regulation of cell cycle,” “energy homeostasis,” “response to estradiol,” “fructose 2,6-bisphosphate metabolic process,” “regulation of glycolytic and glycogen biosynthetic process” in biological process, “nucleus,” “transcription factor complex,” “cytoplasm,” “cytosol” in cellular component, and “protein heterodimerization activity,” “transcriptional activator activity,” “glucocorticoid receptor binding,” “Hsp90 protein binding,” “steroid hormone receptor activity,” and “glucose binding” in molecular function [Figure 3]. KEGG pathway analysis was performed next, and 11 pathways were identified significantly enriched among the DEGs between the HIF-1α-KO and WT mice (all P < 0.05), including “HIF-1 signaling pathway,” “starch and sucrose metabolism,” and “insulin resistance.”
Figure 3.
Enrichment analysis of GO terms and pathways for DEGs. (a-c) GO analysis according to biological process, cellular component, and molecular function, respectively. (d) Pathway analysis based on the KEGG database. GO: Gene ontology; KEGG: Kyoto Encyclopedia of Genes Pathway; DEG: Differentially expressed gene.
Validation of the RNA-sequencing results using quantitative reverse transcription-polymerase chain reaction
From the DEGs, 6 differentially expressed mRNAs (Jun B proto-oncogene [JUNB], metallothionein 1, nuclear receptor subfamily 4, group A, member 2 [NR4A2], adaptor-related protein complex AP-4, potassium inwardly-rectifying channel, toll-interleukin 1 receptor domain-containing adaptor protein) were randomly selected between HIF-1α-KO and WT mice to confirm their expression levels by qRT-PCR. All the 6 mRNAs showed the same change patterns as shown in microarray analysis [Figure 4].
Figure 4.
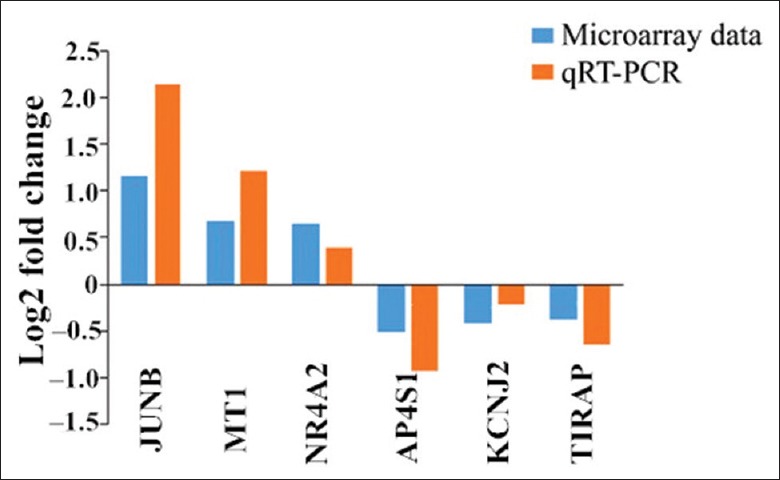
Validation of RNA-seq data by qRT-PCR. RNA-seq: RNA-sequencing; qRT-PCR: quantitative reverse transcription-polymerase chain reaction.
Discussion
It was found that the physiologically local muscle hypoxia induced by low-intensity resistance exercise combined with moderate vascular occlusion could effectively increase the cross-sectional muscle area and enhance the isokinetic contraction strength of skeletal muscle.[10] In our previous study, however, we have found that long-term CIH could alter the physical properties of genioglossus toward a more fatigable phenotype in rats.[8] In addition, our previous researches have demonstrated that estrogen accentuates the contractility of genioglossus in rats, and downregulation of HIF-1α might be a pivotal explanation in the protective effects.[8,11,12] Skelly et al.[13] reported that chronic antioxidant supplementation ameliorated free radical-mediated UA muscle dysfunction induced by CIH. Although the mechanisms underlying UA muscle dysfunction are not explicit yet, there is strong evidence suggesting a link between increased production of HIF-1α and altered muscle function.
In the present study, we applied NGS-based RNA-seq method to study the whole transcriptional changes associated to HIF-1α in genioglossus of skeletal muscle of HIF-1α-KO mice and WT mice, aiming at getting a more global and accurate picture of HIF-1α with regard to genioglossus muscle function. A total of 22,619 mRNAs were detected. Among these expression-changed genes, we identified 77 upregulated and 65 downregulated genes that were significantly differentially expressed in genioglossus of HIF-1α-KO mice in comparison to littermate WT mice (all P < 0.05) and summarized their general characteristics and functional annotations using GO analysis and KEGG pathway. The discovery of genes affected by HIF-1α will provide important guidance for future study of molecular mechanisms underlying OSAHS.
It is well known that a lot of target genes activated by HIF-1α are involved in cell proliferation, survival, and differentiation, including erythropoietin, vascular endothelial growth factor (VEGF), basic fibroblast growth factor, and others.[14] Ono et al.[15] constructed HIF-1α knockdown C2C12 cells to analyze the role of HIF-1α in myoblast differentiation and found that HIF-1a knockdown effectively blocked myotube formation and myosin heavy chain (MHC) expression. We got different results in our previous study. Accompanied with overexpression of HIF-1α, the myotube formation and the expressions of MyoD, myogenin, and MHC were inhibited by hypoxia, while no differences were observed between HIF-1α knockdown myoblasts and the control ones under normoxia at day 6 of myogenesis.[12] The effects of HIF-1α on myoblast differentiation or myogenesis varied in these experiments and need to be further proved. In the present study, HIF-1α was deleted in skeletal muscle and a significant number of upregulated genes were found to map to tissue differentiation and development. Notable upregulated genes were involved in neuron differentiation (BTG2, NTNG2, DLL1, HES1, NR4A2, STAT3, and VEGF-A), myeloid cell and osteoclast differentiation (KLF10, TOB2, and ZFP36), cellular and macromolecular biosynthetic process (KLF13, MKL1, HES1, MYOG, NR4A1, NR4A2, STAT3, and MAF), and skeletal system development (JUND, KLF10, NAB2, MYOG, and TIPARP). Functional analysis and enrichment of GO terms revealed that biological processes were closely correlated to skeletal muscle cell differentiation, fat cell differentiation, muscle organ development, and nervous system development. Furthermore, molecular functions such as protein heterodimerization activity, transcriptional activator and factor activities, and DNA binding were affected. GO analysis indicated an important role of HIF-1α in the myogenesis and muscle development or repair.
It is clear that the transcription factor HIF-1α is an essential factor in changing the way of energy metabolism. A switch from mitochondrial aerobic respiration to glycolysis induced by HIF-1 is considered critical for metabolic adaptation to hypoxia.[16,17] There are also many studies identifying the mechanisms by which HIF-1 suppresses oxidative metabolism, including inhibition of fatty acid oxidation, which also generates acetyl-CoA and entry into the tricarboxylic acid (TCA) cycle,[18] induction of mitochondrial-selective autophagy,[19,20] and inhibition of electron transport chain complex I activity.[21,22,23] In fact, although HIF-1 is typically considered as activation during hypoxia, its loss has an effect on both normoxic and hypoxic ATP levels in a number of tissue types,[24,25] and this implicates the factor in the regulation of metabolic function even during conditions of normal physiologic oxygenation. In the study, a lot of notable downregulated genes were mapped to carbohydrate metabolism, including glucose metabolic process (ACACB and pyruvate dehydrogenase kinase 1 [PDK1]), glycogen biosynthesis and metabolic process (AGL, GSK3B, and GCK), and starch and sucrose metabolism (AMY1, AGL, and GCK). As for glycogen metabolism, Mason et al.[26] reported that HIF-1α targeted to delete mice stored and metabolized more glycogen in response to stimulation than do WT mice. Their research group also analyzed the role of HIF-1α in carbohydrate metabolism and endurance training. HIF-1α null mice have a decreased level of PDK1, increased capillary to fiber ratio, elevated oxidative enzyme activities, and enhanced endurance of skeletal muscles.[27]
This study was to deal with the potential regulation of HIF-1α and skeletal muscle cell proliferation, differentiation, and carbohydrate metabolism through transcriptome analysis in skeletal muscle conditional HIF-1α-KO mice. As reported in some studies, the expression of HIF-1α varied distinctly in different hypoxic conditions, such as sustained hypoxia and intermittent hypoxia.[28] In fact, both fatigability and HIF-1α level were different in skeletal muscle in short-term and prolonged intermittent hypoxia.[29,30] Viganò et al.[31] reported that HIF-1α in human skeletal muscle was at the prehypoxia levels after exposure to hypobaric hypoxia for 7–9 days, despite that proteins involved in TCA cycle, oxidative phosphorylation, and oxidative stress responses were significantly decreased. Considering that the HIF-1α expression changed with different hypoxia conditions or exposure time, we chose HIF-1α-KO mice in normoxia to study the role of HIF-1α, instead of mice in CIH model. The transgene mice were obtained through Cre-loxP site-specific recombination system. HIFflox/- mice were generated from HIF-1a loxP-flanked allele mouse stocks backcrossed into a C57Bl6/J background, and then crossed into MCK-Cre+ hemizygous mice [Figure 1]. MCK-Cre recombinase, which also expressed in a low level in cardiac muscle, was commonly used in creating skeletal muscle conditional activation or inactivation of genes in mice.[32] In this study, cardiac deletion (34.6%) of HIF-1α was detected. Fortunately, no developmental or behavior abnormality was found.
In summary, the present study might provide evidence that HIF-1α affected the expression of multiple genes involved in the myogenesis, muscle development, and carbohydrate metabolism through transcriptome analysis in skeletal muscle conditional HIF-1α-KO mice. The data paved the way for future study of molecular mechanism underlying altered contractile properties of genioglossus induced by CIH.
Financial support and sponsorship
This study was supported by grants from the National Natural Science Foundation of China (No. 81271192 and No. 81600897), and the Shanghai Science and Technology Committee of China (No. 15140903500).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Xin Chen
References
- 1.Xie J, Liu S, Wei YX. Obstructive Sleep Apnea Hypopnea Syndrome: An Incognito Player Contributing to Repeated Pulmonary Embolism? Chin Med J. 2016;129:252. doi: 10.4103/0366-6999.173560. doi: 10.4103/0366-6999.173560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malhotra A, White DP. Obstructive sleep apnoea. Lancet. 2002;360:237–45. doi: 10.1016/S0140-6736(02)09464-3. doi: 10.1016/S0140-6736(02)09464-3. [DOI] [PubMed] [Google Scholar]
- 3.Cai Z, Manalo DJ, Wei G, Rodriguez ER, Fox-Talbot K, Lu H, et al. Hearts from rodents exposed to intermittent hypoxia or erythropoietin are protected against ischemia-reperfusion injury. Circulation. 2003;108:79–85. doi: 10.1161/01.CIR.0000078635.89229.8A. doi: 10.1161/01.CIR.0000078635.89229.8A. [DOI] [PubMed] [Google Scholar]
- 4.Pae EK, Wu J, Nguyen D, Monti R, Harper RM. Geniohyoid muscle properties and myosin heavy chain composition are altered after short-term intermittent hypoxic exposure. J Appl Physiol. 2005;98:889–94. doi: 10.1152/japplphysiol.00978.2004. doi: 10.1152/japplphysiol.00978.2004. [DOI] [PubMed] [Google Scholar]
- 5.Yuan G, Nanduri J, Bhasker CR, Semenza GL, Prabhakar NR. Ca2+/calmodulin kinase-dependent activation of hypoxia inducible factor 1 transcriptional activity in cells subjected to intermittent hypoxia. J Biol Chem. 2005;280:4321–8. doi: 10.1074/jbc.M407706200. doi: 10.1074/jbc.M407706200. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Zhang J, Gan TX, Chen-Izu Y, Hasday JD, Karmazyn M, et al. Left ventricular dysfunction and associated cellular injury in rats exposed to chronic intermittent hypoxia. J Appl Physiol. 2008;104:218–23. doi: 10.1152/japplphysiol.00301.2007. doi: 10.1152/japplphysiol.00301.2007. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–37. doi: 10.1074/jbc.271.51.32529. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 8.Jia SS, Liu YH. Down-regulation of hypoxia inducible factor-1alpha: A possible explanation for the protective effects of estrogen on genioglossus fatigue resistance. Eur J Oral Sci. 2010;118:139–44. doi: 10.1111/j.1600-0722.2010.00712.x. doi: 10.1111/j.1600-0722.2010.00712.x. [DOI] [PubMed] [Google Scholar]
- 9.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takarada Y, Takazawa H, Sato Y, Takebayashi S, Tanaka Y, Ishii N. Effects of resistance exercise combined with moderate vascular occlusion on muscular function in humans. J Appl Physiol. 2000;88:2097–106. doi: 10.1152/jappl.2000.88.6.2097. [DOI] [PubMed] [Google Scholar]
- 11.Hou YX, Jia SS, Liu YH. 17beta-Estradiol accentuates contractility of rat genioglossal muscle via regulation of estrogen receptor alpha. Arch Oral Biol. 2010;55:309–17. doi: 10.1016/j.archoralbio.2010.02.002. doi: 10.1016/j.archoralbio.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Liu Y. Effects of genistein and estrogen on the genioglossus in rats exposed to chronic intermittent hypoxia may be HIF-1α dependent. Oral Dis. 2013;19:702–11. doi: 10.1111/odi.12060. doi: 10.1111/odi.12060. [DOI] [PubMed] [Google Scholar]
- 13.Skelly JR, Edge D, Shortt CM, Jones JF, Bradford A, O’Halloran KD. Tempol ameliorates pharyngeal dilator muscle dysfunction in a rodent model of chronic intermittent hypoxia. Am J Respir Cell Mol Biol. 2012;46:139–48. doi: 10.1165/rcmb.2011-0084OC. doi: 10.1165/rcmb.2011-0084OC. [DOI] [PubMed] [Google Scholar]
- 14.Kunz M, Ibrahim SM. Molecular responses to hypoxia in tumor cells. Mol Cancer. 2003;2:23. doi: 10.1186/1476-4598-2-23. doi: 10.1186/1476-4598-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono Y, Sensui H, Sakamoto Y, Nagatomi R. Knockdown of hypoxia-inducible factor-1alpha by siRNA inhibits C2C12 myoblast differentiation. J Cell Biochem. 2006;98:642–9. doi: 10.1002/jcb.20804. doi: 10.1002/jcb.20804. [DOI] [PubMed] [Google Scholar]
- 16.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Hardie DG, Hawley SA, Scott JW. AMP-activated protein kinase – Development of the energy sensor concept. J Physiol. 2006;574(Pt 1):7–15. doi: 10.1113/jphysiol.2006.108944. doi: 10.1113/jphysiol.2006.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang D, Li T, Li X, Zhang L, Sun L, He X, et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014;8:1930–42. doi: 10.1016/j.celrep.2014.08.028. doi: 10.1016/j.celrep.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–903. doi: 10.1074/jbc.M800102200. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–84. doi: 10.1016/j.cmet.2009.08.015. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tello D, Balsa E, Acosta-Iborra B, Fuertes-Yebra E, Elorza A, Ordóñez Á, et al. Induction of the mitochondrial NDUFA4L2 protein by HIF-1α decreases oxygen consumption by inhibiting complex I activity. Cell Metab. 2011;14:768–79. doi: 10.1016/j.cmet.2011.10.008. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, et al. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol. 2001;21:3436–44. doi: 10.1128/MCB.21.10.3436-3444.2001. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–57. doi: 10.1016/s0092-8674(03)00154-5. doi: 10.1016/S0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mason SD, Howlett RA, Kim MJ, Olfert IM, Hogan MC, McNulty W, et al. Loss of skeletal muscle HIF-1alpha results in altered exercise endurance. PLoS Biol. 2004;2:e288. doi: 10.1371/journal.pbio.0020288. doi: 10.1371/journal.pbio.0020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason SD, Rundqvist H, Papandreou I, Duh R, McNulty WJ, Howlett RA, et al. HIF-1alpha in endurance training: Suppression of oxidative metabolism. Am J Physiol Regul Integr Comp Physiol. 2007;293:R2059–69. doi: 10.1152/ajpregu.00335.2007. doi: 10.1152/ajpregu.00335.2007. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen DD, Kim G, Pae EK. Modulation of muscle fiber compositions in response to hypoxia via pyruvate dehydrogenase kinase-1. Front Physiol. 2016;7:604. doi: 10.3389/fphys.2016.00604. doi: 10.3389/fphys.2016.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powers SK, Ji LL, Kavazis AN, Jackson MJ. Reactive oxygen species: Impact on skeletal muscle. Compr Physiol. 2011;1:941–69. doi: 10.1002/cphy.c100054. doi: 10.1002/cphy.c100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozakowska M, Pietraszek-Gremplewicz K, Jozkowicz A, Dulak J. The role of oxidative stress in skeletal muscle injury and regeneration: Focus on antioxidant enzymes. J Muscle Res Cell Motil. 2015;36:377–93. doi: 10.1007/s10974-015-9438-9. doi: 10.1007/s10974-015-9438-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viganò A, Ripamonti M, De Palma S, Capitanio D, Vasso M, Wait R, et al. Proteins modulation in human skeletal muscle in the early phase of adaptation to hypobaric hypoxia. Proteomics. 2008;8:4668–79. doi: 10.1002/pmic.200800232. doi: 10.1002/pmic.200800232. [DOI] [PubMed] [Google Scholar]
- 32.Miwa T, Koyama T, Shirai M. Muscle specific expression of Cre recombinase under two actin promoters in transgenic mice. Genesis. 2000;26:136–8. doi: 10.1002/(SICI)1526-968X(200002)26:2<136:AID-GENE11>3.0.CO;2-J. [PubMed] [Google Scholar]