Abstract
APETALA2 (AP2) is best known for its role in the regulation of flower meristem and flower organ identity and development in Arabidopsis. We show here that AP2 also plays an important role in determining seed size, seed weight, and the accumulation of seed oil and protein. We demonstrate genetically that AP2 acts through the maternal sporophyte and endosperm genomes to control seed weight and seed yield. Thus, AP2 functions outside the boundaries of flower meristem and flower organ development to affect agronomically relevant traits in Arabidopsis.
Keywords: Apetala2, Arabidopsis, seed protein and oil contents, seed size
Genetic and molecular studies have revealed an evolutionarily conserved network of regulatory genes that orchestrate flower development in Arabidopsis and in other plant species. Included in this core group of flower-promoting and floral organ identity genes is the homeotic regulatory gene APETALA2 (AP2). AP2 is the founding member of a large family of transcription factors in Arabidopsis characterized by the presence of a 68-aa repeated domain referred to as the AP2 domain (1–7). Loss of AP2 activity has been shown to effect qualitative and quantitative changes not only in floral meristem and floral organ identity and development (1, 8–11) but also in seed coat development (1, 12, 13). Molecular studies have shown that the ap2 flower phenotypes result in part as a consequence of ectopic expression of the MADS domain containing transcription factor AGAMOUS (AG) during flower development (14, 15), indicating that one important function of AP2 is to negatively regulate AG transcription.
In flower development, the expression of most of the core regulatory genes is both temporally and spatially restricted to developing flowers (14, 16–18). These include the MADS domain-containing transcription factors AG, APETALA1 (AP1), PISTILLATA (PI), and APETALA3 (AP3). In contrast, AP2 transcripts have been detected at every stage of Arabidopsis development and in virtually every organ type examined, including young seedlings, vegetative leaf, stem and root, the inflorescence meristem, all four types of floral organs, ovules, and developing seeds and embryos (1, 19), suggesting that AP2 may be active at other times in development. Recent studies, however, have shown that the detection of AP2 transcripts may not be a sufficient indicator of AP2 activity. For example, AP2 activity can be controlled at the translational level by the microRNA mi172 (20, 21) or subject to posttranslational modification by phosphorylation, similar to that observed for other AP2 domain-containing proteins (ref. 22; R. Khush and J.K.O., unpublished data). We demonstrate here that AP2 has functions outside the boundaries of floral meristem identity, floral organ identity and the control of floral organ number. Our analysis of AP2 activity and its effects on seed size and seed mass in mutant and transgenic Arabidopsis identify AP2 as a significant player in the control of seed mass and seed yield.
Materials and Methods
Plant Material and Growth Conditions. Arabidopsis thaliana Landsberg erecta (Ler), Columbia (Col), and Columbia C24 were used as control seeds. ap2-1 and ap2-9 seeds (Ler) were provided by M. Koornneef (Wageninen University, Wageninen, The Netherlands) and G. Drews (University of Utah, Salt Lake City), respectively; ap2-3 and ap2-4 seeds (Ler) were provided by K. Okada (Kyoto University, Kyoto); and ap2-5 and ap2-6 seeds (Col) were provided by G. Haughn (University of British Columbia, Vancouver). ap2-10 is in the C24 genetic background (1). malesterile1 (ms1) seeds (Ler) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Plants were grown in either a greenhouse or a growth chamber in a 1:1:1 mixture containing vermiculite, perlite, and peat moss. Plants grown in the growth chamber were exposed to 8 h of darkness and 16 h of light in a Conviron E15 chamber (Controlled Environments, Asheville, NC) as described (23). Plants were watered with a one-quarter strength Peter's solution (Grace-Sierra Co., Milpitas, CA). Mature brown seeds were harvested, dried for 7 days at 25°C, and stratified for 7 days at 7°C before being analyzed.
Plant Transformation. Wild-type Arabidopsis root explants were transformed with Ti plasmid vectors pPW9, pPW14.4, and pPW15 according to standard procedures (24). pPW9, pPW14.4, and pPW15 contain the 1.68-kb AP2 gene coding region (1) cloned in a transcriptional fusion in the sense (pPW9) and antisense (pPW14.4 and pPW15) orientations with the cauliflower mosaic virus 35S constitutive promoter. The Ti plasmid vector used for these constructs pGSJ780A (Plant Genetic Systems N.V., Ghent, Belgium) contains the plant selectable marker gene neomycin phosphotransferase (NPTII) that confers kanamycin resistance to transformed plant cells. Independently transformed Arabidopsis lines were selected for kanamycin resistance and the presence of flowers displaying the ap2 mutant phenotype.
Seed Size and Seed Mass Analysis. Average seed mass was determined by weighing mature dry seeds in batches of 100. The weights of at least five sample batches were measured for each seed lot. Size distributions of wild-type and mutant seed populations were analyzed by separating batches of ≈0.1–0.2 g of seeds by using a series of fine wire sieves. Sieve mesh sizes 35, 40, 45, 50, 60, 70, and 80 (Fisher Scientific) with exclusion sizes of 500, 425, 355, 300, 250, 212, and 180 μm, respectively, were used for each analysis. Seeds retained by each sieve were weighed by using an analytical balance (Mettler-Toledo AG, Greifensee, Switzerland) with mass expressed as a percent of the total weight of the seed sample analyzed.
Transmission Electron Microscopy and Image Processing. Plastic sections of Arabidopsis seeds were generated as described by Yadegari et al. (25) and were examined in a JEOL JEM-100B transmission electron microscope operating with an accelerator voltage of 80 kV. All images were scanned and digitized by using a Polaroid Sprintscan 35 or an AGFA Arcus II flatbed scanner (AGFA Division, Miles). Contrast and brightness were adjusted by using photoshop 8.0 (Adobe Systems, Mountain View, CA).
Seed Yield Analysis. Plants were grown in the greenhouse in individual 3.5-inch square pots filled to the top with a 1:1:1 mixture containing vermiculite, perlite, and peat moss. Plants were bottom-watered with a one-quarter strength Peter's solution. When the seedlings reached the four-leaf stage, each seedling was fitted with an 11-× 11-inch “collar” consisting of an inverted plastic humidity tray to catch all seeds released by dehiscing siliques. All seeds from a single plant were harvested when the plant was mature and the last siliques produced on the inflorescence had elongated, turned brown, dried, and were ready to dehisce. Mature plants were cut at the crown and crushed by hand to release seeds from all remaining siliques. These hand-harvested seeds were combined with those caught in the humidity tray, sieved to remove plant debris, dried at 25°C for 7 days, and weighed.
Determination of Seed Protein and Fatty Acid Content. Total seed protein extracts were prepared as described (26). Protein concentrations were determined by using a colorimetric assay (Bio-Rad). Total seed fatty acids from wild-type and mutant Arabidopsis seeds produced and harvested under similar growth conditions were extracted and converted to methyl esters by using a 1:2 dilution of methanolic-3N HCL (Supelco) in absolute methanol at 85°C for 16 h. The free fatty acid C15:0 (Sigma) was used as an internal control to assess efficiency of extraction. Seed fatty acid methyl esters were analyzed, and total seed fatty acid composition was determined by gas chromatography with an Omega Wax 250 column (Supelco) and a Sigma 300 gas chromatograph (PerkinElmer). Total fatty acid content was determined by comparing the estimated total fatty acid methyl ester peak area to that of the C15:0 internal standard.
Results
APETALA2 Contributes to the Determination of Seed Weight and Seed Size in Arabidopsis. Previous studies showed that ap2 mutant seeds lack a distinctive seed coat epidermal cell structure called the columella and are more irregular in shape than wild-type seeds (1, 12, 13), indicating that AP2 is required for normal seed coat development. We found that AP2 is also involved in seed mass control. Table 1 shows that all seven ap2 mutants examined produced seeds that displayed significant increases in average seed mass, ranging from 27% to 104% greater than wild-type controls. Seeds produced by the weak partial loss-of-function mutants ap2-1 (27) and ap2-5 (10) showed the smallest gains in average seed mass ranging from 27% to 39% greater than that of parental wild-type Ler and Col, respectively (Table 1). In contrast, seeds produced by the strong ap2 mutants ap2-4 (8), ap2-6 (10), and ap2-10 (1) showed 69–104% gains in average seed weight compared with wild type (Table 1).
Table 1. Genetic control of Arabidopsis seed weight.
| Plant | Average seed weight | % increase over wild type |
|---|---|---|
| ap2-1 (Ler) | 2.1 (0.1) | +27 |
| 2.2 (0.1) | +33 | |
| 2.1 (0.2) | +31 | |
| 2.8 (0.2) | +33 | |
| ap2-3 (Ler) | 2.6 (0.1) | +27 |
| ap2-4 (Ler) | 3.5 (0.3) | +69 |
| 3.5 (0.2) | +69 | |
| ap2-5 (Col) | 2.9 (0.1) | +39 |
| ap2-6 (Col) | 3.5 (0.2) | +69 |
| ap2-9 (Ler) | 2.9 (0.1) | +40 |
| ap2-10 (C24) | 3.7 (0.4) | +79 |
| 3.9 (0.3) | +90 | |
| 4.2 (0.5) | +104 | |
| ms1 × Ler | 2.9 | +22 |
| Col | 1.8 (0.1) | |
| 2.1 (0.1) | ||
| Ler | 1.6 (0.1) | |
| 2.1 (0.1) | ||
| 2.3 (0.1) | ||
| C24 | 2.0 (0.1) | |
| 2.3 (0.1) |
Average seed weight is given in mg per 100 seeds. Standard deviation values are given in parentheses. Percent increases in seed weight were calculated based on comparison with that of wild-type seeds produced under similar growth conditions.
It has been suggested that seed fill, and therefore seed weight, is determined in part by the availability of assimilates to the developing seed. If true, then a decrease in total seed number may result in an increase in average seed weight. Because total seed number depends on many factors, including fertility, and because strong ap2 mutants are reduced in male fertility (1, 8–11) and total seed yield, we could not rule out the possibility that the observed increase in ap2 seed weight was produced at the expense of total seed number and yield. To test this hypothesis, we assessed the extent to which male infertility can affect Arabidopsis seed weight. ms1 flowers produce no seeds unless hand pollinated with wild-type pollen (28, 29). We therefore hand pollinated five flowers on five ms1 plants with wild-type pollen and then determined the average weight of the seeds produced. Seeds resulting from this forced pollination develop in a background of unfertilized ovules and therefore, according to the hypothesis, should show an increase in average seed weight. As predicted, the average weight of F1 [ms1 (-/-) × wild type] seeds was increased, but the average gain was only 22% greater than that of wild-type seeds, far lower than the 69–104% gains observed for severe ap2 mutants (Table 1). Even weak ap2 mutants with good fertility and seed set such as ap2-1 showed statistically higher increases in weight (27–31%) (Table 1). Thus, although male infertility can enhance seed weight slightly, it is not solely responsible for the dramatic increase in seed weight displayed by weak or strong ap2 mutants. Consistent with this result, Alonso-Blanco et al. (30) concluded that reduced fertility does not dramatically impact Arabidopsis seed size and weight.
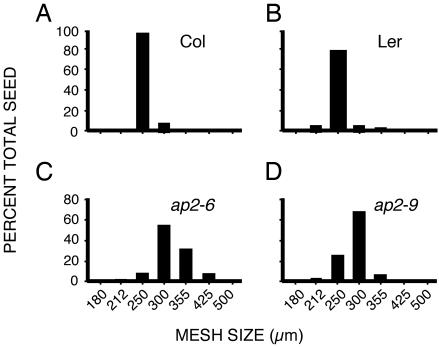
We examined ap2 mutant seed size by fractionating seeds produced by individual wild-type and mutant plants by using a series of wire mesh screens. Fig. 1 shows that seeds produced by a wild-type plant typically displayed a narrow range of exclusion sizes compared with ap2 mutant seeds. For example, wild-type Col seeds ranged in exclusion size from 250 μm to 300 μm (Fig. 1 A), whereas wild-type Ler seeds showed a slightly broader range of 212 μm to 355 μm (Fig. 1B). Both seed types have an average exclusion size between 212 and 250 μm (Fig. 1 A and B). In contrast, both ap2-6 (Col) (Fig. 1C) and ap2-9 (Ler) (Fig. 1D) mutant seeds displayed a shift in range to larger exclusion sizes. In the case of ap2-6, mutant seeds ranged in size from 250 μm to as high as 425 μm (Fig. 1C). In addition, both ap2 mutant seeds showed an increase in average exclusion size compared with wild type. Taken together, these results demonstrate that reducing AP2 gene activity consistently increases both seed size and seed weight in Arabidopsis.
Fig. 1.
ap2 seeds are larger in size than wild-type seeds. Preweighed batches of wild-type Col (A), Ler (B), and ap2-6 (Col) (C) and ap2-9 (Ler) (D) mutant seeds from single plants were passed through a series of wire sieves of decreasing mesh size (in μm) as described in Materials and Methods. ap2-6 (10) and ap2-9 (11) are severe ap2 mutants. Boxes designate the percent total seeds by weight retained by each sieve.
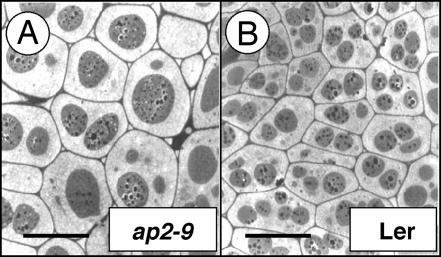
To determine the cellular basis for the difference in seed size between wild-type and ap2 mutant seeds, we examined cotyledon cell size in mature ap2-9 and Ler seeds by transmission electron microscopy. Fig. 2 shows that ap2-9 cotyledon cells are larger than those of wild type. Similar results were obtained for ap2-6 (data not shown). We conclude that the increase in seed size and seed weight observed for ap2 seeds are due in part to an increase in embryo cell size.
Fig. 2.
The ap2 mutation affects embryo cell size in Arabidopsis. Transmission electron micrographs of mature ap2-9 (A) and Ler (B) Arabidopsis seeds show cotyledon cell size and morphology. ap2-9 represents a severe loss-of-function mutant in the Ler background (11). Images were taken at the same magnification. (Bar: 12 μM.)
AP2 Activity Can Be Manipulated to Control Seed Mass in Transgenic Arabidopsis Plants. We used antisense and sense cosuppression strategies to suppress AP2 activity in planta to test whether seed mass could be manipulated in transgenic wild-type Arabidopsis plants. Nine independent lines of transgenic plants containing a chimeric AP2 antisense gene construct and eight lines containing an AP2 sense gene construct tested positive for kanamycin resistance and the presence of one or more copies of T-DNA (data not shown). Seven of the nine AP2 antisense and two of the eight sense cosuppression lines produced ap2-like flowers and seeds that were significantly larger when compared with seeds produced by control transgenic plants. Table 2 shows that the gains in seed weight ranged from 22% for antisense line PW15-2 (C24) to 89% for PW15-3 (C24) compared with controls. Increased seed weight was observed for T1,T2, and T3 generation seeds (Table 2), indicating that the seed size phenotypes are heritable. Similarly, seeds produced by the two AP2 sense cosuppression mutant lines PW9-1 (Ler) and PW9-1 (C24) showed gains in seed weight that ranged from 26% and 86% higher than that of control transgenic seeds, respectively (Table 2). Together, these results demonstrate that AP2 gene sequences can be used to genetically engineer significant increases in Arabidopsis seed weight.
Table 2. Suppression of AP2 activity in transgenic Arabidopsis plants results in increased seed mass.
| Transgenic line | Average seed weight | % increase over wild type |
|---|---|---|
| AP2 antisense lines | ||
| PW14.4-1 (C24) (T1-15) | 3.1 | +35 |
| PW14.4-1 (C24) (T1-15) T3 seeds | 3.4 (0.3) | +47 |
| PW15-1 (C24) T1 seeds | 3.6 (0.1) | +76 |
| PW15-2 (C24) (T1-2) | 2.6 (0.1) | +25 |
| PW15-2 (C24) (T1-7) | 2.5 (0.2) | +22 |
| PW15-3 (C24) T1 seeds | 3.9 (0.1) | +89 |
| PW15-1 (Ler) T1 seeds | 2.4 (0.1) | +42 |
| PW15-2 (Ler) (T1-3) | 2.8 (0.0) | +33 |
| PW15-2 (Ler) (T1-17) | 2.7 (0.0) | +28 |
| AP2 cosuppression lines | ||
| PW9-1 (C24) T1 seeds | 3.8 (0.0) | +86 |
| PW9-1 (Ler) (T1-19) | 2.7 (0.2) | +26 |
| PW9-1 (Ler) (T1-24) | 2.7 (0.1) | +26 |
| pGSJ780A vector control lines | ||
| PW3-1 (C24) T1 seeds | 2.2 (0.1) | +9 |
| PW3-2 (C24) T1 seeds | 2.3 (0.0) | +13 |
| PW3-1 (Ler) T1 seeds | 2.3 (0.1) | +7 |
| PW3-2 (Ler) (T1-4) | 2.4 (0.2) | +12 |
| PW3-2 (Ler) (T1-6) | 2.3 (0.0) | +9 |
| PW3-2 (Ler) (T1-8) | 2.1 (0.0) | -0.5 |
Transgenic lines were in the C24 or Landsberg erecta (Ler) backgrounds. Average seed weight reflects that of T2 seeds produced by a single kanamycin-resistant T1 transgenic plant unless otherwise stated. Standard deviation values are given in parentheses. Percent increases in seed weight were calculated based on comparison to that of wild-type seeds produced under similar growth conditions.
AP2 Controls Arabidopsis Seed Mass in Part Through the Maternal Sporophytic and Endosperm Genomes. As in all angiosperms, seed development in Arabidopsis depends on the interaction between the triploid endosperm and the diploid sporophytic and embryonic genomes to orchestrate morphogenesis and the deposition of seed reserves in the developing seed (31). Previously, we and others (1, 12, 13) showed that AP2 functions through the sporophytic genome to regulate seed coat development. We carried out reciprocal crosses by using ap2-10 and wild-type plants to determine whether AP2 functions through the sporophytic genome to control seed mass. Table 3 shows that ap2-10 (+/-) seeds produced by ap2 mutant flowers pollinated with wild-type pollen were larger by weight than wild-type seeds but still smaller than ap2-10 (-/-) seeds produced by mutant flowers pollinated with ap2 pollen. As expected, seeds produced by an ap2 mother, regardless of the genotype of the pollen donor, displayed the distinctive ap2 seed coat phenotype (data not shown; refs. 1, 12, 13). In contrast, ap2-10 (+/-) seeds produced by wild-type flowers pollinated with ap2-10 pollen were comparable in weight to wild-type seeds (Table 3) and had normal seed coats (data not shown). Together, these results suggest that AP2 controls seed mass in part through its activity in the maternal sporophyte and endosperm and not in the embryo.
Table 3. Genetic control of seed mass and yield by AP2.
| Parental genotypes
|
F1 seed genotype
|
Average seed mass
|
|||||
|---|---|---|---|---|---|---|---|
| ♀ | ♂ | SP | EN | EM | F1 | F2 | Average F2 seed yield |
| -/- | -/- | -/- | -/-/- | -/- | 4.75 (0.07) | ND | ND |
| -/- | +/+ | -/- | -/-/+ | -/+ | 3.65 (0.14) | 2.84 (0.21) | 2.70 (0.35) |
| +/+ | -/- | +/+ | +/+/- | +/- | 2.60 (0.04) | ND | ND |
| +/+ | +/+ | +/+ | +/+/+ | +/+ | 2.30 (0.10) | 2.46 (0.15) | 2.00 (0.22) |
| +/- | -/- | +/- | +/+/- | +/- | 3.13 (0.12) | ND | ND |
| +/- | -/-/- | -/- | |||||
| +/+ | AS/AS | +/+ | +/+/AS | +/AS | 3.50 (0.26) | ND | ND |
The + and - designations indicate the presence of the wild-type AP2 and the recessive mutant ap2-10 alleles, respectively. AS indicates the AP2 antisense transgene in pollen donor PW15-3 (C24) that confers an ap2 seed size phenotype as described in Table 2 Standard deviation values are shown in parentheses. Seed mass is given in units of mg per 100 seeds. Seed yield is the average weight (g) of the total number of F2 seeds produced per plant for WT (n = 7) and ap2-10(+/-) (n = 9) F1 plants. SP, EN, and EM refer to the maternal sporophyte, endosperm, and embryo genotypes, respectively. ND, not determined.
To further test the genetic contribution of AP2 activity in the maternal sporophyte and endosperm, we generated seeds with ap2 mutant endosperm (-/-/-) by crossing ap2-10 (+/-) flowers with ap2-10 mutant pollen. If AP2 activity in the endosperm is required for seed mass control, then the prediction is that 50% of the seeds produced from this cross will be similar in mass to those produced by a homozygous ap2 mutant. Table 3 shows that the average weight of seeds produced by this cross was intermediate between that observed for seeds produced by a homozygous ap2-10 mutant and by a wild-type plant. However, these seeds did not segregate as predicted for ap2-10 mutant seed size when assayed by microscopic and seed sieve analyses (data not shown). We conclude that AP2 activity in the maternal sporophyte is critical for the control of Arabidopsis seed mass.
To definitively test for AP2 seed mass control activity in the endosperm, we generated seeds by crossing wild-type flowers with pollen containing a 35S-AP2 antisense transgene [PW15-3 (C24); Table 2]. We reasoned that the transgene would suppress AP2 gene activity in the endosperm and embryo of the developing seeds but not in the maternal sporophyte. Table 3 shows that the seeds produced by this cross were significantly greater in weight than control seeds (Table 3), but less than those produced by ap2 mutants grown under similar conditions. Because AP2 activity in the embryo does not appear to be critical for seed size control (Table 3), these results indicate that AP2 activity in the endosperm is also critical for the control of seed mass. Taken together, these studies demonstrate that AP2 functions in at least two of the three genomes necessary for Arabidopsis seed production.
ap2 Can Increase Seed Weight Without a Compensatory Decrease in Total Seed Yield. To test whether increased seed weight negatively affects total seed yield, we took advantage of the fact that AP2 controls seed mass in part through the sporophytic and endosperm genomes (Table 3) and compared average seed weight and total seed yield for ap2-10 (+/-) and control plants. As shown previously, F1 [ap2-10 (-/-) × C24 (+/+)] seeds displayed increased seed weight (Table 3). These seeds were germinated, and the resulting plants were found to be fully fertile; they produced F2 seeds that were larger than those of wild type by 15% (P < 0.01) and produced 35% more seed by weight per plant than wild-type C24 (P < 0.01) (Table 3). These results suggest that AP2 controls both seed mass and total seed yield in Arabidopsis.
Increase in ap2 Seed Weight Is Due in Part to Increases in Total Seed Protein and Oil Contents. We extracted total protein from wildtype and ap2 mutant seeds to determine whether increases in ap2 seed mass are due in part to increases in seed reserves. Table 4 shows that total ap2 mutant seed protein content increased by 13–78% when compared with wild type. In addition, the spectrum of soluble proteins extracted from mature wild-type and ap2 mutant seeds under denaturing conditions were qualitatively indistinguishable, with no detectable difference in the relative representation of the two major classes of Arabidopsis seed storage proteins, the 12S cruciferins (32, 33) or the 2S albumins (ref. 34 and data not shown). Similar results were obtained by using wild-type and ap2 seed protein extracts generated by aqueous salt extraction procedures (data not shown). Taken together, these results indicate that the observed increases in seed protein content in ap2 mutant seeds are not due to selective increases in seed storage protein reserves.
Table 4. Genetic control of total seed protein and fatty acid contents in Arabidopsis by AP2.
| % FA methylesters (area % by GC)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Total protein, μg per 100 seeds | Total FA, μg per 100 seeds | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 22:0 | 22:1 | 24:0 |
| ap2-4 | 729 (107) (+33%) | 910 (70) (+25%) | 6.6 | 3.7 | 16.5 | 26.9 | 23.7 | 2.8 | 19.6 | >0.5 | >0.5 | 2.8 |
| Ler | 545 (22) | 730 (20) | 6.0 | 4.1 | 22.7 | 26.7 | 17.4 | 2.2 | 19.3 | >0.5 | >0.5 | 1.6 |
| ap2-5 | 617 (24) (+13%) | 920 (70) (+35%) | 6.9 | 4.1 | 14.8 | 30.7 | 20.8 | 2.2 | 17.0 | >0.4 | >0.4 | 2.2 |
| Col | 548 (42) | 680 (70) | 8.1 | 4.0 | 13.6 | 33.7 | 20.9 | 2.1 | 16.6 | >0.5 | >0.5 | 1.4 |
| ap2-10 | 836 (15) (+78%) | 1450 (90) (+113%) | 7.9 | 4.6 | 12.1 | 25.2 | 24.3 | 3.3 | 19.5 | >0.4 | >0.4 | 2.5 |
| ap2-10(+/-) | ND | 870 (70) (+32%) | 7.0 | 5.2 | 15.1 | 26.5 | 23.2 | 3.1 | 18.2 | >0.5 | >0.4 | 1.8 |
| C24 | 469 (19) | 680 (10) | 8.0 | 5.7 | 15.1 | 26.5 | 22.5 | 3.0 | 17.6 | >0.5 | >0.5 | 1.6 |
Standard deviation values are given in parentheses, and percent increases with respect to wild-type controls are shown. FA, fatty acids; ND, not determined.
We also compared total seed fatty acid content and composition in wild-type and ap2 mutant seeds. Like its close relative oilseed rape (Brassica napus), Arabidopsis seeds produce a small number of fatty acid species that collectively represent as much as 80% of the total seed fatty acids (35–37). These fatty acids include palmitic (C16:0), stearic (C18:0), oleic (C18:1), linoleic (C18:2), linolenic (C18:3), eicosenoic (C20:1), and erucic (C22:1) acids. Table 4 shows that there is ≈680–730 μg of these fatty acid species per 100 Arabidopsis wild-type seeds. In contrast, ap2 mutant seeds as well as seeds produced by ap2-10 (+/-) plants showed increases in fatty acid content of 25–113% when compared with wild type, increases that are roughly proportional to the observed increases in ap2 seed weight and total seed protein content (Tables 1 and 4 and data not shown). In addition, we detected no statistically relevant differences in fatty acid composition between wild-type and ap2 mutant seeds with some notable exceptions. For example, Table 4 shows that ap2 mutant seeds showed a 37–57% increase in the levels of lignoceric acid (C24:0) when compared with that in parental wild-type seeds. In the case of ap2-4 and ap2-10 seeds, these observed increases were accompanied by compensatory 27% and 20% decreases in oleic acid (C18:1) levels, respectively (Table 4). We conclude that reducing AP2 gene activity increases both seed size and contents, and consequently seed yield, without substantial changes in seed protein and fatty acid composition.
Discussion
In many plant species, seed size and seed mass have been implicated as important determinants in seedling survival and vigor upon germination (38). Field studies have shown that large seeds can confer enhanced seedling establishment and survival (39–42), increased tolerance to flooding (43), and tolerance to insect predation (44, 45). Because of the advantages associated with larger seeds and because of the potential of increasing total seed yield through seed size, it is of agronomic importance to identify the genes involved in the determination of seed size and seed mass. We demonstrate here that AP2 plays an important role in the control of seed mass and seed yield in Arabidopsis. The loss of ap2 activity resulted in seed mass increases as much as 100% greater than that of wild-type seeds (Table 1). The increases in seed mass observed for ap2 mutant seeds are not solely due to the homeotic nature and reduced fertility of ap2 flowers because the mass of seeds produced by severe ap2 mutants like ap2-10 greatly surpassed that observed for seeds produced by ms1 mutant plants (Table 1).
How does AP2 carry out its functions to affect seed size, embryo size, seed weight, and the accumulation of seed reserves? Our conclusion that AP2 is acting in the maternal sporophyte and endosperm (Table 3) is supported by RNA gel blot and RT-PCR analyses that show that AP2 is expressed in all vegetative organs examined and in the developing seed (1, 19). The data presented here also suggest that AP2 activity affects source–sink relations. Consistent with this hypothesis, Ohto et al. (46) have shown that ap2 mutants are altered in sucrose sensing, flowering time, leaf number, and soluble sucrose metabolism in developing seeds. Genetic and physiological studies suggest that AP2 acts in part by suppressing gibberellin signaling to ensure uniformity of flower development against normal physiological fluxes (23). Gibberellins are plant growth regulators that have been shown to promote flowering, seed development, and growth in Arabidopsis and in many other plant systems (47, 48). Our observation that ap2 seeds are consistently more heterogeneous in size whereas wild-type seeds are consistently uniform (Fig. 1) suggests that AP2 could play a similar role in the maintenance of seed size uniformity. Although we show here that the major effect of ap2 on mature seeds is increased cell size (Fig. 2), Ohto et al. (46) have recently shown that ap2 also increases embryo cell number. Consistent with these data, the data of Alonso-Blanco et al. (30) show that at least 11 seed size and seed weight quantitative trait loci contribute to seed size variation in Arabidopsis by affecting both cell number and cell size, with one major quantitative trait locus mapping to the region of chromosome IV that contains AP2. We propose that AP2 may negatively affect the ability of gibberellins to regulate metabolism in both source and sink tissues and in so doing, affect cell size and cell number during seed growth.
Efforts to increase agronomic traits like seed protein or seed oil content in Brassica, soybean, and other seed crops have shown that changes in total seed protein levels are often inversely proportional to changes in seed oil levels in the presence of a fixed supply of assimilates (49–51). That is, total seed protein or total seed oil levels per seed can increase but usually at the expense of the other. In contrast, we observed that seed weight increases were accompanied by increases in both total seed protein and total seed oil contents (Tables 1 and 4). To date the potential impact of homeotic genes like ap2 on agronomic traits like seed weight, seed yield, and seed composition have received little attention from traditional plant breeding efforts because of their strong negative effects on fertility, plant growth, and development. The discovery that such traits can be enhanced in Arabidopsis by a genetic reduction in AP2 copy number may be a useful strategy for identifying new yield-limiting genes in Arabidopsis for analysis in crop plants.
Acknowledgments
We thank Chris Robles for technical assistance and Drs. Tony Kinney and Angus Murphy for assistance with the fatty acid analysis. We also express our appreciation to Drs. John Harada, Jane Silverthorne, and Lincoln Taiz for their encouragement during the course of this study. P.K.O. was supported by a University of California Systemwide Biotechnology Training Grant. This study was supported by the National Institutes of Health (Grant GM46309) and the National Science Foundation (Grant 9604507) and by a gift from Ceres, Inc., to J.K.O.
Abbreviations: AP2, APETALA2; Col, Columbia; Ler, Landsberg erecta; ms1, malesterile1.
References
- 1.Jofuku, K. D., den Boer, B. G., Van Montagu, M. & Okamuro, J. K. (1994) Plant Cell 6, 1211-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M. & Jofuku, K. D. (1997) Proc. Natl. Acad. Sci. USA 94, 7076-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohme-Takagi, M. & Shinshi, H. (1995) Plant Cell 7, 173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson, K., Long, D., Swinburne, J. & Coupland, G. (1996) Plant Cell 8, 659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockinger, E. J., Gilmour, S. J. & Thomashow, M. F. (1997) Proc. Natl. Acad. Sci. USA 94, 1035-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou, J., Tang, X. & Martin, G. B. (1997) EMBO J. 16, 3207-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magnani, E., Sjolander, K. & Hake, S. (2004) Plant Cell 16, 2265-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komaki, M. L., Okada, K., Nishino, E. & Shimura, Y. (1988) Development (Cambridge, U.K.) 104, 195-203. [Google Scholar]
- 9.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1989) Plant Cell 1, 37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunst, L., Klenz, J. E., Martinez-Zapater, J. & Haughn, G. W. (1989) Plant Cell 1, 1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1991) Development (Cambridge, U.K.) 112, 1-20. [DOI] [PubMed] [Google Scholar]
- 12.Léon-Kloosterziel, K. M., Keijzer, C. J. & Koornneef, M. (1994) Plant Cell 6, 385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Western, T. L., Burn, J., Tan, W. L., Skinner, D. J., Martin-McCaffrey, L., Moffatt, B. A. & Haughn, G. W. (2001) Plant Physiol. 127, 998-1011. [PMC free article] [PubMed] [Google Scholar]
- 14.Drews, G. N., Bowman, J. L. & Meyerowitz, E. M. (1991) Cell 65, 991-1002. [DOI] [PubMed] [Google Scholar]
- 15.Mizukami, Y. & Ma, H. (1992) Cell 71, 119-131. [DOI] [PubMed] [Google Scholar]
- 16.Jack, T., Brockman, L. L. & Meyerowitz. E. M. (1992) Cell 68, 683-697. [DOI] [PubMed] [Google Scholar]
- 17.Mandel, M. A., Gustafson-Brown, C., Savidge, B. & Yanofsky, M. F. (1992) Nature 360, 273-277. [DOI] [PubMed] [Google Scholar]
- 18.Goto, K. & Meyerowitz, E. M. (1994) Genes Dev. 8, 1548-1560. [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita, T., Miura, A., Choi, Y., Kinoshita, Y., Cao, X., Jacobsen, S. E., Fischer, R. L. & Kakutani, T. (2004) Science 303, 521-523. [DOI] [PubMed] [Google Scholar]
- 20.Aukerman, M. J. & Sakai, H. (2003) Plant Cell 15, 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, X. (2004) Science 303, 2022-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, Y. Q., Yang, C., Thara, V. K., Zhou, J. & Martin, G. B. (2000) Plant Cell 12, 771-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamuro, J. K., Szeto, W., Lotys-Prass, C. & Jofuku, K. D. (1997) Plant Cell 9, 37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valvekens, D., Van Montagu, M. & Van Lijsebettens, M. (1988) Proc. Natl. Acad. Sci. USA 85, 5536-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadegari, R., Paiva, G., Laux, T., Koltunow, A. M., Apuya, N., Zimmerman, J. L., Fischer, R. L., Harada, J. J. & Goldberg, R. B. (1994) Plant Cell 6, 1713-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naito, S., Dubè, P. H. & Beachy, R. N. (1988) Plant Mol. Biol. 11, 109-123. [DOI] [PubMed] [Google Scholar]
- 27.Koornneef, M., de Bruine, J.-H. & Goettsch, P. (1980) Arab. Inf. Serv. 17, 11-18. [Google Scholar]
- 28.Van der Veen, J. H. & Wirtz, P. (1968) Euphytica 17, 371-377. [Google Scholar]
- 29.Dawson, J., Wilson, Z. A., Aarts, M. G. M., Braithwaite, A. F., Briarty, L. G. & Mulligan, B. J. (1993) Can. J. Bot. 71, 629-638. [Google Scholar]
- 30.Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C. J. & Koornneef, M. (1999) Proc. Natl. Acad. Sci. USA 96, 4710-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esau, K. (1997) Anatomy of Seed Plants (Wiley, New York), 2nd Ed.
- 32.Heath, J. D., Weldon, R., Monnot, C. & Meinke, D. W. (1986) Planta 169, 304-312. [DOI] [PubMed] [Google Scholar]
- 33.Pang, P. P., Pruitt, R. E. & Meyerowitz, E. M. (1988) Plant Mol. Biol. 11, 805-820. [DOI] [PubMed] [Google Scholar]
- 34.van der Klei, H., Van Damme, J., Casteels, P. & Krebbers, E. (1993) Plant Physiol. 101, 1415-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Downey, R. K. (1983) in High and Low Erucic Acid Rapeseed Oils, eds. Kamer, J., Sauer, F. & Pigden, W., (Academic, New York).
- 36.James, D. W., Jr., & Dooner, H. K. (1990) Theor. Appl. Genet. 80, 241-245. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, D. C., Giblin, E. M., Reed, D. W., Hogge, L. R., Olson, D. J. & MacKenzie, S. L. (1995) J. Am. Oil Chem. Soc. 72, 305-308. [Google Scholar]
- 38.Westoby, M., Leishman, M. & Lord, J. (1996) Philos. Trans. R. Soc. London B. 351, 1309-1318. [Google Scholar]
- 39.Armstrong, D. P. & Westoby, M. (1993) Ecology 74, 1092-1100. [Google Scholar]
- 40.Kolawole, G. O. & Kang, B. T. (1997) Commun. Soil Sci. Plant Anal. 28, 223-1235. [Google Scholar]
- 41.Milberg, P. & Lamont, B. B. (1997) New Phytol. 137, 665-672. [Google Scholar]
- 42.Vera, M. L. (1997) Plant Ecol. 133, 101-106. [Google Scholar]
- 43.Fenster, C. B. (1997) Aquat. Bot. 56, 215-231. [Google Scholar]
- 44.Moegenburg, S. M. (1996) Oecologia 106, 539-543. [DOI] [PubMed] [Google Scholar]
- 45.Lopes, E. C. A., Destro, D., Montalvan, R., Ventura, M. U. & Perez-Guerra, E. (1997) Euphytica 97, 161-166. [Google Scholar]
- 46.Ohto, M.-a., Fischer, R. L., Goldberg, R. B., Nakamura, K. & Harada, J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 3123-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeevaart, J. A. D. (1983) in The Biochemistry and Physiology of Gibberellins, ed. Crozier, A. (Praeger Publishers, New York), Vol. 2.
- 48.Taiz, L. & Zeiger, E. (2002) Plant Physiology (Sinauer Assoc., Sunderland, MA), 3rd Ed.
- 49.Denis, L., Dominguez, J., Baldini, M. & Vear, F. (1994) Euphytica 79, 29-38. [Google Scholar]
- 50.Eriksson, I., Westerlund, E. & Aman, P. (1994) J. Sci. Food Agric. 66, 233-240. [Google Scholar]
- 51.Hartwig, E. E., Kuo, T. M. & Kenty, M. M. (1997) Crop Sci. 37, 770-773. [Google Scholar]