Abstract
Natural killer (NK) cells are an important component of the innate immune response against viral infections. NK cell-mediated cytolytic activity is defective in HIV-infected individuals with high levels of viral replication. In the present study, we examined the phenotypic and functional characteristics of an unusual CD56–/CD16+ (CD56–) NK subset that is greatly expanded in HIV-viremic individuals. The higher level of expression of inhibitory NK receptors and the lower level of expression of natural cytotoxicity receptors observed in the CD56– NK fraction compared with that of CD56+ NK cells was associated with extremely poor in vitro cytotoxic function of this subset. In addition, the secretion of certain cytokines known to be important in initiating antiviral immune responses was markedly reduced in the CD56–, as compared with the CD56+ NK cell subset. These data suggest that the expansion of this highly dysfunctional CD56– NK cell subset in HIV-viremic individuals largely accounts for the impaired function of the total NK cell population.
Keywords: cytokines, cytotoxicity, inhibitory NK receptors, killer immunoglobulin-like receptors, natural cytotoxicity receptors
Natural killer (NK) cells, which account for up to 15% of peripheral blood lymphocytes, are well known as important effectors of the innate immune system (1, 2). In addition, NK cells are an important source of cytokines that regulate hematopoiesis and link the innate to the adaptive immune response through a bidirectional cross-talk with dendritic cells (3, 4).
NK cells are able to lyse tumor and virally infected cells without prior sensitization while sparing normal cells that express adequate levels of MHC of class I molecules (MHC-I). This cytolytic function is under the dominant control of a heterogeneous family of inhibitory NK receptors (iNKRs) that bind specifically to certain allelic forms of HLA of class I (HLA-I). In humans, the iNKRs are divided into two different groups: (i) killer immunoglobulin-like receptors (KIRs); (ii) LIR1/ILT2 and NKG2A/CD94, which belong to the family of the C-type lectin proteins. Diminution or absence of expression of HLA-I molecules on the cell surface because of viral infection or tumor transformation results in reduced engagement of iNKRs; loss of this dominant-negative signal in turn allows a large group of activating NK receptors and coreceptors to trigger cytolytic activity (5–7).
The role of NK cells in the course of HIV-1 infection remains to be fully elucidated; however, the effect of HIV infection on NK cell function has been increasingly delineated in recent years. It is well documented that NK cells isolated from HIV-infected individuals are impaired in their ability to kill HIV-1-infected autologous cells, as well as tumor cell lines (8–11). More recently, numerous studies have characterized the effects of HIV viremia on NK cell phenotype and function. In this regard, it has been demonstrated that the expression and function of most major iNKRs are either maintained or significantly increased on the surface of NK cells in viremic patients (12, 13), and a direct correlation has been reported between these findings and levels of HIV viremia (14). Moreover, the expression of natural cytotoxicity receptors (NCRs), NKp46, NKp30, and NKp44, is markedly decreased among viremic individuals, along with a concomitant decrease in NK cytolytic activity (13, 15). In addition, HIV viremia affects the capacity of NK cells to secrete CC-chemokines, which are well known as suppressors of HIV replication ex vivo (16, 17). Finally, the expansion of an unusual CD56–/CD16+ (CD56–) NK cell subset has been associated with high HIV viral loads (10, 13, 18). The successful suppression of HIV replication below detectable levels after treatment with effective antiretroviral therapy results in considerable improvement of NK cell cytotoxicity, cytokine secretion, NK cell receptor expression and function (13), and the restoration of normal CD56 expression (19).
The present study characterizes the phenotypic and functional characteristics of the CD56– NK subset isolated from HIV-1-viremic individuals. This population exhibited significantly lower cytolytic activity and ability to secrete cytokines, as well as more dramatic abnormalities in the expression and function of NK activating and inhibiting receptors when compared with the CD56+ NK cell subset from the same individuals. These data suggest that impairments observed in the total NK population of HIV-viremic individuals are largely due to elevated frequencies of this highly dysfunctional CD56– NK cell subset.
Methods
Study Subjects. Forty-eight HIV-1-infected viremic individuals who were not receiving antiretroviral therapy were studied. The median CD4+ cell count was 552 per μl (SD = ±251) and the CD4/CD8 ratio was <1 in all subjects. The median plasma viremia was 36,249 HIV RNA copies (SD =±70,952) per ml of plasma as detected by an ultrasensitive branched DNA (bDNA) assay (Chiron) with a lower limit of detection of 50 copies per ml (Table 1, which is published as supporting information on the PNAS web site). Leukapheresis was performed after signed informed consent was given as part of clinical protocols approved by Institutional Review Boards of the University of Toronto (21 patients) and of the National Institute of Allergy and Infectious Diseases (27 patients). As negative controls, peripheral blood mononuclear cells (PBMCs) from 30 healthy donors seronegative for HIV were obtained by apheresis generously provided by the Transfusion Medicine Department of the Warren Grant Magnuson Clinical Center, National Institutes of Health. The normal donors were enrolled under a protocol approved by the Institutional Review Board. The median CD4+ T cell count was 1,620 cells per μl (SD = ±299) and the CD4/CD8 ratio was >1 in all of the subjects.
Isolation and Culture of NK Cells. PBMCs were isolated over Ficoll–Hypaque gradients (LSM, ICN), and NK cells were isolated by negative selection using column-based cell separation techniques (Stem Cell Technologies, Vancouver), as described in refs. 13 and 17. Purified NK cells contained ≤3% contamination of other PBMC subsets (CD3, CD4, TCRα/β, TCRγ/δ, CD19, or CD14). CD56+ and CD56– NK cell subsets were separated by using a magnetic cell-sorting technique (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany) according to the protocol provided by the manufacturer. The purities of the CD56+ and CD56– NK cell fractions, as assessed by flow cytometry, were consistently more than 95% and 90%, respectively. CD56– and CD56+ NK cell subsets were placed in 96-well round-bottom plates at a concentration of 106 cells per ml and activated for 6 days in medium (RPMI medium 1640 with penicillin/streptomycin, l-glutamine (GIBCO), and 10% FCS (HyClone) containing recombinant IL-2 (rIL-2) at 200 units/ml (Roche Molecular Biochemicals).
Monoclonal Antibodies (mAbs). The mAbs 289 (IgG2a anti-CD3), C218 and FS280 (IgG1 and IgG2a anti-CD56, respectively), KD1 (IgG2a anti-CD16), Gl183 (IgG1 anti-p58.2/KIR2DL2), 11pb6 (IgG1 anti-p58.1/KIR2DL1), Z27 (IgG1 anti-p70/KIR3DL1), F278 (IgG1 anti-LIR1/ILT2), Z270 (IgG1 anti-NKG2A), Xa185 (IgG1 anti-CD94), Q66 (IgM anti-p140/KI3DL2), Qa79 (IgG1 anti-p75/AIRM1), Bab281 (IgG1 anti-NKp46), Az20 (IgG1 antiNKp30), Z231 (IgG1 anti-NKp44), On72 and Bat221 (IgG1 antiNKG2D), pp35 (IgG1 anti-2B4), Ma152 (IgG1 anti-NKp80), and Ma127 (IgG1 anti-NTB-A) were provided by one of the authors (A.M.) and D. Pende (Istituto Scientifico Tumori, Genoa, Italy). Anti-CD4 (IgG1), anti-TCRα/β (IgG1), anti-TCR γ/δ (IgG1), anti CD19 (IgG1), anti-CD14 (IgG1), anti-CD69 (IgG1), and anti CD38 (IgG1) were purchased from BD Pharmingen.
Flow-Cytometric Analysis and Cytolytic Activity. For one or two-color cytofluorimetric analysis, freshly isolated and rIL-2-activated NK cells were stained with the appropriate mAbs followed by phycoerythrin (PE)or FITC-conjugated isotype-specific goat antimouse second reagent (Southern Biotechnology Associates). Second appropriate anti-isotypic mAbs stained with PE or/and FITC were used as negative controls. The data were analyzed by using cellquest software (Becton Dickinson). After 6 days of activation with rIL-2, NK cells were tested for cytolytic activity at various effector-to-target (E/T) ratios in a 4-h 51Cr-release assay, as described in ref. 13.
Cellular Proliferation and Apoptosis of Activated NK Cells. After 6 days of activation with rIL-2, proliferation of CD56– and CD56+ NK cell subsets was measured by [3H]thymidine uptake (16 h) in triplicate. Early apoptotic cells were detected by using a flow-cytometric assay with FITC-conjugated annexin V according to the supplier's instructions (BD Pharmingen).
IFN-γ, Granulocyte/Macrophage Colony-Stimulating Factor (GM-CSF), and TNF-α Secretion by Activated NK Cells. After 6 days of activation with rIL-2, total unfractionated NK cells, as well as CD56+ and CD56– NK subsets, were stimulated overnight with phytohemagglutinin (0.5%), and the levels of supernatant-associated IFN-γ, GM-CSF, and TNF-α were measured by ELISA according to the protocol provided by the supplier (R & D Systems).
Statistical Analysis. The distributions of each immune response variable were compared between uninfected and HIV-infected viremic individuals by using the Mann–Whitney test. For each individual, immune response variables were calculated for CD56– and CD16+ NK subsets, and the difference in these two variables was evaluated by using the Wilcoxon signed-ranks test. All P values are two-sided and unadjusted.
Results
Frequency of CD56– Cells in Freshly Isolated NK Cells from HIV-Viremic Individuals. We have previously shown that the CD56– NK cell subset, which is rarely observed in uninfected individuals, is expanded in HIV-infected subjects in association with high levels of plasma viremia (13). In the present study, freshly isolated NK cells purified from PBMCs of healthy donors and HIV-infected viremic individuals were analyzed by flow cytometry for the expression of CD56 and CD16 cell surface markers. Forty-three of 48 (89%) infected patients with high viral load (Table 1), and 1 of 30 normal volunteers manifested a CD56– NK cell subset frequency of >10%. The median percentage of the CD56– subset of NK cells isolated from HIV-viremic versus HIV-negative control subjects was significantly different (P < 0.0001) (Fig. 4, which is published as supporting information on the PNAS web site). As previously described (13, 20), the frequency of CD56bright/CD16– NK cells in PBMCs was not significantly different between healthy donors and viremic patients (data not shown). HIV-infected individuals whose viremia was suppressed by antiretroviral therapy for 2 years or longer showed a NK cell phenotype similar to that of healthy donors for CD56 and CD16 expression (13).
We and others have reported that the expression and function of inhibiting and activating NK receptors are dysregulated in NK cells of HIV-infected viremic individuals (12–15). To determine whether this abnormal expression of NK receptors was preferentially associated with a particular NK subset, we analyzed their expression on CD56– and CD56+ populations of freshly isolated NK cells by using flow cytometry. The CD56+ analysis gate included both the CD56dim/CD16+, sharply decreased in viremic individuals (13), and the CD56bright/CD16– NK subpopulations. The expression of two iNKRs (p58.2/KIR2DL2 and LIR/ILT2) was significantly higher on CD56–, as compared with CD56+, NK cells (P = 0.016 and P < 0.0001, respectively). There was no significant difference in the levels of expression of the other two major iNKRs (p58.1/KIR2DL1 and p70/KIR3DL1) on CD56– versus CD56+ NK cells; however, some HIV-viremic subjects exhibited an unusually high frequency of CD56– cells positive for these KIRs (up to 73%). To better appreciate the extent of the expansion of the CD56–/iNKRs+ NK subset in viremic patients, the above-mentioned four iNKRs were labeled all together and analyzed in a double cytofluorometric analysis with the CD56 marker. This dysfunctional CD56– NK subset showed a statistically significant higher expression (range 52–83%) of iNKRs when compared with those seen in the CD56+ cells (range 24–76%) (P < 0.0001). NKG2A is the only iNKR that has been previously reported to be decreased in both expression and function in NK cells from viremic individuals (13). Of interest, NKG2A expression was significantly lower in the CD56–, as compared with the CD56+, subset of NK cells (P < 0.0001). CD94, which, if associated with NKG2A, confers iNKR activity to the complex, was not differentially expressed on CD56+ or CD56– subpopulations (Fig. 5 A and B, which is published as supporting information on the PNAS web site). No differences were detected in surface expression between the two different subsets of CD56– and CD56+ NK cells for other iNKRs such as p140/KIR3DL2 and p75/AIRM1 (data not shown).
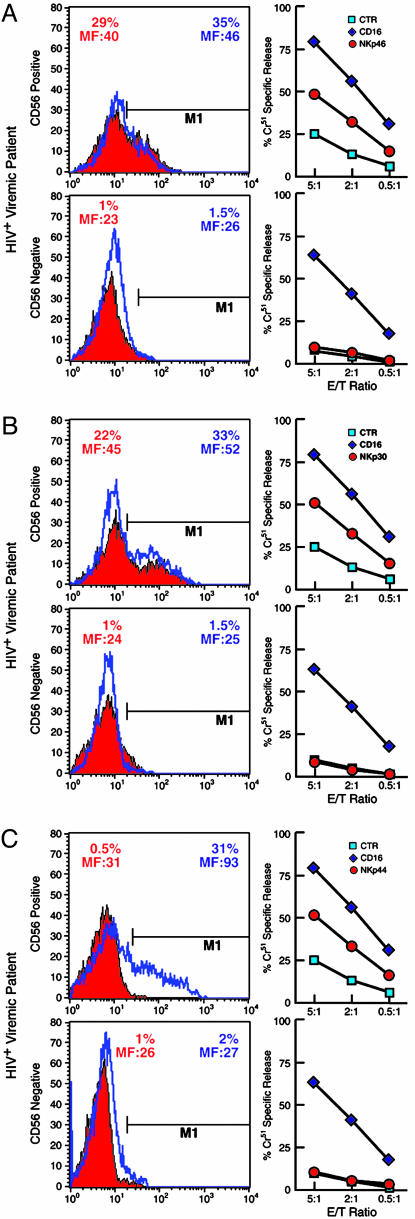
We then analyzed the cell surface expression of NKp46, NKp30, and NKG2D, the main activating receptors that are present constitutively on fresh NK cells. The NCRs NKp46 and NKp30 are specifically expressed on NK cells, whereas NKG2D is also expressed on a subset of CD3+/CD8+ T cells (6). NKp46 and NKp30 were barely detectable on the CD56– NK subset, and this frequency was significantly lower than that observed in the CD56+ NK cell subset (P < 0.0001). NKG2D expression was comparable in the two subsets) (Fig. 5 C and D). No differences were observed between CD56– and CD56+ NK cells in surface expression of the activating coreceptors NKp80, 2B4, and NTB-A (data not shown).
Phenotypic and Functional Profiles of CD56– NK Cells after Activation with rIL-2. Purified CD56– and CD56+ NK cells from HIV-infected viremic individuals were activated in vitro with rIL-2. Expression of the CD56 molecule was fully recovered upon activation of the CD56– subset over the course of 2–3 weeks (Fig. 1A). Despite the partial reversion of the CD56– subset to a CD56+ phenotype after 6 days of rIL-2 stimulation, the cytolytic activity of these cells against the NK-susceptible K562 erythroleukemia cell line target was dramatically reduced compared with that of the activated CD56+ NK cell subset of the same HIV donor (P < 0.05) (Fig. 1B). K562-directed killing by this population did not significantly improve even after 21 days of activation although cells expressed normal levels of classical markers of activation, such as CD69 or CD38 (data not shown) and a complete recovery of CD56 as shown in Fig. 3A. Moreover, the frequency of the entire CD56– and CD56+ freshly derived NK subsets that were undergoing apoptosis (annexin V-positive cells) during the course of activation was low (Fig. 1C). In addition, the level of cellular proliferation of the CD56– subset after 6 days of culture with rIL-2 did not differ significantly from that of the CD56+ subset (P > 0.05) (Fig. 1D). Finally, there were no differences between these subsets with regard to the secretion of perforin or granzyme (data not shown).
Fig. 1.
CD56 phenotype and function of activated CD56– NK cells. (A) Separation of CD56– and CD56+ subsets in total freshly isolated NK cells obtained from a viremic HIV-infected patient and recovery of CD56 expression upon stimulation with rIL-2 in vitro. (B) Spontaneous cytolytic activity against the K562 cell line of NK cells activated with rIL-2 for 6 days at various effector-to-target (E/T) ratios. Data represent the average lytic activity obtained by using CD56+ (green squares) versus CD56– (red circles) NK cells from 48 HIV-viremic patients. (C) Representative example of cytofluorometric analysis with CD56-PE/annexin V-FITC of CD56+ (left green plot) and CD56– (right red plot) NK cells after 6 days of stimulation with rIL-2. (D) Proliferation of CD56+ (green diamond) and CD56– (red circle) NK cell subsets at different cell numbers per well after stimulation with rIL-2 for 6 days. Data are represented as the average [3H]thymidine cpm of experiments conducted with cells from 48 patients.
Fig. 3.
Expression and function of NCRs on CD56+ and CD56– NK cell subsets. CD56+ (Upper) and CD56– (Lower) NK cell subsets were analyzed for NCR expression (Left) and function (Right). Histograms indicate the percent of freshly isolated (solid red) or rIL-2-activated (blue lines) CD56+ and CD56– NK cells expressing NKp46 (A), NKp30 (B), and NKp44 (C). The percentage of NK cells expressing NCRs with their geometric mean fluorescence intensity (MF) are indicated. Functional evaluation of NCRs by a redirected killing assay using an FcγR+ P815 target cell line and IL-2-activated NK cell subsets are adjacent to each histogram. Every graph shows the baseline lysis (blue squares), the maximal lysis triggered by anti-CD16 IgG mAb (blue diamonds), and killing driven by the relevant activating NK receptors (red circles). Data are from a single experiment and are representative of data obtained by using cells isolated from 48 HIV-viremic patients whose expression and function of NCRs were similar.
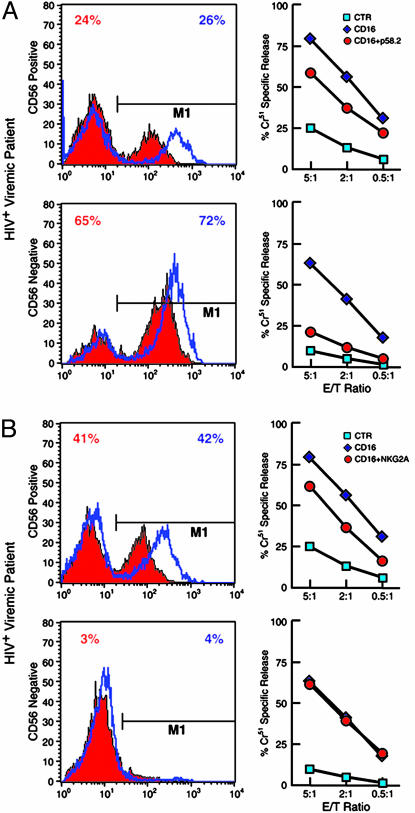
Functional Correlates of Inhibiting and Activating NK Receptor Expression on Activated CD56– and CD56+ NK Cells. Using mAbs with activating properties directed against specific iNKRs and NCRs (redirected killing assays), we sought to determine whether cytolytic activity could be correlated with NK receptor expression on activated CD56–- or CD56+-derived NK cells. After 6 days of stimulation with rIL-2, the frequency of expression of iNKRs on the CD56-positive- and negative-derived NK cells was very similar to that seen in freshly isolated subsets, and the differences observed between fresh CD56+ and CD56– NK subsets in receptor expression remained statistically significant (Fig. 6 A and B, which is published as supporting information on the PNAS web site). To evaluate whether the level of each inhibiting NK receptor was related to its function, CD56+ and CD56– activated NK subsets were analyzed in a redirected killing assay against the FcγR+ P815 target cell line in the presence of anti-CD16 mAb, used alone or in combination with mAb specific for each individual iNKR. The increased expression of one or another inhibitory NK cell receptor on the activated CD56– NK subset was always associated with a greater inhibition of the CD16-induced cytotoxicity when compared with that of the CD56+ NK cells, which had a lower expression of iNKRs. The higher levels of the iNKRs p58.2/KIR2DL2 (Fig. 2A) and LIR1/ILT2 (not shown) on the CD56– NK subset were associated with significantly greater inhibition of lytic activity by this population, as compared with that of the CD56+ NK subset, in CD16-induced redirected killing assays (P < 0.05). This result also indicated that the KIR molecules detected by Gl183 mAb (which reacts with both KIR2DL2 and KIR2DL3) within the CD56– NK subset are essentially represented by inhibitory rather than activating forms of KIRs. Suppression of lytic activity by p58.1/KIR2DL1 and p70/KIR3DL1 was similar between the CD56– and CD56+ NK subsets, reflecting the comparable levels of expression of these iNKRs on the two NK populations (data not shown). Finally, the addition of anti-NKG2A mAb had a significant (P < 0.05) inhibitory effect on the lytic activity of CD56+-, but not CD56–-, activated NK cells (Fig. 2B), a result that correlates with the very low or negative expression of NKG2A on the CD56– cells.
Fig. 2.
Expression and function of iNKRs p58.2/KIR2DL2 and NKG2A on CD56+ and CD56– NK cell subsets. CD56+ (Upper) and CD56– (Lower) NK cell subsets were analyzed for iNKRs expression (Left) and function (Right). Histograms indicate the percent of freshly isolated (solid red) or rIL-2-activated (blue lines) CD56+ and CD56– NK cells expressing p58.2/KIR2DL2 (A) or NKG2A (B). Functional evaluation of iNKRs by a redirected killing assay using an FcγR+ P815 target cell line and IL-2-activated NK cell subsets are adjacent to each histogram. Every graph shows the baseline lysis (blue squares), the maximal lysis triggered by anti-CD16 IgG mAb (blue diamonds), and the inhibition of killing driven by the cotriggering of relevant receptors with anti-CD16 IgG mAb (red circles). Data are from a single experiment and are representative of data obtained by using cells isolated from 48 HIV-viremic patients.
Similar to the expression of iNKRs, the differential expression of the activating NK receptors, NKp46 and NKp30, on fresh CD56– and CD56+ cells did not change significantly over the course of activation (Fig. 3 A and B). NKp44, a NK cell-specific NCR usually expressed only upon activation (6), resulted in up-regulation on the surface of CD56+, but not CD56–, NK cells, after 6 days of stimulation with rIL-2 (P = 0.0003) (Fig. 3C). To evaluate the functional capability of NCRs to induce NK-mediated cytotoxicity, activated NK subsets derived from CD56– and CD56+ cells were analyzed in a redirected killing assay in the presence or absence of mAbs specific for NKp46, NKp30, and NKp44. In cultures containing activated CD56– NK cells none of the three NCR mAbs were able to induce NK-mediated cytotoxicity, consistent with the NCRsdim or NCRsneg phenotype of these cells. In contrast, CD56+ NK cells, which express these NCRs to various degrees, exhibited enhanced cytolytic activity (P < 0.05) after mAb-mediated activation of NKp46, NKp30, and NKp44 (Fig. 3).
Therefore, despite the recovery of CD56 expression on the CD56– NK subset in a time-dependent manner upon stimulation with rIL-2, this unusual population kept the identical profile of expression of activating and inhibiting NK receptors over the time period in question. This conserved phenotype allowed us to exclude any major contamination of the CD56– cell subset with CD56+ NK cells during the growth and proliferation of the CD56– population. In this regard, in the NK subset separation shown as a representative example in Fig. 1A, the CD56 expression on the freshly purified CD56– NK cells was only of 2.5%. On activation with rIL-2, the CD56–-derived NK cells, whose rate of proliferation and apoptosis did not differ from that of the CD56+-derived NK cells, had similar levels of p58.2/KIR2DL2 as well as a similar lack of expression of NKG2A/CD94 and NCRs noted in the original fresh CD56– NK cells. In a similar manner, the activated CD56+-derived NK cells kept the same phenotype of the freshly purified CD56+ cells (Figs. 2 and 3). Moreover, the same striking differences in NK receptor levels between fresh CD56– and CD56+ cells were conserved on the activated cells, reflecting the extreme dichotomy of cytolytic activities of the two subsets (Fig. 6).
IFN-γ, TNF-α, and GM-CSF Secretion. In addition to lytic activity, NK cells release several cytokines that can modulate other effector functions of the immune system (3, 21). The levels of a large panel of NK-related cytokines were assessed in the supernatants from cultures of total unfractionated, CD56–, and CD56+ NK cells after 6 days of stimulation with rIL-2 and 12 h with phytohemagglutinin. As reported by several studies (13, 22–24), activated NK cells from HIV-viremic patients secrete significantly lower levels of IFN-γ, TNF-α, and GM-CSF as compared with NK cells isolated from healthy donors. Furthermore, within the cohort of viremic subjects, activated NK cells derived from fresh CD56– cells produced significantly lower levels of IFN-γ, TNF-α, and GM-CSF as compared with the levels secreted by activated CD56+ NK cells (Fig. 7, which is published as supporting information on the PNAS web site).
Discussion
NK cell functions are significantly impaired in HIV-infected individuals with high levels of ongoing viral replication. In healthy HIV-seronegative individuals, as well as in HIV-infected individuals who are receiving antiretroviral therapy and whose viremia was suppressed to below detectable level for 2 years or longer, the majority of NK cells (85–95%) are CD56dim/CD16+, whereas a minority are CD56bright and CD16dim/neg cells (13, 25). When the HIV viral load is high, there is a dramatic expansion of CD56–/CD16+ (CD56–) NK cells, a subset that is represented at a very low frequency in healthy HIV-negative and aviremic HIV-infected individuals (10, 13, 18). Of note, HIV-1 viremia does not seem to affect the CD56bright NK cells; however, it is associated with a statistically significant decrease of the CD56dim/CD16+ NK subpopulation (13, 20), and it has also been related to a rapid and early progression to AIDS (26). The presence of an expanded CD56– NK cell population among HIV viremic individuals has been associated with significantly reduced spontaneous NK cytolytic activity as compared with that of aviremic individuals or healthy donors (13). The present study delineates the potential mechanisms of these abnormalities by demonstrating that the expression of NCRs is low or negative whereas that of certain iNKRs is elevated on the CD56–, as compared with the CD56+, NK cell subset isolated from HIV-infected viremic subjects. In addition, the impaired cytolytic function of CD56– NK cells reflects the aberrant expression of certain NK receptors on this subset. Furthermore, the production of numerous cytokines known to modulate the function of other immune effectors was reduced in the CD56–, as compared with the CD56+ NK cell subset. Taken together, these data suggest that impaired NK cell function in HIV-infected viremic individuals may be largely attributable to the expansion of this highly dysfunctional CD56– NK cell subset.
Significant differences in spontaneous cytolytic activity between the CD56+ and CD56– NK cells isolated from HIV-infected individuals have been previously reported (27). The present study confirmed that the CD56– NK subset isolated from HIV-viremic individuals exhibits very low cytotoxicity against K562 targets, even after the complete recovery of the expression of CD56 upon activation with rIL-2. We were unable to identify significant differences between rIL-2-activated CD56– and CD56+ NK subsets in terms of cellular proliferation or perforin/granzyme expression; in addition, rates of apoptosis were low in both subsets. However, a striking correlation was observed between the cytolytic dysfunction of CD56– NK cells and the expression of activating and inhibiting NK receptors on this subset.
In a previous study we noted that the expression of certain iNKRs, such as the inhibitory forms of KIRs and LIR1/ILT2, are either conserved or increased on NK cells from HIV viremic patients, as compared with HIV-negative or aviremic HIV-infected subjects who had been receiving antiretroviral therapy for 2 years or more, with the exception of NKG2A, which is decreased (13). In the present study, we have demonstrated that the level of expression of iNKRs was much higher on the CD56– NK cell subset compared with the CD56+ NK cell subset isolated from the same HIV-1-viremic individuals. Furthermore, functional analysis confirmed the greater receptor-specific inhibition of CD16-mediated cytolytic activity in CD56– NK cells consistent with the increased expression of iNKRs on their cell surface. The relevance of elevated iNKR expression/function in the context of HIV-infected target cells is suggested by the observation that, despite the selective decrease of MHC-I expression that generally occurs in HIV infection in vitro (28), NK cells are not able to lyse autologous infected T cell blasts (11) unless the interaction between specific HLAs and the corresponding iNKR is blocked (29). Clearly, the elevated expression of certain, but not all, iNKRs on CD56– NK cells in parallel with HIV-mediated down-regulation of MHC-I molecules presents a complex scenario.
If the dominant-negative effects of iNKR-self MHC-I interactions are overcome as would be expected by the virus-induced down-regulation of MHC-1 molecules, the triggering of activating NCRs by their ligands induces NK cytolytic activity. In this regard, the low or absent expression of NK-specific NCRs on CD56– NK cells versus CD56+ NK cells was more striking than the higher expression of iNKRs on CD56– versus CD56+ NK cells. A significant proportion of fresh and/or activated CD56+ NK cells expressed the three NCRs, NKp46, NKp30, and NKp44, and this subset of NK cells was able to lyse target cells through these activating receptors. In contrast, CD56– NK cells were negative for or had a very low surface density of these NCRs and almost a complete lack of ability to respond to NCR antibody-mediated triggering of cytotoxicity. While it remains unclear what the relative role of NK cells is in controlling ongoing HIV replication, the marked diminution of activating NCRs on the CD56– population could certainly contribute to a reduced ability to eliminate virus-infected cells. In this regard, it has been shown that immature dendritic cells of HIV-infected individuals are able to escape NK cell-mediated killing (30), an activity that in healthy individuals has been reported to be mainly dependent on the engagement of NKp30 receptor (31).
The mechanism(s) whereby the expression of certain NK receptors is dysregulated on CD56– NK cells of HIV-viremic individuals is not clear. Several cytokines have been shown to modulate the expression of NK receptors and some, such as TGF-β, which down-regulates the levels of NKp30 (32), and IL-10, which increases the expression of several iNKRs (33), have been found to be present at elevated levels in the serum of AIDS patients (34–36). We did not find significant differences in the levels of TGF-β (13) or IL-10 (data not shown) secreted by NK cells from viremic individuals compared with those of healthy donors, suggesting that at least an autocrine mechanism of receptor modulation among NK cells is not likely. However, NK cells are not the only cell types capable of producing these cytokines. Alternatively, CD56 down-regulation and altered expression of NCRs and iNKRs may reflect persistent antigenic exposure and a general state of chronic cellular activation seen with the HIV viremic state (37, 38). However, the expansion of the CD56–/CD16+ NK subset in HIV-infected individuals appears to be strictly associated with HIV viremia and has not been described in any other viral infections or immunological diseases that are associated with an aberrant state of immune activation. We have previously demonstrated that the NK subsets isolated in our studies are negative for HIV DNA, ruling out possible effects of direct viral infection (13). However, it remains possible that direct physical interaction between HIV virions or viral products and NK cells by means of chemokine receptors or C-lectin type receptors may result in dysregulation of NK receptors.
NK cells are an important component of the innate immune response to viral infection; however, they also secrete several cytokines and chemokines that play a role in the recruitment and activation of the adaptive immune response (39, 40). IFN-γ, TNF-α, and GM-CSF can play a role in activating antiviral activities of other cell types (41); however, they can also promote the maturation and migration of dendritic cells, allowing effective antigen-presentation for the adaptive immune response (3, 21). A significant reduction in levels of IFN-γ, TNF-α, and GM-CSF produced by NK cells from viremic patients versus healthy donors has been previously reported (13, 22–24). The present study demonstrates that the CD56– NK subset is particularly impaired in its ability to produce these cytokines. Thus, it remains to be determined whether this defect in cytokine secretion is related to a defect in triggering of NCRs and whether this defect also has an impact on dendritic cell function in HIV disease.
In conclusion, the present study describes the phenotypic and functional characteristics of the CD56–/CD16+ subset of NK cells, a population that is present at high frequencies in viremic individuals but not in aviremic HIV-infected individuals or healthy donors (13). As the CD56+ NK subset from these individuals appeared to be relatively intact with regard to NK receptor expression and function, we believe that the dramatic expansion of highly dysfunctional CD56– NK cells likely accounts for the defects previously reported for total NK cell populations from HIV-viremic individuals. Additional studies will be needed to further verify the HIV specificity of CD56–/CD16+ NK cell expansion and to elucidate the molecular mechanism(s) that drive the dysregulation of NK receptor expression and function in this subset.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Angelo Mavilio. We thank the patients for their participation in this study. We also thank Shyam Kottilil for his invaluable assistance in reviewing the manuscript.
Abbreviations: NK, natural killer; iNKRs, inhibitory NK receptors; KIRs, killer immunoglobulin-like receptors; NCRs, natural cytotoxicity receptors; PBMC, peripheral blood mononuclear cell; rIL-2, recombinant IL-2; GM-CSF, granulocyte/macrophage colony-stimulating factor.
References
- 1.Karre, K., Ljunggren, H. G., Piontek, G. & Kiessling, R. (1986) Nature 319, 675–678. [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri, G. (1989) Adv. Immunol. 47, 187–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moretta, A. (2002) Nat. Rev. Immunol. 2, 957–964. [DOI] [PubMed] [Google Scholar]
- 4.Raulet, D. H. (2004) Nat. Immunol. 5, 996–1002. [DOI] [PubMed] [Google Scholar]
- 5.Moretta, A., Bottino, C., Vitale, M., Pende, D., Biassoni, R., Mingari, M. C. & Moretta, L. (1996) Annu. Rev. Immunol. 14, 619–648. [DOI] [PubMed] [Google Scholar]
- 6.Moretta, A., Bottino, C., Vitale, M., Pende, D., Cantoni, C., Mingari, M. C., Biassoni, R. & Moretta, L. (2001) Annu. Rev. Immunol. 19, 197–223. [DOI] [PubMed] [Google Scholar]
- 7.Cerwenka, A. & Lanier, L. L. (2001) Nat. Rev. Immunol. 1, 41–49. [DOI] [PubMed] [Google Scholar]
- 8.Ullum, H., Gotzsche, P. C., Victor, J., Dickmeiss, E., Skinhoj, P. & Pedersen, B. K. (1995) J. Exp. Med. 182, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad, A. & Menezes, J. (1996) AIDS 10, 143–149. [DOI] [PubMed] [Google Scholar]
- 10.Scott-Algara, D. & Paul, P. (2002) Curr. Mol. Med. 2, 757–768. [DOI] [PubMed] [Google Scholar]
- 11.Bonaparte, M. I. & Barker, E. (2003) AIDS 17, 487–494. [DOI] [PubMed] [Google Scholar]
- 12.Ahmad, R., Sindhu, S. T., Tran, P., Toma, E., Morisset, R., Menezes, J. & Ahmad, A. (2001) J. Med. Virol. 65, 431–440. [PubMed] [Google Scholar]
- 13.Mavilio, D., Benjamin, J., Daucher, M., Lombardo, G., Kottilil, S., Planta, M. A., Marcenaro, E., Bottino, C., Moretta, L., Moretta, A. & Fauci, A. S. (2003) Proc. Natl. Acad. Sci. USA 100, 15011–15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kottilil, S., Shin, K., Planta, M., McLaughlin, M., Hallahan, C. W., Ghany, M., Chun, T. W., Sneller, M. C. & Fauci, A. S. (2004) J. Infect. Dis. 189, 1193–1198. [DOI] [PubMed] [Google Scholar]
- 15.De Maria, A., Fogli, M., Costa, P., Murdaca, G., Puppo, F., Mavilio, D., Moretta, A. & Moretta, L. (2003) Eur. J. Immunol. 33, 2410–2418. [DOI] [PubMed] [Google Scholar]
- 16.Oliva, A., Kinter, A. L., Vaccarezza, M., Rubbert, A., Catanzaro, A., Moir, S., Monaco, J., Ehler, L., Mizell, S., Jackson, R., et al. (1998) J. Clin. Invest. 102, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kottilil, S., Chun, T. W., Moir, S., Liu, S., McLaughlin, M., Hallahan, C. W., Maldarelli, F., Corey, L. & Fauci, A. S. (2003) J. Infect. Dis. 187, 1038–1045. [DOI] [PubMed] [Google Scholar]
- 18.Hu, P. F., Hultin, L. E., Hultin, P., Hausner, M. A., Hirji, K., Jewett, A., Bonavida, B., Detels, R. & Giorgi, J. V. (1995) J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10, 331–340. [PubMed] [Google Scholar]
- 19.Sondergaard, S. R., Aladdin, H., Ullum, H., Gerstoft, J., Skinhoj, P. & Pedersen, B. K. (1999) J. Acquir. Immune Defic. Syndr. 21, 376–383. [PubMed] [Google Scholar]
- 20.Tarazona, R., Casado, J. G., Delarosa, O., Torre-Cisneros, J., Villanueva, J. L., Sanchez, B., Galiani, M. D., Gonzalez, R., Solana, R. & Pena, J. (2002) J. Clin Immunol. 22, 176–183. [DOI] [PubMed] [Google Scholar]
- 21.Cooper, M. A., Fehniger, T. A., Fuchs, A., Colonna, M. & Caligiuri, M. A. (2004) Trends Immunol. 25, 47–52. [DOI] [PubMed] [Google Scholar]
- 22.Scott-Algara, D., Vuillier, F., Cayota, A. & Dighiero, G. (1992) Clin. Exp. Immunol. 90, 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azzoni, L., Papasavvas, E., Chehimi, J., Kostman, J. R., Mounzer, K., Ondercin, J., Perussia, B. & Montaner, L. J. (2002) J. Immunol. 168, 5764–5770. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, R., Weber, K., Wendt, K., Heiken, H. & Schmidt, R. E. (2004) J. Clin. Immunol. 24, 281–286. [DOI] [PubMed] [Google Scholar]
- 25.Cooper, M. A., Fehniger, T. A., Turner, S. C., Chen, K. S., Ghaheri, B. A., Ghayur, T., Carson, W. E. & Caligiuri, M. A. (2001) Blood 97, 3146–3151. [DOI] [PubMed] [Google Scholar]
- 26.Bruunsgaard, H., Pedersen, C., Skinhoj, P. & Pedersen, B. K. (1997) Scand. J. Immunol. 46, 91–95. [DOI] [PubMed] [Google Scholar]
- 27.Sondergaard, S. R., Ullum, H. & Pedersen, B. K. (2000) APMIS 108, 831–837. [DOI] [PubMed] [Google Scholar]
- 28.Cohen, G. B., Gandhi, R. T., Davis, D. M., Mandelboim, O., Chen, B. K., Strominger, J. L. & Baltimore, D. (1999) Immunity 10, 661–671. [DOI] [PubMed] [Google Scholar]
- 29.Bonaparte, M. I. & Barker, E. (2004) Blood 104, 2087–2094. [DOI] [PubMed] [Google Scholar]
- 30.Tasca, S., Tambussi, G., Nozza, S., Capiluppi, B., Zocchi, M. R., Soldini, L., Veglia, F., Poli, G., Lazzarin, A. & Fortis, C. (2003) AIDS 17, 2291–2298. [DOI] [PubMed] [Google Scholar]
- 31.Ferlazzo, G., Tsang, M. L., Moretta, L., Melioli, G., Steinman, R. M. & Munz, C. (2002) J. Exp. Med. 195, 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castriconi, R., Cantoni, C., Della Chiesa, M., Vitale, M., Marcenaro, E., Conte, R., Biassoni, R., Bottino, C., Moretta, L. & Moretta, A. (2003) Proc. Natl. Acad. Sci. USA 100, 4120–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parato, K. G., Kumar, A., Badley, A. D., Sanchez-Dardon, J. L., Chambers, K. A., Young, C. D., Lim, W. T., Kravcik, S., Cameron, D. W. & Angel, J. B. (2002) AIDS 16, 1251–1256. [DOI] [PubMed] [Google Scholar]
- 34.Lotz, M. & Seth, P. (1993) Ann. N.Y. Acad. Sci. 685, 501–511. [DOI] [PubMed] [Google Scholar]
- 35.Stylianou, E., Aukrust, P., Kvale, D., Muller, F. & Froland, S. S. (1999) Clin. Exp. Immunol. 116, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Havlir, D. V., Torriani, F. J., Schrier, R. D., Huang, J. Y., Lederman, M. M., Chervenak, K. A. & Boom, W. H. (2001) J. Clin. Microbiol. 39, 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun, T. W., Justement, J. S., Sanford, C., Hallahan, C. W., Planta, M. A., Loutfy, M., Kottilil, S., Moir, S., Kovacs, C. & Fauci, A. S. (2004) Proc. Natl. Acad. Sci. USA 101, 2464–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fogli, M., Costa, P., Murdaca, G., Setti, M., Mingari, M. C., Moretta, L., Moretta, A. & De Maria, A. (2004) Eur. J. Immunol. 34, 2313–2321. [DOI] [PubMed] [Google Scholar]
- 39.Cooper, M. A., Fehniger, T. A. & Caligiuri, M. A. (2001) Trends Immunol. 22, 633–640. [DOI] [PubMed] [Google Scholar]
- 40.Robertson, M. J. (2002) J. Leukocyte Biol. 71, 173–183. [PubMed] [Google Scholar]
- 41.Ahmad, A. & Ahmad, R. (2003) Curr. HIV Res. 1, 295–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.