Abstract
Haploinsufficiency of tumor suppressor genes (TSGs) indicates that the reduced levels of proteins in cells that lack one allele of the genomic locus results in the inability of the cell to execute normal cellular functions contributing to tumor development. Representative cases of haploinsufficient TSGs are p27Kip1, p53, DMP1, NF1, and PTEN. Tumor development is significantly accelerated in both mice with homozygous and heterozygous gene deletion, with expression of the wild type allele in the latter. Newly characterized TSGs such as AML1, EGR1, TGFβR1/2, and SMAD4 have also shown haploid insufficiency for tumor suppression. This phenotype has typically been demonstrated in gene knockout mouse models, but analyses of human samples have been conducted in some cases. Recent studies suggest collaboration of multiple haploinsufficient TSGs in 5q-, 7q-, and 8q- syndromes, which is called compound haploinsufficiency. Although ARF is a classical TSG, it also belongs to this category since Arf+/− accelerates tumor development when both alleles for Ink4a are inactivated. Haploid insufficiency of Arf was also reported in myeloid leukemogenesis in the presence of inv(16). In case of p53, p53+/− cells achieve only ~25% of p53 mRNA and protein levels as compared to those in wild type, which could explain the mechanism. TGFβR1+/− collaborates with ApcMin+/− in colorectal cancer development; TGFβR2+/− and Smad4+/− collaborates with K-Ras mutation in pancreatic ductal adenocarcinomagenesis, demonstrating the synergism of haploinsufficient TSGs and other oncogenic events. These TSGs can be targets for activation therapy in cancer since they retain a functional allele even in tumor cells.
Keywords: haploinsufficiency, tumor suppressor gene, p27Kip1, p53, DMP1 (DMTF1), ARF, AML1, EGR1, TGFβ/TGFβR/SMAD4, mouse model
Introduction
Cancer is a complex genetic disorder caused by alterations for both gene coding and non-coding regions. Two major classes of genes have been identified, namely oncogenes and tumor suppressor genes (TSGs). For instance, overexpression of a proto-oncogene, such as c-Myc, and/or inactivation of a tumor suppressor, such as TP53 or RB induces tumors. Oncogene activation in tumors is relatively straightforward since they are overexpressed or activated by mutation(s) while inactivation of a TSG is a complicated process performed by different mechanisms, e.g. gene deletion, mutation, epigenetic silencing, abnormal activity of microRNAs (miRNAs), and/or aberrant splicing. Although complete loss of TSG is common in human cancers as predicted by the Knudson’s two-hit hypothesis (1), recent studies indicate that an incomplete process for TSG function termed ‘haploinsufficiency’ that contributes to the development and progression of many cancers (Fig. 1). In this category of TSG, one functional allele of a gene is lost by mutation or deletion while the remaining normal allele is still retained in tumors, but the activity is not enough to execute its physiological function to prevent abnormal cell proliferation. Promoter hypermethylation or point mutation does not happen in the retained locus in haploinsufficient TSGs, thus tumor cells still express the wild type mRNA. We use this definition of haploinsufficient TSG throughout this review.
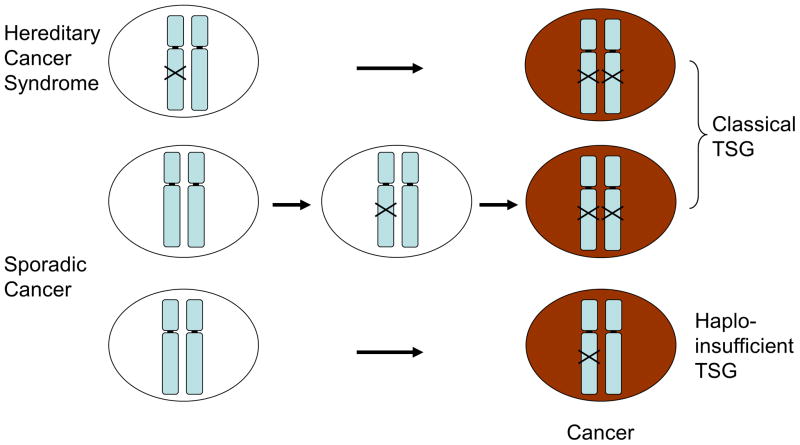
Figure 1. Tumor development caused by classical and haploinsufficient tumor suppressor genes.
Top panel: In case of hereditary cancer syndrome (e.g. Li-Fraumeni syndrome), one TSG locus is mutated in all the chromosomes, and inactivation of the intact allele leads to cancer following the two-hit hypothesis by Dr. Knudson (1). Tumor development is much accelerated because only a single hit on the TSG locus leads to total inactivation of the gene leading to carcinogenesis.
Middle panel: Tumor development in classical TSGs. It will take longer than the above for the tumor to develop since biallelic inactivation of the locus is needed.
Lower panel: Tumor development in haploinsufficient TSGs. It will take more time for tumor(s) to develop than classical TSGs since collaboration of mono-allelic loss with other genetic alteration(s) are required.
The Cdk inhibitor p27kip1 (2) was the first gene to be characterized as haploinsufficient for tumor suppression, followed by Dmp1 (3, 4; reviewed in 5) through observation of tumor development in homozygous and heterozygous knockout mice. Pten and Nf1 are also haploinsufficient TSGs since mice with heterozygous genomic DNA deletion often develop tumors without deletion of the wild type allele; however, comparative tumor development assays were not possible using global knockout mice due to the lethality of homozygous gene deletion (6–8; reviewed in 9). Creation of prostate-specific gene deletion in mice showed that complete loss of Pten expression activates a p53-dependent cellular senescence that can act as a brake on tumor formation (10). Hence the genomic locus for PTEN needs to be mono-allelic loss to promote prostate carcinogenesis, raising the concept of ‘obligate haploinsufficiency’ (9). TGFβ was shown to be haploinsufficient for tumor suppression (11); however the situation is different from others since it is a secreted protein. Although p53 has been classified as a classical TSG, haploid insufficiency of p53 has also been demonstrated in mice (12) as well as in humans (13) from analyses of tumor samples of Li-Fraumeni syndrome patients.
In case of classical TSGs (e.g. RB, p53, and INK4a/ARF), both loci are inactivated by i) point mutation, ii) gene deletion, iii) promoter hypermethylation, or iv) any combinations of these during tumor development (Fig. 2A). Knockout mouse models for such TSGs usually exhibit obvious tumor-prone phenotype without carcinogenic challenge than those for haplo-insufficient TSGs (14–16; 17, 18 for reviews). Conversely, a study from the Dr. Pandolfi’s lab showed that even a 20% reduction of PTEN protein level contributed to the development of cancer (19). When targeted by shRNAs, p53 knockdown mice showed distinct phenotypes ranging from hyperplasia to malignancy depending on the expression levels in its protein (20). Based on such observations, Berger et al. proposed a continuum model that accounts for subtle dosage effects of tumor suppressors including their regulation by miRNAs (21). The dosage and function of haploinsufficient TSGs are critical, and understanding the impact of haploinsufficiency is important for assessment of inter-individual genetic variation as well as the molecular mechanism of haploinsufficient disorders.
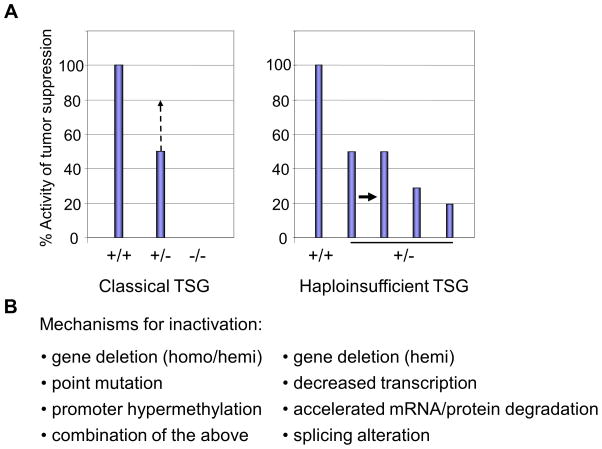
Figure 2. The activity of tumor suppressor genes/proteins in gene knockout mice (A) and possible mechanisms for inactivation of TSGs (B).
A. The wild type activity represents 100% diploid gene function and null (−/−) represents complete loss of function of the TSG. In classical TSGs, both loci are inactivated by i) point mutation, ii) gene deletion, iii) promoter hypermethylation, or iv) any combinations of these during tumor development. In this case, loss of one locus leads to compensatory activation of the remaining allele, leading to more than 50% of the tumor suppressor activity (left columns, middle, discontinued line; ref. 124). In haploinsufficient TSGs, tumors from mice with heterozygous gene deletion express the mRNA/protein at levels ~50% of wild type cells. The levels may be lower if other inactivation mechanisms co-exist to lower the levels of the tumor suppressive mRNA/protein (right columns). In p53+/− cells, both mRNA and protein levels are reduced to ~25% of those in wild type cells (48).
B. Possible mechanisms for inactivation of classical and haploinsufficient TSGs. Classical TSGs are inactivated by homozygous or heterozygous gene deletion plus point mutation or promoter hypermethylation(s). Conversely, haploinsufficient TSGs are inactivated by hemizygous gene deletion or point mutation at the single locus without promoter methylation or mutation of the remaining wild type allele. Recent studies show that other mechanisms, such as decreased transcription, accelerated RNA degradation (20), enhanced protein degradation, or aberrant splicing (72, 74, 119, 125–127) can coexist to down-regulate the TSG mRNA/protein expression to less than the 50% level of that in wild type cells in subsets of malignancies.
Haploinsufficiency of multiple genes cooperate to promote tumorigenesis, a phenomenon called ‘compound haploinsufficiency’. The 5q deletion (5q-) and 7q deletion (7q-) syndromes are typical examples of compound haploinsufficiency (22, 23), which demonstrate the importance of combinatorial interactions to elicit specific phenotypes. Deletion of chromosome 8p is also very common in human cancer. Using a murine model of hepatocellular carcinoma and in vivo RNAi, Xue et al. silenced the genes frequently deleted on human 8p22 and showed that multiple genes on chromosome 8p (Dlc1, Vps37a, Fgl1) could cooperatively inhibit tumorigenesis in mice predicting poor survival (24).
Although haploinsufficient TSGs do not have promoter hypermethylation or mutation(s) in the coding region for the retained locus, the mRNA and/or protein expression levels may decrease to less than 50% (usually 20–30%) of those in wild type cells (Fig. 2A). We include these TSGs into the category of haplo-insufficient TSGs since the protein levels in TSG+/− mice never reach 0%. The possible mechanisms for partial inactivation of the wild type locus include 1) decreased transactivation of the promoter due to autoregulation, 2) accelerated degradation of mRNAs or proteins, and 3) splicing alterations (Fig. 2B). For 2), recent research indicated that single nucleotide polymorphisms (SNPs) in the 3′ untranslated region of mRNAs and miRNAs seed sequences that may cause haploinsufficiency at the level of mRNAs through altered binding specificity of miRNAs (20). Networking analysis suggested that the haploinsufficient TSGs strongly interacted with one another, and any subtle alterations in this network could contribute to tumorigenesis. A typical case for 3) is a transcription factor Dmp1 (3, 4), which will be discussed in this review.
In this chapter, we focus on the identification and characterization of early generation haploinsufficient TSGs, such as p27Kip1, p53, DMP1, TGFβ; and then on recently characterized TSGs, such as 53BP1, AML1, EGR1, and TGFβR/SMAD4. Although ARF is a classical TSG, it behaves like a haploinsufficient TSG under specific circumstances. Finally, we discuss future directions and therapeutic values for the research on haploinsufficient TSGs.
p27Kip1
Entry, progression, and exit from the G1 phase of the mammalian cell cycle in response to mitogens are governed by cyclin-dependent kinases (Cdks) regulated by the D- and E-type cyclins (25, 26). p27Kip1 was first discovered as a key regulator of cell proliferation (canonical function; ref. 27, 28). In addition to its initial identification as a CDK inhibitor, p27Kip1 has also emerged as a multifunctional protein with numerous non-canonical, CDK-independent functions that exert influence on key processes such as cell cycle regulation, cytoskeletal dynamics and cellular plasticity, cell migration, and stem-cell proliferation and differentiation (29). While both p21Cip1 and p27Kip1 bind directly to cyclin/Cdk complexes to inhibit their Cdk activity and block cell proliferation, they are essential activators of cyclin D-dependent kinases in murine fibroblasts, thus are called ‘assembly factors’ (‘AF’ in Fig. 3; ref. 30). Abnormally low levels of the p27Kip1 protein are frequently found in human carcinomas, and these low levels correlate directly with both histological aggressiveness and patient mortality (2). The p27KIP1 gene was mapped to chromosome band 12p13 that is hemizygously deleted in leukemias without mutations, which may confer a growth advantage to leukemic cells (31). Fero et al. showed that that both p27Kip1+/− and p27Kip1−/− mice were predisposed to tumors in multiple tissues (intestinal and lung adenomas, pituitary tumors) when challenged with γ-irradiation or a potent DNA alkylating agent N-ethyl-nitrosourea (ENU, Table 1; ref. 2). Therefore p27Kip1 is a multiple-tissue tumor suppressor in mice. Molecular analyses of tumors in p27Kip1+/− mice show that the remaining wild type allele is neither mutated nor silenced. Hence, p27Kip1 is haploinsufficient for tumor suppression (2).
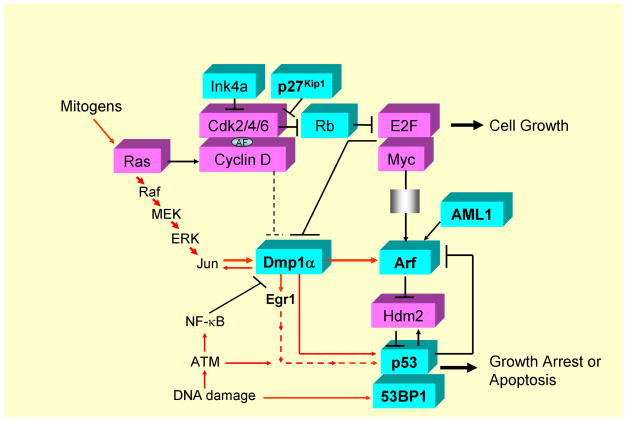
Figure 3. Intracellular signaling pathways involving p27Kip1, Dmp1, Arf, p53, 53BP1, and AML1.
Cyclin D/Cdks are activated in response to mitogenic signals and initiate phosphorylation of Rb, a process that is completed by cyclin E - Cdk2. Once cells enter the S phase, cyclin E is degraded and cyclin A enters into complexes with Cdk2. Proteins of the Ink4 family (p15, p16, p18, p19) bind only to Cdk4 and inhibit its activity while those in the Kip/Cip family (p21, p27, p57) inhibit cyclin-bound Cdks (25–28). Both p21Cip1 and p27Kip1 act as an assembly factor (AF; ref. 30). Arf is induced by potentially oncogenic signals stemming from overexpression of oncogenes such as c-Myc, E2F1, and activated Ras, which quenches inappropriate mitogenic signaling by diverting incipient cancer cells to undergo p53-dependent growth arrest or cell death (76, 77). Dmp1 is activated by oncogenic Ras or HER2, which, in turn, binds and activates the Arf promoter and induces cell cycle arrest (61, 65). Three different splicing variants have been reported for human DMP1 (72, 73). Dmp1 (Dmp1α) directly binds to the CD13 (128) and Arf (56) promoters to transactivate the gene expression, thereby accelerates myeloid cell differentiation by interfering cell cycle progression. Interestingly the CD13 promoter activation is inhibited by D-type cyclins in a Cdk-independent fashion (54, 128; see the dotted line for inhibition) while the Arf promoter activation is stimulated by cyclin D1 (129) in a Cdk-dependent fashion (57, 61). Other transcriptional targets for Dmp1α include Areg, Thbp-1, JunB, and Egr1 (118), suggesting that it is also involved in signal transduction related to angiogenesis and/or metastasis. Dmp1α physically interacts with p53 and neutralizes all the activities of Mdm2 to activate the p53 pathway (59, 60). Both Dmp1−/− and Dmp1+/− mice show hypersensitivity to develop tumors in response to carcinogen or γ-irradiation. E2F1-3a (130, 131) directly binds to the Dmp1 promoter and causes transcriptional repression (62). The Dmp1 promoter is repressed by NF-κB through direct binding of the promoter to RelA (63). Conversely, the Dmp1 promoter is activated by the oncogenic Ras-Raf-MEK-ERK-Jun pathway (61) and HER2-Pi3k-Akt-NF-κB (65), and thus Ras or HER2-driven carcinogenesis is dramatically accelerated in Dmp1-null mice. The transcription factor AML1 locus is frequently translocated to create hybrid molecules in human acute leukemias, and plays essential roles in normal hematopoiesis through dimerization of its partner CBFβ (91, 92). Direct transcriptional activation of the ARF promoter by AML1 and its inhibition by AML1-ETO has been reported (96). 53BP1 is a p53-binding protein, and is involved in DNA-damage response by choosing recombination and end joining at DNA double-strand breaks (50–52). Haploid insufficiency of p27Kip1, p53, 53BP1, Dmp1α, Arf, AML1, and Egr1 TSGs are discussed in this review. These genes are shown in bold. The DMP1 locus generates other two splice variants, namely DMP1β and γ, and the oncogenic role of DMP1β has recently been demonstrated in vivo (72–74). Aberrant splicing of TSGs and their roles in carcinogenesis are currently extensively studied (119, 125–127), which can further affect the function of the remaining allele of haploinsufficient TSGs. Proteins that have mitogenic/oncogenic functions are shown in dark while those with tumor-suppressive activities are shown in white.
Table 1. The summary of haploinsufficient TSGs explained in this review.
Genes for which evidence of haploinsufficiency comes from mouse and/or human studies are shown. LFS: Li-Fraumeni syndrome; FAMMM: familial atypical multiple mole melanoma syndrome; FPD: familial platelet disorder; Homo: homozygous knockout; Het: heterozygous knockout; ENU: N-ethyl-nitrosourea; T-PLL: T-cell prolymphocytic leukemia; MTCP1: mature T cell proliferation-1; MPD: myeloproliferative disorder; t-MN: therapy-related myeloid neoplasm; AML: acute myelogenous leukemia; PDAC: pancreatic ductal adenocarcinoma; MCN: mucinous cystic neoplasm.
| Genes | Human syndromes | Mouse Model | Cancer types | References |
|---|---|---|---|---|
| p27Kip1 | homo, het with γ-irradiation, ENU | intestinal adenoma, lung adenoma, pituitary tumor female reproductive tract tumor |
2 | |
| het;MMTV-neu TG, het;MTCP1 TG, het;Ptc+/− | mammary tumor, T-PLL, medulloblastoma | 32, 33, 36, 37 | ||
| p53 | homo | T-cell lymphoma, sarcoma, hemangiosarcoma | 14 | |
| LFS | het | soft tissue sarcoma, osteosarcoma, carcinoma | 12 | |
| 53BP1 | homo, het | glioblastoma multiforme | 50, 53 | |
| DMP1 | 7q- syndrome | homo, het with DMBA, γ-irradiation | lung cancer, T cell lymphoma, ovarian tumor, liver tumor | 3, 4 |
| Eμ-Myc, K-RasLA, MMTV-neu, cyclin D1 | B cell lymphoma, lung cancer, mammary tumor | 4, 57, 64, 65 | ||
| ARF | homo | sarcoma, lymphoma, carcinoma, glioma | 16, 78 | |
| het | lymphoma, hemangioma, sarcoma, carcinoma | 136 | ||
| FAMMM | het;Ink4a−/− | melanoma and other tumors | 79 | |
| AML1 | FDP/AML | het (homo lethal) | predispposition to myeloid leukemia | 93–98 |
| EGR1 | 5q- syndrome | het, homo with ENU | MPD, T cell lymphoma | 103 |
| het;Apc+/−, het;p53+/− | t-MNs, AML | 106, 107 | ||
| TGFβRI | het;ApcMin/+ (homo lethal) | intestinal tumor | 110 | |
| het;KrasG12D | PDAC | 113 | ||
| SMAD4 | Juvenile Polyposis | het (homo lethal) | gastrointestinal polyposis, intestinal tumor | 114, 115 |
| het;KrasG12D | MCNs developing into invasive PDAC | 116 |
ErbB2/neu destabilizes p27Kip1 and increases the expression of cyclin D1. Muraoka et al. studied the roles of p27Kip1 and cyclin D1 in ErbB2-mediated mammary epithelial cell transformation (32). Overexpression of ErbB2 or cyclin D1 in p27Kip1+/− primary murine mammary epithelial cells resulted in increased proliferation, cyclin D1 nuclear localization, and colony formation in soft agar as compared to those in p27Kip1+/+ cells. In contrast, ErbB2- or cyclin D1-overexpressing p27Kip1−/− cells displayed reduced proliferation, anchorage-independent growth, Cdk4 activity, and nuclear cyclin D1 expression compared to wild type cells. Mammary glands from MMTV (mouse mammary tumor virus)-neu;p27Kip1+/− mice exhibited alveolar hyperplasia, enhanced proliferation, decreased apoptosis, and accelerated tumor formation compared to MMTV-neu;p27Kip1+/+ glands. Interestingly, MMTV-neu;p27Kip1−/− glands showed decreased proliferation, cyclin D1 expression, and Cdk4 activity, as well as markedly prolonged tumor latency, compared to MMTV-neu;p27Kip1+/+ glands. Therefore p27Kip1+/− mammary epithelium are more susceptible to HER2/neu-induced tumorigenesis while p27Kip1-null glands, due to severely impaired cyclin D1/Cdk4 function, are more resistant to transformation (32), suggesting another case of obligate haploinsufficiency.
Haploid insufficiency of p27KIP1 was also demonstrated in T-cell prolymphocytic leukemia (T-PLL, 33) although hematopoietic malignancies were not reported in p27Kip1−/− mice. T-PLL is consistently associated with inactivation of the ATM gene and chromosomal rearrangements leading to an overexpression of the MTCP1/TCL1 oncoprotein that interact with AKT1. These alterations are present at the earliest stage of malignant transformation, suggesting that additional events are required for overt malignancy. Toriellec et al. studied the 12p13 deletion that occurs in approximately half of T-PLLs where found that the p27KIP1 gene was hemizygously deleted in leukemic cells without mutation (33). Consistently, in a p27Kip1+/− background, MTCP1-transgenics (TGs) had multiple emergences of preleukemic clones not observed in control cohorts. The remaining p27Kip1 allele was maintained and expressed in these preleukemic clones, strongly implicating that p27Kip1 was haploinsufficient in the pathogenesis of T-PLL (33). Haploid insufficiency of p27Kip1 for tumor suppression was also reported in human acute myeloid leukemia with complex karyotype (34), human small intestinal neuroendocrine tumors (35), mouse models for medulloblastoma (36) and pancreatic cancer (37), thus acceleration of tumorigenesis has been considered to be a generalized phenotype for heterozygous loss of p27Kip1.
p53 and 53BP1
p53 is an established tumor suppressor that is activated on cellular stresses such as DNA damage, oncogene activation, hypoxia, which transactivates sets of genes that induce cell cycle arrest, apoptosis, DNA repair, or autophagy, playing essential roles in the prevention of tumor development (38–40). It is mutated or deleted in over ~50% of human cancers (41), but is inactivated in others with wild type p53 through different mechanisms, indicating that functional inactivation of the p53 pathway is essential for tumor development. Loss of heterozygosity (LOH) of p53 often occurs in tumors having a p53 mutation indicating that it behaves as a classical TSG that meets the Knudson’s two-hit hypothesis (1). However, p53 is also a representative example of haploinsufficient TSG (12). The first evidence that p53 may exhibit haploinsufficiency comes from the analysis of Li-Fraumeni syndrome (LFS) patients (13, 42). LFS is a hereditary cancer predisposition syndrome in whom a variety of cancers are found in affected families, having a higher risk of developing cancer, and the relatively early age of cancer development (42). The majority of LFS cases are caused by germline missense mutations for p53 (42, 43). The most common types of cancer found in families with LFS include osteosarcoma, soft tissue sarcoma, acute leukemia, breast/lung/gastrointestinal cancer, brain tumor, and adrenocortical tumor. When tumors from LFS patients were analyzed for LOH of p53, only half of the tumors exhibited LOH at the locus (13), raising the question of whether p53 mutation acted as a dominant-negative inhibitor of the wild type allele (44) or if in fact single-copy loss of p53 could contribute to tumorigenesis. In case of a dominant-negative mutation for p53, the wild type allele does not need to be lost to develop tumors since proteins from the mutant locus subvert the function of the wild type allele by making non-functional complexes because i) p53 proteins form tetramers to act as a transcription factor (45), ii) the level of Mdm2 is very low in p53-mutant cells, which results in accumulation of the protein to extremely high levels (46), and 3) degradation of mutant p53 is selectively compromised in tumor cells (46). As a consequence, tetramers from p53wt/mut cells will almost exclusively be composed of mutant p53 that are non-functional. In case of a haploinsufficient mutation, the wild type p53 allele is retained because half the normal wild type protein is insufficient to maintain the functionality. Since p53 mutations from LFS patients often affect the oligomerization domain, the proteins remain unfolded and unable to interact with wild type p53; hence the locus is considered to be haploinsufficient than having a dominant-negative activity (47).
To address this question of haploid insufficiency of p53 for tumor suppression, Venkatachalam et al. analyzed tumors from both p53+/− and p53−/− mice (12; Table 1) for the latency and tumor spectra. The survival of p53+/− mice (median, 70 weeks) was somewhere in between that of the p53+/+, >100 weeks) and p53−/− mice (18 weeks). Of note, tumors that developed in p53+/− mice without loss of the remaining allele (lymphomas, sarcomas) occurred at later time points than those exhibiting LOH of the locus, suggesting that the wild type p53 allele was partially effective for tumor suppression in p53+/− mice. A high proportion of tumors from the p53+/− mice retained a functional wild type p53 allele. Thus a mere reduction in p53 levels was sufficient to promote tumorigenesis. Lynch and Milner determined both basal p53 mRNA and protein levels, and compared the p53 stress response in p53+/+, p53+/− and p53−/− isogenic clones derived from human colon adenocarcinoma cell line HCT116 (48). Basal expression of p53 in p53+/− cells was only 25% in comparison to that in p53+/+ cells, and this difference was maintained following oncogenic stress. The p53 stress responses were attenuated in p53+/− cells, in particular for p21CIP1 upregulation, G1 arrest, and apoptosis (48). This is the first study that demonstrated the molecular basis for haploinsufficiency of p53, which explains the attenuated tumor-suppressive phenotype observed in p53+/− mice and humans with LFS. In hematopoietic cells, p53-dependent cell cycle control, senescence, and apoptotic functions are actively involved in maintaining homeostasis under normal and stress conditions (49). Whereas loss of p53 function promotes leukemia/lymphoma development, increased p53 activity results in myelodysplasia. Since genetic alterations in TP53 is rather low in hematopoietic malignancies (10% – 20%, ref. 49), it is expected that p53 is functionally inactivated through other mechanisms e.g. ARF or DMP1 deletion (see below).
Haploid insufficiency was also reported in the p53-binding protein 1, 53BP1 (50). When DNA double-strand breaks (DBSs) occur, the cell cycle stage has a major influence on the choice of the repair pathway employed (51). Specifically, non-homologous end joining is the predominant mechanism used in the G1 phase of the cell cycle, while homologous recombination becomes fully activated in the S phase. Studies over the past 2 decades have revealed that the aberrant joining of replication-associated breaks leads to catastrophic genome rearrangements, revealing an important role of DNA break repair pathway choice in the preservation of genome integrity. 53BP1, a putative DNA-damage sensor that accumulates at sites of double-strand breaks (DSBs) dependent on histone H2AX, and BRCA1, a well-known breast cancer tumor suppressor, are at the center of this choice (51).
Genomic profiling of human cancers has provided insights into the mutational landscape of genes that are involved in the response to radiation-induced DNA-damage. The sub-nuclear accumulation of proteins (i.e. foci) in the DNA-damage response (DDR), such as gamma-H2AX, 53BP1, or RAD51, has been studied as a substitute of treatment sensitivity (52). Recent preclinical studies have demonstrated the predictive potential of DDR foci with clinically relevant end points, such as tumor control probability. This will yield functional insight that may complement or even supersede genomic information, thereby giving radiation oncologists unique opportunities to individualize cancer treatments in the near future (52).
Loss of a single 53BP1 allele compromised genomic stability and DSB repair, which could explain the susceptibility of 53BP1+/− mice for tumorigenesis (50). 53BP1 was heterozygously deleted in ~20% of human glioblastoma multiform specimens, and low 53BP1 expression was associated with worse prognosis. 53BP1 behaved as a haploinsufficient TSG in a mouse model of platelet-derived growth factor-induced gliomagenesis (53). Since human glioma cell lines in which 53BP1 was robustly silenced by shRNA showed higher sensitivity to ionizing radiation, 53BP1 can be pharmacologically targeted in glioblastoma multiforme in combination with standard therapies (53).
Dmp1 (Dmtf1)
Dmp1, a cyclin D binding myb-like protein 1 (also called Dmtf1), was originally isolated in a yeast two-hybrid screen of a murine T-lymphocyte library with cyclin D2 as bait (54, 55). Dmp1 transactivates the myeloid differentiation marker CD13 (128) and shows its activity as a tumor suppressor by directly binding to the Arf promoter to activate its gene expression and, thereby, induces Arf- and p53-dependent cell cycle arrest (56). Recent studies show that it binds to endogenous cyclin D1 and transactivates both p19Arf and p16Ink4a (57, 58). In Arf-null cells, Dmp1 physically interacts with p53 to induce senescence or apoptosis for tumor suppression (59; reviewed in 60). The Dmp1 promoter is efficiently activated by oncogenic Ras as well as by constitutively active MEK, ERK (61), but is repressed by E2Fs and NF-κB (62, 63). Hence Dmp1 has been shown to be a key mediator between Ras-Raf-MEK-ERK mitogenic signaling and the Arf-p53 tumor suppressor pathway.
The activity of the Arf-53 pathway is significantly attenuated in Dmp1-deficient cells since those cells can easily give rise to immortalized cell lines that retain wild type p19Arf and functional p53 and are transformed by oncogenic Ras alone (3). Dmp1-deficient mice are prone to tumor development (3, 4; Table 1). Tumors induced by dimethylanthracene, Eμ-Myc (Fig. 4A; ref. 4) or K-RasLA (64) transgene were greatly accelerated in both Dmp1+/− and Dmp1−/− backgrounds, with no differences between groups lacking one or two Dmp1 alleles (4; reviewed in 5). Indeed, all tumors from Dmp1+/− mice retained and expressed the wild type Dmp1 allele, and most expressed wild type Dmp1 mRNA and protein without inactivating mutation in both dimethylanthracene - treated mice and oncogene - transgenic cross, suggesting typical haploinsufficiency of Dmp1 in tumor suppression (4, 64, 65).
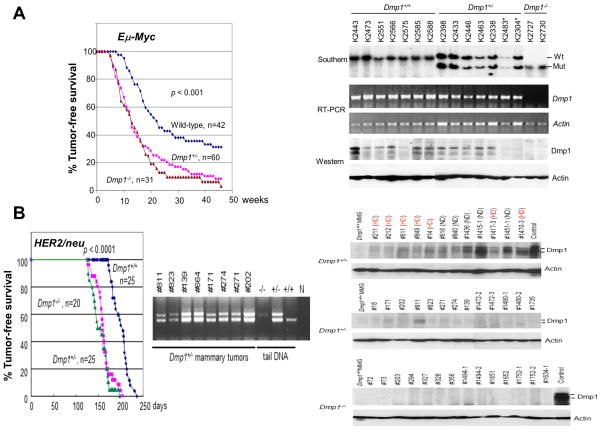
Figure 4. Haploid insufficiency of Dmp1 in tumor suppression.
A. Tumor development in Eμ-Myc TG mice (modified with permission from ref. 4).
Left: Tumor-free survival of Eμ-Myc TG animals of the indicated Dmp1 genotypes. The disease-free survival was significantly shortened in both Dmp1+/− [box] and Dmp1−/− [triangle] mice (both p <0.001 as compared to that of Dmp1+/+;Eμ-Myc [diamond] mice) without significant difference between the two cohorts.
Right: Both Dmp1 RNA and protein were detected by RT-PCR and immunoblotting, respectively, using β-actin as an internal control. Sequencing analyses showed no mutations within the DNA-binding domain for Dmp1. The Dmp1 protein was expressed in tumors from Dmp1+/− mice (~50% of that in Dmp1+/+), but was undetectable in some of them (K2483* and K 2304*).
B. Tumor development in MMTV-neu mice (modified with permission from ref. 65).
Left: Tumor-free survival of MMTV-neu (mutant) mice of the indicated Dmp1 genotypes (65). The ErbB2/neu gene is activated by point mutation in rat neu but an aberrant splicing in human breast cancer. The disease-free survival was significantly shortened in both Dmp1+/− [box] and Dmp1−/− [triangle] in comparison to Dmp1+/+ [diamond] mice (p < 0.001) without significant difference between the two cohorts. Genomic DNA PCR shows retention of the wild type allele in all mammary tumors from Dmp1+/− mice, showing haploinsufficiency.
Right: Western blotting analyses of MMTV-neu mammary tumors for Dmp1 using actin as an internal control. The Dmp1 protein expression was 2–16 folds higher in tumors from Dmp1 wild type mice than normal mammary glands (Dmp1+/+ MMG from a 15-week-old non-lactating female), reflecting the Dmp1 promoter activation by HER2/neu. HD means spontaneous hemizygous deletion of the Dmp1 locus in tumors from Dmp1+/+;MMTV-neu* mice. ND: no deletion. The protein expression levels were ~50% in HD tumors as compared with of those in ND tumors. The Dmp1 protein expression levels in Dmp1+/−;MMTV-neu* mice were ~25% of that in Dmp1+/+;MMTV-neu* ND (no deletion) mice suggesting additional mechanism of destabilization of the protein in these tumors; however this is not important for tumor development since the survival curve for Dmp1+/−; MMTV-neu* mice was overlapping with that of Dmp1−/−; MMTV-neu* mice (65). See ref. 132 for a general review for MMTV-driven mouse models.
We recently characterized the signaling pathway between HER2/neu (Fig. 4B) or cyclin D1 and Dmp1 using MMTV-neu or MMTV-cyclin D1 (T286A) mice as a model (57, 65). Both Dmp1 and p53 were induced in pre-malignant hyperplastic lesions from MMTV-neu* (mutant) mice, and mammary carcinogenesis was significantly accelerated in both Dmp1+/− and Dmp1−/− animals (65). We also showed that constitutive expression of Dmp1α delayed HER2/neu-induced mammary tumorigenesis (58). All of the mammary tumors from Dmp1+/−;neu* mice retained the wild type Dmp1 allele (Fig. 4B, left), and expressed the protein at levels less than 30% of those in Dmp1wtND;neu* (i.e. wild type neu tumors without Dmp1 deletion) (Fig. 4B, right, ref. 65). We also observed selective deletion of Dmp1 in >50% of wild type neu* mammary tumors (hemizygous Dmp1 deletion: HD) while the involvement of Arf, Mdm2, or p53 was rare (65). Tumors from Dmp1-deficient mice showed significant downregulation of Arf and p21Cip1, showing p53 inactivity and more aggressive phenotypes than tumors without Dmp1 deletion (65). Selective Dmp1 deletion was also found in 21% of the MMTV-D1 and D1T286A mammary tumors and the Dmp1-heterozygous status significantly accelerated mouse mammary tumorigenesis with reduced apoptosis and increased metastasis (57). Thus our study shows the pivotal roles of Dmp1 in HER2/neu-p53 and cyclin D1-p16Ink4a-Rb signaling and mammary tumor development.
Haplo-insufficient TSGs have been studied mainly in mice using gene-knockout mice than humans simply because it is easier to create artificial gene deletion models (homozygous or heterozygous deletion) in mice and conduct tumor development assays than analyzing numerous human cancer specimens where tissue availability can be an issue. Another hurdle to demonstrate the haploinsufficiency of a particular TSG in human cancer is a technical issue. The human DMP1 (hDMP1; hDMTF1) gene is located on chromosome 7q21, a region often deleted in human breast/lung cancers and hematopoietic malignancies (66–68). Bodner et al. reported that the hDMP1 locus was hemizygously deleted in all of 7q- leukemia/lymphoma specimen regardless of the results for the detailed cytogenetic analysis (68). We analyzed 51 human non-small cell lung carcinoma samples and found that LOH of hDMP1 was present in ~35% of lung cancers, which happened in a mutually exclusive fashion with that of INK4a/ARF and/or p53 in the same samples (64, 69). LOH here means loss of one of the two genomic signals present in normal cells (64, 69); thus our definition of LOH is different from loss of the mutant allele in tumor cells in classical TSGs. This raised the possibility that hemizygous hDMP1 deletion might define a new disease entity with different response to therapy (70). We then analyzed 110 pairs of normal and tumor tissues from breast cancer patients for LOH of hDMP1, INK4a/ARF, p53 and gene amplification of Hdm2 to study the frequency and prognostic value for each genetic alteration. LOH of the hDMP1 locus was found in 42% of human breast carcinomas, while those of INK4a/ARF and p53 were found in 20% and 34%, respectively. Hdm2 amplification was found in 13% of the same sample, which was found independently of LOH for hDMP1. Again, LOH for the hDMP1 locus was found in mutually exclusive fashion with that of INK4a/ARF and p53, and was associated with low Ki67 index and diploid karyotype (71). Consistently, LOH for hDMP1 was associated with luminal A category and longer relapse-free survival, while that of p53 was associated with non-luminal A and shorter survival. Detailed LOH analyses with 10 independent primer sets showed that selective deletion of hDMP1 was found in 79% of breast cancer (71) and 94% of lung cancer (64) indicating haploid insufficiency of hDMP1 in suppressing human cancers.
The human DMP1 locus encodes two other splice variants, namely DMP1β and γ, and our recent study indicates that the splice variant DMP1β protein is overexpressed in more than half of human breast cancers, and has oncogenic activity in vivo (72). Although it blocks the activity of DMP1α (73), it has a p53-independent functions as well (72; reviewed in 74, 75). Since half of the DMP1β protein overexpression occurred simultaneously with LOH of the hDMP1 locus in breast cancer (72), it was expected that these two events might synergize to lower the tumor-suppressive activities of DMP1α to less than 50% of those in wild type cells, which should be analyzed in detail. The function of the third transcript DMP1γ should also be analyzed in vivo through creation of transgenic mouse models.
ARF
CDKN2A (INK4a/ARF) is frequently disrupted in various types of human cancers, and germline mutations of this locus can confer susceptibility to melanoma and other tumors (76, 77). Genetic disruption of p19Arf alone predisposes mice to tumorigenesis, demonstrating that Arf is a tumor-suppressor gene in mice (16). Arf is a classical TSG in an Eμ-Myc B cell lymphoma model because tumor development is extremely accelerated in Arf−/− mice (7 weeks) in comparison to Arf+/− (11 weeks) or Arf+/+ (30 weeks) mice, and because 80% of lymphomas from Arf+/− mice lost the wild type allele (78). Krimpenfort et al. specifically mutated p16Ink4a and demonstrated that these mice, designated Ink4a*/*, did not show a significant predisposition to spontaneous tumor formation within 17 months (79). Murine embryonic fibroblasts (MEFs) derived from them proliferate normally, were mortal, and were not transformed by oncogenic H-Ras alone (79, 80). The very mild phenotype of the Ink4a*/* mice indicated that the very strong phenotypes of the original Ink4a/ArfΔ2,3 mice (79) were primarily due to loss of Arf. Of note, Ink4a*/Δ2,3 mice that were deficient for Ink4a and heterozygous for Arf spontaneously developed a wide spectrum of tumors, including melanoma (79). Those tumors expressed Arf demonstrating haploinsufficiency of Arf in the absence of Ink4a (Table 1). Arf haplo-insufficiency was also demonstrated in Eμ-Myc lymphomas where loss of one allele of Arf rescued protracted lymphoma development in Mdm2+/−;Eμ-Myc TGs (81).
The inv(16) is one of the most frequent chromosomal translocations associated with acute myeloid leukemia (AML) and creates a chimeric fusion protein CBFβ/MYH11 (82). The ARF tumor suppressor is transactivated by the AML1 protein (see below), suggesting that the inv(16) fusion protein may repress ARF expression. Moreno-Miralles et al. established a murine bone marrow transplant model of the inv(16) in which wild type, Arf+/−, and Arf−/− bone marrow cells were engineered to express the inv(16) fusion protein (82). Its expression was sufficient to induce a myelomonocytic AML even in wild type bone marrow, yet deletion of only a single Arf allele greatly accelerated the disease, indicating that Arf is haploinsufficient for the induction of AML in the presence of the inv(16) (82). These reports indicate that even a classical tumor suppressor behaves like a haploinsufficient TSG when combined with other genetic alterations, which can happen frequently in human malignancies.
AML1:CBFβ
The core-binding factor (CBF) is a heterodimeric transcription factor complex that consists of 3 distinct DNA-binding CBFα subunits (RUNX1, 2, and 3), and a common CBFβ subunit, which does not bind to DNA (83–87). RUNX1 was the first CBF gene to be isolated and has been known as AML1, PEBPA2B, or CBFA2. The binding affinity of AML1 subunit to the DNA promoter sequences is significantly increased by its association with CBFβ, protects AML1 subunit from proteolysis. Core-binding factor acute myeloid leukemia (AML) is cytogenetically defined by the presence of t(8;21)(q22;q22) or inv(16)(p13q22)/t(16;16)(p13;q22), producing AML1-ETO (83–86) and CBFB/MYH11 (87) chimeric proteins, respectively, both contributing to leukemogenesis (85–87). In both subtypes, the cytogenetic rearrangements disrupt genes that encode subunits of core-binding factor, a transcription factor that functions as an essential regulator of normal hematopoiesis. The t(3;21)(q26;q22) translocation, which is one of the consistent chromosomal abnormalities found in blastic crisis of chronic myelocytic leukemia, is thought to play an important role in the leukemic progression of CML to an acute blastic crisis phase. The AML1 gene was also rearranged by the t(3;21)(q26;q22) translocation producing the AML1-Evi1 fusion protein (88), which leads to dysplastic hemopoiesis in vivo (89). Lineage determinant Runx proteins organize and assemble multi-protein complexes at sites of transcription within the nucleus and regulate both RNA polymerase II- and I-mediated gene expression (90). In addition, Runx proteins epigenetically control lineage determining transcriptional programs including: 1) architectural organization of macromolecular complexes in interphase, 2) regulation of gene expression through bookmarking during mitosis, and 3) microRNA-mediated translational control in the interphase nucleus. These mechanisms are compromised in the onset and progression of leukemias with altered RUNX proteins (90).
Null mutations in mouse AML1 or CBFβ results in mid-gestational lethality with a complete lack of fetal liver hematopoiesis (91, 92). Thus the AML1:CBFβ transcription factor complex is essential for hematopoiesis. Cai et al. examined the consequences of AML1-loss in hematopoietic stem cells (HSC) of the mouse embryo, and demonstrated an absolute requirement for AML1 in functional HSCs (93). Moreover, single allelic loss of AML1 resulted in a dramatic change in the distribution of HSCs, leading to their early appearance in the aorta-gonad-mesonephros region and the yolk sac. The effect of AML1 dosage on adult hematopoiesis was studied by comparing the hematopoietic systems of AML1+/− and AML1+/+ mice (94). Sun et al. reported that AML1+/− bone marrow had an increase in multilineage and lineage-restricted progenitors (94). They also showed that even though AML1+/− mice had a decrease in the number of long-term repopulating hematopoietic stem cells, the engraftment levels were increased. These data demonstrate a dosage-dependent effect for AML1 in regulating the quantity of HSCs and their downstream lineage-committed hematopoietic progenitors (94).
Although AML1+/− mice are genetically comparable models with human FPD/AML, they do not develop spontaneous leukemia. To induce additional genetic alterations, retroviral insertional mutagenesis was performed with the use of BXH2-TG mice, which develop myeloid leukemia because of the random integration of retrovirus (95). Heterozygous disruption of AML1 in BXH2-TG mice resulted in a shortening of the latency period for developing leukemia. Moreover, the c-Kit gene that is frequently mutated in human RUNX leukemias was recurrently activated in BXH2-TG;AML1+/− mice, and a colony forming assay revealed synergism between monoallelic loss of AML1 and c-Kit overexpression. Thus, the BXH2-TG;AML1+/− system is an excellent mouse model to study the mechanism of leukemogenesis in familial platelet disorder (FPD)/AML patients (Table 1).
Linggi et al. identified the p14ARF tumor suppressor as a direct transcriptional target of AML1-ETO, the product of t(8;21) frequently found in AML (96). Of note, AML1 stimulated p14ARF transcription (Fig. 3) and induced phenotypes consistent with cellular senescence, which was antagonized by AML1-ETO (96). This explains why p53 is not mutated in t(8;21)-containing leukemias.
FPD that predisposes to AML is an autosomal-dominant disease characterized by platelet defects, which tends to result in AML (97). Mutational analysis of candidate genes on chromosome 21q revealed non-sense mutations or intragenic deletion of one allele of AML1. Analysis of blood cells from affected FPD/AML patients showed a decrease in megakaryocyte colony formation, demonstrating that AML1 dosage affects megakaryopoiesis (98). Hence haploinsufficiency of AML1 is a pre-neoplastic condition that leads to acute leukemia in humans as well (98).
EGR1 and 5q
The Early growth response 1 (Egr1) gene encodes a zinc-finger transcription factor of 59,000 Daltons that activates transcription by binding to DNA as a monomer (99–101). Depending on the cell type and the stimuli, EGR1 induces the expression of growth factors, growth factor receptors, proteins involved in the regulation of cell growth, differentiation, apoptosis, and extracellular matrix proteins. Because the consensus sequences for SP1- and EGR1-binding overlap, EGR1 often displaces SP1 from gene promoters; thus EGR1 competes with SP1 that is involved in the constitutive expression of housekeeping genes [so called ‘SP box’] and others.
The deletion of part of the long arm of chromosome 5 [del(5q)], is the most common chromosomal abnormality in primary myelodysplastic syndromes (MDS) and therapy-related myeloid neoplasms (t-MNs; ref. 21). Mouse models have implicated heterozygous loss of APC, DIAPH1, EGR1, and NPM1 in the pathophysiology of MDS with del(5q) (102). To study whether loss of Egr1 is an initiating event in the pathogenesis of AML/MDS, Egr1-knockout mice were treated with ENU to induce cooperating mutations (103). Both Egr1+/− and Egr1−/− mice treated with ENU developed T-cell lymphoma or a myeloproliferative disorder (MPD) at increased rates and with shorter latencies than those of wild type littermates (Table 1). Of note, biallelic inactivation of Egr1 was not observed in MPDs in Egr1+/− mice, suggesting that haploinsufficiency for Egr1 plays a role in murine leukemogenesis, and also in the development of AML/MDS with the alteration of chromosome 5.
It has been reported that loss of p53 activity, through mutation or deletion, is highly associated with t-MNs with a del(5q) (104–106). Stoddart et al. reported that loss of one copy of Egr1 or p53 in an Apcdel/+ background accelerated the development of a macrocytic anemia with monocytosis, early features of t-MN (106). The development of Apcdel/+-induced anemia was significantly accelerated by treatment of mice with the alkylating agent ENU regardless of the genomic status of Egr1 and Tp53 demonstrating that the anemia was cell-extrinsic (i.e. caused by microenvironmental change). These data emphasized the synergistic role of cell intrinsic and extrinsic factors in the pathogenesis of t-MN, and raised awareness of the deleterious effects of cytotoxic therapy on the stromal microenvironment (106). Stoddart et al. also showed that loss of p53 in combination with mono-allelic loss of Egr1 increased the rate of development of hematologic malignancies, but not overt myeloid leukemias (107). Cell intrinsic loss of p53 in Egr1+/−;Apc+/− hematopoietic cells led to the development of AML in 17% of mice (107; Table 1). Thus, loss of p53 activity in cooperation with Egr1 and Apc haploinsufficiency creates an environment that predisposes to the development of AML.
TGFβ, TGFβR, and SMAD4
Transforming growth factor-β (TGFβ) is a cytokine that controls cell proliferation and differentiation. It is involved in both inflammatory disorders and cancer. TGFβ acts as an anti-proliferative factor in normal epithelial cells at early stages of oncogenesis while it stimulates cell proliferation at advanced stages (108, 109). Haploinsufficiency of TGFβ1 in tumor suppression was shown by the tumor predisposition of TGFβ+/− mice (11). However, because TGFβ is a secreted protein, its tumor-inhibitory effect depends on its expression in the whole animal, thus the mechanism for haploinsufficiency cannot be discussed within a single cell.
TGFβ dimers bind to a type II receptors which recruits and phosphorylates a type I receptor (Fig. 5). The type I receptor then recruits and phosphorylates a receptor-regulated SMAD (R-SMAD; Smad 1, 2, 3, 5, 8/9). The R-SMAD then binds to the common SMAD:SMAD4 (also called DPC4) and forms a heterodimeric complex (Fig. 5). This complex then enters the cell nucleus where it acts as a transcription factor for various genes to activate the mitogen-activated protein kinase 8 pathway, which triggers apoptosis.
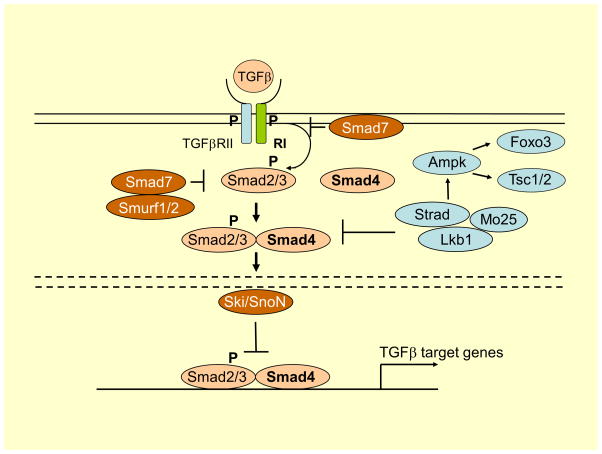
Figure 5. Signaling pathways involving TGFβ, Smad4, and Lkb1.
The solid lines represent the plasma membrane, and the discontinuous lines show the nuclear membrane. TGFβ binds to two distinct receptor types, known as type I and type II. Both type I and type II receptors contain serine/threonine kinase domains in their intracellular portions. The type II receptor kinases are constitutively active. When they bind to the ligand, they form hetero-tetrameric complexes composed of two molecules each of type I and type II receptors. In the hetero-tetrameric receptor I and II complexes, type II receptor kinases transphosphorylate the GS-domain of type I receptor, which, in turn, phosphorylate intracellular substrates. Smad proteins are major signaling molecules acting downstream of the serine/threonine kinase TGFβ receptors. Smads are classified into three subclasses, i.e. receptor-regulated Smads (R-Smads), common-partner Smad (co-Smad: Smad 4), and inhibitory Smads (I-Smads).
After activation of TGFβRI, the signal activates Smad2 and Smad3 proteins bound to the receptors, by phosphorylation of their C-terminal (SXS motif) residues. The next step is formation of a functional trimeric complex by phosphorylated R-Smad and Smad4, and then this complex is translocated to the nucleus, where it regulates the transcription of TGFβ1-dependent genes; thus Smads have the activity of transcription factors. The activity of TGFβ signaling pathway is regulated by a negative regulatory feedback loop mediated by I-Smads (Smads inhibitors): Smad 6 and 7. They are able to interact with membrane receptors by forming stable complex with activated TGFβRI, and thus impairing their interaction with the R-Smad (inhibition of their phosphorylation). Smad7 expression is induced by TGFβ, leading to inhibition of the cellular response to this cytokine. Smad7 has been shown to promote recruitment of E3 ubiquitin ligases, including Smad ubiquitin regulatory factors (Smurf1/2) into the receptor complex. Binding of Smad7 and Smurf to the receptor complex also results in competitive inhibition of Smad2/3 binding to TGFβRI. Direct Interaction of Ski with either Smad3 or Smad4 is necessary and sufficient for Ski-mediated repression of TGFβ signaling (133).
Lkb1 is a well-studied tumor suppressor kinase that regulates cell growth and polarity. It encodes a serine-threonine kinase that directly phosphorylates and activates AMPK, a central metabolic sensor (134). Lkb1 is capable of phosphorylating Smad4 on its DNA-binding domain, inhibiting Smad4 from binding to either TGFβ- or bone morphogenetic protein-specific promoter sequences, thereby Smad4-dependent transcription (135). Lkb1 activates AMPK and send signals to Foxo3, Tsc1/2, p53, fatty acid synthase, and other molecules, which enforces metabolic checkpoints for regulating cell growth and metabolism.
The TGFβ signaling pathway is frequently altered in colorectal cancer. Zheng et al. crossed Tgfbr1+/− mice with ApcMin/+ mice, and showed that Tgfbr1+/−;ApcMin/+ mice developed twice as many intestinal tumors as Tgfbr1+/+;ApcMin/+ mice without losing the wild type Tgfbr1 locus (110). Decreased Smad2/3 phosphorylation, increased cyclin D1 expression, and cellular proliferation were observed in the colonic epithelium crypts of Tgfbr1+/−;ApcMin/+ mice. Thus Tgfbr1 is haploinsufficient for tumor suppression and provides a molecular mechanism for colorectal cancer development in individuals with constitutively altered TGFβR1 expression (110; Table 1).
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in the United States with a median survival of <6 months (111). KRAS mutation is a hallmark of PDAC but remains an intractable pharmacologic target (112). TGFβ signaling plays critical role in PDAC progression since SMAD4 is deleted or mutated in 55% and the TGFβR2 gene is altered in a smaller subset of human PDAC (113). Pancreas-specific Tgfβr2 knockout mice have been generated, alone or in combination of active Kras (KrasG12D) expression driven by the pancreatic transcription factor 1a. Pancreas-specific KrasG12D activation alone generated only intraepithelial neoplasia within 1 year (113). Conversely, both Tgfβr2−/−;KrasG12D and Tgfβr2+/−;KrasG12D developed PDAC, which demonstrated a haploinsufficiency of TGFβ signaling in PDCA prevention (Table 1).
Smad4+/− mice begin to develop polyposis in the fundus at 6–12 months old, and in the duodenum and cecum in older animals. With increasing age, polyps in the antrum show sequential changes from hyperplasia to full adenocarcinoma. However, loss of the remaining wild type allele was detected only in later stages of tumor progression, suggesting that mono-allelic loss of Smad4 is sufficient for tumor progression (114). Alberici et al. (115) subsequently reported that haploinsufficiency of Smad4 underlies tumor initiation in the gastrointestinal tract, which was sufficient to affect tumor progression both prior to and upon loss of Apc function (Table 1).
Oncogenic Kras initiates pancreatic tumorigenesis, while subsequent genetic events shape the resultant disease. Izeradjene et al. showed that concomitant expression of KrasG12D and haploinsufficiency of the Smad4/Dpc4 TSG induced a distinct class of pancreatic tumors, mucinous cystic neoplasms (MCNs), which developed into invasive ductal adenocarcinomas (116). Progression of MCNs was accompanied by LOH of Smad4 and mutation of either p53 or p16Ink4a. Thus, these distinct phenotypic routes to invasive adenocarcinoma nevertheless share the same overall mutational spectra (116).
Research in progress and future directions
Haploinsufficiency of TSGs has been demonstrated mostly in mouse knockout models (18, 117), but analyses of human cancer specimen have also been performed in some cases. For instance, hemizygous loss of hDMP1 was reported in 35–42% of lung and breast cancers (64, 71) with proof in mouse models (64, 65). Since the wild type allele is retained without mutation or promoter hypermethylation in haploinsufficient TSGs, characterization of human cancer specimen for candidate haploinsufficient TSG, esp. for mutational analyses and the expression of the wild type mRNA from the wild type locus, will be essential before an attempt to re-activate the wild type locus for novel cancer therapies.
The outstanding question for haploinsufficient TSG research is the molecular genetic mechanisms that explain the phenotype. In case of p27Kip1, tumors from p27Kip1+/− mice express ~50% of the protein in comparison to p27+/+ tumors, indicating that it is a typical haploinsufficient TSG (2). In both p27 and Pten, tumor development was accelerated in TSG+/− mice than in TSG−/− indicating that the tumor-prone phenotype was obligatory (9). In case of Eμ-Myc tumors in Dmp1 knockout mice, Dmp1+/− lymphomas expressed ~50% of the wild type protein, suggesting that it a typical case of haploid insufficiency (4). However, it is not an obligatory haploinsufficiency since there was no significant difference in tumor-free survival between Dmp1+/− and Dmp1−/− mice (4; see 64, 65 for different models). In case of p53 knockout mice, it was shown that p53+/− cells express only 25% of wild type mRNA and protein, which could explain the acceleration of tumor development in p53+/− mice without losing the wild type allele (12, 48). Likewise, the protein expression level in Dmp1+/−;MMTV-neu tumors was ~25% as compared to those in Dmp1+/+;MMTV-neu tumors (65). Thus these are representative cases of ‘one and a half’ inactivation of the TSG loci. We have put both of these TSGs in the category of non-classical, haploinsufficient TSGs.
In the latter case of ‘one and a half inactivation’, it is reasonable to speculate that some unknown mechanisms may co-exist to lower, but they do not completely eliminate the expression of wild type tumor suppressive protein in TSG+/− tumors. Indeed, our recent study showed that the levels of Dmp1 and Arf mRNAs in Dmp1+/− lungs were only 30 and 15%, respectively, of those in Dmp1+/+ tissues (118). One possible mechanism for these findings is that the transcription for the Dmp1 promoter is decreased in Dmp1+/− tissues through disruption of auto-regulation since the genomic locus has multiple Dmp1-binding consensus sequences. The second possibility for the decreased expression (i.e. less than 50%) of mRNA/protein in haploinsufficient TSG+/− tissues is accelerated degradation of the mRNA/protein, the former of which may be mediated by microRNAs that bind to the 3′ non-coding region. In fact, bioinformatics analysis of haploinsufficient genes for variations in their 3′UTR showed that the occurrence of SNPs result in the creation of new binding sites for miRNAs (20). Thus haploinsufficiency of TSGs can be driven by the cumulative effect of miRNAs, miRNA-binding-site polymorphisms, and miRNA polymorphisms. The third possibility is aberrant splicing of the locus that generates transcripts that block the activities the wild type TSG, which can co-exist with mono-allelic loss of the wild type allele locus of the TSG (72, 74). In p63/p73, oncogenic splicing variants that lack the N-terminal sequences are often overexpressed in tumors and are associated with poor prognosis (119). The mechanisms for degradation of haploinsufficient TSG protein products should be investigated since reversal for such process(es) can lead to a novel therapy of cancer.
This chapter focused on TSGs that are close to our research fields. We could not review other important haploinsufficient TSGs (e.g. PTEN, LKB1, NF1, DOK2, NPM, RPS14, or NKX2-1/TTF-1) due to limitations. Please refer to other reviews (9, 120–123) if you are interested in these molecules.
Acknowledgments
We thank all other members of Dr. Inoue’s laboratory for sharing unpublished research findings.
Financial Support
K. Inoue was supported by NIH/NCI 2R01CA106314, ACS RSG-07-207-01-MGO, and KG080179.
Footnotes
Conflicts of interest
The authors have no conflicts of interest related to this work.
Introduction
- 1.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ. The murine gene p27Kip1 is haplo-insufficient for tumor suppression. Nature. 1998;396:177–180. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue K, Wen R, Rehg JE, Adachi M, Cleveland JL, Roussel MF, Sherr CJ. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, ras transformation, and tumorigenesis. Genes Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- 4.Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001;15:2934–2939. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15:2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 6.Di Cristofano A, Kotsi P, Peng YF, Cordon-Cardo C, Elkon KB, Pandolfi PP. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 7.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 8.Gutmann DH, Loehr A, Zhang Y, Kim J, Henkemeyer M, Cashen A. Haploinsufficiency for the neurofibromatosis 1 (NF1) tumor suppressor results in increased astrocyte proliferation. Oncogene. 1999;18:4450–4459. doi: 10.1038/sj.onc.1202829. [DOI] [PubMed] [Google Scholar]
- 9.Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J Pathol. 2011;223:137–146. doi: 10.1002/path.2800. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumourigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang B, Böttinger EP, Jakowlew SB, Bagnall KM, Mariano J, Anver MR, Letterio JJ, Wakefield LM. Transforming growth factor-B1 is a new form of tumor suppressor with true haploid insufficiency. Nat Med. 1998;4:802–807. doi: 10.1038/nm0798-802. [DOI] [PubMed] [Google Scholar]
- 12.Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, Donehower LA. Retention of wild type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varley JM, Evans DG, Birch JM. Li-Fraumeni syndrome--a molecular and clinical review. Br J Cancer. 1997;76:1–14. doi: 10.1038/bjc.1997.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Current Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 15.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 16.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 17.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 18.Taneja P, Zhu S, Maglic D, Fry EA, Kendig RD, Inoue K. Transgenic and knockout mice models to reveal the functions of tumour suppressor genes. Clin Med Insights Oncol. 2011;5:235–257. doi: 10.4137/CMO.S7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, Richardson AL, Zhang J, Pandolfi PP. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manikandan M, Raksha G, Munirajan AK. Haploinsufficiency of tumor suppressor genes is driven by the cumulative effect of microRNAs, microRNA binding site polymorphisms and microRNA polymorphisms: an insilico approach. Cancer Inform. 2012;11:157–171. doi: 10.4137/CIN.S10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumor suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boultwood J, Pellagatti A, McKenzie AN, Wainscoat JS. Advances in the 5q- syndrome. Blood. 2010;116:5803–5811. doi: 10.1182/blood-2010-04-273771. [DOI] [PubMed] [Google Scholar]
- 23.Honda H, Nagamachi A, Inaba T. -7/7q- syndrome in myeloid-lineage hematopoietic malignancies: attempts to understand this complex disease entity. Oncogene. 2015;34:2413–25. doi: 10.1038/onc.2014.196. [DOI] [PubMed] [Google Scholar]
- 24.Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, Revill K, Weissmueller S, Rappaport AR, Simon J, Zhang J, Luo W, Hicks J, Zender L, Wang XW, Powers S, Wigler M, Lowe SW. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
p27Kip1
- 25.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 26.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 27.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 28.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 29.Sharma SS, Pledger WJ. The non-canonical functions of p27Kip1 in normal and tumor biology. Cell Cycle. 2016:1–13. doi: 10.1080/15384101.2016.1157238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietenpol JA, Bohlander SK, Sato Y, Papadopoulos N, Liu B, Friedman C, Trask BJ, Roberts JM, Kinzler KW, Rowley JD, Vogelstein B. Assignment of the human p27Kip1 gene to 12p13 and its analysis in leukemias. Cancer Res. 1995;55:1206–1210. [PubMed] [Google Scholar]
- 32.Muraoka RS, Lenferink AE, Law B, Hamilton E, Brantley DM, Roebuck LR, Arteaga CL. ErbB2/Neu-induced, cyclin D1-dependent transformation is accelerated in p27-haploinsufficient mammary cells but impaired in p27-null cells. Mol Cell Biol. 2002;22:2204–2219. doi: 10.1128/MCB.22.7.2204-2219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Toriellec E, Despouy G, Pierron G, Gaye N, Joiner M, Bellanger D, Vincent-Salomon A, Stern MH. Haploinsufficiency of CDKN1B contributes to leukemogenesis in T-cell prolymphocytic leukemia. Blood. 2008;111:2321–2328. doi: 10.1182/blood-2007-06-095570. [DOI] [PubMed] [Google Scholar]
- 34.Feurstein S, Rücker FG, Bullinger L, Hofmann W, Manukjan G, Göhring G, Lehmann U, Heuser M, Ganser A, Döhner K, Schlegelberger B, Steinemann D. Haploinsufficiency of ETV6 and CDKN1B in patients with acute myeloid leukemia and complex karyotype. BMC Genomics. 2014;15:784. doi: 10.1186/1471-2164-15-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crona J, Gustavsson T, Norlén O, Edfeldt K, Åkerström T, Westin G, Hellman P, Björklund P, Stålberg P. Somatic Mutations and Genetic Heterogeneity at the CDKN1B Locus in Small Intestinal Neuroendocrine Tumors. Ann Surg Oncol. 2015 Dec;22(Suppl 3):S1428–35. doi: 10.1245/s10434-014-4351-9. Epub 2015 Jan 14. [DOI] [PubMed] [Google Scholar]
- 36.Ayrault O, Zindy F, Rehg J, Sherr CJ, Roussel MF. Two tumor suppressors, p27Kip1 and patched-1, collaborate to prevent medulloblastoma. Mol Cancer Res. 2009;7:33–40. doi: 10.1158/1541-7786.MCR-08-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diersch S, Wenzel P, Szameitat M, Eser P, Paul MC, Seidler B, Eser S, Messer M, Reichert M, Pagel P, Esposito I, Schmid RM, Saur D, Schneider G. Efemp1 and p27(Kip1) modulate responsiveness of pancreatic cancer cells towards a dual PI3K/mTOR inhibitor in preclinical models. Oncotarget. 2013;4:277–288. doi: 10.18632/oncotarget.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
p53, 53BP1
- 38.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 40.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 41.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merino D, Malkin D. p53 and hereditary cancer. Subcell Biochem. 2014;85:1–16. doi: 10.1007/978-94-017-9211-0_1. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 44.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 45.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–8295. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davison TS, Yin P, Nie E, Kay C, Arrowsmith CH. Characterization of the oligomerization defects of two p53 mutants found in families with Li-Fraumeni and Li-Fraumeni-like syndrome. Oncogene. 1998;17:651–656. doi: 10.1038/sj.onc.1202062. [DOI] [PubMed] [Google Scholar]
- 48.Lynch CJ, Milner J. Loss of one p53 allele results in four-fold reduction of p53 mRNA and protein: a basis for p53 haploinsufficiency. Oncogene. 2006;25:3463–3470. doi: 10.1038/sj.onc.1209387. [DOI] [PubMed] [Google Scholar]
- 49.Pant V, Quintás-Cardama A, Lozano G. The p53 pathway in hematopoiesis: lessons from mouse models, implications for humans. Blood. 2012;120:5118–5127. doi: 10.1182/blood-2012-05-356014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward IM, Difilippantonio S, Minn K, Mueller MD, Molina JR, Yu X, Frisk CS, Ried T, Nussenzweig A, Chen J. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol. 2005;25:10079–10086. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34:1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willers H, Gheorghiu L, Liu Q, Efstathiou JA, Wirth LJ, Krause M, von Neubeck C. DNA damage response assessments in human tumor samples provide functional biomarkers of radiosensitivity. Semin Radiat Oncol. 2015;25:237–250. doi: 10.1016/j.semradonc.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Squatrito M, Vanoli F, Schultz N, Jasin M, Holland EC. 53BP1 is a haploinsufficient tumor suppressor and protects cells from radiation response in glioma. Cancer Res. 2012;72:5250–526. doi: 10.1158/0008-5472.CAN-12-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
DMP1 (DMTF1)
- 54.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene. 2007;26:4329–4335. doi: 10.1038/sj.onc.1210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci USA. 1999;96:3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu S, Mott RT, Fry EA, Taneja P, Kulik G, Sui G, Inoue K. Cooperation between cyclin D1 expression and Dmp1-loss in breast cancer. Am J Pathol. 2013;183:1339–1350. doi: 10.1016/j.ajpath.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fry EA, Taneja P, Maglic D, Zhu S, Sui G, Inoue K. Dmp1α inhibits HER2/neu-induced mammary tumorigenesis. PLoS One. 2013;8:e77870. doi: 10.1371/journal.pone.0077870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frazier DP, Kendig RD, Kai F, Maglic D, Sugiyama T, Taneja P, Morgan RL, Fry EA, Lagedrost SJ, Sui G, Inoue K. Dmp1 physically interacts with p53 and positively regulates p53’s stabilization, nuclear localization, and function. Cancer Res. 2012;72:1740–1750. doi: 10.1158/0008-5472.CAN-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inoue K, Fry EA, Frazier DP. Transcription factors that interact with p53 and Mdm2. Int J Cancer. 2016;138:1577–1585. doi: 10.1002/ijc.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its repression by E2Fs. Oncogene. 2006;25:7703–7713. doi: 10.1038/sj.onc.1209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taneja P, Mallakin A, Matise LA, Frazier DP, Choudhary M, Inoue K. Repression of Dmp1 and Arf promoter by anthracyclins: critical roles of the NF-κB subunit p65. Oncogene. 2007;26:7457–7466. doi: 10.1038/sj.onc.1210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mallakin A, Sugiyama T, Taneja P, Matise LA, Frazier DP, Choudhary M, Hawkins GA, D’Agostino RB, Jr, Willingham MC, Inoue K. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007;12:381–394. doi: 10.1016/j.ccr.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taneja P, Maglic D, Kai F, Sugiyama T, Kendig RD, Frazier DP, Willingham MC, Inoue K. Critical role of Dmp1 in HER2/neu-p53 signaling and breast carcinogenesis. Cancer Res. 2010;70:9084–9094. doi: 10.1158/0008-5472.CAN-10-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cornelisse CJ. Loss of heterozygosity, chromosome 7q, and breast cancer. Lancet. 1992;339:1423–1424. doi: 10.1016/0140-6736(92)91248-7. [DOI] [PubMed] [Google Scholar]
- 67.Kristjansson AK. Loss of heterozygosity at chromosome 7q in human breast cancer: association with clinical variables. Anticancer Res. 1997;17:93–98. [PubMed] [Google Scholar]
- 68.Bodner SM, Naeve CW, Rakestraw KM, Jones BG, Valentine VA, Valentine MB, Luthardt FW, Willman CL, Raimondi SC, Downing JR, Roussel MF, Sherr CJ, Look AT. Cloning and chromosomal localization of the gene encoding human cyclin D-binding Myb-like protein (hDMP1) Gene. 1999;229:223–228. doi: 10.1016/s0378-1119(98)00591-5. [DOI] [PubMed] [Google Scholar]
- 69.Inoue K, Sugiyama T, Taneja P, Morgan RL, Frazier DP. Emerging roles of DMP1 in lung cancer. Cancer Res. 2008;68:4487–4490. doi: 10.1158/0008-5472.CAN-07-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugiyama T, Frazier DP, Taneja P, Morgan RL, Willingham MC, Inoue K. The role of Dmp1 and its future in lung cancer diagnostics. Expert Rev Mol Diagn. 2008;8:435–448. doi: 10.1586/14737159.8.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maglic D, Zhu S, Fry EA, Taneja P, Kai F, Kendig RD, Sugiyama T, Miller LD, Willingham MC, Inoue K. Prognostic value of the hDMP1-ARF-Hdm2-p53 pathway in breast cancer. Oncogene. 2013;32:4120–4129. doi: 10.1038/onc.2012.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maglic D, Stovall DB, Cline JM, Fry EA, Mallakin A, Taneja P, Caudell DL, Willingham MC, Sui G, Inoue K. DMP1β, a splice isoform of the tumor suppressor DMP1 locus, induces proliferation and progression of breast cancer. J Pathol. 2015;236:90–102. doi: 10.1002/path.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tschan MP, Federzoni EA, Haimovici A, Britschgi C, Moser BA, Jin J, Reddy VA, Sheeter DA, Fischer KM, Sun P, Torbett BE. Human DMTF1β antagonizes DMTF1α regulation of the p14ARF tumor suppressor and promotes cellular proliferation. Biochim Biophys Acta. 2015;1849:1198–1208. doi: 10.1016/j.bbagrm.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inoue K, Fry EA. Aberrant splicing of the DMP1-INK4a/ARF-MDM2-p53 pathway in cancer. Int J Cancer. 2016 Jan 23; doi: 10.1002/ijc.30003. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Inoue K, Fry EA. Novel biomarkers for breast cancer. Biomark Cancer. 2016;8:1–18. doi: 10.4137/BIC.S38394. [DOI] [PMC free article] [PubMed] [Google Scholar]
ARF
- 76.Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer. 2010;127:2239–2247. doi: 10.1002/ijc.25511. [DOI] [PubMed] [Google Scholar]
- 77.Maggi LB, Jr, Winkeler CL, Miceli AP, Apicelli AJ, Brady SN, Kuchenreuther MJ, Weber JD. ARF tumor suppression in the nucleolus. Biochim Biophys Acta. 2014;1842:831–839. doi: 10.1016/j.bbadis.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 78.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16INK4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 80.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 81.Eischen CM, Alt JR, Wang P. Loss of one allele of ARF rescues Mdm2 haploinsufficiency effects on apoptosis and lymphoma development. Oncogene. 2004;23:8931–8940. doi: 10.1038/sj.onc.1208052. [DOI] [PubMed] [Google Scholar]
- 82.Moreno-Miralles I, Pan L, Keates-Baleeiro J, Durst-Goodwin K, Yang C, Kim HG, Thompson MA, Klug CA, Cleveland JL, Hiebert SW. The inv(16) cooperates with ARF haploinsufficiency to induce acute myeloid leukemia. J Biol Chem. 2005;280:40097–40103. doi: 10.1074/jbc.M506855200. [DOI] [PubMed] [Google Scholar]
AML1
- 83.Sangle NA, Perkins SL. Core-binding factor acute myeloid leukemia. Arch Pathol Lab Med. 2011;135:1504–1509. doi: 10.5858/arpa.2010-0482-RS. [DOI] [PubMed] [Google Scholar]
- 84.Ichikawa M, Yoshimi A, Nakagawa M, Nishimoto N, Watanabe-Okochi N, Kurokawa M. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int J Hematol. 2013;97:726–734. doi: 10.1007/s12185-013-1347-3. [DOI] [PubMed] [Google Scholar]
- 85.Nakazawa T, Harada Y, Tatsumi N, Tsuboi A, Kawakami M, Oka Y, Oji Y, Aozasa K, Kawase I, Sugiyama H. AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1. Blood. 2006;107:3303–3312. doi: 10.1182/blood-2005-04-1656. [DOI] [PubMed] [Google Scholar]
- 86.Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, Chen IM, Chen Z, Rowley JD, Willman CL, Zhang DE. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- 87.Lukasik SM, Zhang L, Corpora T, Tomanicek S, Li Y, Kundu M, Hartman K, Liu PP, Laue TM, Biltonen RL, Speck NA, Bushweller JH. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol. 2002;9:674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- 88.Mitani K, Ogawa S, Tanaka T, Miyoshi H, Kurokawa M, Mano H, Yazaki Y, Ohki M, Hirai H. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 1994;13:504–510. doi: 10.1002/j.1460-2075.1994.tb06288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maki K, Yamagata T, Asai T, Yamazaki I, Oda H, Hirai H, Mitani K. Dysplastic definitive hematopoiesis in AML1/EVI1 knock-in embryos. Blood. 2005;106:2147–2155. doi: 10.1182/blood-2004-11-4330. [DOI] [PubMed] [Google Scholar]
- 90.Zaidi SK, Trombly DJ, Dowdy CR, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Epigenetic mechanisms in leukemia. Adv Biol Regul. 2012;52:369–376. doi: 10.1016/j.jbior.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 91.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 92.Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, Dzierzak E. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 2000;13:423–431. doi: 10.1016/s1074-7613(00)00042-x. [DOI] [PubMed] [Google Scholar]
- 94.Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3672. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- 95.Yamashita N, Osato M, Huang L, Yanagida M, Kogan SC, Iwasaki M, Nakamura T, Shigesada K, Asou N, Ito Y. Haploinsufficiency of Runx1/AML1 promotes myeloid features and leukaemogenesis in BXH2 mice. Br J Haematol. 2005;131:495–507. doi: 10.1111/j.1365-2141.2005.05793.x. [DOI] [PubMed] [Google Scholar]
- 96.Linggi B, Müller-Tidow C, van de Locht L, Hu M, Nip J, Serve H, Berdel WE, van der Reijden B, Quelle DE, Rowley JD, Cleveland J, Jansen JH, Pandolfi PP, Hiebert SW. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–750. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 97.Minelli A, Maserati E, Rossi G, Bernardo ME, De Stefano P, Cecchini MP, Valli R, Albano V, Pierani P, Leszl A, Sainati L, Lo Curto F, Danesino C, Locatelli F, Pasquali F. Familial platelet disorder with propensity to acute myelogenous leukemia: genetic heterogeneity and progression to leukemia via acquisition of clonal chromosome anomalies. Genes Chromosome Canc. 2004;40:165–171. doi: 10.1002/gcc.20030. [DOI] [PubMed] [Google Scholar]
- 98.Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
EGR1
- 99.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pagel JI, Deindl E. Early growth response 1--a transcription factor in the crossfire of signal transduction cascades. Indian J Biochem Biophys. 2011;48:226–235. [PubMed] [Google Scholar]
- 101.Zwang Y, Oren M, Yarden Y. Consistency test of the cell cycle: roles for p53 and EGR1. Cancer Res. 2012;72:1051–1054. doi: 10.1158/0008-5472.CAN-11-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ebert BL. Molecular dissection of the 5q deletion in myelodysplastic syndrome. Sem Oncol. 2011;38:621–626. doi: 10.1053/j.seminoncol.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cleven AH, Nardi V, Ok CY, Goswami M, Dal Cin P, Zheng Z, Iafrate AJ, Abdul Hamid MA, Wang SA, Hasserjian RP. High p53 protein expression in therapy-related myeloid neoplasms is associated with adverse karyotype and poor outcome. Mod Pathol. 2015;28:552–563. doi: 10.1038/modpathol.2014.153. [DOI] [PubMed] [Google Scholar]
- 105.Wang SA, Bueso-Ramos CE. MLL gene amplification in acute myeloid leukemia and myelodysplastic syndromes is associated with characteristic clinicopathological findings and TP53 gene mutation. Hum Pathol. 2015;46:65–73. doi: 10.1016/j.humpath.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 106.Stoddart A, Wang J, Fernald AA, Karrison T, Anastasi J, Le Beau MM. Cell intrinsic and extrinsic factors synergize in mice with haploinsufficiency for Tp53, and two human del(5q) genes, Egr1 and Apc. Blood. 2014;123:228–238. doi: 10.1182/blood-2013-05-506568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stoddart A, Fernald AA, Wang J, Davis EM, Karrison T, Anastasi J, Le Beau MM. Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood. 2014;123:1069–1078. doi: 10.1182/blood-2013-07-517953. [DOI] [PMC free article] [PubMed] [Google Scholar]
TGFβ/TGFβR/SMAD4
- 108.Inman GJ. Switching TGFβ from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev Cold Spring Harb Perspect Biol. 2011;21:93–99. doi: 10.1016/j.gde.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 109.Principe DR, Doll JA, Bauer J, Jung B, Munshi HG, Bartholin L, Pasche B, Lee C, Grippo PJ. TGF-β: Duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst. 2014;106:djt369. doi: 10.1093/jnci/djt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zeng Q, Phukan S, Xu Y, Sadim M, Rosman DS, Pennison M, Liao J, Yang GY, Huang CC, Valle L, Di Cristofano A, de la Chapelle A, Pasche B. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009;69:678–686. doi: 10.1158/0008-5472.CAN-08-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Collisson EA. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu X, Brodie SG, Yang X, Im YH, Parks WT, Chen L, Zhou YX, Weinstein M, Kim SJ, Deng CX. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]
- 115.Alberici P, Jagmohan-Changur S, De Pater E, Van Der Valk M, Smits R, Hohenstein P, Fodde R. Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene. 2006;25:1841–1851. doi: 10.1038/sj.onc.1209226. [DOI] [PubMed] [Google Scholar]
- 116.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, Deng CX, Hruban RH, Adsay NV, Tuveson DA, Hingorani SR. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
Research in progress and outstanding questions
- 117.Inoue K, Fry EA, Taneja P. Recent progress in mouse models for tumor suppressor genes and its implications in human cancer. Clin Med Insights Oncol. 2013;7:103–122. doi: 10.4137/CMO.S10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mallakin A, Sugiyama T, Kai F, Taneja P, Kendig RD, Frazier DP, Maglic D, Matise LA, Willingham MC, Inoue K. The Arf-inducing transcription factor Dmp1 encodes transcriptional activator of amphiregulin, thrombospondin-1, JunB and Egr1. Int J Cancer. 2010;126:1403–1416. doi: 10.1002/ijc.24938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Inoue K, Fry EA. Alterations of p63 and p73 in human cancers. Subcell Biochem. 2014;85:17–40. doi: 10.1007/978-94-017-9211-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumour suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 121.Larizza L, Gervasini C, Natacci F, Riva P. Developmental abnormalities and cancer predisposition in neurofibromatosis type 1. Curr Mol Med. 2009;9:634–653. doi: 10.2174/156652409788488801. [DOI] [PubMed] [Google Scholar]
- 122.Yamaguchi T, Hosono Y, Yanagisawa K, Takahashi T. NKX2–1/TTF-1: an enigmatic oncogene that functions as a double edged sword for cancer cell survival and progression. Cancer Cell. 2013;23:713–723. doi: 10.1016/j.ccr.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 123.Pellagatti A, Boultwood J. Recent Advances in the 5q- Syndrome. Mediterr J Hematol Infect Dis. 2015;7:e2015037. doi: 10.4084/MJHID.2015.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Figures and the table
- 124.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Godell MA, Weinberg RA. Effects of Rb mutation in the mouse. Nature. 2002;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 125.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34:1–14. doi: 10.1038/onc.2013.570. [DOI] [PubMed] [Google Scholar]
- 127.Inoue K, Fry EA. Aberrant splicing of estrogen receptor, HER2, and CD44 in breast cancer. Genet Epigenet. 2015;7:19–32. doi: 10.4137/GEG.S35500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Inoue K, Sherr CJ, Shapiro LH. Regulation of the CD13/aminopeptidase N gene by DMP1, a transcription factor antagonized by D-type cyclins. J Biol Chem. 1998;273:29188–21994. doi: 10.1074/jbc.273.44.29188. [DOI] [PubMed] [Google Scholar]
- 129.Inoue K, Fry EA. Aberrant expression of Cyclin D1 in cancer. Signal Transduction Insights. 2015;4:1–13. doi: 10.4137/STI.S30306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taneja P, Frazier DP, Sugiyama T, Lagedrost SJ, Inoue K. In: Control of cellular physiology by transcription factors E2F and their roles in ‘Carcinogenesis in Control of Cellular Physiology by E2F transcription factors’. Yoshida Ken-ichi., editor. Chapter 10. Research Signpost; Trivandrum, Kerala, India: 2008. pp. 179–197. [Google Scholar]
- 131.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Taneja P, Frazier DP, Kendig RD, Maglic D, Sugiyama T, Kai F, Taneja NK, Inoue K. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev Mol Diagn. 2009;9:423–440. doi: 10.1586/ERM.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ueki N, Hayman MJ. Direct interaction of Ski with either Smad3 or Smad4 is necessary and sufficient for Ski-mediated repression of transforming growth factor-beta signaling. J Biol Chem. 2003;278:32489–32492. doi: 10.1074/jbc.C300276200. [DOI] [PubMed] [Google Scholar]
- 134.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Morén A, Raja E, Heldin CH, Moustakas A. Negative regulation of TGFβ signaling by the kinase LKB1 and the scaffolding protein LIP1. J Biol Chem. 2011;286:341–353. doi: 10.1074/jbc.M110.190660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]