Abstract
We describe an 85-yr-old male of Ashkenazi Jewish descent with biopsy-proven locally advanced pancreatic ductal adenocarcinoma (PDA). The patient underwent a modified course of gemcitabine and stereotactic body radiation therapy and survived for 42 mo with a stable pancreatic head mass and no evidence of metastatic disease before death due to complications from a stroke. Whole-exome sequencing of his tumor revealed a simple genome landscape with no evidence of mutations, copy-number changes, or structural alterations in genes most commonly associated with PDA (i.e., KRAS, CDKN2A, TP53, or SMAD4). An analysis of his germline DNA revealed no pathogenic variants of significance. Whole-exome and whole-genome sequencing identified a somatic mutation of RNF213 and an inversion/deletion of CTNNA2 as the genetic basis of his PDA. Although PDA is classically characterized by a predictable set of mutations, these data suggest that alternate genetic paths to PDA may exist, which can be associated with a more indolent clinical course.
Keywords: neoplasm of the pancreas, pancreatic adenocarcinoma
INTRODUCTION
The mountains within the genomic landscape of pancreatic ductal adenocarcinoma (PDA) have been well documented and indicate that four genes—KRAS, CDKN2A, TP53, and SMAD4—are the most frequent targets of somatic alteration (Jones et al. 2008). Many other genes are also recurrently altered, albeit at frequencies of ∼10% or less, that constitute the hills of this landscape. Collectively, these high- and low-frequency genes are believed to target a distinct set of core signaling pathways that, when dysregulated, contribute to pancreatic carcinogenesis.
Despite the typical aggressive clinical course of PDA, a subset of these tumors can demonstrate favorable clinical outcomes after therapy. These include the ∼10% of PDAs associated with germline or somatic mutations in BRCA1/2, which are sensitive to DNA cross-linking agents such as cisplatin (Holter et al. 2015; Waddell et al. 2015). Whole-genome sequencing has further revealed that 14% of PDAs display excessive numbers of structural rearrangements (>200) indicative of a defective DNA-damage phenotype and greater response to platinum chemotherapy (Waddell et al. 2015). Less commonly, PDAs contain mutations in mismatch repair genes, which can lead to excessive mutational burden and sensitivity to immune checkpoint therapy (Le et al. 2015).
Outside of these examples, PDA patients with long-term survival after standard therapy are rare. We describe a patient with locally advanced PDA who survived 42 mo following gemcitabine and stereotactic body radiotherapy (SBRT) and ultimately died of causes unrelated to PDA. Sequencing of this patient's tumor revealed the putative basis for this indolent clinical course was its genomically “flat” landscape that, although targeting the core pathways of this disease, did so in an uncommon manner.
RESULTS
Clinical Presentation
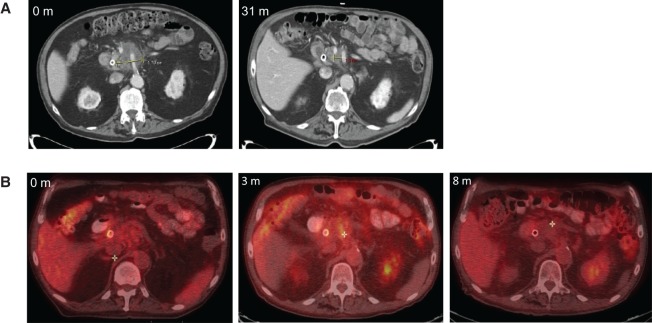
An 85-yr-old male with a past medical history of chronic obstructive pulmonary disease and chronic stable angina and a remote history of early-stage lung adenocarcinoma presented with weight loss and jaundice, with a bilirubin of 2.7 mg/dl. Computed tomography (CT) showed a 2.0-cm pancreatic head mass compressing the bile duct and encasing the superior mesenteric artery and celiac axis (Fig. 1A). Endoscopic retrograde cholangiopancreatography was performed with successful placement of a bile duct stent. A fine-needle aspirate of the mass confirmed the diagnosis of adenocarcinoma. The specimen was too acellular for molecular or genetic studies. Eastern Cooperative Oncology Group (ECOG) performance status at presentation was two and carbohydrate antigen 19-9 (CA 19-9) was measured to be 64.5 U/ml.
Figure 1.
(A) Representative computed tomography (CT) images from the time of diagnosis and ∼10 mo before autopsy. A 2.0-cm diameter pancreatic head mass was identified at diagnosis. Before autopsy and after treatment with gemcitabine and stereotactic body radiotherapy (SBRT), the mass was stable to decreased size. (B) Fluorodeoxyglucose positron emission tomography (PET)-CT scans demonstrating a decrease in size and avidity of the pancreatic lesion by 8 mo after SBRT.
After multidisciplinary review, the patient was deemed to have locally advanced disease and was offered enrollment on a clinical trial combining gemcitabine with fractionated SBRT (Herman et al. 2015). Per protocol, he received three doses of gemcitabine followed by 5 d of SBRT to 33 Gy. Single-agent gemcitabine was then resumed for 3 mo until poor tolerability. A positron emission tomography (PET)-CT performed 3 mo after completion of SBRT demonstrated persistent fluorodeoxyglucose (FDG) avidity, which resolved at 8 mo (Fig. 1B). Carbohydrate antigen (CA) 19-9 also normalized, consistent with a favorable response to treatment.
The patient was then clinically stable until he developed cholangitis and a rising CA 19-9 to 146.6 U/ml ∼31 mo after SBRT. At that time, a CT revealed stable to decreased size of the pancreatic head mass (Fig. 1A). Ascites was also identified, but paracentesis revealed negative cytology. The patient then suffered a stroke and passed away 42 mo after SBRT as a result of complications related to this event.
Genomic Analyses
No metastases were identified at autopsy based on gross and histologic examination of all organs, including the lungs and liver. The primary tumor was submitted in its entirety and contained a 2.3 cm focus of infiltrating well to moderately differentiated duct carcinoma in a background of prominent fibrosis and radiation changes (Fig. 2A). Perineural invasion was identified. Immunolabeling for CK7 and CK19 was positive, and CK20 and CDX2 was negative in the neoplastic cells, consistent with a pancreatic origin (Fig. 2B). There was no histologic evidence of intrapancreatic cysts lined by mucinous epithelial cells to suggest the invasive carcinoma arose in a cystic precursor lesion. Immunolabeling patterns for p53, p16, and SMAD4 proteins were consistent with wild-type status of their respective genes in the neoplastic cells (Fig. 2C).
Figure 2.
(A) Tumor samples obtained at the time of autopsy were hemotoxylin and eosin (H&E) stained and showed extensive fibrosis with focal areas of well-to-moderately differentiated duct carcinoma. (B) Immunolabeling of this same tissue for CK7, CK19, CK20, and CDX2. Staining patterns are consistent with a tumor of pancreatic origin. (C) Immunolabeling patterns for p53, p16, and SMAD4 proteins, all of which are consistent with wild-type status of the genes encoding each of these proteins.
The diagnostic fine-needle aspirate for this patient was largely acellular and thus not amenable to sequencing analysis. To characterize the genetics of this cancer, we therefore processed four tissue samples from the pancreatic tumor collected at the time of autopsy, each derived from a geographically distinct region of the neoplasm. Genomic DNA was extracted from each sample and whole-exome sequencing was performed. Somatic nonsynonymous single-nucleotide variants (SNVs) were identified in 16 different genes (Table 1). None of these 16 missense mutations involved high- or low-frequency somatic targets described for the PDA genome nor were any identified as significantly mutated by bioinformatics algorithms in five different PDA whole-exome or whole-genome sequencing studies (Jones et al. 2008; Biankin et al. 2012; Waddell et al. 2015; Witkiewicz et al. 2015; Bailey et al. 2016). However, one gene, RNF213, was reported to contain nonsilent SNVs in multiple patients in each of four prior PDA studies (Biankin et al. 2012; Waddell et al. 2015; Witkiewicz et al. 2015; Bailey et al. 2016). Overall, the frequency of RNF213 mutation was 1.7% across all 531 PDAs previously analyzed (Fig. 3A). In the present case, the G>C variant within RNF213 at hg19 genome coordinate Chr17:78338242 is predicted to result in a nonconservative amino acid substitution of cysteine for tryptophan at codon 3920. Previously described SNV functional algorithms Polymorphism Phenotyping (PolyPhen) (Adzhubei et al. 2010), Sorting Intolerant From Intolerant (SIFT) (Ng and Henikoff 2001), and Very Efficient Substitution Transposition (VEST) (Carter et al. 2013; Douville et al. 2016) predicted this substitution to be highly damaging at the protein level (Table 1).
Table 1.
Single-nucleotide variants identified by whole-exome sequencing
| Gene symbol | Chr | HGVS DNA reference | HGVS protein reference | Variant type | SIFT prediction | SIFT score | PolyPhen-2 prediction | PolyPhen-2 score | VEST pathogenicity P-value | VEST pathogenicity FDR | dbSNP ID | Genotype | Mutated in PDA studies | Read count per sample (wild-type/mutant) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal | T1 | T2 | T3 | T4 | ||||||||||||||
| HDAC4 | 2 | c.100C>T | p.V34M | Missense | Damaging | 0.001 | Benign | 0.412 | 0.19687 | 0.3 | Heterozygous | No | 188/0 | 517/9 | 171/18 | 439/0 | 377/10 | |
| RGS12 | 4 | c.3940G>A | p.A1314T | Missense | Tolerated | 1.000 | Benign | 0.000 | 0.83625 | 0.85 | rs148763505 | Heterozygous | No | 134/0 | 345/1 | 125/17 | 304/5 | 178/6 |
| HNRNPA0 | 5 | c.196C>A | p.A66S | Missense | Tolerated | 0.494 | Possibly damaging | 0.659 | 0.30144 | 0.4 | Heterozygous | No | 431/1 | 915/4 | 228/30 | 1095/0 | 687/12 | |
| COBL | 7 | c.833G>A | p.T278M | Missense | Damaging | 0.047 | Probably damaging | 1.000 | 0.47385 | 0.6 | Heterozygous | No | 201/0 | 360/7 | 307/37 | 307/0 | 313/18 | |
| FREM1 | 9 | c.4963C>T | p.V1655M | Missense | Tolerated | 0.24 | Probably damaging | 0.963 | 0.18193 | 0.3 | rs114410606 | Heterozygous | No | 95/0 | 166/0 | 126/12 | 187/0 | 170/2 |
| TNNI2 | 11 | c.27G>C | p.R9S | Missense | Tolerated | 0.062 | Benign | 0.218 | 0.22691 | 0.35 | Heterozygous | No | 92/0 | 478/4 | 103/12 | 337/0 | 325/11 | |
| LRRC10B | 11 | c.91C>T | p.R31W | Missense | Damaging | 0.012 | Possibly damaging | 0.926 | 0.09989 | 0.25 | Heterozygous | No | 42/0 | 201/2 | 59/25 | 164/0 | 169/14 | |
| MMP17 | 12 | c.1066C>T | p.R356W | Missense | Damaging | 0.000 | Probably damaging | 1.000 | 0.00751 | 0.1 | Heterozygous | No | 217/1 | 757/3 | 203/27 | 600/5 | 504/10 | |
| PGAM5 | 12 | c.362C>A | p.T121N | Missense | Damaging | 0.000 | Probably damaging | 0.997 | 0.03917 | 0.15 | Heterozygous | No | 68/0 | 358/2 | 124/13 | 379/0 | 434/1 | |
| CDH3 | 16 | c.40C>G | p.L14V | Missense | Tolerated | 0.144 | Benign | 0.003 | 0.51275 | 0.6 | Heterozygous | No | 42/0 | 200/2 | 70/9 | 176/0 | 102/5 | |
| MYO19 | 17 | c.1640T>A | p.N547I | Missense | Damaging | 0.000 | Probably damaging | 1.000 | 0.01905 | 0.1 | Heterozygous | No | 170//0 | 323/3 | 142/17 | 310/0 | 289/3 | |
| RNF213 | 17 | c.11760G>C | p.W3920C | Missense | Damaging | 0.000 | Probably damaging | 1.000 | 0.01688 | 0.1 | Heterozygous | Yes | 104/0 | 235/3 | 83/11 | 197/2 | 167/8 | |
| HIPK4 | 19 | c.91G>A | p.R31W | Missense | Damaging | 0.002 | Probably damaging | 1.000 | 0.01520 | 0.1 | Heterozygous | No | 361/0 | 905/5 | 356/37 | 823/0 | 756/24 | |
| PIWIL3 | 22 | c.2462G>A | p.T821M | Missense | Damaging | 0.013 | Probably damaging | 1.000 | 0.19481 | 0.3 | Heterozygous | No | 64/0 | 156/1 | 97/11 | 229/1 | 145/5 | |
| PLXNB2 | 22 | c.4291G>C | p.Q1431E | Missense | Damaging | 0.002 | Probably damaging | 0.999 | 0.02196 | 0.1 | Heterozygous | No | 347/0 | 1121/14 | 374/33 | 1049/2 | 909/12 | |
| CFAP47 | X | c.2524G>A | p.A842T | Missense | Damaging | 0.000 | Probably damaging | 1.000 | 0.02964 | 0.1 | Heterozygous | No | 46/0 | 73/0 | 82/8 | 110/0 | 77/3 | |
Chr, chromosome; HGVS, Human Genome Variation Society; SIFT, Sorting Tolerant From Intolerant; PolyPhen-2, Polymorphism Phenotyping v2; VEST, Very Efficient Substitution Transposition; dbSNP, Database for Short Genetic Variations; PDA, pancreatic ductal carcinoma.
Figure 3.
(A) A lollipop plot of all reported mutations in RNF213 among a total of 531 sequenced pancreatic ductal adenocarcinomas (PDAs) (green, missense mutation; black, frameshift mutation). These include the present case (large green lollipop with a star) that contains a potentially damaging W3920C mutation. (B) Bayesian analysis of mutant allele frequencies for all missense mutations identified by whole-exome sequencing. The probability of a mutation being in a given sample ranges from likely absent (>99.99% confidence, dark red) to likely present (>99.99% confidence, dark blue). Based on these results, sample T3 was excluded from further analysis. (C) Proposed phylogenetic tree based on sequencing data of samples T1, T2, and T4. Numbers represent quantity of mutations gained in predicted subclones (SC1, SC2) or tumor samples. (D) Circos plot detailing the structural alterations found by whole-genome sequencing, including CTNNA2. Inversions are shown in red, deletions in blue, and duplications in green. (E) Phylogenetic tree of the present case drawn to scale compared with the average trunk length of PDA based on published data (Yachida et al. 2010). Dashed lines of branches in the standardized tree are simulated lengths and have no quantitative value. SNVs, single-nucleotide variants.
Gene level copy-number analysis identified six genes with amplifications of five or more copies in at least one sample, none of which affected previously reported PDA driver genes (Supplemental Table S2). Given the small number of somatic mutations and copy-number alterations found, we performed an analysis of the patient's germline DNA to assess for deletions or other pathogenic variants of significance. None were found, suggesting that this PDA arose sporadically.
Analysis of mutant allele frequencies in individual tumor samples revealed that all 16 missense mutations were present with high confidence in sample T2 (Table 1; Fig. 3B). In contrast, sample T3 had no mutations that could be called with high confidence (Reiter et al. 2017), likely because of low tumor cellularity. A phylogenetic reconstruction of this PDA based on samples T1, T2, and T4 indicated an extremely short trunk with only three founder mutations (Fig. 3C). We next hypothesized that one or more structural events may have occurred in this patient's tumor that were not detectable by whole-exome sequencing. Genomic DNA from the most cellular tissue sample of the primary tumor (sample T2, estimated tumor cellularity 40%) was therefore further analyzed for structural rearrangements by whole-genome sequencing. A total of 10 intrachromosomal structural rearrangements were identified that involved seven different genes and six chromosomes (Fig. 3D; Supplemental Table S3). One of these, catenin alpha 2 (CTNNA2), has previously been described as significantly mutated or structurally disrupted in PDA (Jones et al. 2008; Bailey et al. 2016). It has also been proposed to function as a tumor-suppressor gene in head and neck and gastric cancers (Fanjul-Fernandez et al. 2013; Wang et al. 2014). In addition, a 19.9-Mb deletion of Chromosome 1q that included RAB GTPase-activating protein 1-like (RABGAP1L) was identified. This gene has also been identified as disrupted by a structural alteration in PDA, although its functional significance remains uncertain (Jones et al. 2008; Bailey et al. 2016).
DISCUSSION
This elderly patient presented with an unresectable PDA and modest performance status, a common clinical scenario for this disease. After treatment with SBRT and gemcitabine, a therapeutic regimen associated with a median survival of 13.9 mo (Herman et al. 2015), the patient had a marked involution of his tumor that persisted until he died of other causes almost 4 yr later. Although this PDA is quite classical based on routine histology, a thorough genetic analysis revealed a genomically “flat” landscape meeting minimal genetic requirements for PDA carcinogenesis.
Previous studies of PDA phylogenetic trees have revealed trunks that contain an average of 40 founder mutations (Yachida et al. 2010). Phylogenetic analysis of this tumor showed a much smaller trunk more analogous to that of a sapling rather than a tree (Fig. 3E), suggesting that tumorigenesis occurred following a rare event rather than from a mitotically active precursor lesion. This tree not only had a short trunk but also one long and one short branch, suggesting a relative lack of genomic instability within the tumor. Interestingly, our thorough genetic characterization revealed no mutations in classical PDA driver genes. Because genomic studies often begin with screening of samples for the presence of a common PDA driver gene mutation as a surrogate for quality, examples of PDA as presented here are often excluded. In doing so, studies can be biased against patients with these unique genetic subtypes of PDA that may be treatable with standard therapies. This patient is an ideal example—despite being elderly and having a suboptimal performance status, he experienced a favorable outcome after an aggressive yet tolerable course of chemotherapy and radiation.
In this patient, we hypothesize that the indolent clinical course after therapy was the result of a remarkable lack of mutations that are typically associated with aggressive PDA biology. Without classical PDA driver gene mutations in this case, p53 function would be expected to be intact, KRAS signaling would remain in the normal physiologic range, and transforming growth factor β (TGF-β) signaling would maintain a degree of tissue homeostasis. Although the possibility remains that additional genetic alterations were present in the primary tumor at diagnosis that were eradicated by treatment, previous analyses of treated PDAs have demonstrated persistence of driver gene mutations after chemoradiation (Yachida et al. 2010).
Several alternate driver gene candidates were identified in our analysis of this unique tumor. CTNNA2, a known tumor suppressor that has been identified in other PDA studies, likely played a role in this tumor. RNF213 is also a compelling candidate for a rare but important PDA driver gene given its role in noncanonical Wnt signaling and the presence of inactivating mutations in a small number of PDAs from four other independent studies. The significance of the RABGAP1L deletion is less clear. Its GTPase function may overlap with aspects of KRAS, yet few data are available describing the function of this protein product.
Although PDA is classically characterized by a predictable set of mutations, these data suggest that alternate genetic paths to PDA are sometimes associated with a more indolent clinical course. As comprehensive genomic studies of PDA become more common, the frequency with which such cases are identified will likely increase. By better characterizing these less common genetic mechanisms, we can perhaps gain a more thorough understanding of the genomic features of PDA most sensitive to standard of care therapies and of the “minimal genetic requirements” for PDA tumorigenesis.
Methods
Tissue Samples and Processing
Tissue samples of the entire primary tumor and representative normal tissues collected at autopsy were snap-frozen and stored at −80°C. Frozen tissue was embedded in Tissue-Tek O.C.T. compound and 5–10 µM sections cut using a Leica cryostat. Sections were stained with hematoxylin and eosin and visually assessed for neoplastic cellularity using a two-headed microscope. Areas with tumor cellularity >30% were macrodissected from serial unstained sections.
Immunohistochemistry
Samples of the primary carcinoma were fixed and paraffin-embedded. Unstained 5-μm sections were cut and deparaffinized by routine techniques followed by incubation in 1× target retrieval solution (Dako) and steaming for 25 min at 70°C. Slides were cooled for 20 min, washed with 1× wash buffer (Dako), treated with 3% H2O2 for 10 min, washed again, and incubated with antibodies to CK7 (1:500 dilution, cat no. m7018; DAKO), CK19 (1:50, clone RCK108, Dako), CK20 (1:500 dilution, cat. no. m7019; DAKO), CDX2 (1:50 dilution, MU392A-UC; BioGenex), p16 (CINtec p16 histology, cat. no. 705-4713, Ventana), p53 (CONFIRM DO-7, cat. no. 790-2912, Ventana), or SMAD4 (1:200 dilution, clone EB618Y, Abcam).
Whole-Exome Sequencing
Genomic DNA was extracted using standard phenol–chloroform extraction methods. Whole-exome sequencing was performed to a mean depth of 292× with mean distinct coverage of 189× (Supplemental Table S1). Sequencing was performed by Personal Genome Diagnostics using methods previously described (Jones et al. 2015). Briefly, DNA was sheared using a Covaris sonicator and Illumina TruSeq library construction (Illumina) was performed using 50 ng to 3 µg of gDNA according to manufacturer's instructions. Exonic regions were captured using the Agilent SureSelect system (Agilent) per manufacturer's instructions. Paired-end sequencing was performed using HiSeq 2500 instrumentation (Illumina) to achieve sequencing of 100 bases from each end of the fragments. Analysis of somatic variants and copy-number alterations was performed by Personal Genome Diagnostics using methods previously described in detail (Jones et al. 2015). Somatic variants detected by these methods were further validated using MuTect (Cibulskis et al. 2013).
Whole-Genome Sequencing
Genomic DNA extracted from tissue was subjected to whole-genome sequencing by Human Longevity Inc. Whole-genome sequencing was performed to a depth of 83× with a distinct coverage of 78×. Briefly, genomic DNA was quantified with a Quant-iT fluorescence assay (Life Technologies) and an eight-point standard curve. The genomic DNA was then normalized and sheared with a Covaris LE220 instrument. Next-generation sequencing library preparation was carried out using the TruSeq Nano DNA HT kit (Illumina), essentially following the manufacturer's recommendations. Individual DNA libraries were characterized in regard to size and concentration using a LabChip DX Touch (PerkinElmer) and Quant-iT (Life Technologies), respectively. Libraries were normalized to 2–3.5 nM and stored at −20°C until used. Normalized DNA libraries were combined into multisample pools and clustered on cBot cluster stations following the manufacturer's recommendations. Flow cells were subsequently sequenced on the HiSeqX (Illumina) utilizing a 150 base paired-end single index format. Analysis of structural variants was then performed by Personal Genome Diagnostics using methods previously described in detail (Sausen et al. 2013; Jones et al. 2015).
ADDITIONAL INFORMATION
Data Deposition and Access
Sequence data have been deposited at the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/search/site), which is hosted by the European Bioinformatics Institute (EBI) and the Centre Regulació Genòmica (CRG), under accession number EGAS00001002192. Interpreted variants have been submitted to the Catalogue of Somatic Mutations in Cancer (COSMIC) database under accession number COSP43200.
Ethics Statement
The patient provided written informed consent to participate in the clinical trial of SBRT and gemcitabine. In the time during which his clinical status deteriorated, his legal next of kin provided written informed consent for a research autopsy upon his death. Both protocols were approved by the Johns Hopkins Institutional Review Board. Approval for publication was obtained from next of kin.
Author Contributions
J.M.H., J.P., and C.A.I.-D. participated in treatment of the patient and tissue collection. L.M.R., J.M.H., and C.A.I.-D. collected and analyzed clinical data. Z.A.K., J.P., and C.A.I.-D. performed the experiments. Z.A.K., J.H., M.A.A., A.M.-M., and C.A.I.-D. analyzed experimental data. Z.A.K. and C.A.I.-D. wrote the manuscript. All coauthors assisted in preparing and reviewing the manuscript.
Funding
This work was supported by National Institutes of Health grants CA140599, CA179991, and P30 CA008748.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, et al. 2016. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531: 47–52. [DOI] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. 2012. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. 2013. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics 14Suppl 3: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville C, Masica DL, Stenson PD, Cooper DN, Gygax DM, Kim R, Ryan M, Karchin R. 2016. Assessing the pathogenicity of insertion and deletion variants with the variant effect scoring tool (VEST-Indel). Hum Mutat 37: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanjul-Fernandez M, Quesada V, Cabanillas R, Cadinanos J, Fontanil T, Obaya A, Ramsay AJ, Llorente JL, Astudillo A, Cal S, et al. 2013. Cell–cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat Commun 4: 2531. [DOI] [PubMed] [Google Scholar]
- Herman JM, Chang DT, Goodman KA, Dholakia AS, Raman SP, Hacker-Prietz A, Iacobuzio-Donahue CA, Griffith ME, Pawlik TM, Pai JS, et al. 2015. Phase 2 multi-institutional trial evaluating gemcitabine and stereotactic body radiotherapy for patients with locally advanced unresectable pancreatic adenocarcinoma. Cancer 121: 1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holter S, Borgida A, Dodd A, Grant R, Semotiuk K, Hedley D, Dhani N, Narod S, Akbari M, Moore M, et al. 2015. Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J Clin Oncol 33: 3124–3129. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E, et al. 2015. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 7: 283ra253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. 2015. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2001. Predicting deleterious amino acid substitutions. Genome Res 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JG, Makohon-Moore AP, Gerold JM, Bozic I, Chatterjee K, Iacobuzio-Donahue CA, Vogelstein B, Nowak MA. 2017. Reconstructing phylogenies of metastatic cancers. Nat Commun 8: 14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA Jr, Papadopoulos N, et al. 2013. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet 45: 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. 2015. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. 2014. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 46: 573–582. [DOI] [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. 2015. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 6: 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467: 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.