SUMMARY
Antibiotic-mediated microbiota destruction and the consequent loss of colonization resistance can result in intestinal domination with vancomycin-resistant Enterococcus (VRE), leading to bloodstream infection in hospitalized patients. Clearance of VRE remains a challenging goal that, if achieved, would reduce systemic VRE infections and patient-to-patient transmission. Although obligate anaerobic commensal bacteria have been associated with colonization resistance to VRE, the specific bacterial species involved remain undefined. Herein we demonstrate that a precisely defined consortium of commensal bacteria containing the Clostridium cluster XIVa species Blautia producta and Clostridium bolteae restores colonization resistance against VRE and clears VRE from the intestines of mice. While C. bolteae did not directly mediate VRE clearance it enabled intestinal colonization with B. producta, which directly inhibited VRE growth. These findings suggest that therapeutic or prophylactic administration of defined bacterial consortia to individuals with compromised microbiota composition may reduce inter-patient transmission and intra-patient dissemination of highly antibiotic-resistant pathogens.
eTOC Blurb
Vancomycin-resistant Enterococcus (VRE) can densely colonize intestines and cause bloodstream infections. The intestinal microbiota provides resistance against VRE colonization. Caballero and colleagues demonstrate in mice that Blautia producta and Clostridium bolteae restore resistance against VRE. Administration of specific consortia of commensal bacteria can re-establish colonization resistance against highly antibiotic-resistant pathogens.
INTRODUCTION
Vancomycin-resistant Enterococcus (VRE) is one of the leading causes of nosocomial infection and has been declared a serious public health threat due to its high level of antibiotic resistance (Antibiotic resistant threats in the United States, 2013; Arias and Murray, 2008, 2012). VRE can colonize and dominate the intestine following antibiotic-mediated disruption of the indigenous microbiota (Donskey et al., 2000; Ubeda et al., 2010). Dense intestinal colonization can lead to systemic VRE infection and patient-to-patient transmission of VRE within healthcare facilities (Arias and Murray, 2008; Taur et al., 2012). Patients undergoing allogeneic hematopoietic stem cell transplantation are at especially high risk for VRE colonization and subsequent bloodstream infection due to prophylactic and empiric antibiotic treatment and development of transient transplant-related intestinal mucositis with loss of epithelial integrity (Taur et al., 2012).
Upon colonization of the gastrointestinal (GI) tract, VRE can persist at high densities for months despite discontinuation of antibiotic treatment (Ubeda et al., 2010). Importantly, VRE can be efficiently cleared from the gut via transplantation of an antibiotic-naïve, diverse fecal microbiota, a procedure known as FMT (Caballero et al., 2015; Ubeda et al., 2013). The genus Barnesiella has been associated with resistance to VRE colonization (Ubeda et al., 2013), however, it remains unclear whether Barnesiella species mediate VRE clearance in vivo. Thus, although administration of a complex yet uncharacterized microbiota can establish resistance to VRE, the commensal bacterial species that mediate protection against VRE remain undefined.
The identification of bacterial strains that provide colonization resistance is challenging because they are, for the most part, obligate anaerobes, difficult to culture and sensitive to antibiotics that impair colonization resistance (Reeves et al., 2012; Lawley et al., 2012; Ubeda et al., 2013; Buffie et al., 2015). Furthermore, most commensal bacterial species have evolved as members of complex ecosystems, with metabolic interdependencies that are just beginning to be defined (Rakoff-Nahoum et al., 2016), suggesting that the species that provide resistance against VRE may be incapable of functioning independently.
To simplify the analysis of colonization resistance in mice made susceptible to VRE colonization by ampicillin treatment, we established a mouse model whereby ampicillin administration could be maintained by using the ampicillin-resistant microbiota (ARM) isolated from a mouse strain that has been treated continuously with ampicillin for over 15 years. Efficient ARM colonization in the presence of ampicillin minimized the recovery of indigenous ampicillin-sensitive bacteria, allowing us to dissect the impact of ARM-derived strains on VRE colonization.
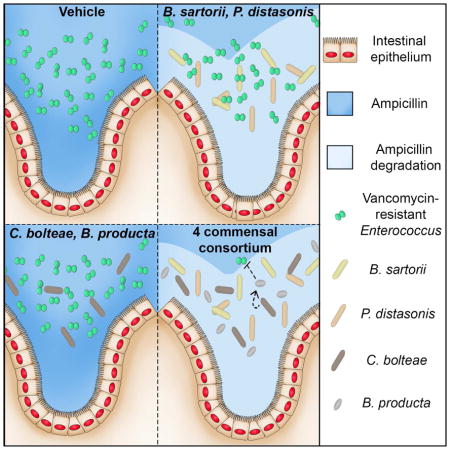
Using metagenomic sequencing and a bank of commensal bacterial isolates, we assembled a consortium of four bacterial species that, upon administration to ampicillin-treated mice, restored complete resistance to VRE. The four-bacteria consortium, consisting of Bacteroides sartorii, Parabacteroides distasonis and Clostridium cluster XIVa members Clostridium bolteae and Blautia producta, prevents and eliminates VRE colonization in mice. While ex vivo studies revealed that B. producta alone is able to suppress VRE growth, we found that B. sartorii and P. distasonis produce high levels of β-lactamase that allows C. bolteae and B. producta to colonize the intestine and clear VRE. Our results demonstrate inter-species cooperativity that is essential for colonization resistance against an important nosocomial pathogen.
RESULTS
Bacterial composition of intestinal microbiota resistant to ampicillin
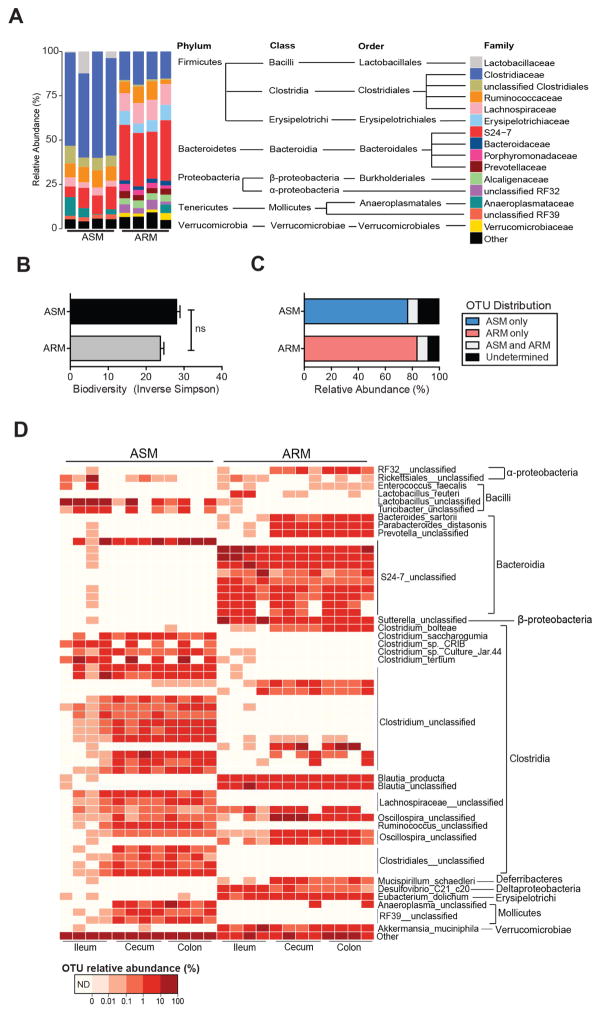
To identify bacterial species that promote resistance to VRE, we characterized the fecal microbiota of a colony of MyD88−/− mice on the C57BL/6 background that evolved an ampicillin-resistant microbiota (ARM) as a result of long-term ampicillin treatment. Taxonomic comparison of the fecal microbiota of MyD88−/− mice and C57BL/6 mice from Jackson laboratories, which harbor an ampicillin-sensitive microbiota (ASM), revealed differences in the representation of Firmicutes and Bacteroidetes, the two major intestinal bacterial phyla. In ASM, over 75% of bacterial taxa belonged to the Firmicutes phylum while in ARM the representation of Firmicutes and Bacteroidetes was approximately equal (Figure 1A and Figure S1A). Bacterial families associated with the murine intestinal microbiota, including Clostridiaceae, Ruminococcaceae, S24-7 and Lachnospiraceae (Dethlefsen et al., 2008; Eckburg et al., 2005; Ubeda et al., 2013) were well represented in ASM and ARM, although the frequency of S24-7 was roughly 40% in ARM compared to 5–10% in ASM (Figure 1A). Conversely, the Clostridiaceae family represented approximately 50% of ASM taxa in contrast to 15% abundance in ARM. Other bacterial families, such as the Erysipelotrichiaceae, Bacteroidaceae, Prevotellaceae, Alcaligenaceae and Verrucomicrobiaceae, were more highly represented in ARM than ASM (Figure 1A). Despite these differences, ARM and ASM fecal microbial communities were similarly diverse, as quantified by the inverse Simpson index (Figure 1B). In the ileum, the microbiota of antibiotic-naïve animals had lower diversity than in long-term ampicillin-treated mice, with predominant representation of the Lactobacillaceae and Clostridiaceae families in ASM while S24-7, Alcaligenaceae and Lachnospiraceae families predominated in the ileum of ARM-harboring mice. (Figures S1B and S1C).
Figure 1. Characterization of ampicillin-resistant and ampicillin-sensitive microbiota.
(A) Fecal microbiota composition of the ampicillin-resistant microbiota (ARM) harbored by MyD88−/− mice and the ampicillin-sensitive microbiota (ASM) of C57BL/6 animals. Each bar corresponds to an individual mouse. Operational taxonomic units (OTUs) at > 0.01% relative abundance are shown. (B) Fecal microbiota diversity (ns = non-significant, Student’s t test; n = 4 mice per group). Data are means ± SEM. (C) Distribution of fecal OTUs (≥ 97% similarity, > 0.01% relative abundance). Data shown is the average of 4 mice per group. (D) Relative abundance of the top 60 OTUs (horizontal bars) in the ileum, cecum and feces of ARM-harboring animals. Each column corresponds to an individual mouse. ARM, antibiotic-resistant microbiota; ASM, antibiotic-sensitive microbiota. Data are means ± SEM. See also Figure S1.
Analysis of operational taxonomic units (OTUs) in ileum, cecum and feces revealed that 75–85% of detected OTUs were present in either ARM or ASM but not both (Figure 1C and 1D). ARM contained Bacteroides sartorii, Parabacteroides distasonis and multiple S24-7 OTUs within the Bacteroidia class while the Clostridia class consisted mainly of cluster XIVa members Clostridium bolteae and Blautia producta in addition to several uncharacterized Clostridium spp. (Figure 1D). These bacterial taxa, many of which are present in the microbiota of humans (Atarashi et al., 2013; Qin et al., 2010), were not detected in ASM. Although several studies have linked microbiota composition to host genotype, previous work from our laboratory revealed that compositional differences among mice lacking distinct arms of innate immunity are a consequence of long-term breeding in isolation and vertical transmission rather than innate immune signaling deficiency (Ubeda et al., 2012). Therefore, it is likely that husbandry conditions such as chronic exposure to ampicillin and long-term segregated breeding have resulted in disparate bacterial compositions of ASM and ARM.
ARM transplantation restores intestinal homeostasis and resistance to VRE during antibiotic treatment
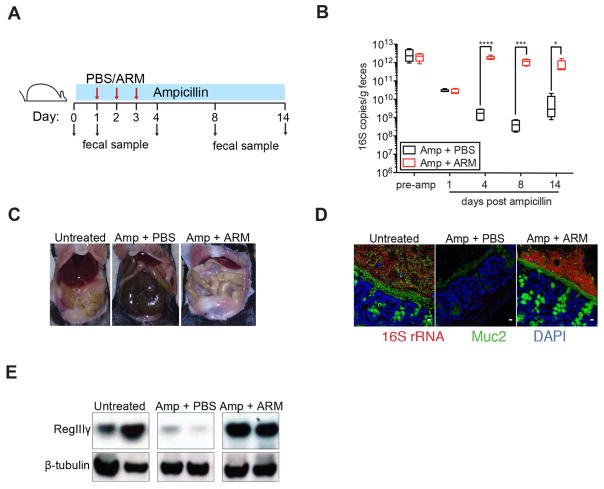
To functionally characterize ARM, we investigated whether adoptive ARM transfer into C57BL/6 mice could reverse the impact of short-term (2-week) ampicillin treatment (Figure 2A). Consistent with previous studies (Ubeda et al., 2010), treatment with ampicillin reduced fecal bacterial density between 100 and 1000 fold (Figure 2B). ARM administration to ampicillin-treated mice increased bacterial density, as measured by 16S rRNA gene copy numbers, within 24 hours to levels detected in untreated mice (Figure 2B). Sequencing of 16S rRNA genes demonstrated complete and stable transplantation of ARM OTUs (Figures S2A–S2C). Ampicillin-treated mice that received PBS had reduced bacterial diversity and a microbiota composition distinct from untreated and ARM-transplanted animals (Figures S2A–S2C).
Figure 2. ARM transplantation prevents the loss of intestinal homeostasis caused by antibiotic treatment.
(A) Experimental scheme for ARM administration. Three doses of PBS or ARM were administered daily by oral gavage to C57BL/6 ampicillin-treated mice beginning on day 2 of ampicillin treatment. Fecal samples were collected before antibiotics (d0) and on days 1 (prior to transplant), 4, 8 and 15 after antibiotic initiation. (B) Density of 16S rRNA gene copies in fecal samples over time (n = 5 mice per group). Box plots show the median, 25th percentile and 75th percentile; whiskers represent minimum and maximum values (*P = 0.0137, ***P = 0.0009, ****P > 0.0001, Student’s t test). (C) Difference in cecum size among the different groups. (D) Visualization of the inner mucus layer and goblet cells (Muc2) as well as all bacteria (16S rRNA gene) in colon cross-sections. Hoechst dye was used to visualize nuclei. Original magnification, 63X. Scale bars, 10μm. (E) Protein extracts from ileal tissues were analyzed by Western blotting with RegIIIγ-specific antiserum and anti-β-tubulin as loading control. (D–F) Representative images and samples from 5 mice per group are shown. See also Figure S2.
Physiological changes associated with antibiotic treatment such as cecal enlargement (Reikvam et al., 2011; Savage and Dubos, 1968), thinning of the colonic mucus layer (Wlodarska et al., 2011) and down-regulation of the bactericidal C-type lectin RegIIIγ (Brandl et al., 2008; Kinnebrew et al., 2010) were reversed by ARM administration to ampicillin-treated C57BL/6 mice, indicating that ARM corrects defective fiber degradation and provides the stimuli to trigger mucin production and innate immune responses in the presence of ampicillin. (Figures 2C–2E).
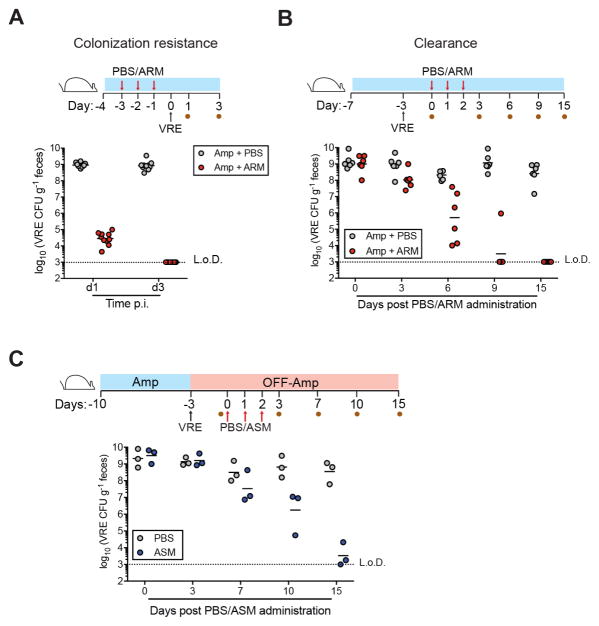
Previous studies demonstrated that antibiotic-treated mice become densely colonized with VRE upon oral inoculation (Caballero et al., 2015; Ubeda et al., 2010). To determine whether ARM could provide colonization resistance against VRE, we challenged ampicillin-treated mice that had received ARM or PBS. While VRE reached a density of 109 colony-forming units (CFU) per gram in the feces of PBS-treated mice 24 hrs post challenge, VRE levels in ARM-treated animals were lower (104 CFU) and became undetectable by day 3 post inoculation (Figure 3A). Thus, ARM administration to ampicillin-treated C57BL/6 mice prevented intestinal VRE colonization. Once VRE has colonized and dominated the intestine it can persist for months, even in the absence of antibiotic pressure, and clearance can be achieved by transplantation of a healthy and diverse microbiota (Arias and Murray, 2012; Caballero et al., 2015; Taur et al., 2012; Ubeda et al., 2010). Administration of ARM to ampicillin-treated, VRE-dominated mice resulted in progressive reduction in VRE levels and clearance within 15 days (Figure 3B). Notably, the rate of VRE clearance by ARM was similar to that obtained by transplantation of feces from antibiotic-naïve C57BL/6 mice (Figure 3C and Ubeda et al., 2013), suggesting that ARM and ASM, despite significant compositional differences, are functionally similar.
Figure 3. ARM transplantation restores resistance against VRE during antibiotic treatment.
(A) Prevention and (B) clearance of VRE colonization following PBS or ARM administration to ampicillin-treated mice. Three doses of ARM or PBS were administered daily by oral gavage to C57BL/6 ampicillin-treated mice before VRE challenge (A) or starting on the third day of VRE colonization (B). Fecal pellets were collected at the indicated time points for VRE quantification (n = 6–12 mice per group). Blue bar denotes continuous ampicillin treatment in the drinking water. (C) Clearance of VRE colonization following PBS or ASM administration measured in feces. C57BL/6 mice were challenged with VRE on the seventh day of ampicillin treatment, at which point treatment was stopped. The first of 3 doses of PBS or ASM was administered on day 3 post VRE challenge. L.o.D., limit of detection. CFU, colony-forming units.
Ampicillin-sensitive bacterial strains within ARM mediate colonization resistance against VRE
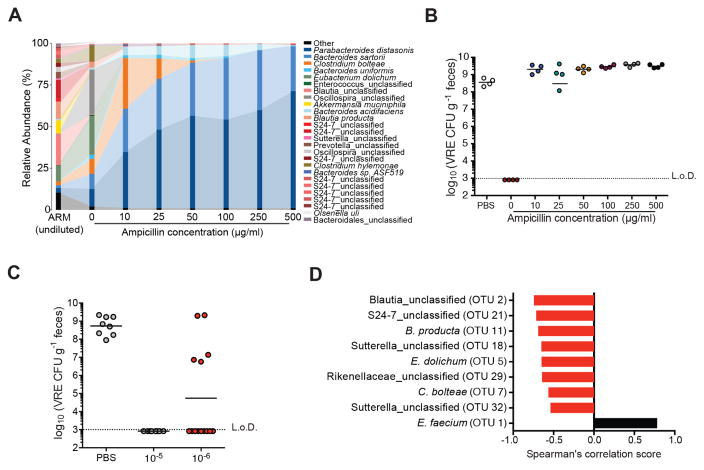
In vitro studies have demonstrated that antibiotic-resistant bacteria within complex bacterial populations can enable neighboring sensitive species to survive antibiotic-treatment, either by secretion of antibiotic-degrading enzymes (i.e. β-lactamases) or by inducing susceptible cells to withstand antibiotic stress (Lee et al., 2010; Medaney et al., 2016; Yurtsev et al., 2013). To determine whether bacterial species within ARM differed in terms of antibiotic resistance, we plated a diluted suspension of ARM on media containing concentrations of ampicillin ranging from 0 to 500 μg/ml and determined bacterial composition by 16S rRNA gene sequencing. Approximately 40% of the bacterial species detected in uncultured ARM were detected in anaerobic cultures grown in the absence of ampicillin (Figure 4A). However, bacterial diversity was dramatically reduced upon addition of ampicillin, with concentrations as low as 10 μg/ml yielding predominantly Parabacteroides distasonis, Clostridium bolteae and several Bacteroides spp. (B. sartorii, B. uniformis, B. acidifaciens). Increasing ampicillin concentrations further reduced bacterial diversity and enriched for P. distasonis and B. sartorii (Figure 4A and Figure S3A). Assays for β-lactamase, which inactivates ampicillin (Blair et al., 2015), were positive with ARM but not ASM (Figure S3B), and cultures of P. distasonis, B. sartorii and B. acidifaciens isolated from ARM were strongly positive for β-lactamase activity, while an isolate of B. producta was negative (Figure S3C). These findings suggest that most bacterial species within ARM are ampicillin-sensitive and that a small number of resistant strains provide population-wide ampicillin resistance.
Figure 4. Ampicillin-sensitive bacterial strains within ARM confer resistance to VRE.
(A–B) Fecal suspensions of ARM were diluted 10−5-fold and grown on plates containing 0, 10, 50, 100 and 500 μg/ml of ampicillin. (A) Bacterial composition of cultured fecal fractions. Each bar corresponds to pooled cultures from 3 plates per group. (B) Ampicillin-treated mice were administered PBS or cultured fecal fractions from each group by oral gavage on three consecutive days starting on day 2 of ampicillin treatment and challenged with VRE the day following the third gavage. VRE density in fecal samples was determined 3 days after infection (n = 4 mice per group). (C) VRE levels 3 days post challenge of ampicillin-treated mice inoculated with 10−5 and 10−6 plate cultures as described in (B) (n = 8–14 mice per group). (D) Spearman correlation of OTUs associated with resistance to VRE colonization. OTUs with P values < 0.05 are plotted. L.o.D., limit of detection. See also Figures S3 and S4 and Table S1.
To determine whether ampicillin-resistant strains contribute to VRE-specific colonization resistance, we transplanted ARM cultures grown in the presence or absence of ampicillin into ampicillin-treated mice prior to VRE challenge. Mice that received untreated cultures were highly resistant to VRE while colonization resistance was abrogated in mice inoculated with ARM cultured with even the lowest concentration of ampicillin (Figure 4B). Thus, the bacterial species within ARM suppressing VRE intestinal expansion are culturable under anaerobic conditions and are ampicillin-sensitive. Interestingly, mice that received bacterial cultures grown in the presence of ampicillin, which consisted mostly of Bacteroides and Parabacteroides spp., exhibited higher VRE colonization levels than control animals (Figure 4B). Bacteroides spp. have been shown to enhance intestinal expansion of C. difficile and Salmonella enterica serovar Typhimurium by increasing free sialic acid levels in the gut (Ng et al., 2013). Therefore, it is possible that in our model, Bacteroides and Parabacteroides colonization alter the carbohydrate pool in the intestine, favoring VRE growth.
To identify the bacterial species that mediate VRE resistance, we used a reductionist approach and adoptively transferred bacterial fractions from plates of 10−5 or 10−6 dilutions of ARM into ampicillin-treated mice before VRE challenge. While un-transplanted, PBS-treated mice became densely colonized with VRE and mice transplanted with the 10−5 ARM dilution were completely protected, mice that received cultures from the 10−6 ARM dilution varied in terms of resistance to VRE colonization (Figure 4C). Low bacterial diversity increases the likelihood of intestinal colonization and infection (Taur et al., 2012; Ubeda et al., 2010). Importantly, administration of the 10−5 and 10−6 ARM dilutions variably restored microbiota diversity in antibiotic-treated mice (Figure S4A). However, some mice within the 10−6 dilution group were resistant to VRE colonization despite exhibiting low bacterial diversity (Figure S4B). The finding that low biodiversity does not always correlate with colonization susceptibility was previously reported (Buffie et al., 2015) and suggests that the presence of specific bacterial species, rather than the degree of diversity, is critical for colonization resistance. To identify candidate bacteria protective against VRE, we stratified mice from the 10−6 dilution group by VRE colonization density and plotted the abundance of the 20 most highly represented OTUs (Figure S4C). While most OTUs were present in resistant mice, albeit at various frequencies, several, including members of the Clostridia (Blautia spp., Oscillospira spp. and Clostridium bolteae), Mollicutes (Eubacterium dolichum), Bacteroidia (Bacteroides spp., Barnesiellaceae and Rikenellaceae families) and β-proteobacteria (Sutterella spp.) classes were undetectable in susceptible mice (Figure S4C). Of these, 8 bacterial OTUs correlated significantly with resistance to VRE colonization by Spearman’s rank correlation test (Figure 4D). As previously reported, S24-7 (Barnesiella) was strongly associated with VRE protection (Ubeda et al., 2013).
Adoptive transfer of an assembled and precisely defined bacterial consortium reestablishes resistance against VRE
In order to isolate bacterial strains responsible for VRE suppression, we cultured 250 individual colonies from ARM and sequenced their 16S rRNA genes. Bacterial species belonging to the Bacteroidaceae, Clostridiaceae, Porphyromonadaceae, Coriobacteriaceae, Rikenellaceae, Lachnospiraceae, unclassified Clostridiales and Verrucomicrobiaceae families were cultured (Table S1). Among the cultured bacterial isolates, 4 shared 100% sequence homology with strains associated with ARM-induced resistance to VRE colonization: Blautia_unclassified, Blautia producta, Clostridium bolteae and Eubacterium dolichum (whose 16S sequence is identical to that of Clostridium innocuum). Of note, OTUs corresponding to Akkermansia muciniphila, Bacteroides sartorii and Parabacteroides distasonis were highly abundant in susceptible and resistant animals alike (Figure S4C). These organisms are known for their ability to digest polysaccharides into less complex sugars, enhancing the nutrient pool for other members of the community (Derrien et al., 2004; Flint et al., 2012; Marcobal et al., 2011; Ng et al., 2013). In addition, B. sartorii and P. distasonis are highly resistant to ampicillin. Therefore, even though these OTUs did not reach statistical significance in our correlation analysis, we hypothesized that they could play an important role in the establishment and maintenance of resistance-associated strains by providing a source of energy and/or inactivating ampicillin.
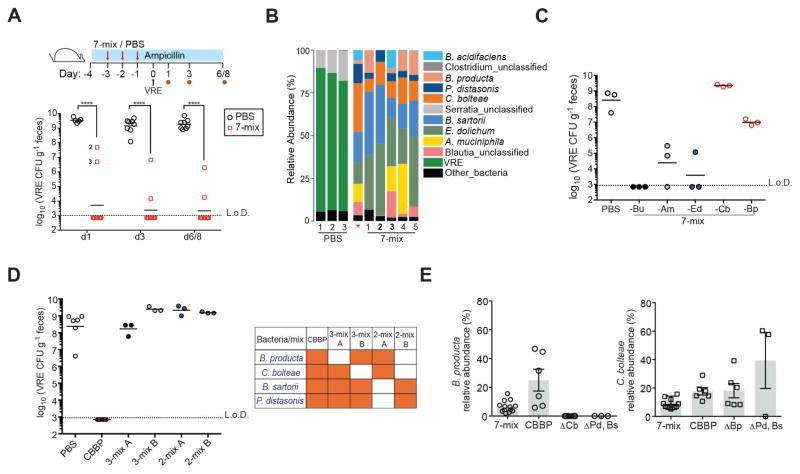
We assembled a consortium of 7 ARM isolates: Blautia_unclassified, B. producta, C. bolteae, E. dolichum, A. muciniphila, B. sartorii and P. distasonis and assessed its ability to prevent VRE colonization. Eight of ten mice that received a mixture of the 7 strains (7-mix) had undetectable VRE 1 day after challenge and two had a VRE burden of 106–108 CFU that had decreased by 2-logs on day 6/8 post infection. In contrast, VRE levels in PBS-treated animals ranged between 109–1010 CFU at all time points (Figure 5A). Taxonomic analysis demonstrated that while control mice were VRE-dominated, the microbiota of protected animals was comprised almost entirely of bacteria from the 7-mix. Importantly, B. producta was present at low to undetectable levels in the two mice from the 7-mix group with reduced and delayed VRE-resistance, suggesting a role for this strain in protection against VRE (Figure 5B).
Figure 5. Adoptive transfer of bacterial consortia containing C. bolteae and B. producta prevents VRE colonization.
(A–B) Ampicillin-treated mice were orally gavaged with PBS or a mixture of C. bolteae, B. producta, Blautia_unclassified, E. dolichum, A. muciniphila, P. distasonis and B. sartorii (7-mix) for three consecutive days beginning on day 2 of antibiotic treatment. Mice were challenged with VRE the day following the third gavage and fecal samples were collected at the indicated time points. (A) VRE CFUs in stool samples 1, 3 and 6/8 days post inoculation (p.i.) (****P < 0.0001, Mann-Whitney test; n = 10 mice per group). (B) Fecal microbiota composition of treated mice on day 1 p.i. *, 7-mix input. (C) VRE stool burden 3 days p.i. in mice treated with 7-mix consortia lacking individual bacterial strains as described in (A–B). Bu, Blautia_unclassified; Am, A. muciniphila; Ed, E. dolichum; Cb, C. bolteae; Bp, B. producta (n = 3 mice per group). (D) VRE density 3 days p.i. in mice treated with a consortium of four bacterial strains (CBBP) and combinations thereof. Experimental approach described in (A–B) was followed (n = 3–6 mice per group). (E) Relative abundance levels of B. producta and C. bolteae on day 3 p.i. in feces from mice administered the 7-mix, CBBP or bacterial mixtures lacking C. bolteae, B. producta or P. distasonis (Ps) and B. sartorii (Bs) (n = 3–14 mice per group). Δ indicates the absence of corresponding strain (s).
To assess the extent to which B. producta and the remaining strains in the 7-mix contributed to colonization resistance, we individually excluded A. muciniphila, E. dolichum, Blautia_unclassified, C. bolteae or B. producta from the consortia but kept P. distasonis and B. sartorii in every group for their role in providing community-wide antibiotic resistance. Exclusion of Blautia_unclassified or E. dolichum did not impact resistance to VRE colonization whereas an intermediate effect was detected in the absence of A. muciniphila (Figure 5C). Remarkably, loss of colonization resistance against VRE occurred in mice treated with bacterial mixtures deficient in either C. bolteae or B. producta (Figure 5C). These findings demonstrate that Blautia_unclassified, E. dolichum and A. muciniphila are dispensable for VRE-specific colonization resistance while C. bolteae and B. producta are essential. Indeed, adoptive transfer of C. bolteae and B. producta together with B. sartorii and P. distasonis (CBBP) fully prevented VRE expansion while various subsets lacking either C. bolteae or B. producta were ineffective (Figure 5D). Furthermore, administration of C. bolteae and B. producta in the absence of P. distasonis and B. sartorii or vice-versa had no impact on VRE colonization (Figure 5D). These results indicate that the ampicillin-resistant strains do not, on their own, inhibit VRE colonization but are required to act in a cooperative manner for the engraftment or activity of the ampicillin-sensitive strains. Consistent with this, B. producta was undetectable in ampicillin-treated mice administered C. bolteae and B. producta alone, whereas C. bolteae colonization was detected in two of three animals (Figure 5E). Further supporting the notion of bacterial cooperation, we found that B. producta did not colonize the gut when administered in the absence of C. bolteae even in the presence of the β-lactamase-producing strains, suggesting a second cooperative interaction during engraftment of the CBBP consortium (Figure 5E). On the other hand, C. bolteae relative abundance levels were similar in mice treated with mixtures of the 7- and 4-bacteria consortia containing or lacking B. producta (Figure 5E). Taken together, our observations suggest that reestablishment of colonization resistance in the ampicillin-treated mouse model requires two cooperative interactions. First, B. sartorii and P. distasonis inactivate ampicillin, allowing engraftment of ampicillin-sensitive C. bolteae, which in turn supports engraftment of B. producta.
A four-bacteria consortium effectively eliminates established VRE colonization
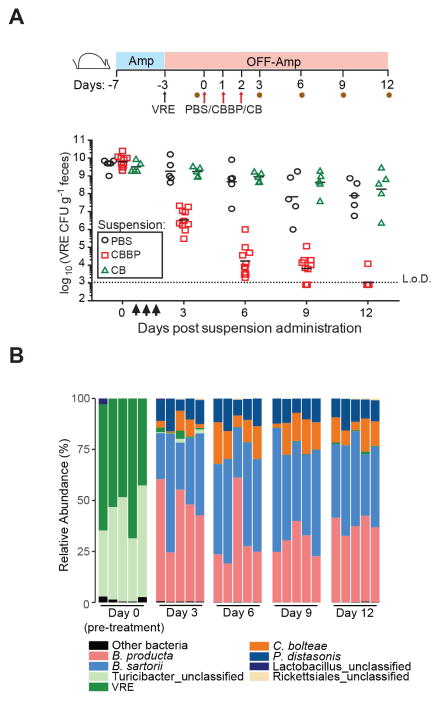
Although reestablishing colonization resistance in vulnerable patients represents an important potential approach to reduce VRE acquisition, reducing the density of VRE in colonized patients would also reduce infection and transmission rates within healthcare settings (Arias and Murray, 2008; Taur et al., 2012). Therefore, we tested whether the CBBP consortium or C. bolteae together with B. producta (CB) could clear VRE from the gut of densely colonized mice. Because B. producta and C. bolteae are not resistant to ampicillin, we challenged ampicillin-treated mice with VRE and, following discontinuation of antibiotics, administered PBS, CBBP or CB and quantified the density of VRE in fecal samples at various time points post challenge. (Figure 6A). All mice were densely colonized with VRE prior to treatment, and while PBS-treated animals remained colonized, mice inoculated with CBBP exhibited a reduction in VRE colonization that progressed for 12 days, at which point VRE was no longer detected (Figure 6A). VRE was also cleared from the ileum and cecum (Figure S5A), demonstrating that decolonization of the small and large intestines can be achieved with CBBP.
Figure 6. Administration of a four-bacteria consortium clears VRE from densely colonized mice.
(A) VRE clearance with CBBP. Experimental scheme, mice were treated with ampicillin (Amp) for 4 days prior to VRE inoculation Antibiotic-treatment was discontinued at the time of challenge and three days later, the first of three daily doses of PBS, CBBP or CB (C. bolteae and B. producta) was administered orally. Fecal samples were collected on day 3 p.i. prior to PBS, CBBP or CB administration (d0) and on days 3, 6, 9 and 12 after the first gavage. VRE burden in stool samples from the indicated time points (n = 5-10 mice per group) is shown. (B) Fecal microbiota composition of mice treated with CBBP. Each bar represents an individual mouse. Data is representative of two experiments. L.o.D., limit of detection. See also Figure S5.
Importantly, CB administration had no effect on VRE colonization suggesting that in the absence of ampicillin, P. distasonis and B. sartorii enhance the inhibitory activity of CB by mechanism(s) in addition to β-lactam degradation (Figure 6A). Consistent with these observations, VRE levels remained unchanged following administration of either C. bolteae or B. producta (Figure S5B). Sequence analysis demonstrated that CBBP strains represented ≥90% of the fecal microbiota in transplanted mice, and remained remarkably stable over the 12-day duration of the experiment despite discontinuation of ampicillin treatment (Figure 6B). Therefore, in addition to providing VRE-specific colonization resistance, reconstitution of VRE-dominated mice with this minimal bacterial consortium promotes nearly one million-fold reduction in VRE density.
B. producta directly inhibits VRE growth
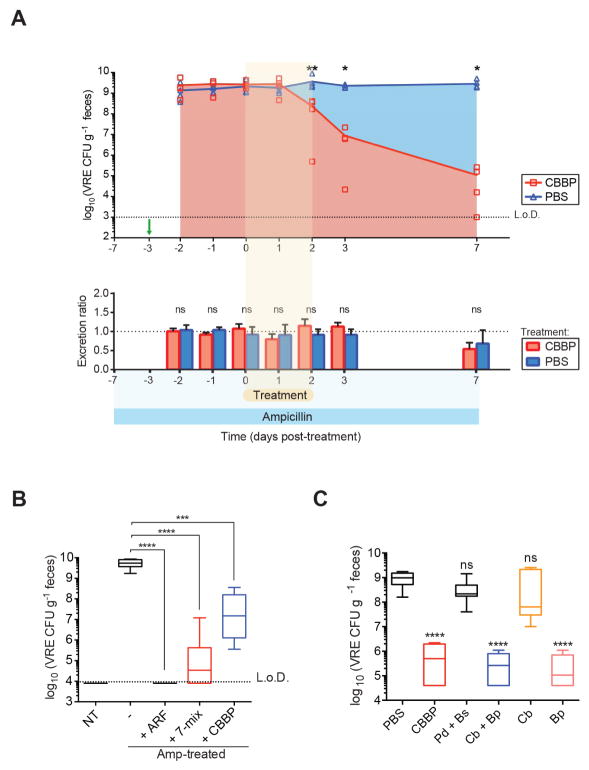
To begin to identify the mechanism of enhanced VRE clearance from the gut, we first determined whether administration of CBBP increased the rate of VRE excretion in feces. Administration of CBBP to ampicillin-treated, VRE-dominated mice did not increase either the amount of feces produced per day or density of excreted VRE (Figure 7A and Figures S6A and S6B), suggesting that the four-bacteria consortium does not increase bowel transit rates to simply flush VRE out.
Figure 7. Blautia producta directly inhibits VRE growth.
(A) Ampicillin-treated mice were challenged with VRE on the fourth day of ampicillin treatment (day -3, green arrow) and administered CBBP or PBS for three consecutive days starting on the third day of VRE colonization (day 0). Fecal pellets were collected at the indicated time points to determine VRE CFUs (top graph). For quantification of total fecal VRE excretion (bottom graph), all fecal pellets per cage were collected and weighed at 24-hour intervals and normalized to the initial time point (day -2). An excretion ratio of 1 indicates no change in the excretion rate (*P = 0.0159, **P = 0.0079, ns = non-significant by the Mann-Whitney test; n = 5 mice per group). (B) Ex vivo quantification of VRE cultured in fecal suspensions from antibiotic-naïve mice (NT), antibiotic-treated mice, and antibiotic-treated mice colonized with ARM, 7-mix or CBBP (***P = 0.0007, ****P < 0.0001, Mann-Whitney test; n = 5–10 samples per group). (C) VRE growth in cecal content supplemented with cultures of the indicated CBBP strains prior to seeding with VRE (***P < 0.0001, ns = non-significant, Mann-Whitney test with respect to PBS control; n = 12 samples per group). L.o.D., limit of detection; Amp, ampicillin; Cb, C. bolteae; Bp, B. producta; Pd, P. distasonis; Bs, B. sartorii. See also Figures S6 and S7.
To determine whether CBBP inhibits VRE growth, we developed an ex vivo assay that approximates in vivo conditions to test VRE growth and survival. Colonization resistance against VRE has been modeled ex vivo using fecal material from antibiotic-naïve and/or antibiotic-treated animals (Pultz et al., 2005). Using this approach, we detected 106-fold VRE expansion upon culture in a fecal suspension generated from ampicillin-treated mice whereas no growth was detected in fecal suspensions from antibiotic-naïve mice (Figure 7B). Consistent with our in vivo findings, VRE growth was completely inhibited or significantly suppressed in fecal suspensions from ampicillin-treated mice colonized with ARM, the 7-mix or CBBP (Figure 7B). Confirming that viable, anaerobic bacteria are responsible for inhibiting VRE growth, heating to 55°C or treatment with metronizadole, an antibiotic with potent anti-anaerobe activity, markedly reduced the ability of ARM to inhibit VRE expansion (Figures S7A–S7C).
To confirm that B. producta is the key species within the consortium providing VRE-specific colonization resistance, we added CBBP strains individually or in various combinations to fecal suspensions from ampicillin-treated mice (two days after stopping ampicillin) prior to VRE culture. Similar to our in vivo findings, CBBP suppressed VRE growth while P. distasonis and B. sartorii or C. bolteae did not. In contrast, B. producta alone or in combination with C. bolteae suppressed VRE expansion to the same extent as CBBP (Figure 7C). Importantly, CBBP also inhibited the growth of two other VRE clinical strains isolated from colonized patients, suggesting that CBBP could be effective in defense against a wide range of VRE strains (Figure S7D). Altogether, our results indicate that a consortium of four anaerobic strains is required for in vivo colonization resistance to VRE, and that B. producta is the key contributor to VRE inhibition.
DISCUSSION
Reducing the density of intestinal VRE colonization and shedding can be anticipated to benefit patients harboring VRE and reduce patient-to-patient transmission. Our study demonstrates that administration of a defined bacterial consortium containing B. producta and C. bolteae provides colonization resistance against VRE and clears persistent VRE colonization in mice. Quantification of VRE fecal shedding in reconstituted mice suggests that the mechanism by which B. producta and C. bolteae reduce intestinal VRE colonization is not simply a matter of increased excretion. Instead, this bacterial consortium either reduces VRE replication or increases VRE killing in the gut. Although previous studies from our laboratory have shown that microbiota-induced production of RegIIIγ reduces VRE colonization in the ileum (Abt et al., 2016; Brandl et al., 2008; Kinnebrew et al., 2010), the finding that B. producta directly inhibits VRE growth ex vivo suggests that the mechanism(s) by which B. producta suppresses VRE expansion likely involve direct bacterial interactions, such as nutrient competition or production of inhibitory factors (e.g. metabolites or bacteriocins).
Although FMT can clear VRE from the mouse intestine, it remains unclear whether microbiota replenishment can eliminate VRE from the human GI tract. However, a recent, uncontrolled, retrospective analysis of patients who received a commercial microbiota suspension for recurrent Clostridium difficile infection suggested that microbiota transplantation might reduce colonization with VRE (Dubberke et al., 2016). Given the high frequency of VRE colonization and intestinal domination within specific patient populations, controlled clinical trials of FMT or treatment with defined bacterial consortia will be necessary to move microbiota enhancement for the prevention of VRE infection and transmission into the clinical mainstream. While FMT has been used very effectively and safely in patients with recurrent C. difficile infection, patients with VRE colonization are often immunocompromised, making the incompletely defined composition of feces, even from healthy and screened donors, potentially problematic (Pamer, 2014). Precisely defined and characterized bacterial consortia, such as the four-mix described in this study, offer the advantage of being of known, consistent composition and testable safety.
Our study reveals two levels of cooperativity between bacterial species providing resistance against intestinal VRE domination in ampicillin-treated mice. At one level, we demonstrate that two bacterial species expressing a secreted antibiotic resistance enzyme, β-lactamase, enable a diverse but antibiotic-sensitive population to survive in the gut in the presence of antibiotics. The second level of cooperativity is the in vivo requirement of C. bolteae, P. distasonis and B. sartorii to support B. producta intestinal colonization, which is required for VRE clearance. The mechanisms underlying this second level of cooperativity are likely complex and require additional study. Nevertheless, identifying cooperative interactions between commensal bacteria and determining their respective contributions to colonization resistance represent important steps towards the eventual generation of therapeutic microbial consortia (McKenney and Pamer; 2015). While commensal bacteria constituting the intestinal ecosystem compete for physical and metabolic niches within the gut and are influenced by the diet’s fiber content (Lee et al., 2013; Earle et al., 2015; Desai et al., 2016), other studies have demonstrated cooperation between different commensal bacterial species. For example, extracellular degradation of inulin, a complex carbohydrate, by Bacteroides ovatus benefits Bacteroides vulgatus, which, in the context of a complex microbiota, reciprocates by undefined mechanisms and enhances B. ovatus growth (Rakoff-Nahoum et al., 2016).
The ability of CBBP to clear VRE colonization is similar to that of FMT with ASM or ARM, suggesting that we have identified a minimal, but likely not unique, consortium to support in vivo VRE clearance. Indeed, ASM is effective at clearing VRE but did not contain detectable levels of B. producta or C. bolteae, suggesting that other combinations of commensal bacterial species can mediate clearance of VRE from the intestine. Nevertheless, our finding that four commensal bacterial species can mediate VRE-specific colonization resistance demonstrates the feasibility of assembling bacterial consortia for the prevention and clearance of VRE colonization.
The ability of defined bacterial consortia to provide resistance against C. difficile infection is well established (Lawley et al., 2012; Petrof et al., 2013; Buffie et al., 2015; Tvede et al., 2015). More recently, a mouse-derived bacterial consortium consisting of 15 strains was shown to protect mice against Salmonella enterica serovar Typhimurium colonization as efficiently as FMT (Brugiroux et al., 2016). In this study, we demonstrate that a four-member consortium can, in a mouse model, provide high level protection against another clinically-relevant pathogen, VRE. While VRE is one of the most important antibiotic-resistant bacterial pathogens causing difficult-to-treat infections, the complete list of antibiotic resistant bacterial pathogens is long and contains many species that originate from the gut. An important challenge, therefore, is to identify commensal bacterial species that provide colonization resistance against the wide range of antibiotic-resistant opportunistic pathogens, including bacteria belonging to the Enterobacteriaceae family, such as Escherichia coli, Klebsiella pneumoniae and Enterobacter aerogenes among many others. Given the continued increase in antibiotic resistance and the paucity of new antibiotics, microbiota enhancement or augmentation with defined commensal bacterial species provides an important potential avenue to markedly reduce intestinal colonization with and transmission of antibiotic-resistant pathogens.
STAR METHODS TEXT
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact, Eric Pamer (pamere@mskcc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
A colony of MyD88−/− mice that has been continuously treated for 15 years with ampicillin in the Memorial Sloan Kettering Cancer Center (MSKCC) animal facility was used to obtain ampicillin-resistant microbiota (ARM). Age/gender-matched C57BL/6 mice harboring an ampicillin-sensitive microbiota (ASM) were purchased from Jackson Laboratories. MyD88−/− and C57BL/6 mice from separate cages were sampled for microbiota analysis. Mice were female and 6–8 weeks of age at the beginning of experiments. All mouse handling, cage changes and tissue collection were performed in a biosafety level 2 facility wearing sterile gowns, masks and gloves. All animal procedures were approved by the Institutional Animal Care and Use Committee of MSKCC.
METHOD DETAILS
Ampicillin treatment and VRE colonization
For VRE challenge, 6–8 week old female C57BL/6 mice from Jackson Laboratories were treated with 0.5 g/L ampicillin (Fisher) in their drinking water and inoculated with 5x104 VRE CFUs (E. faecium, ATCC 700221) in 200μl by gavage. Animals were maintained in a specific pathogen-free facility at MSKCC.
VRE colonization with diluted ARM fractions
Fecal samples from ARM-harboring mice were collected and resuspended in 1ml pre-reduced PBS under anaerobic conditions. Fecal suspensions were titrated by culturing 100μl from five 10-fold serial dilutions (10−5) on Columbia agar supplemented with 5% sheep blood and ampicillin concentrations in the 0–500 μg/ml range. 10−6 dilutions were cultured similarly but without ampicillin. Plate cultures were incubated anaerobically at 37°C for 3 –5 days, bacterial colonies were scraped off individual plates and resuspended in 5 ml pre-reduced PBS. For inocula preparation, glycerol was added to 10−5 and 10−6 bacterial suspensions at a concentration of 15% and aliquoted before freezing at −80°C. For VRE colonization resistance experiments, 200μl was administered per mouse daily for 3 days. ARM and ASM fecal transplants were prepared by resuspending fresh stool pellets from MyD88−/− (SFB-positive) or C57BL/6 (SFB-negative) mice, respectively in 1 ml pre-reduced PBS and 200μl of the fecal suspension was administered to mice by oral gavage daily for three days . For colonization resistance experiments, mice were administered three daily doses of ARM or bacterial suspensions beginning the second day of ampicillin treatment up to one day before VRE challenge. For clearance experiments, three daily doses of ARM, ASM or bacterial suspensions were administered starting on the third day following VRE inoculation. Mice were single-housed from the time of challenge and kept on ampicillin unless otherwise noted.
VRE culture
For VRE quantification, tenfold dilutions of weighed intestinal samples resuspended in PBS were plated on Difco Enterococcosel agar (supplemented with 8 μg/ml vancomycin; Novaplus and 100 μg/ml streptomycin; Fisher) and incubated at 37°C for 48 hr.
16S rRNA gene sequencing
Intestinal samples were frozen in a dry ice/ethanol slurry immediately after collection and stored at −80°C and DNA extraction w as performed as previously described (Buffie et al., 2015). Briefly, samples were resuspended in 500 μl of extraction buffer (200 mM Tris pH 8.0/200 mM NaCl/20 mM EDTA), 200 μl of 20% SDS, 500 μl of phenol:chloroform:isoamyl alcohol (24:24:1) and 500 μl of 0.1-mm diameter zirconia/silica beads (BioSpec Products). Cells were lysed by bead beating (2 min). DNA was extracted in a phenol/chloroform/isoamyl alcohol solution twice and precipitated with ethanol and sodium acetate. DNA was resuspended in 200 μl of TE buffer containing 100 μg/ml RNase, purified with QIAmp Mini Spin Columns (Qiagen) and eluted in 100 μl water. The V4–V5 region of the 16S rRNA gene was amplified following DNA extraction, using the primers 563F (5’-nnnnnnnn-NNNNNNNNNNNN-AYTGGGYDTAAAGNG-3′) and 926R (5′-nnnnnnnn-NNNNNNNNNNNN-CCGTCAATTYHTTTRAGT-3′). The PCR reaction consisted of 50 ng of purified DNA, 0.2 mM dNTPs, 1.5 μM MgCl2, 1.25 U Platinum TaqDNA polymerase, 2.5 μl of 10X PCR buffer and 0.2 μM of each primer. A unique 12-base Golay barcode (Ns) preceded the primers for sample identification after pooling amplicons. The cycling conditions were: 94°C (3 min), 27 cycles of 94°C (50 s), 51°C (30 s) and 72°C (1 min) followed by a final elongation step at 72 °C (5 min). Replicate PCRs we re pooled and purified using the Qiaquick PCR Purification Kit (Qiagen) and Qiagen MinElute PCR Purification Kit. PCR products were quantified using the Illumina TruSeq Sample Preparation procedure and combined at equimolar amounts before Illumina barcodes and adaptors were added. The completed library was sequenced on an Ilumina Miseq platform as per Illumina specifications.
Sequencing data analysis
Sequence analysis was performed using the MOTHUR pipeline (Schloss et al., 2009), version 1.33.3. Sequences were aligned using the Silva reference alignment as a template and chimeric sequences were eliminated using UCHIME (Edgar et al., 2011). Sequences with a distance-based similarity of ≥97% were binned into operational taxonomic units (OTUs) using the average-neighbor algorithm. OTUs were classified using a modified Greengenes 16S rRNA reference database (DeSantis et al., 2006). OTU-based biodiversity was calculated by the Inverse Simpson index. A phylogenetic tree was generated using the Clearcut command in MOTHUR (Sheneman et al., 2006). Unweighted UniFrac was run on the resulting tree and principal component analysis was performed on the resulting distance matrix between each group of samples (Lozupone et al., 2011; Ubeda et al., 2012). OTUs at relative abundance >0.01% were plotted.
Quantification of bacterial density by qPCR
Copies of the 16S rRNA gene were determined by performing quantitative PCR on total DNA extracted from fecal samples using primers specific to the V4 region of the 16S gene, 517F (5’-GCCAGCAGCCGCGGTAA-3’) and 798R (5’-AGGGTATCTAATCCT-3’), at 0.2 μM concentration with the DyNAmo SYBR green RT-PCR kit (Finnzymes). Standard curves were prepared by serial dilution of the PCR blunt vector (Invitrogen) containing a single copy of the 16S rRNA gene. The cycling protocol was: 95°C for 10 mi n, followed by 40 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min.
FISH and Muc2 Immunofluorescence
Colonic tissue was fixed in Methacarn solution (60% methanol, 30% chloroform and 10% acetic acid). Costaining for Muc2 and 16S rRNA gene was carried out as previously described (Caballero et al., 2015). Briefly, for FISH, tissue sections were deparaffinized with xylene (twice, 10 min each) and rehydrated through an ethanol gradient (95%, 10 min; 90%, 10 min) to water. Sections were incubated with a universal bacterial probe directed against the 16S rRNA gene or with probes specific to K. pneumoniae and Enterococcus at 50°C for 3 hours. Probes were diluted to 5ng/μl in 0.9M NaCl, 20mM Tris-HCl at pH7.2 and 0.1% SDS prior to use. Sections were later washed twice in 0.9M NaCl, 20mM Tris-HCl at pH7.2 (wash buffer) for 10 min and counterstained with Hoechst (1:3000 in wash buffer) for nuclear staining. The following FISH probes were used: universal bacterial probe EUB338: [Cy3]-GCTGCCTCCCGTAGGAGT-[AmC7~Q+Cy3es]. For MUC2 immunofluorescence, deparaffinized sections were incubated in 0.9M NaCl, 20mM Tris-HCl at pH7.2 and 0.1% SDS at 50°C f or 3 hours, rinsed in PBS and blocked with 5% goat serum in PBS for 30 min at room temperature to minimize non-specific binding. Sections were then washed in PBS for 10 min prior to overnight incubation at 4°C with an anti-M uc2 rabbit polyclonal antibody (H300, Santa Cruz; 1:200 in PBS). Following incubation with primary antibody, tissues were washed 3 times in PBS for 10 min and incubated with goat-anti-rabbit Alexa 488 secondary antibody (Life Technologies, 1:1000 in PBS) for 1 hour at room temperature. Sections were washed twice in PBS for 10 min and counterstained with Hoechst (1:3000 in PBS). For FISH-Muc2 dual staining, sections were briefly rinsed in wash buffer after FISH hybridization and incubated directly with the anti-Muc2 primary antibody diluted in wash buffer. Incubation with secondary antibody was carried out at 4°C for 2 hours. A single 10 min PBS wash was performed after incubation with the primary and secondary antibodies before Hoechst nuclear staining and mounting with Mowiol solution.
Image acquisition was performed with a Leica TCS SP5-II upright confocal microscope using a 63x oil immersion lens as a series of short Z-stacks. Fiji (ImageJ) software was used for maximum intensity Z-stack projection.
Western blot analysis
A 2-cm segment from the distal portion of the small intestine was excised and stored in RNA stabilization solution (RNAlater) at 4°C prior to homogenization with Trizol reagent (Invitrogen). Total protein was extracted according to the manufacturer’s specifications and resuspended in a solution of 8 mol/L urea, 1% sodium dodecyl sulfate (SDS) and 0.15 mol/L Tris-HCl at pH 7.5. Equal amounts of protein were denatured, loaded onto a 4–12% SDS-polyacrylamide electrophoresis gel (Nupage Bis-Tris gel, Invitrogen) and transferred to a nitrocellulose membrane. Protein blots were incubated with rabbit polyclonal RegIIIγ specific-antiserum and mouse anti-β-tubulin (Santa Cruz Biotechnology) antibodies, followed by horseradish peroxidase-conjugated anti-rabbit (GE Healthcare) and anti-mouse (Santa Cruz technology) antibodies. Protein bands were detected using chemiluminescence (GE Healthcare).
β-lactamase detection assay
Fecal pellets from MyD88−/− (ARM) and C57BL/6 (ASM) animals were collected and resuspended in PBS at 25mg/ml. Samples were left undisturbed for 5 minutes to allow particulate matter to sediment. 100μl of the suspension was pipetted into a 96-well plate with 50μl of nitrocefin (0.2 mg/ml, Fisher) and incubated for 30 minutes at room temperature while protected from light. For detection of β-lactamase activity from individual isolates, 100μl from a 3-day anaerobic culture corresponding to 106 CFU was used.
Quantification of VRE fecal shedding
Fecal pellets from ampicillin-treated, VRE-colonized mice administered PBS or CBBP were resuspended in PBS to a final concentration of 100 mg/ml for VRE CFU quantification. For fecal weight kinetics, fecal pellets were collected, weighed, incubated at room temperature for 24 and 48 hours and weighed again. Dryness of the pellets due to evaporation after 24 hours reduced the excretion rate by half. As a result, quantification of fecal pellets incubated for 24 hours at room temperature were adjusted accordingly by a factor of 2 when resuspending in PBS to compensate for the difference between initial fecal weight and the weight of the fecal pellet after 24 hours. To determine VRE viability, fecal pellets were collected and divided in half. Half of the pellet was immediately plated to quantify VRE CFU. The remaining pellet was incubated at room temperature for 24 hours and subsequently plated to assess changes in VRE CFU that may have resulted from drying. The excretion ratio for a given mouse at time x is expressed as: Excretion ratio = f (x)/f (0), where f (x) corresponds to the fecal weight at any given time and f (0) is the fecal weight at time = 0. For in vivo VRE clearance, sterile, customized cage grates (Thoren Caging Systems, Inc.) were placed in each cage to prevent coprophagy. At each 24-hour time point, all fecal pellets per cage were collected and mice were placed into a new cage. The fecal pellets were incubated at room temperature for 24 hours prior to quantification for VRE CFU.
Bacterial isolation and adoptive transfer
To isolate ARM bacteria, colonies were picked from 10−5 dilution cultures and streaked onto fresh agar to ensure purity. Isolated clones were resuspended in PBS plus glycerol (15%) and stored at −80°C. Full-l ength 16S rRNA genes were amplified by colony PCR using primers 8F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’). The resulting PCR product was Sanger sequenced with primers spanning the full 16S gene (8F and 1492R) as well as primers specific to the V4–V5 region (517F, 5’-GCCAGCAGCCGCGGTAA-3’) and classified using BLAST (98–100% sequence identity). For colonization experiments, bacteria were individually cultured on Columbia plus 5% sheep blood agar (BD Biosciences) for 3 days at 37°C under anaerobic conditions, plate cultures were scraped off, mixed in a 1:1 ratio (107–108 CFU per isolate) in PBS plus glycerol (15%) and stored at −80°C. For CBBP and 2-mix experiments, the bacterial inoculum administered to mice was normalized to total CFU. Bacterial mixtures were administered to mice by oral gavage in 200μl daily for 3 days.
VRE competition/suppression assay
For ex vivo assays, fecal samples from C57BL/6 ampicillin-treated mice colonized with different bacterial mixtures were freshly-collected, transferred to an anaerobic chamber and resuspended in pre-reduced PBS at a concentration of 25 mg/ml. Fecal suspensions were inoculated with 5x103 VRE CFU and incubated overnight at 37°C . For in vitro assays, B. producta , C. bolteae , P. distasonis and B. sartorii were cultured in BHI medium supplemented with L-cysteine (1.0 g/L) and yeast-extract (5.0 g/L) for 2 days. Bacterial cultures were normalized to the same optical density and 50–150 μl of each strain(s) was added to suspensions of cecal content collected from mice two days after stopping antibiotic treatment and incubated anaerobically at 37°C overnight. The following day, VRE (5x103 CFU) was seeded into cecal cultures and grown anaerobically at 37°C. For antibiotic treatment of fecal samples, fecal pellets from ampicillin-treated mice that had or had not received ARM were resuspended in pre-reduced PBS containing metronidazole (Sigma-Aldrich), gentamicin (Fisher) or streptomycin (Fisher) at 0.5 mg/ml or no antibiotics and seeded with 5x103 VRE CFU. For in vitro CBBP-mediated VRE suppression, VRE isolates derived from the stool of two VRE-colonized patients at Memorial Hospital were tested in addition to the ATCC strain. These patients were enrolled in a fecal collection protocol where feces were collected during hospitalization and stored in a biospecimen bank. The study was approved by the institutional review board at MSKCC and patients provided informed consent for biospecimen collection and analysis. This study was conducted in accordance with the Declaration of Helsinki. Each CBBP member was grown in BHI medium supplemented with L-cysteine (1.0 g/L) and yeast-extract (5.0 g/L) for 2 days. Bacterial cultures were normalized to the same optical density and 50–150 μl of each strain was added to fresh BHI medium in conjunction with 5x103 VRE CFU. VRE quantification was performed by CFU plating 16 hours post inoculation.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Graph-Pad Prism (version 6.0). Data are expressed as mean ± SEM. The statistical parameters for each experiment are detailed in the Figure legends. Two-tailed unpaired Student’s t test was used to determine statistical significance in biodiversity scores and 16S bacterial density. The Mann-Whitney nonparametric test was used to determine if VRE levels were statistically significant among different groups. Spearman’s rank correlation test (two-tailed) was used to determine statistical correlations between two variables. The Benjamini-Hochberg method was applied to control for false discovery rate. For the Student’s t test, Mann-Whitney test and Spearman’s correlation test, P values < 0.05 were considered significant. No assumption on the distribution of the data were made, and only nonparametric statistical tests were used.
Supplementary Material
Highlights.
Microbiota damage and VRE infection are corrected by fecal transplantation.
Fecal fractionation identified four commensal species that prevent VRE infection.
Colonization resistance against VRE requires commensal bacterial cooperation.
Acknowledgments
This work was supported by grants RO1 AI42135, RO1 AI95706, UO1 AI124275 and P30 CA008748 from the US National Institutes of Health and the Tow Foundation and Lucille Castori Center for Microbes, Inflammation and Cancer to E.G.P. S.C. was supported by the Gilliam pre-doctoral fellowship from the Howard Hughes Medical Institute. We thank members of the Pamer laboratory for helpful discussions and comments on the manuscript. We also thank the Molecular Cytology Facility at MSKCC for tissue processing, sectioning and imaging.
Footnotes
AUTHOR CONTRIBUTIONS
S.C. and E.G.P. designed the experiments and wrote the manuscript. S.C. performed experiments and most analyses. S.K. designed and performed experiments to quantify fecal VRE shedding and assess the impact of CBBP on VRE clinical isolates. G.J.K., L.M. and L.L. performed DNA extractions and 16S MiSeq Illumina sequencing and analyses. R.A.C. and B.S. assisted in bacterial culturing and Western blot analysis and contributed to experimental design. I.M.L. and R.A.C. maintained and screened mouse strains and contributed to experimental design.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antibiotic Resistance Threats in the United States. 2013 [PubMed] [Google Scholar]
- Abt MC, Buffie CG, Susac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8:327ra325. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CA, Murray BE. Emergence and management of drug-resistant enterococcal infections. Expert Rev Anti Infect Ther. 2008;6:637–655. doi: 10.1586/14787210.6.5.637. [DOI] [PubMed] [Google Scholar]
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiroux S, Beutler M, Pfann C, Garzetti D, Ruscheweyh HJ, Ring D, et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol. 2016;2:16215. doi: 10.1038/nmicrobiol.2016.215. [DOI] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero S, Carter R, Ke X, Susac B, Leiner IM, Kim GJ, Miller L, Ling L, Manova K, Pamer EG. Distinct but Spatially Overlapping Intestinal Niches for Vancomycin-Resistant Enterococcus faecium and Carbapenem-Resistant Klebsiella pneumoniae. PLoS Pathog. 2015;11:e1005132. doi: 10.1371/journal.ppat.1005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med. 2000;343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubberke ER, Mullane KM, Gerding DN, Lee CH, Louie TJ, Guthertz H, Jones C. Clearance of Vancomycin-Resistant Enterococcus Concomitant With Administration of a Microbiota-Based Drug Targeted at Recurrent Clostridium difficile Infection. Open Forum Infect Dis. 2016;3:ofw133. doi: 10.1093/ofid/ofw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, et al. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe. 2015;18(4):478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, et al. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. PLoS Pathog. 2012;8(10):e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Pamer EG. From hype to hope: the gut microbiota in enteric infectious disease. Cell. 163(6):1326–1332. doi: 10.1016/j.cell.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medaney F, Dimitriu T, Ellis RJ, Raymond B. Live to cheat another day: bacterial dormancy facilitates the social exploitation of beta-lactamases. ISME J. 2016;10:778–787. doi: 10.1038/ismej.2015.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG. Fecal microbiota transplantation: effectiveness,complexities, and lingering concerns. Mucosal Immunology. 2014;7:210–214. doi: 10.1038/mi.2013.117. [DOI] [PubMed] [Google Scholar]
- Petrof EO, Gloor GB, Vanner SJ, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. 2005;191:949–956. doi: 10.1086/428090. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533(7602):255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the Gastrointestinal Tracts of Germfree Mice Inoculated with a Murine Isolate from the Family Lachnospiraceae. Infect and Immun. 2012;80(11):3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen FE. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, Dubos R. Alterations in the mouse cecum and its flora produced by antibacterial drugs. J Exp Med. 1968;128:97–110. doi: 10.1084/jem.128.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheneman L, Evans J, Foster JA. Clearcut: a fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–2824. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvede M, Tinggaard M, Helms M. Rectal bacteriotherapy for recurrent Clostridium difficile-associated diarrhoea: results from a case series of 55 patients in Denmark 2000–2012. Clin Microbiol Infect. 2015;21:48–53. doi: 10.1016/j.cmi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurtsev EA, Chao HX, Datta MS, Artemova T, Gore J. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol Syst Biol. 2013;9:683. doi: 10.1038/msb.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.