Abstract
Objectives
Exosomes are important mediators in intercellular communications and play a role in cancer progression and metastasis. Exosomal membranes are enriched in endosome-specific tetraspanins (CD9 and CD63). Here, we explored the expression of CD63 and CD9 utilizing immunohistochemistry in malignant and non-malignant cells in 29 resected pancreatic specimens (RPS) of mixed racial background.
Methods
The pathologic tissues (PTs) and adjacent normal tissues (ANTs) in each RPS were stained for CD63 and CD9. Two pathologists independently scored the expression of CD63 and CD9. Staining intensity was graded from 1–3. Staining percentage was estimated in 10% increments. An average Q score (Intensity X Percentage of staining) was calculated. Unpaired t test was used for statistical analysis.
Results
The mean multiplicative Quick-score (Q-score) for CD63 and CD9 expression is higher in PTs (209 and 72) compared to ANTs (154 and 24) (p= 0.0041; p=0.0018). The Mean Q score for CD63 and CD9 expression is higher in the malignant PTs (231 and 85) compared to ANTs (129 and 25) (p<0.0001 and p < 0.0124).
Conclusions
Exosomal markers (CD63 and CD9) expression assessment using IHC is feasible in RPS. The expression of CD63 and CD9 is higher in PTs and malignant PTs compared to their ANT.
Keywords: Exosomes, Exosomal Markers, CD63, CD9, Pancreatic Tissues
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of death from cancer in the United States. 53,070 new cases and 41,780 deaths are estimated to occur in 2016. 1 The prognosis of PDAC is notoriously poor, with a 5-year overall survival of 5%. 2 Early diagnosis of PDAC has been and continues to be a challenge. Metastasis to distant organs, invasion of surrounding vasculature, and resistance to available chemotherapy are major causes of treatment failure and poor prognosis. The progress in the treatment of PDAC is unsatisfactory and that immensely motivated researchers to better understand tumor interaction with microenvironment, the process of metastasis to distant organs and resistance to chemotherapy.
Extracellular vesicles (EVs) are secreted membrane-enclosed vesicles that include exosomes, microvesicles, apoptotic bodies and other EV subsets. The biogenesis of exosomes is not clear; therefore the term EVs is often used. 3 Exosomes are membraneous nanovesicles (EVs of 30–150 nm diameter) of endocytic origin released by most cells types from diverse organisms. 4 They contain functional biomolecules including nucleic acids, lipids and proteins and are released into the extracellular space and enter the circulation. 4–7 Exosomal membranes are enriched in endosome-specific tetraspanins (CD9, CD63, CD81). Various other proteins have been identified to play a role in exosomes biogenesis (Alix, TSG101) and membrane transport and fusion (flotillin, GTPase). 8
Exosomes are secreted from cancer cells at higher rates compared to healthy cells. 9 Tumor exosomes are considered as an important mediator of intercellular communication, and play pivotal roles in facilitating cancer progression and metastasis. 4 Tumor exosomes represent a central mediator of the tumor microenvironment and can promote angiogenesis, stromal remodeling, pathways activation, chemoresistance and genetic intercellular exchange. 8 In PDAC cells, exosomes provided remarkable survival benefits to PDAC cells against gemcitabine treatment. Moreover, tumor exosomes can exert a broad array of detrimental effects on the immune system and mediate cancer-associated immunosuppressive microenvironment and help initiate pre-metastatic niche. 10,11 We, therefore, set out to explore the expression pattern of exosomal markers, CD63 and CD9, in malignant and non-malignant (Premalignant, inflammatory and normal) cells in resected pancreatic specimens by immunohistochemical (IHC) staining.
Materials and Methods
Pancreatic tissue collection
This study was conducted at the University of South Alabama Mitchell Cancer Institute in Mobile, Alabama. Patients included were identified through searching the pancreatic surgeon specific registry at the University of South Alabama Medical Center. The University of South Alabama Institutional Review Board (IRB) approved this study and the IRB-approved database provided a waiver of the requirement for informed consent and allowed for publication of de-identified data.
Immunohistochemical analysis
Two 5μm sections were obtained from every RPS. One was obtained from the pathologic tissues (PTs) and one was obtained from the adjacent normal tissues (ANTs). To study the expression pattern of the exsosomal markers (CD63 and CD9), immunohistochemical staining was performed. 12 In brief, the unstained slides were first deparaffinaized by using xylene (X1-10 min, X2-10 min) and subsequently hydrated by sequential incubation in ethanol (100% EtOH-5 min, 70% EtOH-5min, 50% EtOH-5 min, 30% EtOH-5 min, rinse the slides in running water-5 min). Thereafter, antigen retrieval was performed using Declokar chamber (Biocare Medical, Concord, CA) with 1X Declokar buffer (Biocare Medical) followed by blocking of the endogenous peroxidase by incubation with peroxidase-1 (Biocare Medical). Later tissue sections were blocked with Background Sniper (Biocare Medical) for 10 min and incubated with the primary antibodies against CD63 and CD9 (1:50 and 1:25, respectively) (mouse monoclonal; Abcam, Cambridge, MA) overnight at 4°C. Post-incubation, sections were washed and incubated with recommended polymer and probe (Biocare Medical) according to the manufacturer’s protocol. Immunoreactivity was visualized by DAB Chromogen followed by haematoxylin counterstain. Tissue sections incubated with normal mouse IgG (Santa Cruz Biotechnology Inc, Dallas, TX) served as negative control.
Scoring and statistical analysis
Two pathologists, independently, scored the staining of CD63 and CD9. The intensity of cytoplasmic staining was graded from 1 to 3: 1 (weak), 2 (moderate) and 3 (strong). The percentage of stained cells was estimated on each section in 10% increments. For each tissue section, a multiplicative Quick-score (Q-score) was calculated by multiplying the percentage of positive cells by the intensity of the staining. 13 The average Q-score was calculated for each section. Unpaired t test was used for statistical analysis. Statistical significance was defined as p < 0.05, and all tests were two-sided. Tests were performed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA).
Results
Between October 2013 and December 2015, searching the pancreatic surgeon specific registry at the University of South Alabama Medical Center identified twenty-nine patients that underwent pancreatic resection. The baseline characteristics of the patients are summarized in Table 1. The Median age was 59 years (range 35–80). 17 patients (59%) were white and 12 patients (41%) were African Americans. 16 patients (55%) were females, while 13 patients (45%) were males. 10 patients (34.5%) had PDAC, 3 patients (10.3%) had ampullary adenocarcinoma, 4 patients (13.8%) had pancreatic neuroendocrine tumors, 6 patients had premalignant conditions. 3 patients (10.3%) had Intraductal papillary mucinous neoplasm (IPMN) and 3 patients (10.3%) had pancreatic intraepithelial neoplasm, 6 patients (21.6%) had acute/chronic pancreatitis.
TABLE 1.
Patients Baseline Characteristics (N = 29)
| Age, median (range), y | 59 (35–80) |
|---|---|
|
| |
| Sex, n (%) | |
| Male | 13 (45) |
| Female | 16 (55) |
|
| |
| Ethnicity, n (%) | |
| White | 17 (59) |
| Black | 12 (41) |
|
| |
| Diagnosis, n (%) | |
| Malignant | |
| Pancreatic Adenocarcinoma | 10 (34.5) |
| Ampullary Adenocarcinoma | 3 (10.3) |
| Neuroendocrine tumor | 4 (13.8) |
| Premalignant | |
| IPMN | 3 (10.3) |
| Intraepithelial Neoplasm | 3 (10.3) |
| Inflammatory | |
| Acute/Chronic Pancreatitis | 6 (21.6) |
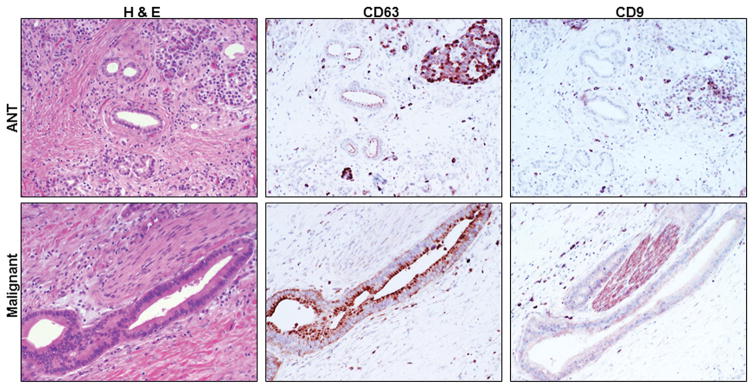
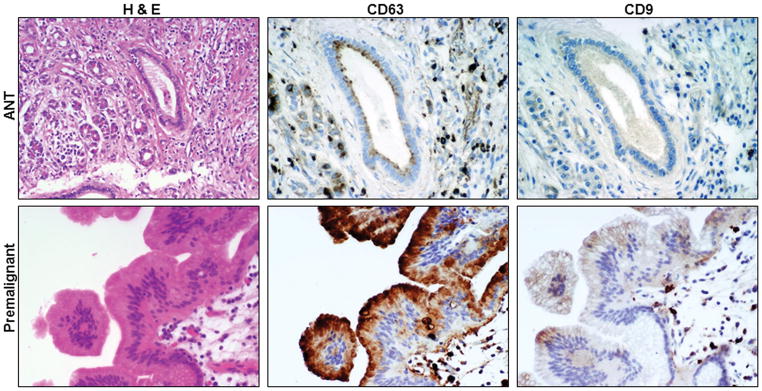
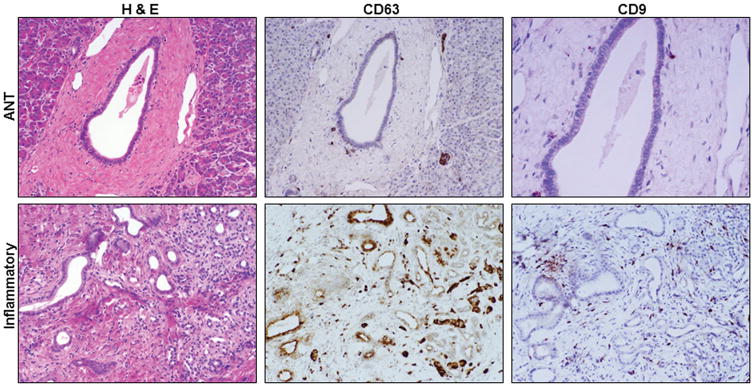
The H&E staining and the expression of CD63 and CD9 in the malignant PTs and their ANTs are shown respectively in figure 1. The H&E staining and the expression of CD63 and CD9 in the premalignant PTs and their ANTs are shown respectively in figure 2. The H&E staining and the expression of CD63 and CD9 in the inflammatory PTs and their ANTs are shown respectively in figure 3.
Figure 1.
H&E staining and CD63 and CD9 expression in malignant pathologic tissues (PTs) and their adjacent normal tissues (ANTs). (200X)
Figure 2.
H&E staining and CD63 and CD9 expression in premalignant pathologic tissues and their adjacent normal tissues (ANTs). (200X)
Figure 3.
H&E staining and CD63 and CD9 expression in inflammatory pathologic tissues and their adjacent normal tissues (ANTs). 200X
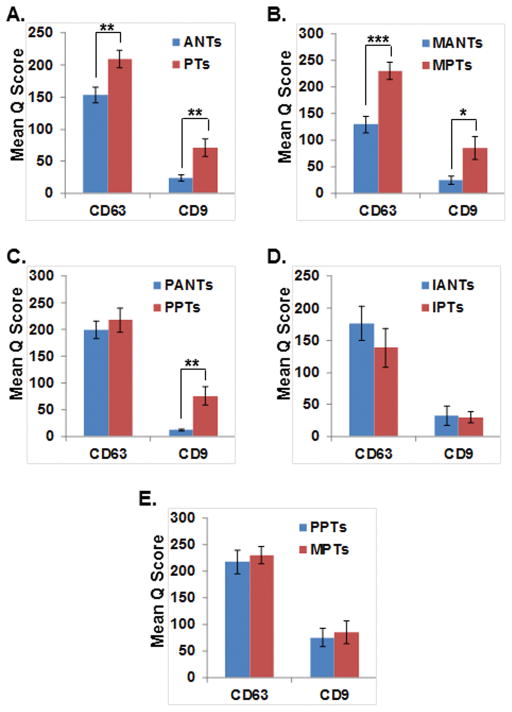
The mean multiplicative Quick scores for CD63 and CD9 are summarized in Table 2. The mean Q score in the PTs compared to their ANTs is higher in CD63 (209 Vs 154; p=0.0041) and CD9 (72 vs 24; p=0.0018) (Figure 4A). The mean Q score in the malignant PTs (MPTs) compared to their ANTs (MATs) is higher in CD63 (231 Vs 129; p<0.0001) and CD9 (85 vs 25; p=0.0132) (Figure 4B). The mean Q score in the premalignant PTs (PPTs) compared to their ANTs (PANTs) is higher in CD63 (218 Vs 100; p=0.52) and CD9 (76 vs 12; p=0.0048) (Figure 4C). The mean Q score in the inflammatory PTs (IPTs) compared to their ANTs (IANTs) is lower in CD63 (138 Vs 177; p=0.36). The mean Q score is equal in CD9 (30 vs 30; p=0.88) (Figure 4D). The mean Q score in the malignant PTs (MPTs) compared to premalignant ANTs (PANTs) is higher in CD63 (231 Vs 218; p=0.68) and CD9 (85 vs 76; p=0.812) (Figure 4E).
Table 2.
CD63 and CD9 mean Q scores
| Tissue | N | CD63 Mean Q score | CD9 Mean Q Score |
|---|---|---|---|
|
| |||
| Pathologic Tissues (PTs) | 29 | 209 | 72 |
| Malignant PTs | 17 | 231 | 85 |
| Premalignant PTs | 6 | 218 | 76 |
| Inflammatory PTs | 6 | 138 | 30 |
|
| |||
| Adjacent Normal Tissues (ANTs) | 29 | 154 | 24 |
| Malignant ANTs | 17 | 129 | 25 |
| Premalignant ANTs | 6 | 200 | 12 |
| Inflammatory ANTs | 6 | 177 | 30 |
Figure 4.
Exosomal markers CD9 and CD63 expression pattern in resected pancreatic tissues. Pathologic Tissues (PTs); Adjacent Normal Tissues (ANTs); Malignant PTs (MPTs); Malignant ANTs (MANTs); Premalignant PTs (PPTs); Premalignant ANTs (PANTs); Inflammatory PTs (IPTs); Inflammatory ANTs (IANTs); ***P <0.001; **P <0.01; *P <0.05.
The patients included in our study represent heterogeneous patient population including patients with malignant (N=17), premalignant (N=6) and inflammatory (n=6) conditions. Among the patients diagnosed with malignant conditions, we explored the pathologic characteristics and survival data of patients with PDAC (N=10). Given the small sample size, the presented data represent a descriptive summary. The data on patients with ampullary adenocarcinoma (N= 3) and pancreatic neuroendocrine tumors (N=4) were not further explored due to very small sample size. The relationship between the Q score of CD9 and CD63 and Tumor T stage, number of involved lymph nodes (LNs), tumor grade, the presence of lymphovascular invasion (LVI), perineural invasion (PNI), progression free survival (PFS) and overall survival (OS) is shown in Table # 3. Two patients (case # 1 and # 8) have received neoadjuvant chemotherapy prior to pancreatic resection. Four patients (case # 1, 5, 9 and 10) have not progressed yet and are still alive.
Table 3.
The relationship between the mean Q score of CD9 and CD63 and Tumor T stage, number of involved lymph nodes (LNs), tumor grade, the presence of lymphovascular invasion (LVI), perineural invasion (PNI), progression free survival (PFS) and overall survival (OS). NA: Not applicable.
| Case # | Mean CD63 Q score | Mean CD9 Q score | T | Involved LNs # | Grade | LVI | PNI | PFS | OS |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 255 | 140 | ypT3 | 0 | G2 | No | No | NA | NA |
| 2 | 255 | 10 | pT3 | 23 | G3 | Yes | Yes | 2 | 7 |
| 3 | 270 | 210 | pT3 | 0 | G2 | No | Yes | 16 | 16 |
| 4 | 270 | 90 | pT3 | 7 | G2 | Yes | Yes | 9 | 9 |
| 5 | 210 | 10 | pT3 | 3 | G3 | Yes | Yes | NA | NA |
| 6 | 150 | 65 | pT3 | 4 | G3 | Yes | Yes | 3 | 5 |
| 7 | 270 | 120 | pT3 | 1 | G2 | Yes | Yes | 9 | 19 |
| 8 | 270 | 25 | ypT3 | 0 | G2 | No | Yes | 20 | 20 |
| 9 | 110 | 0 | pT3 | 0 | G2 | No | No | NA | NA |
| 10 | 270 | 180 | pT3 | 0 | G2 | Yes | Yes | NA | NA |
Discussion
Extracellular Vesicles are now being widely studied especially in cancer research. They include apoptotic bodies (500nm–3mm), microvesicles (100nm–1mm) and exosomes (30–150 nm). 14–16 Exosomes appear to contribute to a diverse range of biological processes, depending on the cell of origin and the conditions for secretion. Current evidence suggests that exosomes fuse with the plasma membrane of the recipient cell and release their contents into the target cell. 17,18
Recent studies have shown that exosomes can be isolated in vivo in bodily fluids such as blood, urine, breast milk, amniotic fluid, malignant ascites, bronchoalveolar lavage fluid and synovial fluid. Exosomes isolation and identification is strongly based on electron microscopy (size and shape) and western blotting for the existence of proteins markers that are involved in exosomal biogenesis such as biogenesis-related proteins (Alix, TSG101), and/or exosomal membrane tetraspanins (CD9, CD63, CD81). 19–25 In conventional histological sections, recognition of EVs is substantially limited by the resolving power of the light microscope, as their diameter usually falls below the limit of resolution. Furthermore, not only histological assessment but also conventional cell biology techniques including laser confocal microscopy or flow cytometry have substantial limitations when used for analysis of EVs. 26
PDAC cells-derived exosomes play important roles in their pathobiology. They were shown to be involved in pre-metastatic niche formation by inducing fibrotic environment in the liver of naïve mice. Their uptake by kupffer cells caused transforming growth factor β secretion and upregulation of fibronectin production by hepatic stellate. Macrophage migration inhibitory factor (MIF) was highly expressed in PDAC-derived exosomes, and its blockade prevented pre-metastatic niche formation and metastasis. In Patients with stage I PDAC, macrophage MIF was higher in exsosomes from patients who later developed liver metastasis compared to other patients who didn’t. 10 Clinical data indicated that exosomal integrins could be used to predict organ-specific metastasis. Exosomes proteomic revealed distinct integrin expression patterns and shed light on metastatic organotropism. The αvβ5 binds to Kupffer cells and is linked to liver metastasis. On the other hand, α6β1 and α6β4 bind to lung fibroblasts and epithelial cells and is linked to lung metastasis. Targeting those integrins decreased exosomes uptake and consequently liver and lung metastasis. 11 PDAC cells-derived exosomes facilitate the survival and proliferation of disseminated cells at metastatic sites through inhibition of RFXAP (transcription factor) expression via miR-212-3p resulting in MCH II down regulation and induction of tolerance of dendritic cells. 27
The development of resistance to cytotoxic chemotherapy and targeted therapy in patients with PDAC is inevitable. Cross talk between tumor and microenvironment is one recognized mechanism of acquired resistance. 28,29 In one experiment, dynamic Light Scattering based-size distribution identified EVs of three different size, large (>1500 nm), medium (500–1500 nm) and small (100–300 nm). Treatment with large and medium sized vesicular fraction of conditioned media from Gemcitabine-treated PDAC cells didn’t show any chemoprotective effect in PDAC cells, while smaller sized vesicular fraction provided remarkable survival benefits to PDAC cells against gemcitabine treatment. Immunoblotting was performed to examine the specific markers associated with EVs. Immunoblot data revealed that these active EVs are enriched with exosomal markers CD9 and CD63.
Our work indicates that the transformation of normal pancreatic tissues into pathologic and malignant pathologic tissues is associated with increased expression of both exosomal markers (CD63 and CD9) (Figure 4A and 4B). The transformation of normal pancreatic tissues into premalignant pathologic tissue is associated with increased expression of the exosomal marker CD9 (Figure 4C). There is a trend toward increased expression of CD63 but the difference in CD63 expression between premalignant PTs and their ANTs didn’t reach statistical significance (probably due to small sample size). The transformation of premalignant pathologic tissues into malignant pathologic tissues was associated with a trend toward increased expression of exosomal markers (CD63 and CD9) (figure 4E) but the difference in expression didn’t reach statistical significance (probably due to small sample size).
The exosomal markers CD9 and CD63 have been recognized as differentially expressed proteins that can be detected using IHC among other modalities.30,31 The relationship between CD9 gene expression (using RT-PCR and IHC) and tumor pathologic characteristics was previously explored. There was no statistically significant relationship between CD9 gene expression and metastatic status. However, CD9 gene expression was associated with lymph node status (p=0.028), tumor stage (p=0.024) and histopathologic grading (p=0.041). 36.8% of the N0 stage patients had reduced gene expression compared with 71.4% of N1 stage patients. Moreover, CD9 gene expression increased from 14.3 % in patients with stage I disease to 85.7% in patients with stage IV disease.31 In the same study, the IHC for CD63 was positive in all cases and the mRNA levels for CD63 in almost all pancreatic tumor were preserved and no decreased gene expression was detected. In our study, the relationship between the Q score of CD9 and CD63 and Tumor T stage, number of involved lymph nodes, tumor grade, the presence of lymphovascular invasion, perineural invasion, progression free survival and overall survival is shown in Table # 3. Given the small sample size (N=10), the presented data represents descriptive summary.
The correlation between CD9 gene expression (using RT-PCR and IHC) and survival was previously explored as well. Patients with low CD9 gene expression had a worse 1-year survival and median overall survival compared to patients with high CD9 gene expression (0% vs 25.5%, p=0.0004), (226 days Vs 397 days, p=0.018). In our study, the mean Q score in patients who progressed and died within 12 months of their time of diagnosis compared to those who progressed or died after 12 months of their time of diagnosis was lower in CD63 (225 Vs 236, p=0.91) and in CD9 (55 vs 98; p=0.76). The noticeable trend in our study toward worse survival with lower mean Q score in CD9 is similar to the previously published report. The lack of statistical significance is probably due to small sample size. We are currently working on expanding our cohort to include patients with stage IV pancreatic adenocarcinoma to further explore and validate the correlation between exosomal markers CD63 and CD9 expression and PFS and OS.
The exosome-based therapy is an attractive strategy for targeted drug therapy. Utilizing exosomes to carry therapeutic drugs, proteins, microRNA and other transferable materials is a step forward in fighting cancer. Engineering exosomes to recognize a specific target/organ in the body would improve on our progress with personalized medicine. This will help maximize treatment efficacy and reduce associated toxicity. Moreover, the interaction between the tumor and the immune system via exosomes is an additional target that might further revolutionize immunotherapy. Component for successful exosome-based drug delivery include choice of donor cell, therapeutic cargo, use of targeting peptide, loading method and administration route.32,33
Conclusions
Scientists and clinicians are actively looking for diagnostic and predictive biomarkers in patients with pancreatic adenocarcinoma. Exosomal markers are among the biomarkers that have been and continue to an ongoing interest. The exosomal markers CD9 and CD63 have been recognized as differentially expressed proteins that can be detected using IHC among other modalities. Our work shed more light and provided qualitative assessment using the Q score to the differential expression of CD9 and CD63 among a spectrum of normal, inflammatory, premalignant and malignant pancreatic cells using IHC. We have also demonstrated that using the Q-score, CD63 was differentially expressed in patients with PDAC. Sho et al. reported that the IHC for CD63 was positive in all cases and the mRNA levels for CD63 in almost all pancreatic tumors were preserved and no decreased gene expression was detected. Moreover, the noticeable trend in our study toward worse survival with lower mean Q score in CD9 is similar to the previously published report. The lack of statistical significance is probably due to small sample size.
Abbreviations
- IHC
Immunohistochemistry
- RPS
Resected Pancreatic Specimens (RPS)
- PTs
Pathologic Tissues
- MPTs
Malignant Pathologic Tissues
- PPTs
Premalignant Pathologic Tissues
- IPTs
Inflammatory Pathologic Tissues
- ANTs
Adjacent Normal Tissues
- MANTs
Malignant Adjacent Normal Tissues
- PANTs
Premalignant Adjacent Normal Tissues
- IANTs
Inflammatory Adjacent Normal Tissues
- PDAC
Pancreatic ductal adenocarcinoma
- EVs
Extracellular Vesicles
- IRB
Institutional Review Board
- Q-score
Quick-score
- MIF
Migration Inhibitory Factor
Footnotes
Disclosure: The authors of this manuscript have no conflicts of interest to disclose.
PREVIOUS PRESENTATION
Presented as a poster at the AACR Special Conference on Pancreatic Cancer in Orlando, Florida (May 12–15, 2016)
Presented as a poster at the XXXI International Congress of the International Academy of Pathology (IAP) and 28th Congress of the European Society of Pathology (ESP) to be held in Cologne, Germany from 25 – 29 September 2016.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Stathis A, Moore MJ. Advanced pancreatic carcinoma: current treatment and future challenges. Nat Rev Clin Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 3.Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathivanan S, Fahner CJ, Reid GE, et al. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demory BM, Higginbotham JN, Franklin JL, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan BT, Teng K, Wu C, et al. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skog J, Wurdinger T, van RS, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. 2011;33:441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- 9.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 10.Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng YH, Rome S, Jalabert A, et al. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One. 2013;8:e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detre S, Saclani JG, Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gyorgy B, Szabo TG, Pasztoi M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo SS, Ishibashi O, Ishikawa G, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 17.Morelli AE, Larregina AT, Shufesky WJ, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 18.Segura E, Guerin C, Hogg N, et al. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179:1489–1496. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 19.Admyre C, Grunewald J, Thyberg J, et al. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur Respir J. 2003;22:578–583. doi: 10.1183/09031936.03.00041703. [DOI] [PubMed] [Google Scholar]
- 20.Admyre C, Johansson SM, Qazi KR, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 21.Bard MP, Hegmans JP, Hemmes A, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 22.Caby MP, Lankar D, Vincendeau-Scherrer C, et al. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 23.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skriner K, Adolph K, Jungblut PR, et al. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 25.Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J Immunol. 2006;176:1534–1542. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 26.van der Pol E, Hoekstra AG, Sturk A, et al. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- 27.Ding G, Zhou L, Qian Y, et al. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212–3p. Oncotarget. 2015;6:29877–29888. doi: 10.18632/oncotarget.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charrier A, Chen R, Chen L, et al. Connective tissue growth factor (CCN2) and microRNA-21 are components of a positive feedback loop in pancreatic stellate cells (PSC) during chronic pancreatitis and are exported in PSC-derived exosomes. J Cell Commun Signal. 2014;8:147–156. doi: 10.1007/s12079-014-0220-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava SK, Arora S, Singh S, et al. MicroRNAs in pancreatic malignancy: progress and promises. Cancer Lett. 2014;347:167–174. doi: 10.1016/j.canlet.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gronborg M, Kristiansen TZ, Iwahori A, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Sho M, Adachi M, Taki T, et al. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. Int J Cancer. 1998;79:509–516. doi: 10.1002/(sici)1097-0215(19981023)79:5<509::aid-ijc11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 32.Johnsen KB, Gudbergsson JM, Skov MN, et al. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846:75–87. doi: 10.1016/j.bbcan.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Patel GK, Patton MC, Singh S, et al. Pancreatic Cancer Exosomes: Shedding Off for a Meaningful Journey. Pancreat Disord Ther. 2016;6:e148. doi: 10.4172/2165-7092.1000e148. [DOI] [PMC free article] [PubMed] [Google Scholar]