Abstract
Iron is an essential nutrient for bacteria but the reactivity of Fe2+ and the insolubility of Fe3+ present significant challenges to bacterial cells. Iron storage proteins contribute to ameliorating these challenges by oxidizing Fe2+ using O2 and H2O2 as electron acceptors, and by compartmentalizing Fe3+. Two types of iron-storage proteins coexist in bacteria, the ferritins (Ftn) and the heme-containing bacterioferritins (Bfr), but the reasons for their coexistence are largely unknown. P. aeruginosa cells harbor two iron storage proteins (FtnA and BfrB), but nothing is known about their relative contributions to iron homeostasis. Prior studies in vitro have shown that iron mobilization from BfrB requires specific interactions with a ferredoxin (Bfd), but the relevance of the BfrB:Bfd interaction to iron homeostasis in P. aeruginosa is unknown. In this work we explore the repercussions of (i) deleting the bfrB gene, and (ii) perturbing the BfrB:Bfd interaction in P. aeruginosa cells by either deleting the bfd gene or by replacing the wild type bfrB gene with a L68A/E81A double mutant allele in the P. aeruginosa chromosome. The effects of the mutations were evaluated by following the accumulation of iron in BfrB, analyzing levels of free and total intracellular iron, and by characterizing the ensuing iron homeostasis dysregulation phenotypes. The results reveal that P. aeruginosa accumulate iron mainly in BfrB, and that the nutrient does not accumulate in FtnA to detectable levels, even after deletion of the bfrB gene. Perturbing the BfrB:Bfd interaction causes irreversible flow of iron into BfrB, which leads to the accumulation of unusable intracellular iron while severely depleting the levels of free intracellular iron, which drives the cells to an acute iron starvation response despite harboring “normal” levels of total intracellular iron. These results are discussed in the context of a dynamic equilibrium between free cytosolic Fe2+ and Fe3+ compartmentalized in BfrB, which functions as a buffer to oppose rapid changes of free cytosolic iron. Finally, we also show that P. aeruginosa cells utilize iron stored in BfrB for growth in iron-limiting conditions, and that the utilization of BfrB-iron requires a functional BfrB:Bfd interaction.
Graphical Abstract
Introduction
Iron is essential for all forms of life. In bacteria iron is involved in multiple metabolic processes, including respiration (heme-containing cytochromes, [Fe-S]-containing cytochromes), and in key enzymatic reactions, such as those carried out in the TCA cycle by the [Fe-S]-containing aconitase and fumarase enzymes. Despite the essentiality of iron, the chemical properties of its most common oxidation states (Fe2+ and Fe3+) can present significant challenges to living cells. Fe2+ is soluble in aqueous solution (~0.1 M at pH 7.0) but is readily oxidized by O2 to the extremely insoluble Fe3+ form (~10−18 M at pH 7.0), drastically reducing the nutrient’s bioavailability. In addition, the reactivity of Fe2+ with O2 and H2O2, which forms reactive oxygen species such as superoxide and the highly reactive hydroxyl radical, can be conducive to iron-induced toxicity.1,2 Consequently, bacteria rely on a complex iron homeostatic machinery to ensure iron sufficiency for metabolic needs while preventing iron- induced toxicity. Because iron is essential for bacterial growth and survival, mammals maintain extremely low concentrations of free iron, which are further reduced during infections.3 To overcome the iron starvation caused by the limited bioavailability of iron in the environment, or imposed by the immune system of a mammalian host, bacteria employ redundant iron procuring strategies. These include the production of siderophores (iron chelators capable of binding Fe3+ with association constants that can exceed 1030), heme acquisition systems aimed at utilizing host-heme as an iron source, transferrin and lactoferrin receptors, and Fe2+ importers.4–7 Efficient deployment of these iron uptake strategies depends on intact iron homeostasis machinery, which endows the bacterial cell with the ability to sense intracellular and environmental iron levels, and to coordinate responses to procure iron, or to defend from potential iron-induced toxicity. To maintain iron homeostasis bacteria must balance the synthesis and secretion of iron scavenging systems, the capture and import of iron-scavenger complexes, the incorporation of the nutrient into iron-utilizing proteins, and the storage of iron in iron storage proteins, which can also provide a source of iron when external supplies are limited.
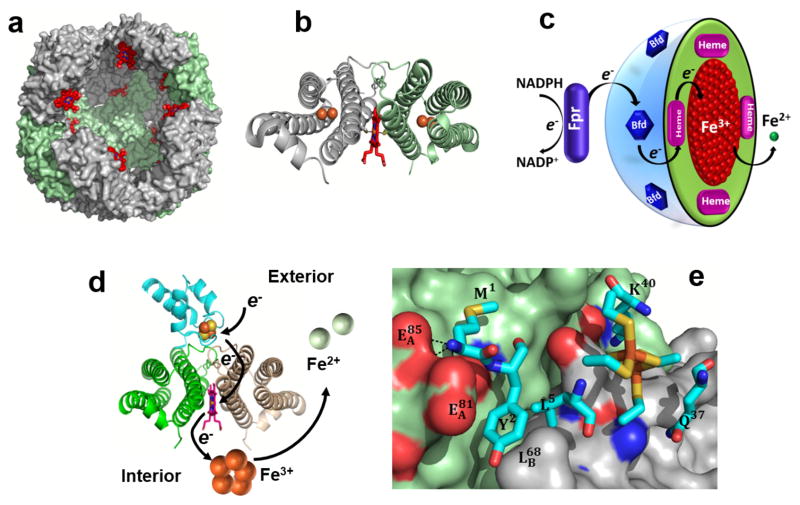
Bacteria store iron in two types of cytosolic ferritin-like molecules, the bacterial ferritins (Ftns) and the bacterioferrins (Bfrs); the latter are found exclusively in bacteria.2 The Ftns and Bfrs are spherical, hollow proteins (~120 Å external diameter and ~80 Å internal diameter) assembled from 24 identical subunits (Figure 1a). Despite their nearly identical subunit fold and quaternary structure, the eukaryotic Ftns, bacterial Ftns and Bfrs share <20% sequence similarity, which strongly influences the charge distribution, packing and function of the 24-mer assemblies.8,9 Eukaryotic Ftns assemble from two types of subunits, H and L, with only the H subunits harboring ferroxidase catalytic centers for oxidizing Fe2+ prior to storage as Fe3+.10,11 Bacterial Ftns and Bfrs are assembled from a single type of subunit and with a ferroxidase catalytic center in each subunit. Only Bfrs bind heme at 2-fold intersubunit sites, where a conserved Met residue from each subunit coordinates the heme-iron (Figure 1b).12
Fig. 1.
Structural organization of P. aeruginosa BfrB and the BfrB:Bfd complex. (a) BfrB is assembled from 24 identical subunits and 12 heme molecules (red) into a nearly spherical molecule with a hollow cavity ~80 Å in diameter where iron is stored in the form of a Fe3+ mineral. (b) Each of the 12 heme molecules (red sticks) binds between two subunits, forming a subunit dimer where the heme-iron is coordinated by a conserved Met residue from each of the subunits. (c) Schematic representation of the reactions delivering electrons originating in NADPH to the core mineral in BfrB. (d) View of a Bfd molecule (cyan) binding BfrB at the interface of two subunits, above each heme, showing schematically how electrons flow from the [2Fe-2S] cluster in Bfd (orange and yellow spheres) to the Fe3+ mineral in the interior cavity of BfrB (orange spheres), which promotes the mobilization of Fe2+ (green spheres). (e) Close-up view of the BfrB:Bfd interface depicting Bfd in cyan sticks and the BfrB surface in green (subunit 1) and gray (subunit 2). The view illustrates the cleft on the BfrB surface formed by residues L68 and E81 where Y2 and L5 from Bfd anchor; the [2Fe-2S] cluster of Bfd is depicted in orange and yellow sticks.
We have focused on understanding the function of iron storage proteins in Pseudomonas aeruginosa,8 a Gram-negative opportunistic pathogen that causes serious infections in immune-compromised individuals, burn victims, and cystic fibrosis patients.13 P. aeruginosa encodes an Ftn-like gene originally termed bfrA, and a Bfr-like gene termed bfrB.14 Structural characterization of the proteins showed that the products of bfrA and bfrB assemble into independent 24-mer proteins with similar subunit fold and quaternary structures. One important difference however, is the fact that the protein coded by bfrB binds heme, whereas that coded by bfrA does not. The structures also revealed that the ferroxidase centers in the protein coded by bfrB are identical to those seen in bacterioferritins, whereas the ferroxidase centers of the protein coded by bfrA are reminiscent of those present in bacterial Ftns. These observations established that the protein coded by bfrA is a bacterial ferritin (the gene has accordingly been re-named ftnA), and indicated the coexistence of two distinct 24-mer iron storage proteins in P. aeruginosa, a bacterial ferritin (FtnA) and a bacterioferritin (BfrB).14
In P. aeruginosa the ftnA gene is adjacent to the katA gene, which codes for a heme catalase (KatA). It is interesting that a ftnA-null mutant of P. aeruginosa exhibits 50% the catalase activity of the wild type cells, an observation that led to the speculation that iron from FtnA (formerly BfrA) is incorporated into protoporphyrin IX to make the heme cofactor for KatA.15 The bfrB gene is adjacent to the bfd gene, which codes for Bfd, a [2Fe-2S] ferredoxin (bacterioferritin-associated ferredoxin). Early in vitro investigations showed that recombinant E. coli Bfd binds to E. coli Bfr, an observation that was interpreted to suggest that Bfd may function either as electron donor to the Fe3+ stored in Bfr for the mobilization of Fe2+, or as electron acceptor to facilitate oxidation of Fe2+ for storage in Bfr.16–18 While mining the genetic response of P. aeruginosa to high and low iron conditions, we noticed that of the ~120 genes up-regulated by low iron conditions, the expression of bfd is stimulated ~200-fold, and that of a gene coding for a ferredoxin reductase (fpr) is increased 3-fold.20, 21 The strong up-regulation of bfd in response to low iron led us to propose a model where Bfd accepts electrons from the ferredoxin reductase (Fpr) and transfers these to the Fe3+ mineral stored in BfrB, enabling the mobilization of Fe2+ from BfrB to the cytosol (Figure 1c).19 We cloned the genes, characterized the BfrB, Bfd and Fpr proteins biochemically and structurally,20–22 and showed that Bfd, Fpr and NADPH are necessary and sufficient to mobilize iron from BfrB in vitro.19,23,24 The X-ray crystal structure of the BfrB:Bfd complex23 showed a biological assembly where 12 Bfd molecules bind a 24-mer BfrB; each Bfd occupies an identical site on BfrB, at the interface of a subunit dimer, above a heme molecule (Figure 1d). Additional characterization of the BfrB:Bfd complex in solution showed that the 12 Bfd-binding sites on BfrB are equivalent and independent, and that the BfrB:Bfd interaction is characterized by a dissociation constant (Kd) of approximately 3 μM.24 Analysis of the interface showed that L68 and E81 in BfrB form a continuous set of interactions with M1, Y2 and L5 in Bfd (Figure 1e). The Kd values for the association between Bfd and the E81A and L68A mutants of BfrB are ~100-fold higher than wt BfrB, and the affinity of Bfd for the L68A/E81A double mutant of BfrB is undetectable.24 In agreement, iron mobilization from the L68A/E81A BfrB mutant is completely inhibited in vitro, which indicates that the L68A/E81A mutation in BfrB blocks the BfrB:Bfd interaction and inhibits iron mobilization from BfrB.24
In this study we capitalized from the insights obtained from our previous investigations in vitro to study the contributions of BfrB and the BfrB:Bfd interaction to iron homeostasis in P. aeruginosa cells. This was carried out by examining the consequences of deleting the bfrB gene or by inhibiting the BfrB:Bfd interaction in P. aeruginosa cells by two methods: (i) deleting the bfd gene or (ii) replacing the wild type bfrB gene with a L68A/E81A double mutant bfrB allele at the native site in the P. aeruginosa chromosome. The results show that P. aeruginosa deposits iron reserves in BfrB, but not in FtnA, and that these reserves are mobilized in late exponential and early stationary growth phases, when iron concentrations in the environment are depleted. Iron stored in BfrB provides a source of iron for P. aeruginosa growth in iron limiting conditions but utilization of the stored nutrient requires a functional BfrB:Bfd interaction. The data also shows that inhibition of the BfrB:Bfd interaction in P. aeruginosa causes the irreversible accumulation of iron in BfrB and severely depletes free intracellular iron levels, which triggers an acute iron starvation response. We interpret the findings to suggest a model whereby the function of BfrB in P. aeruginosa is not simply iron storage; rather, BfrB and its interactions with Bfd function to establish a dynamic equilibrium between Fe3+ compartmentalized in BfrB and cytosolic free iron (Fe2+) that acts as a buffer to regulate cytosolic levels of Fe2+.
Experimental
Media and growth conditions
Chemicals were obtained from Fisher Scientific unless otherwise stated. All strains were kept on Pseudomonas Isolation Agar (PIA) (BD Biosciences). Pseudomonas Isolation (PI) media (20 g/L peptone, 2.99 g/L MgCl2•6H2O, 10 g/L K2SO4, 25 mg/L irgasan (Sigma-Aldrich), and 20 mL/L glycerol, pH 7.0) was used for normal growth conditions. Colorimetric analysis26 showed that PI media contains 1.15 mM phosphate, which originates from the peptone. Unless otherwise indicated PI media was supplemented with iron by addition of a small volume of 20 mM (NH4)2Fe(SO4)2 (pH ~2.0) stock solution to give a final concentration of 10 μM in Fe. Cultures (50 mL PI media in 250 mL Erlenmeyer flasks) were inoculated with a 5 mL overnight culture grown in PI media to an optical density at 600 nm (OD600) of 0.001 and incubated with shaking at 230 rpm and 37 °C. For cultures in low iron conditions, glassware was rinsed with 1% TraceSelect nitric acid (Sigma) and then rinsed 5 times with nanopure water. Media for low iron conditions (1L) was prepared using 24 g HEPES, 0.93 g (NH4)2SO4, 3.245 g succinic acid, 615 mL of 2 mM K2HPO4 (Sigma-Aldrich) and 385 mL of 2 mM KH2PO4 (Sigma-Aldrich) at pH 7.0. The following trace metals were added: 2 mL of 1 M MgSO4, 100 μL of 4 M CaCl2, 10 μL 15 mM ammonium molybdate, 1 mL of 17 mM EDTA, 300 μL of 10 mM CuSO4, 100 μL of 20 mM Co(NO3)2, 100 μL of 94 mM Na2B4O7, and 100 μL of 76 mM ZnSO4.
Strains
Bacterial strains and plasmids used in this study are listed in Table 1 and Table S1, respectively, and primers in Table S2. Pseudomonas aeruginosa PAO1-UW25 was purchased from the University of Washington Genome Center. PAO1-derived strains with unmarked, in-frame deletions of bfrB or bfd or a variant bfrB allele encoding BfrB(L68A/E81A) were made using methods described previously.27 Briefly, the plasmid pEXG228 was used to deliver each of the alleles to the P. aeruginosa chromosome. The pEXG2 plasmids used to construct the bfrB and bfd deletion strains were made using PCR-amplified DNA products flanking each gene. The pEXG2 plasmid used to introduce the gene coding BfrB(L68A/E81A) was made using a synthetic DNA fragment (Genescript) identical to bfrB except with three base substitutions; C202G and T203C to create the L68A mutation and A242C to create the E81A mutation. The pEXG2 derivatives were transformed into E. coli RHO29 and then crossed into PAO1 by mating. Transconjugants were selected on Pseudomonas isolation agar containing gentamicin, and deletion mutants were selected using no-salt LB agar containing 10% (wt/vol) sucrose. To complement the bfrB and bfd mutations, we introduced an IPTG-inducible bfrB or bfd gene to the neutral att site using the pUC18-miniTn7-LAC plasmid as described previously30 and then removed the gentamicin resistance marker using plasmid pFLP2 as described previously.31
Table 1.
P. aeruginosa Strains
| Strains | Description | Reference |
|---|---|---|
| PAO1 | Wild type strain | (25) |
| PAO1 ΔbfrB | PAO1 containing an unmarked, in-frame bfrB deletion | This study |
| PAO1 Δbfd | PAO1 containing an unmarked, in-frame bfd deletion | This study |
| PAO1 bfrB(L68A/E81A) | PAO1 harboring a gene encoding the BfrB L68A/E81A allele at the native bfrB locus | This study |
| PAO1ΔbfrB attn7::PlacbfrB | Made by introducing pUC18-miniTn7T-LAC bfrB to PAO1 ΔbfrB and removing the GentR marker | This study |
| PAO1 Δbfd attn7::Placbfd | Made by introducing pUC18-miniTn7T-LAC bfd to PAO1 Δbfd and removing the GentR marker | This study |
Growth curves
Cells were cultured in 250 mL PI media supplemented with 10 μM Fe as described in Media and Growth Conditions. Samples (100 μL) from 250 mL PI cultures were removed every 2 h, serially diluted in phosphate buffer saline (PBS pH 7.4), and plated on PIA plates. Plates were incubated for 18 h at 37 °C and colony forming units per mL enumerated. Growth curves in low iron media were obtained as follows: Pre-cultures (5 mL) were grown at 37 °C in PI media supplemented with 10, 20, or 30 μM Fe for 24 h, shaking at 230 rpm. Cells were then washed twice by centrifugation (13,000 rpm, 10 min) in ice-cold low iron media. Washed cells were resuspended in 1 mL ice-cold, low iron media, diluted to an OD600 = 0.01 and transferred to 96 well plates (200 μL/well). Cultures were then grown at 37 °C in a BioTek EPOCH2 microplate reader, shaking at 205 cpm and reading OD600 every 15 min.
Imaging iron stored in BfrB
P. aeruginosa cells were cultured in 250 mL PI media supplemented with 10 μM as described in Media and Growth Conditions. Samples (15 mL) were collected at 6, 8, 10, 12, 24, 36, and 48 h post inoculation and centrifuged for 15 min at 4,000 rpm and 4 °C. Cells were resuspended in 1 mL PI media, transferred to 1.7 mL microcentrifuge tubes and centrifuged for 10 min at 13,300 rpm and 4 °C; the cell pellets were then frozen at −80 °C. Frozen cell pellets were thawed at room temperature and lysed by addition of 300 μL of lysis buffer (Tris-base buffer (50 mM, pH 8.0) containing 10% glycerol, lysozyme (20 mg/mL), DNAse (0.2 mg/mL) (Gold Bio), NaCl (0.1 M), 1 mM MgSO4 and 1% Triton-X100 (Sigma)) and incubation at 25 °C (90 min), and then at 37 °C (30 min). The resultant lysates were centrifuged for 10 min at 13,300 rpm, the supernatant filtered through 0.45 μm filtration membrane, and the filtrate (100 μL) mixed with 10 μL of loading dye (5.9 mL water, 0.5 mL glycerol, 0.4 mL β-mercaptoethanol, 0.4 mL 1% bromophenol blue, and 0.5 mL 1 M Tris-HCl pH 6.8). The resultant solutions (100 μL) were loaded onto 2 mm-thick native PAGE gels (4% stacking gel and 8% resolving gel) for separation. Electrophoresis was carried out at 60 V for 9 h in a cold room (4 °C), and the gels were stained with Ferene S32 in the dark by immersion (10 min) in a solution containing 0.049 g Ferene-S, 250 μL thioglycolic acid, 2.4 mL acetic acid and 100 mL water.
Analysis of secreted pyoverdin
The release of pyoverdin from P. aeruginosa strains was studied in plates and in liquid culture. In the case of plates, single colonies were grown overnight (14 h) in 5 mL LB broth. 100 μL of the culture was serially diluted in PBS and ten drops (10 μL each) of the 106 dilution were plated on PIA plates, dried at room temperature and incubated in the dark at 37 °C for 24 h. Pyoverdin secreted by the cells was imaged by photographing the yellow-green fluorescence of pyoverdin stimulated in a UV trans-illuminator. To measure the release of pyoverdin in liquid culture, cells were incubated in 250 mL PI media supplemented with 10 μM Fe. At 12, 24, 36, and 48 h, 1 mL of culture was removed and centrifuged for 3 min at 13,300 rpm and 4 °C. The supernatant was treated with 1 mL of chloroform to extract pyocyanin from the culture supernatants, the aqueous phase was recovered, diluted 1000-fold in 200 mM Tris buffer (pH 7.4) containing 5 mM EDTA, and the fluorescence of pyoverdin was measured with excitation at 400 nm. To account for small differences in cell density among the different strains, fluorescence intensity (FI) at 460 nm was normalized to cell count CFU/mL (FI/CFU/mL).
Cellular iron levels
Total levels of cellular iron were measured at 12 and 18 h post-inoculation using a published method33 with small modifications: Cultures (50 mL PI media supplemented with 10 μM Fe in 250 mL Erlenmeyer flasks) were grown as described in Media and Growth Conditions. The cultures were centrifuged for 45 min at 4,000 rpm and 4 °C. The cell pellets were washed by resuspension and centrifugation, once in 4.5 mL ice-cold PI media containing 10 mM diethylenetriaminepentaacetic acid (DTPA) (Sigma-Aldrich), once in 5 mL ice-cold PBS buffer, and once in 5 mL ice-cold nanopure water, prior to overnight storage at −20 °C. The cell pellets were thawed at ambient temperature, resuspended in 1.0 mL lysis buffer (10 mg/mL lysozyme, 50 mM Tris-base (pH=8.0), 100 mM NaCl, 1 mM EDTA and 1% Triton-X100), and incubated at 37 °C for 1 h. The lysed cells were then treated with 1.0 mL of freshly prepared digestion reagent (5% concentrated HCl and 2.25% w/v KMnO4 in water), thoroughly mixed by vortexing, and then incubated at 65 °C for 6 h. The resultant solutions were cooled to room temperature, treated with 300 μL of iron detection reagent (6.5 mM ferrozine, 15.4 mM neocuproine, 2 M ascorbic acid, and 5 M ammonium acetate (Sigma-Aldrich)), incubated for 20–30 min at room temperature, and centrifuged for 15 min at 4,000 rpm. The iron concentration of the resultant solution was measured from the absorbance of the Fe2+- ferrozine complex at 564 nm (ε564 = 27.9 mM−1cm−1) using a Varian-Cary 50 Bio UV-vis spectrophotometer, normalized by cell population and reported as Fe atoms/CFU. The colorimetric determination of total intracellular iron using ferrozine offers a sensitive, accurate and low cost analytical technique,33,34 which has been shown to produce results similar to those obtained by atomic absorption spectrometry.35
Levels of free intracellular iron were measured at 12 and 18 h post-inoculation using a previously reported whole-cell electron paramagnetic resonance (EPR) spectroscopy method,36–38 with some modifications in sample preparation: Cultures (50 mL PI media supplemented with 10 μM Fe in 250 mL Erlenmeyer flasks) were grown as described in Media and Growth Conditions. The cultures were centrifuged for 45 min at 4,000 rpm and 4 °C. The cell pellets were suspended in 5 mL ice-cold PI media (not reconstituted with Fe) containing 10 mM DTPA and 20 mM of the intracellular iron chelator desferroxamine mesylate (DFO) and incubated with shaking for 10 min at 37 °C and 230 rpm. The cells were then centrifuged and washed twice with 10 mL ice-cold PBS buffer (pH 7.4), suspended in 200 μL of ice-cold PBS containing 10% glycerol, loaded onto 4 mm quartz EPR tubes, and immediately frozen by immersion in a dry-ice/acetone bath. EPR spectra of whole cells were acquired at 5 K in a Bruker EMXplus spectrometer equipped with an Oxford cryostat using the following parameters: field center, 1610 G; field sweep, 400 G; modulation frequency, 100 kHz; modulation amplitude, 8 G; time constant, 20.48 ms; receiver gain, 60 dB; power, 2.000 mW. A standard curve was generated using identical parameters and solutions containing 0, 5, 10, 20, and 30 μM Fe3+ (from Fe2(SO4)3·7H2O) and 2 mM DFO in 20 mM Tris buffer (pH 7.4) and 10% (v/v) glycerol.
Iron in spent media
To determine the concentration of iron in spent media, cultures were grown in PI media supplemented with 10 μM Fe, as described in Media and Growth Conditions. Samples (3 mL) obtained at different times (0 h, 12 h, 18 h, 24 h, 30 h, and 35 h) were centrifuged at 4,000 rpm for 45 min. The cell-free media was freeze-dried (~16 h) using a Savant Speed Vac SC110, treated with 500 μL digestion reagent (see above), and incubated for 6 h at 65 °C. The samples were cooled to room temperature, treated with 100 μL of iron detection reagent (see above), incubated for 30 min, and centrifuged for 5 min at 13,000 rpm. The iron concentration of the resultant solution was measured from the absorbance of the Fe2+-ferrozine complex at 564 nm (ε564 = 27.9 mM−1cm− 1) using a Varian-Cary 50 Bio UV-vis spectrophotometer.39
Results and discussion
BfrB is the main iron storage protein in P. aeruginosa
Our studies in vitro indicated that Bfd is required to mobilize iron stored in BfrB (see Figure 1).19 Additional structural investigations defined the crucial interface of the BfrB:Bfd complex23 and analysis of this interface allowed us to demonstrate that mutation of two hot-spot residues in BfrB (L68A/E81A) is sufficient to block the BfrB:Bfd interaction and inhibit iron mobilization from BfrB.24 To test these ideas in P. aeruginosa cells, and to investigate the consequences of removing BfrB, Bfd or the BfrB:Bfd interaction on iron homeostasis, we constructed mutant P. aeruginosa strains with unmarked, in-frame deletions of the bfrB (ΔbfrB) or bfd (Δbfd) genes, and a strain with the bfrB gene encoding the L68A/E181A mutations (bfrB(L68A/E81A)) (see Experimental). When wild type P. aeruginosa or mutant strains are cultured in iron sufficient media (PI media supplemented with 10 μM Fe), all the strains show nearly identical growth (Figure 2), although, as will be shown below, all three of the mutants grow significantly more slowly than wild type in iron-deplete conditions.
Fig. 2.
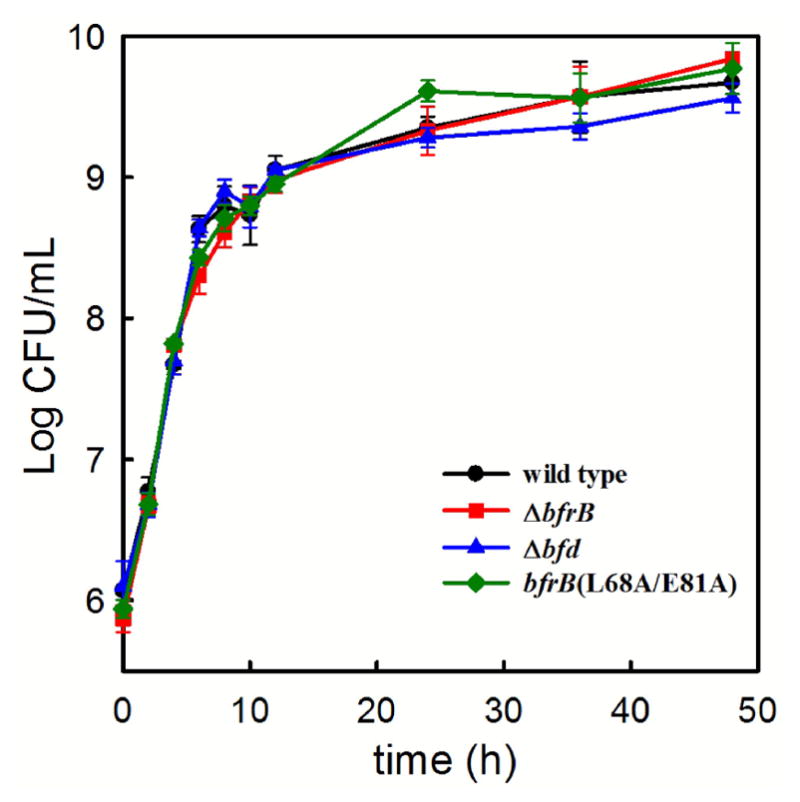
The wild type and mutant cells grow at the same rate and to similar cell density in iron sufficient growth media (PI media supplemented with 10 μM iron). Error bars represent the standard deviation of three independent measurements.
The genomes of many bacteria harbor genes coding for at least two types of 24-mer ferritin-like proteins, the bacterial ferritinis (Ftn) and the bacterioferritins (Bfr). Biochemical and structural characterization has shown that the subunits of Ftns and Bfrs share an identical fold (see Figure 1A) and assemble into very similar quaternary structures consisting of 24 subunits, which are also shared with the eukaryotic ferritins. This shared quaternary structure has contributed to the widespread notion that in bacteria Ftns and Bfrs function as iron storage proteins; this notion has been reinforced by in vitro observations showing that both Ftns and Bfrs can oxidize Fe2+ and deposit a Fe3+-mineral in their interior cavity. Although the reasons why bacteria would require two types of 24-mer iron storage proteins remain enigmatic, attempts to address the issue have led to the suggestion that the specific physiological roles played by Ftns and Bfrs are distinct in different bacteria.40 For example, whereas FtnA is the main iron accumulator in E. coli cells,35 the same role has been attributed to Bfr in Salmonella enterica typhimurium.41
There are two 24-mer ferritin-like molecules in P. aeruginosa, FtnA and BfrB.14 To investigate their relative contributions to iron storage in the cell, we followed the incorporation of iron into FtnA and BfrB by culturing the wild type and each of the three mutant strains in iron sufficient media (PI media supplemented with 10 μM Fe), harvesting and lysing cells at different time points, loading clarified supernatant onto a native PAGE gels for separation, and staining with Ferene S, which stains iron in ferritin but does not stain heme-iron.32 Recombinant FtnA and BfrB, each mineralized with an iron core of approximately 400 iron ions were used as standards to show that the electrophoretic mobility of the two proteins is clearly discernible in the native PAGE gels (Figure 3). Lanes loaded with wild type cell lysate solutions show iron-stained bands migrating with the electrophoretic mobility of BfrB; the bands become clearly detectable in mid-exponential growth phase (> 6 h). Surprisingly, bands corresponding to the electrophoretic mobility of FtnA are not detectable in the P. aeruginosa cell lysates, even if the cells are cultured in iron rich (30 μM Fe) media (Figure S1 in supplementary material). These findings indicate that despite the presence of two potential iron accumulators in P. aeruginosa cells (BfrB and FtnA), the function of iron storage, at least under our laboratory conditions, rests mainly on BfrB. In agreement with the idea that FtnA does not contribute significantly to iron storage, there is no detectable iron accumulation in the ΔbfrB mutant cells, regardless of whether the cells are cultured in iron sufficient (10 μM) (Figure 3) or in iron rich (30 μM) media (Figure S1). We had anticipated that in cultures of the ΔbfrB mutant, the other presumed iron storage protein in P. aeruginosa, FtnA, would compensate for the absence of BfrB and accumulate detectable levels of iron, especially when the cells were cultured in iron rich media. Surprisingly, the native PAGE gels revealed that even when BfrB is absent, P. aeruginosa cells do not accumulate iron in FtnA, or in other ferritin-like molecule, to levels that are detectable by the Ferene-S stain. Consequently, we conclude that BfrB functions as the main iron storage protein in P. aeruginosa, a conclusion that is in agreement with previous observations indicating that whereas the bfrB gene is upregulated in response to high iron levels in the environment, expression of the ftnA gene does not respond to external iron stimuli.42,43 The function of FtnA in P. aeruginosa remains unknown; the only report addressing its possible function suggests that FtnA is somehow important in enabling function of the heme catalase (KatA),15 but the details of how this is accomplished remain to be explored in detail.
Fig. 3.
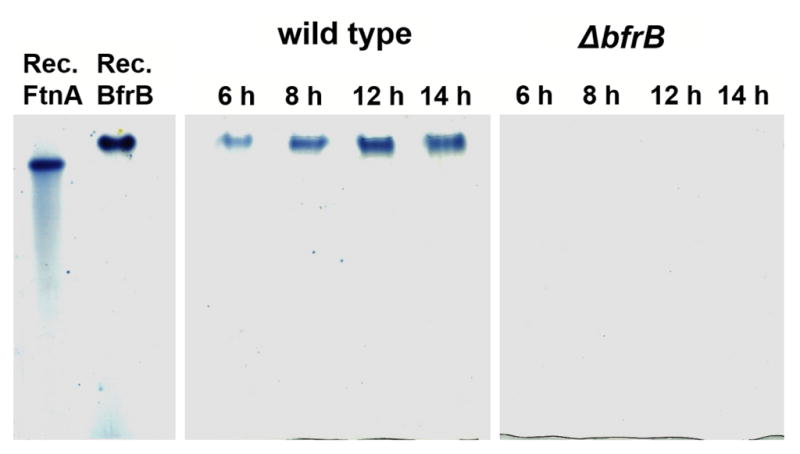
Visualizing iron stored in ferritin-like molecules with the aid of native PAGE gels stained with Ferene-S. Left: Recombinant FtnA (Rec. FtnA) and recombinant BfrB (Rec. BfrB) exhibit different electrophoretic mobility and can be resolved in native PAGE gels. The iron-chelating agent Ferene-S stains iron stored in the central cavity of both recombinant proteins blue. Center: Lanes loaded with lysates of wild type P. aeruginosa cultured in PI media supplemented with 10 μM Fe and harvested at different times post inoculation only show iron-stained bands migrating with the electrophoretic mobility of BfrB. Right: Lanes loaded with lysates of ΔbfrB cells cultured in PI media supplemented with 10 μM Fe and harvested at different times post inoculation show that iron is not accumulated in FtnA, or in any other protein.
E. coli Bfr and P. aeruginosa BfrB are the best characterized bacterioferritins.8,12,40,44 In contrast to our findings with P. aeruginosa, FtnA is the main iron accumulator in E. coli, while the function of Bfr remains unknown.35 The distinct physiological roles played by P. aeruginosa BfrB and E. coli Bfr are fascinating in the context that structural and biochemical characterization of these proteins have revealed important differences, despite very similar quaternary structures. In E. coli Bfr the ferroxidase centers are stable once formed and function as a catalytic site for O2 reduction.40,45,46 In comparison, the seemingly structurally identical ferroxidase centers in BfrB are not stable and function in a dual role of O2 reduction and transit pore for the passage of Fe3+ ions into the interior structural cavity.8,12,20 Additional structural and dynamic characterization of BfrB revealed long-range dynamic communication between pores in the BfrB structure and ferroxidase centers. These concerted motions appear to be crucial to impart the ferroxidase centers in BfrB with the flexibility necessary to capture and oxidize Fe2+, and subsequently allow passage of Fe3+ into the central cavity.8,47,48 It is therefore possible that seemingly subtle amino acid sequence differences between E. coli Bfr and P. aeruginosa BfrB impart the 24-mer proteins with different dynamic properties, which provide the corresponding ferroxidase centers with their distinct chemical properties. These unique properties in turn may be important determinants of the physiological role of the corresponding bacterioferritins. Although these ideas require the structural, biochemical and functional (in cell) characterization of additional systems, comparison of the dynamic properties of eukaryotic and bacterial Ftns and Bfrs has already suggested that amino acid sequence indeed exerts important dynamic and potentially functional properties to 24-mer ferritins.9 Hence, future efforts aimed at the detailed characterization of Ftns and Bfrs from other organisms may prove important to understand how the structure and dynamics of these complicated proteins contribute to fine-tuning their physiological function in bacterial cells.
Bfd is required for the mobilization of iron from BfrB in P. aeruginosa cells
This aspect of the work builds from our previous biochemical and structural work, which showed in vitro that Bfd and Fpr are sufficient to mobilize iron from BfrB, in a process that requires specific interactions between BfrB and Bfd to enable electron transfer from the [2Fe-2S] cluster in Bfd to the Fe3+ mineral stored in the central cavity of BfrB (see Figure 1c).19,23 In addition, the BfrB:Bfd association, which occurs with a Kd of 3 μM in vitro, is completely inhibited when residues L68 and E81 in BfrB are mutated to alanine.24 Consequently, to interrogate the consequences of perturbing the BfrB:Bfd interaction in P. aeruginosa cells we followed two strategies: (i) deleting the bfd gene (Δbfd) and (ii) inhibiting the BfrB:Bfd interaction by introducing a bfrB variant allele encoding the the L68A and E81A mutations to the P. aeruginosa chromosome (bfrB (L68A/E81A)).
To evaluate how iron is mobilized from BfrB in P. aeruginosa we resorted to native PAGE gels stained with Ferene S to examine the accumulation of iron in BfrB in the wild type and in each of the mutant cells harvested at different times post inoculation. The results show that in the wild type cells the amount of iron accumulated in BfrB reaches a maximum ca. 12 h post inoculation (early stationary phase). Beyond this point, the accumulated iron disappears from the cell lysates (Figure 4a). We presume this is because iron is mobilized from BfrB when the nutrient becomes scarce in the medium. Similar experiments carried out with the Δbfd and bfrB(L68A/E81A) strains also show that iron is stored only in BfrB (Figures 4b and 4c). In contrast to the observations made with the wild type cells, however, the iron accumulated in BfrB is not mobilized from BfrB. Therefore, in addition to corroborating the ideas inferred from the in vitro studies regarding the function of Bfd and the BfrB:Bfd interaction in the mobilization of iron reserves from BfrB, the findings also demonstrate that the BfrB:Bfd interaction is the only mechanism that enables P. aeruginosa cells to mobilize iron reserves from BfrB.
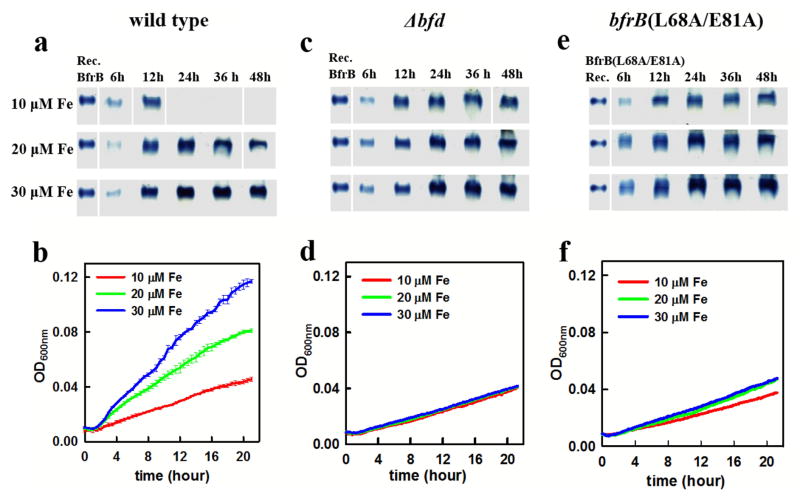
Fig. 4.

Monitoring the deposition and subsequent mobilization of iron in BfrB using native PAGE and Ferene-S staining. Wild type and mutant cells were cultured in PI media supplemented with 10 μM Fe. (a) Wild type cells deposit iron in BfrB during late exponential and early stationary growth phases, which are subsequently mobilized in late stationary phase. In contrast, the deposition of iron in BfrB in cultures of (b) Δbfd and (c) bfrB(L68A/E81A) cells is irreversible. (d) ΔbfrB cells do not accumulate iron, but (e) complementing ΔbfrB cells by expressing BfrB from an IPTG-inducible bfrB gene inserted into neutral site in the genome restores iron accumulation in BfrB followed by its mobilization in late stationary phase. (f) Δbfd cells accumulate iron irreversibly in BfrB, but (g) addition of IPTG at 12 h post inoculation (red arrow) induces the expression of Bfd from an IPTG-inducible bfd gene inserted into a neutral site of the genome and promotes iron mobilization from BfrB).
We were able to restore the phenotypes of the ΔbfrB and Δbfd mutants by expressing the missing gene from an IPTG-inducible lac promoter at another site in the P. aeruginosa chromosome (att). In the absence of IPTG, there was no iron accumulation in the ΔbfrB mutant with the lac-promoter-driven bfrB gene (Figure 4d). However, adding IPTG (1 mM final concentration) to induce BfrB expression resulted in iron accumulation during exponential and early stationary phases, which is later mobilized (Figure 4e), similar to that of wild type cells. The timing of maximum accumulation is not identical to that in the wild type cells, presumably because the IPTG was present from the start of the culture and this might have altered the normal pattern of BfrB expression. Similarly, inducing the expression of a lac-promoter-driven bfd gene in the Δbfd mutant by adding IPTG (1 mM final concentration) to cultures at the 12 h time point (indicated by the red arrow) induces the mobilization of iron stored in BfrB, similar to wild type (Figures 4e and 4g).
Inhibiting the BfrB:Bfd interaction in P. aeruginosa causes bacterial iron deficiency
The results so far support the conclusion that Bfd and the Bfd interaction with BfrB are required to mobilize iron from BfrB. We surmised that the mobilization of iron from BfrB might be critical to maintain iron homeostasis in the cell, and therefore predicted that mutants that fail to mobilize iron from BfrB would have low levels of free iron in the cytosol, which in turn would stimulate the synthesis and secretion of iron scavenging molecules, such as pyoverdin (Pvd) and pyochelin (PCH), the main siderophores secreted by P. aeruginosa to scavenge Fe3+ from the environment.49 Pvd exhibits significantly higher affinity for Fe3+ than PCH,50 which is produced at much lower levels than Pvd, even by strains growing in the lungs of cystic fibrosis patients.51 A striking phenotypic characteristic of P. aeruginosa cells challenged with low iron conditions is the secretion of Pvd, which exhibits a characteristic green-yellow color with absorbance at 400 nm and fluorescence at 460 nm. Therefore, we chose to follow the time-dependent secretion of Pvd as a reporter of the relative levels of iron starvation sensed by the wild type and mutant cells. To initiate testing of these ideas, we compared the relative amounts of Pvd secreted by the wild type and each of the three mutants by spotting cells on PIA plates, culturing for 22 h at 37 °C, and visualizing the fluorescent emission by illuminating the plates from the bottom with UV light. Results from these experiments (Figure S2) show that both the Δbfd and the bfrB(L68A/E81A) strains secrete Pvd more abundantly than wild type P. aeruginosa, observations that are in agreement with the idea that disrupting Bfd or the BfrB:Bfd interaction traps iron irreversibly in BfrB and causes the cells to sense low iron conditions and upregulate the synthesis of siderophores. It is also interesting to note that the ΔbfrB cells secrete less Pvd than the wild type cells, an observation that is addressed below.
To corroborate that similar observations can be made in liquid media, we cultured cells in PI media supplemented with 10 μM Fe and monitored the secreted Pvd using fluorescence spectrophotometry. As indicated above, when cultured under these conditions wild type and mutant cells grow similarly and reach stationary phase in approximately 12 h with comparable cell densities (Figure 2). Plotting the time-dependent secretion of Pvd normalized to cell density (Figure 5a) shows that none of the strains secrete detectable levels of Pvd during exponential growth (<12 h). Differences in the levels of secreted Pvd become evident at 24 h post inoculation and become largely exacerbated at longer culture times, with the Δbfd mutant and the bfrB(L68A/E81A) variant secreting 3- to 4-fold larger amounts of Pvd relative to wild type. These observations suggest that in these mutants iron starvation is sensed more acutely than in identically grown wild type. To understand the relationship between the timing of Pvd release and the availability of iron in the media, we analyzed the concentration of iron in the spent media as a function of time. The results (Figure 5b) show that wild type, the Δbfd, or the bfrB(L68A/E81A) cells all deplete iron in the growth medium at similar rates, reducing the concentration of the nutrient in the growth medium to <1 μM in nearly 20 h, which is approximately the time when Pvd becomes detectable in spent media (Figure 5a). In comparison, ΔbfrB cells deplete iron in the growth medium at a slower rate, such that the concentration of the nutrient is reduced to <1 μM in approximately 30 h, which also corresponds to the approximate time when Pvd becomes detectable in spent media of ΔbfrB cultures. From these observations, it is evident that P. aeruginosa cells sense the need to secrete Pvd for scavenging iron when the concentration of the nutrient in the growth media is ~1 μM.
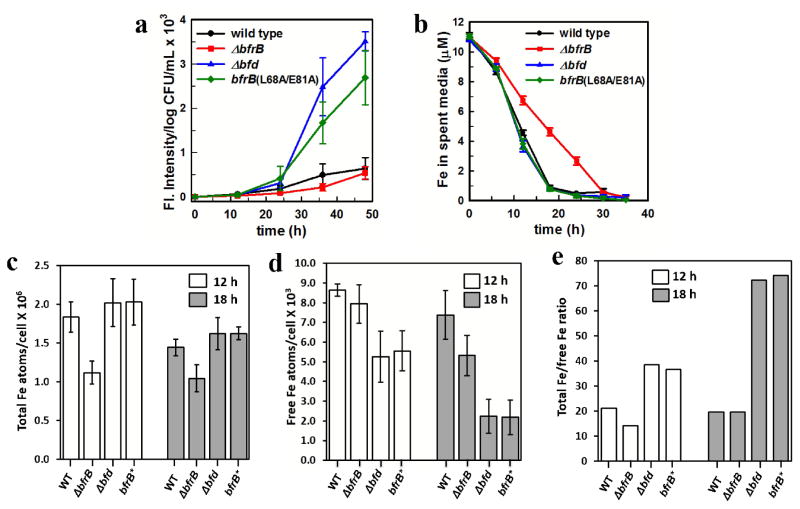
Fig. 5.
Phenotypic manifestations of iron dysregulation in P. aeruginosa strains with deletions in Bfd or BfrB, or a variant with amino acid mutations in BfrB that abolish Bfd binding (bfrB(L68A/E81A)). In (c – e) the bfrB(L68A/E81A) variant is abbreviated BfrB* for simplicity. Wild type and mutant cells were cultured in PI media supplemented with 10 μM Fe. (a) Disrupting the BfrB:Bfd interaction in the Δbfd or bfrB(L68A/E81A) strains elicits secretion of Pvd at levels 3 to 4-fold larger than wild type cells. (b) Analysis of the time-dependent concentration of iron in the culture media show that Δbfd and bfrB(L68A/E81A) cells deplete iron from the media at the same rate as wild type cells, whereas ΔbfrB cells deplete iron from the media at a significantly slower rate. (c) Total iron and (d) free iron levels in wild type and mutant cells harvested in early (12 h) and late (18 h) stationary growth phase. (e) Ratio of total-Fe/free-Fe in cells harvested in early (12 h) and late (18 h) stationary growth phase. Error bars represent the standard deviation of three independent experiments.
Our findings that both the Δbfd and bfrB(L68A/E81A) strains sense iron deprivation more acutely than wild type is probably a consequence of the unidirectional flux of iron into BfrB when Bfd is absent or when the BfrB:Bfd interaction is disrupted. Unidirectional iron flux into BfrB causes irreversible accumulation of iron in BfrB (Figures 4b and 4c), and is expected to depress the concentration of free iron in the cytosol. To establish a direct correlation between cellular iron levels and the observed phenotype of acute iron deprivation, we measured the levels of free and total intracellular iron in the wild type and mutant cells cultured in PI media supplemented with 10 μM iron. Free intracellular iron is conceived as iron not stably incorporated into proteins, enzymes or iron storage proteins (free iron pool),52 whereas total iron measures all iron in the cell, free and stably incorporated into macromolecules. Cells were harvested at 12 h (early stationary phase), when there is significant iron accumulated in BfrB in both, the wild type and in the mutant cells, and at 18 h (late stationary phase) when most of the iron has been mobilized from BfrB in the wild type cells (see Figure 4a). Levels of intracellular total iron were measured using UV-vis spectrophotometry, as indicated in Methods. The levels of intracellular free iron were measured using whole-cell electron paramagnetic resonance (EPR) spectroscopy, as described in Experimental, using previously reported methodology36–38 which utilizes the cell permeable iron chelator desferroxamine mesylate (DFO). Because DFO binds Fe3+ more strongly than Fe2+, it facilitates the quantitative oxidation of free intracellular Fe2+ to a Fe3+- DFO chelate, which exhibits a sharp first derivative EPR signal with a g-value of 4.3 (Figure S3). The iron content can be quantified from the amplitude of this signal and a standard curve prepared from Fe-DFO standards, and normalized to cell population (CFU).
The results show that cells with bfd deleted or with the bfrB(L68A/E81A) allele have similar levels of total iron relative to the wild type cells at 12 and 24 h, which in Figure 5c are expressed as iron atoms/CFU. In this context, it is noteworthy that the relatively common practice of only analyzing total iron levels would seem to indicate nothing abnormal in the iron metabolism of the Δbfd and bfrB(L68A/E81A) strains. Analysis of free iron levels, however, revealed important differences (Figure 5d): Free iron levels in both the Δbfd and the bfrB(L68A/E81A) cells are ~60% (12 h) and ~25% (18 h) that of wild type cells. It is illustrative to present these results as the ratio of total-Fe/free-Fe (Figure 5e) because comparison of the corresponding ratios makes it evident that wild type cells maintain the total-Fe/free-Fe ratio at ~20, during early and late stationary phases. In contrast, the corresponding ratios in the Δbfd and the bfrB(L68A/E81A) cells are much higher; ~40 at 12 h and ~75 at 18 h, which is a clear indication that irreversible accumulation of iron in BfrB causes iron dyshomeostasis by severely depleting free iron levels in the cytosol. In fact, the “nearly normal” levels of total iron observed in the Δbfd and the bfrB(L68A/E81A) strains are probably due to unusable iron irreversibly trapped in BfrB.
The above-described experimental findings are in excellent agreement with the idea that disrupting Bfd or the BfrB:Bfd interaction results in unidirectional iron flux into BfrB, which causes iron to irreversibly accumulate in BfrB, while simultaneously depressing the levels of free iron in the cytosol. The abnormally low levels of free iron in the cytosol of the Δbfd and the bfrB(L68A/E81A) cells, in turn, are expected to lead to the dissociation of the Fe-Fur complex, with concomitant de-repression of iron acquisition genes, as manifested in the very high levels of Pvd secreted by the Δbfd and the bfrB(L68A/E81A) cells. In this context, it is interesting to note that at 12 h, when the growth medium still contains ~5 μM iron (Figure 5b), the free iron levels in the Δbfd and the bfrB(L68A/E81A) cells are less severely compromised (total-Fe/free-Fe ~40) than at 18 h, because the nutrient can still be readily procured from the medium. As the concentration of iron in the medium decreases to < 1 μM (~18 h), the unidirectional flux of iron into BfrB aggravates the depletion of free iron levels in the cytosol (total-Fe/free-Fe ~75), which prompts the cells to respond by aggressively secreting Pvd (Figure 5a). Consequently, an important outcome of inhibiting the BfrB:Bfd interaction is the irreversible compartmentalization of iron in BfrB, which for all practical purposes renders the nutrient inaccessible to cell metabolism, leading to a paradoxical iron deficiency phenotype despite “nearly normal” levels of total iron the cells.
It is also interesting to consider that the BfrB:Bfd interaction functions to enable a dynamic equilibrium between free Fe2+ in the cytosol and Fe3+ stored in BfrB. As suggested in Figure 6a, this dynamic equilibrium probably functions as a buffer that regulates levels of cytosolic free Fe2+, which in turn are sensed via a dynamic equilibrium between the apo-form of the master iron uptake regulator (Fur) and its Fe2+ complex. Hence, when cytosolic Fe2+ concentrations are similar (or above) the Kd for the dissociation of the Fe2+-Fur complex, iron sufficiency is sensed, enabling (i) iron incorporation into iron-utilizing proteins, (ii) iron accumulation in BfrB, and (iii) repression of genes encoding iron acquisition systems. In this context, it is important to appreciate that the buffering capacity of the equilibrium between Fe2+ in the cytosol and Fe3+ compartmentalized in BfrB opposes large changes in the levels of cytosolic Fe2+, and enables a regulated deployment of iron acquisition systems by Fur. Perturbing the equilibrium between free Fe2+ in the cytosol and Fe3+ in BfrB by inhibiting iron flow out of BfrB (Figure 6b) eliminates the Fe2+-buffer capacity of the system and allows drastic lowering of cytosolic Fe2+ levels (see Figure 5), which in turn lead to complete dissociation of the Fe2+-Fur complex and the concomitant unregulated de-repression of iron acquisition systems. The latter is evident in the aggressive secretion of Pvd from the Δbfd and the bfrB(L68A/E81A) cells, which is several-fold larger that of wild type cells.
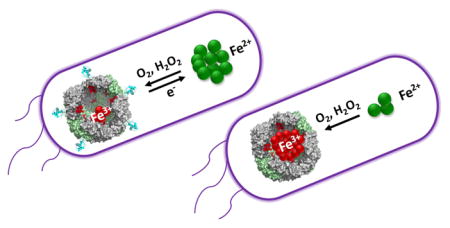
Fig. 6.
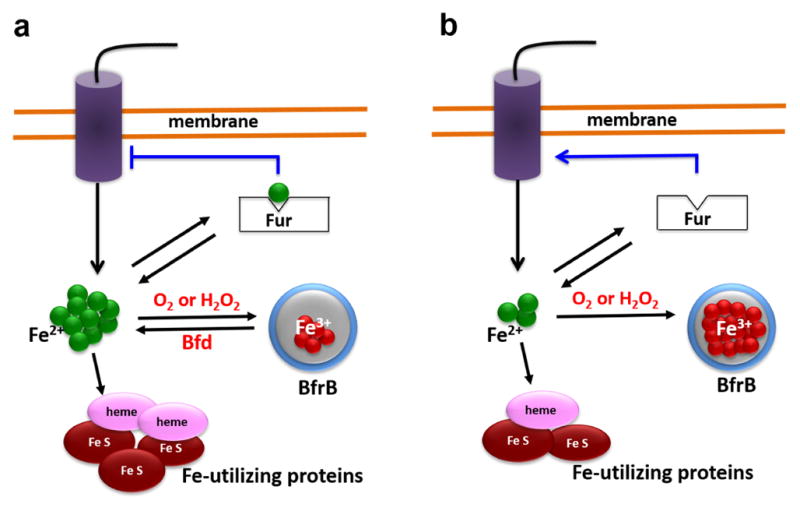
Schematic depiction of iron metabolism in P. aeruginosa. (a) Under conditions of iron sufficiency internalized iron is directed toward the synthesis/repair of iron utilizing proteins and into BfrB. The dynamic equilibrium between free iron (Fe2+) in the cytosol and Fe3+ compartmentalized in BfrB functions as a buffer that maintains the cytosolic Fe2+ concentrations near (above) the value of the dissociation constant (Kd) of the equilibrium between apo-Fur and its Fe-bound form (Fe2+-Fur), thus allowing regulated repression of genes involved in iron scavenging. Under conditions of iron limitation, the upregulation of Bfd synthesis enhances the rate of iron mobilization from BfrB, which contributes to a less precipitous depletion of Fe2+ in the cytosol and to a “measured” de-repression of iron scavenging genes. (b) When the mobilization of iron from BfrB is inhibited, iron that flows into BfrB is irreversibly trapped. Although under conditions of iron sufficiency the cells appear to partially compensate the irreversible trapping of iron in BfrB, under iron limiting conditions, Fe2+ concentrations are rapidly depleted, eliciting an unregulated de-repression of iron scavenging genes. A significant consequence of irreversible iron accumulation in BfrB is loss of the buffering action of the dynamic equilibrium, which causes acute iron deprivation in the cytosol, and non-regulated derepression of iron acquisition genes.
P. aeruginosa cells appear to compensate for the absence of BfrB by depleting iron from the growth media more slowly than wild type cells
The observations made with the ΔbfrB mutant cells are also noteworthy. Total iron levels in ΔbfrB cells are significantly lower at 12 and 18 h post inoculation relative to wild type cells (Figure 5c). The lower levels of total iron observed in ΔbfrB cells is consistent with reports indicating that bacterial strains with null mutations that eliminate a dominant iron storage protein accumulate less iron than the corresponding parental strains.35,41 Surprisingly, analysis of free iron revealed that free iron levels in ΔbfrB cells are similar to wild type at 12 h and somewhat smaller (~75%) at 18 h, and interestingly, the total-Fe/free-Fe ratio in the ΔbfrB cells is similar to that in wild type cells, at 12 and 18h (Figure 5e). These observations suggest that in the absence of a depository (BfrB) where to place iron not immediately utilized in metabolism, P. aeruginosa cells employ yet unidentified mechanisms to prevent accumulation of toxic levels of free iron in the cytosol. The time-dependent depletion of iron from the growth medium (Figure 5b) of ΔbfrB cultures may provide some initial clues regarding this issue: The significantly slow rate of iron depletion from the media compared to wild type cells grown under identical conditions suggests that one mechanism enabling the ΔbfrB cells to fend potential iron-induced toxicity is a slower rate of iron internalization, which could result from slower iron uptake and/or accelerated iron efflux. Regardless, the slower rate at which ΔbfrB cells deplete iron from the growth medium appears to enable the ΔbfrB cells to maintain nearly optimal levels of free intracellular iron, which in turn permit the Fur-Fe2+ complex to maintain iron acquisition genes repressed. In fact, it is important to notice that de-repression of iron acquisition genes in the ΔbfrB mutant, like in wild type cells, also occurs when the concentration of iron in the growth medium reaches ~1 μM, except that in ΔbfrB cultures this value is reached at ~30 h post inoculation (Figure 5b). Beyond this point, Pvd becomes detectable and at 48 h the levels of Pvd secreted by ΔbfrB cells are similar to those seen in cultures of wild type cells. It is therefore possible to conclude that P. aeruginosa cells can compensate for the absence of the main iron storage protein, and it is evident that the compensatory mechanism does not involve iron accumulation in FtnA, which is indicated by the absence of bands in the Ferene S-stained gels of lysates from ΔbfrB cells cultured in the presence of iron sufficient (10 μM) or iron rich (30 μM) growth media (Figures 3 and S1). It is tempting to speculate that the compensatory mechanism involves a slower rate of iron uptake and/or an increased rate of iron efflux. Although iron exporter genes have not yet been identified in P. aeruginosa, its genome encodes several proteins identified as belonging to P-type ATPase and cation diffusion facilitator (CDF) proteins, which exhibit some sequence similarity to corresponding proteins known to function in the transport and tolerance of Fe2+ and Zn2+ in other bacteria.53–55 Investigations directed at probing these questions are currently underway in our laboratory.
Mobilization of iron stored in BfrB promotes the growth of cells challenged with iron limiting conditions
As shown in Figure 7a, wild type P. aeruginosa cells cultured in PI media supplemented with 10 μM iron deposit iron reserves in BfrB, and later mobilize it when environmental iron becomes scarce, such that at 24 h iron stored in BfrB is undetectable in PAGE gels stained with Ferene-S. In comparison, when wild type cells are cultured in PI media supplemented with 20 or 30 μM Fe, the nutrient accumulates in BfrB until ca. 24 h post-inoculation, and although BfrB-iron is gradually mobilized beyond 24 h, it remains readily detectable in PAGE gels at 48 h. The comparison clearly illustrates that whereas P. aeruginosa cells cultured in media supplemented with 10 μM Fe have only small (non-detectable) amounts of iron reserves in BfrB at 24 h, cells cultured in media supplemented with 20 or 30 μM Fe have significant iron reserves at 24 h. We capitalized from these insights to probe whether P. aeruginosa cells utilize iron stored in BfrB to grow when passaged into iron-limiting medium. To this end, we grew pre-cultures of wild type cells in PI media supplemented with 10, 20, or 30 μM iron for 24 h. Cells were washed and then diluted in iron deficient media (< 0.1 μM Fe), transferred to 96-well plates and cultured as indicated in Methods. The growth curves (Figure 7b) show that cultures inoculated with pre cultures grown in 30 μM Fe grow faster and to a higher cell density than cultures inoculated with pre cultures grown in 20 μM Fe, and that cultures inoculated with pre cultures grown in 10 μM Fe grow at the slowest rate and to the lowest cell density. These observations are in excellent agreement with the idea that wild type P. aeruginosa cells utilize iron reserves stored in BfrB to grow when challenged by an iron-limiting environment. The idea is also supported by similar experiments conducted with Δbfd and bfrB(L68A/E81A) cells. As illustrated in Figures 7c, and 7e, these mutants irreversibly accumulate iron in BfrB when cultured in PI media supplemented with 10, 20, or 30 μM Fe. Since iron in BfrB cannot be mobilized for incorporation into metabolism, the growth of Δbfd and bfrB(L68A/E81A) strains is expected to be significantly impaired when challenged with iron deficient conditions. In agreement with these expectations, the corresponding growth curves (Figures 7d and 7f) show that the growth rate of the Δbfd and bfrB(L68A/E81A) cells is significantly slower than that of wild type cells and independent of the concentration of iron used to grow the inoculum. Similar experiments carried out with ΔbfrB cells also show that the growth of the mutant in iron deficient media is significantly slower than that of wild type cells and independent of the concentration of iron used to grow the pre cultures (Figure 8), observations that are in agreement with the above-described findings showing that in absence of BfrB, P. aeruginosa cells do not accumulate iron (Figure 3). Taken together, the findings clearly demonstrate that an important function of BfrB in P. aeruginosa fits within the classical view of iron storage proteins, which accumulate iron for subsequent utilization when the cells are challenged with low iron conditions. Utilization of iron reserves as a source of the nutrient requires Bfd and the BfrB:Bfd interaction, which are necessary to deliver electrons to the ferric mineral stored in BfrB and enable mobilization of Fe2+ into the bacterial cytosol.
Fig. 7.
Iron stored in BfrB promotes the growth of P. aeruginosa cells in iron deficient media. (a) Ferene-stained native PAGE gels of lysates from wild type cells cultured in media supplemented with 10 μM Fe show iron accumulation in BfrB at 12 h and undetectable iron in BfrB at 24 h and beyond, whereas cells cultured in 20 and 30 show significant accumulation of iron in BfrB at 24 h and beyond. (b) The relative growth rates of wild type cells in iron deficient media inoculated with pre cultures grown for 24 h in media supplemented with 10 μM (red), 20 μM (green) and 30 μM (blue) Fe indicate that iron stored in BfrB promotes growth under iron limiting conditions. (c and e) Ferene-stained native PAGE gels of lysates from Δbfd and bfrB(L68A/E81A) cells cultured in media supplemented with 10, 20 or 30 μM Fe show that iron is irreversibly accumulated in BfrB. (d and f) The growth rates of Δbfd and bfrB(L68A/E81A) cells in iron deficient media are significantly slower than those of wild type cells and independent of the iron concentration in the media used to grow the pre cultures. Error bars represent the standard deviation of three independent experimets; in d and f error bars are omitted for simplicity.
Fig. 8.
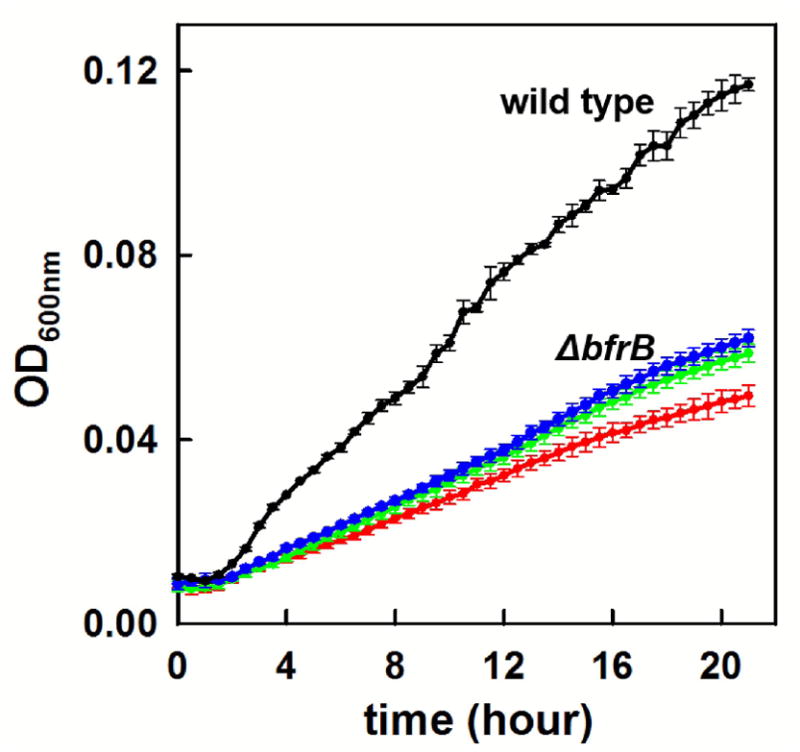
The growth rates of ΔbfrB cells in iron deficient media are significantly slower than those of wild type cells and independent of the iron concentration in the media used to grow the pre cultures; 10 μM (red), 20 μM (green), 30 μM (blue). For comparison the growth rate of wild type cells in iron deficient media inoculated with pre cultures grown in 30 μM Fe is shown in black. Error bars represent the standard deviation of three independent experiments.
Conclusions
Significant progress has been made since Stiefel and Watt first revealed the existence of iron storage proteins in bacteria.56 Despite these advancements, important questions regarding the roles played by iron storage proteins in the bacterial cell remain unknown. Insights from studying iron storage proteins from a select number of organisms in vitro and in bacterial cells indicate that distinct bacteria employ either Ftn or Bfr as the main iron accumulator protein. Importantly, in all cases where the function of iron storage has been experimentally attributed to either Bfr or Ftn, the function of the other Ftn-like molecule remains enigmatic. The work summarized here shows that BfrB is the main iron storage protein in P. aeruginosa, and that the mobilization of iron reserves from BfrB is important to buffer levels of intracellular free iron, and when appropriate, to provide a source of iron to overcome iron limitation in the environment. In the genomes of many bacteria, like in the genome of P. aeruginosa, the bfd and bfr genes are adjacent, which would suggest that the BfrB:Bfd interaction is of widespread importance to bacterial iron homeostasis.23,24 It is not yet known, however, whether in these organisms Bfr is also the main iron accumulator, an issue that underscores the need to investigate several organisms in similar detail. It is also important to stress that nothing is known regarding the mechanism of iron mobilization from bacterial Ftns, an issue that is clearly of importance to understanding iron homeostasis in bacteria where iron is stored in Ftn. Finally, it is interesting to consider that the iron core of Bfr has a high phosphate content (Fe:P ~1:1), which has led to the suggestion that phosphate may be stored in Bfr.57 Hence, it will be interesting to investigate whether inhibition of the BfrB:Bfd interaction also affects phosphate homeostasis in P. aeruginosa.
Supplementary Material
Significance to Metallomics.
Bacteria depend on iron homeostasis to satisfy their nutritional iron requirement while preventing iron-induced toxicity. Iron homeostasis requires iron storage proteins, where Fe3+ can be deposited, and when necessary, mobilized as Fe2+ for incorporation in metabolism. This study demonstrates that P. aeruginosa accumulates iron in bacterioferritin (BfrB) and that upon passage to iron-limiting conditions the cells must mobilize iron from BfrB to sustain growth, a process that requires binding of BfrB to a ferredoxin (Bfd). Finally, our results also suggest that the role of iron storage proteins in bacteria is more sophisticated than simply iron storage: BfrB and the BfrB-Bfd interaction enable an equilibrium between iron stored in BfrB and free iron in the cytosol, which assists in the maintenance of optimal free iron levels in the cytosol, which in turn enable a regulated cellular response to environmental changes in iron availability.
Acknowledgments
This study was supported by a grant from the National Science Foundation (MCB-1615767) and grants from the National Institutes of Health (AI125529 and P20GM103638). A 2014 University of Kansas Strategic grant (Level I) also supported this work. M.R. thanks Dr. Amanda Oglesby-Sherrouse for helpful discussions.
Footnotes
Supplementary Information (ESI) available: [details of supplementary information available should be included here].
References
- 1.Touati D. Iron and oxidative stress in bacteria. Archives of biochemistry and biophysics. 2000;373:1–6. doi: 10.1006/abbi.1999.1518. [DOI] [PubMed] [Google Scholar]
- 2.Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial Iron Homeostasis. FEMS Microbiology Reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 3.Schaible UE, Kaufmann SH. Iron and microbial infection. Nature reviews Microbiology. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 4.Andrews S, Norton I, Salunkhe AS, Goodluck H, Aly WS, Mourad-Agha H, Cornelis P. Control of iron metabolism in bacteria. Metal ions in life sciences. 2013;12:203–239. doi: 10.1007/978-94-007-5561-1_7. [DOI] [PubMed] [Google Scholar]
- 5.Benson DR, Rivera M. Heme Uptake and Metabolism in Bacteria. Met Ions Life Sci. 2013;12:279–332. doi: 10.1007/978-94-007-5561-1_9. [DOI] [PubMed] [Google Scholar]
- 6.Wilks A, Burkhard KA. Heme and Virulence: How Bacterial Pathogens Regulate, Transport and Utilize Heme. Nat Prod Rep. 2007;24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 7.Wilks A, Ikeda-Saito M. Heme utilization by pathogenic bacteria: not all pathways lead to biliverdin. Accounts of chemical research. 2014;47:2291–2298. doi: 10.1021/ar500028n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera M. Bacterioferritin: Structure, Dynamics and Protein-Protein Interactions at Play in Iron Storage and Mobilization. Accounts of chemical research. 2017;50:331–340. doi: 10.1021/acs.accounts.6b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruvinsky AM, Vakser IA, Rivera M. Local packing modulates diversity of iron pathways and cooperative behavior in eukaryotic and prokaryotic ferritins. The Journal of chemical physics. 2014;140:115104. doi: 10.1063/1.4868229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Theil EC. Ferritins: Dynamic Management of Biological Iron and Oxygen Chemistry. Acc Chem Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 11.Honarmand Ebrahimi K, Hagedoorn PL, Hagen WR. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem Rev. 2015;115:295–326. doi: 10.1021/cr5004908. [DOI] [PubMed] [Google Scholar]
- 12.Rivera M. In: Handbook of Porphyrin Science. Kadish KK, Smith KM, Guilard R, editors. Vol. 30. 2014. pp. 136–179. ch. 159. [Google Scholar]
- 13.Murphy TF. The many faces of Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2008;47:1534–1536. doi: 10.1086/593187. [DOI] [PubMed] [Google Scholar]
- 14.Yao H, Jepkorir G, Lovell S, Nama PV, Weeratunga SK, Battaille KP, Rivera M. Two Disctinct Ferritin-Like Molecules in P. aeruginosa: The Product of the bfrA Gene is a Bacterial Ferritin (FtnA) not a bacterioferritin (Bfr) Biochemistry. 2011;50:5236–5248. doi: 10.1021/bi2004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma JF, Ochsner UA, Klotz MG, Nanayakkara VK, Howell ML, Johnson Z, Posey JE, Vasil ML, Monaco JJ, Hassett DJ. Bacterioferritin A Modulates Catalase A (KatA) Activity and Resistance to Hydrogen Peroxide in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews SC, Harrison PM, Guest JR. Cloning, Sequencing, and Mapping of the Bacterioferritin Gene (bfr) of Escherichia coli K-12. J Bacteriol. 1989;171:3940–3947. doi: 10.1128/jb.171.7.3940-3947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg RP, Vargo CJ, Cui X, Kurtz DMJ. A [2Fe-2S] Protein Encoded by an Open Reading Frame Upstream of the Escherichia coli Bacterioferritin Gene. Biochemistry. 1996;35:6297–6301. doi: 10.1021/bi9600862. [DOI] [PubMed] [Google Scholar]
- 18.Quail MA, Jordan P, Grogan JM, Butt JN, Lutz M, Thomson AJ, Andrews SC, Guest JR. Spectroscopic and Voltammetric Characterization of Bacterioferritin- Associated Ferredoxin of Escherichia coli. Biochem Biophys Res Commun. 1996;229:635–642. doi: 10.1006/bbrc.1996.1856. [DOI] [PubMed] [Google Scholar]
- 19.Weeratunga S, Gee CE, Lovell S, Zeng Y, Woodin CL, Rivera M. Binding of Pseudomonas aeruginosa Apobacterioferritin-Associated Ferredoxin to Bacterioferritin B Promotes Heme Mediation of Electron Delivery and Mobilization of Core Mineral Iron. Biochemistry. 2009;48:7420–7431. doi: 10.1021/bi900561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weeratunga S, Lovell S, Yao H, Battaile KP, Fischer CJ, Gee CE, Rivera M. Structural Studies of Bacterioferritin B (BfrB) from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center. Biochemistry. 2010;49:1160–1175. doi: 10.1021/bi9015204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang A, Zeng Y, Han H, Weeratunga S, Morgan BN, Moënne-Loccoz P, Schönbrunn E, Rivera M. Biochemical and Structural Characterization of Pseudomonas aeruginosa Bfd and FPR: Ferredoxin NADP+ Reductase and Not Ferredoxin is the Redox Partner of Heme Oxygenase under Iron-Starvation Conditions. Biochemistry. 2007;46:12198–12211. doi: 10.1021/bi7013135. [DOI] [PubMed] [Google Scholar]
- 22.Wang A, Rodríguez JC, Han H, Schönbrunn E, Rivera M. X-Ray Crystallographic and Solution State Nuclear Magnetic Resonance Spectroscopic Investigations of NADP+ Binding to Ferredoxin NADP Reductase from Pseudomonas aeruginosa. Biochemistry. 2008;47:8080–8093. doi: 10.1021/bi8007356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao H, Wang Y, Lovell S, Kumar R, Ruvinsky AM, Battaile KP, Vakser IA, Rivera M. The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin. J Am Chem Soc. 2012;134:13470–13481. doi: 10.1021/ja305180n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Yao H, Cheng Y, Lovell S, Battaile KP, Middaugh CR, Rivera M. Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Potein-Protein Interactions in Solution and Determination of Binding Energy Hot Spots. Biochemistry. 2015;54:6162–6175. doi: 10.1021/acs.biochem.5b00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Simith K, Spencer D, Wong GKS, Wu Z, Paulsen IT, Reizer J, Saler MH, Hancock REW, Lory S, Olson MV. Complete Genome Sequence of Pseudomonas aeruginosa PA01, an Opportunistic Pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 26.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Analytical biochemistry. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 27.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. Precision- engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nature protocols. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon R, Priefer U, Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nature Biotechnology. 1983;1:784–791. [Google Scholar]
- 30.Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nature protocols. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 31.Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. Genetic tools for select- agent-compliant manipulation of Burkholderia pseudomallei. Applied and environmental microbiology. 2008;74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung MC. A specific iron stain for iron-binding proteins in polyacrylamide gels: application to transferrin and lactoferrin. Anal Biochem. 1985;148:498–502. doi: 10.1016/0003-2697(85)90258-1. [DOI] [PubMed] [Google Scholar]
- 33.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods in enzymology. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 34.Flores SE, Day AS, Keenan JI. Measurement of total iron in Helicobacter pylori-infected gastric epithelial cells. Biometals: an international journal on the role of metal ions in biology, biochemistry, and medicine. 2015;28:143–150. doi: 10.1007/s10534-014-9810-z. [DOI] [PubMed] [Google Scholar]
- 35.Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. Journal of bacteriology. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keyer K, Imlay JA. Superoxide Accelerates DNA- Damage by Elevating Free-Iron Levels. Proc Natl Acad Sci USA. 1996;93:13635–13649. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodmansee AN, Imlay JA. Quantitation of intracellular free iron by electron paramagnetic resonance spectroscopy. Methods in enzymology. 2002;349:3–9. doi: 10.1016/s0076-6879(02)49316-0. [DOI] [PubMed] [Google Scholar]
- 38.Yan Y, Waite-Cusic JG, Kuppusamy P, Yousef AE. Intracellular free iron and its potential role in ultrahigh- pressure-induced inactivation of Escherichia coli. Applied and environmental microbiology. 2013;79:722–724. doi: 10.1128/AEM.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.British Medical Association. British medical journal. 1883;2:1113–1123. [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley JM, Le Brun NE, Moore GR. Ferritins: furnishing proteins with iron. J Biol Inorg Chem. 2016;21:13–28. doi: 10.1007/s00775-016-1336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velayudhan J, Castor M, Richardson A, Main-Hester KL, Fang FC. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Molecular microbiology. 2007;63:1495–1507. doi: 10.1111/j.1365-2958.2007.05600.x. [DOI] [PubMed] [Google Scholar]
- 42.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip Expression Analysis of the Iron Starvation Response in Pseudomonas Aeruginosa: Identification of Novel Pyoverdine Biosynthesis Genes. Mol Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 43.Palma M, Worgall S, Quadri LEN. Transcriptome Analysis of the Pseudomonas aeruginosa Response to Iron. Arch Microbiol. 2003;180:374–379. doi: 10.1007/s00203-003-0602-z. [DOI] [PubMed] [Google Scholar]
- 44.Bradley JM, Moore GR, Le Brun NE. Mechanisms of iron mineralization in ferritins: one size does not fit all. J Biol Inorg Chem. 2014;19:775–785. doi: 10.1007/s00775-014-1136-3. [DOI] [PubMed] [Google Scholar]
- 45.Le Brun NE, Wilson MT, Andrews SC, Guest JR, Harrison PM, Thomson AJ, Moore GR. Kinetic and Structural Characterization of an Intermediate in the Biomineralization of Bacterioferritin. FEBS letters. 1993;333:197–202. doi: 10.1016/0014-5793(93)80404-i. [DOI] [PubMed] [Google Scholar]
- 46.Crow A, Lawson TL, Lewin A, Moore GR, Le Brun NE. Structural Basis for Iron Mineralization by Bacterioferritin. J Am Chem Soc. 2009;131:6808–6813. doi: 10.1021/ja8093444. [DOI] [PubMed] [Google Scholar]
- 47.Rui H, Rivera M, Im W. Protein dynamics and ion traffic in bacterioferritin. Biochemistry. 2012;51:9900–9910. doi: 10.1021/bi3013388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao H, Rui H, Kumar R, Eshelman K, Lovell S, Battaile KP, Im W, Rivera M. Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic. Biochemistry. 2015;54:1611–1627. doi: 10.1021/bi501255r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mislin GL, Schalk IJ. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics: integrated biometal science. 2014;6:408–420. doi: 10.1039/c3mt00359k. [DOI] [PubMed] [Google Scholar]
- 50.Brandel J, Humbert N, Elhabiri M, Schalk IJ, Mislin GL, Albrecht-Gary AM. Pyochelin, a siderophore of Pseudomonas aeruginosa: physicochemical characterization of the iron(III), copper(II) and zinc(II) complexes. Dalton transactions. 2012;41:2820–2834. doi: 10.1039/c1dt11804h. [DOI] [PubMed] [Google Scholar]
- 51.Martin LW, Reid DW, Sharples KJ, Lamont IL. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals: an international journal on the role of metal ions in biology, biochemistry, and medicine. 2011;24:1059–1067. doi: 10.1007/s10534-011-9464-z. [DOI] [PubMed] [Google Scholar]
- 52.Kakhlona O, Cabantchik ZL. The labile iron pool: characterization, measurement, and participation in cellular processes. Free Radic Biol Med. 2002;33:1037–1046. doi: 10.1016/s0891-5849(02)01006-7. [DOI] [PubMed] [Google Scholar]
- 53.Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan G, Pinochet-Barros A, Gaballa A, Patel SJ, Arguello JM, Helmann JD. PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Molecular microbiology. 2015;98:787–803. doi: 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolaj-Robin O, Russell D, Hayes KA, Pembroke JT, Soulimane T. Cation Diffusion Facilitator family: Structure and function. FEBS letters. 2015;589:1283–1295. doi: 10.1016/j.febslet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Stiefel EI, Watt GD. Azotobacter cytochrome b557.5 is a bacterioferritin. Nature. 1979;279:81–83. doi: 10.1038/279081a0. [DOI] [PubMed] [Google Scholar]
- 57.Watt GD, Frankel RB, Jacobs D, Huang H. Fe2+ and Phosphate Interactions in Bacterial Ferritin from Azotobacter vinelandii. Biochemistry. 1992;31:5672–5679. doi: 10.1021/bi00139a035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.