Abstract
A triple-negative breast cancer patient had no hereditary BRCA1, BRCA2, or TP53 risk variants. After exhaustion of standard treatments, she underwent experimental treatments and whole-exome sequencing of tumor, blood, and a metastasis. Well-tolerated experimental bortezomib monotherapy was administered for a progression-free period of 11 mo. After progression, treatments were changed and the exome data were evaluated, expanded with RNA and exome sequencing of a late-stage metastasis. In the final stage, eribulin alone and in combination with anthracyclines were administered. While suffering from grade 3 adverse events, skin metastases progressed. She lived 51 mo after initial diagnosis.
Toxicity from anthracyclines and cisplatin may have been due to associated germline variants CBR3 C4Y and V224M and GSTP1 I105V, respectively. Somatic mutations predicted or reported as pathogenic were detected in 38 genes in tumor tissues. All tumor samples harbored the heterozygous TP53 Y220C variant, known to destabilize p53 and down-regulate p53-mediated apoptosis. The success of bortezomib may be explained by the previously reported up-regulation of caspase-mediated apoptosis, which is p53-independent. Phylogenetic analysis of blood, primary tumor, and two metastases inferred an ancestral tumor cell with 12 expressed tumor mutations from which all three tumors may have evolved.
Although our first urgent analysis could only include 40 genes, postmortem analysis uncovered the aggressiveness and suggested experimental therapies including 16 actionable targets, partly validated by immunohistochemistry. Exome and transcriptome analyses yielded comprehensive therapy-relevant information and should be considered for patients at first diagnosis.
Keywords: ductal carcinoma in situ, multifocal breast carcinoma
INTRODUCTION
About one in five breast cancer patients test negative for the estrogen, progesterone, and Her2 receptors in their tumor tissues. The diagnosis of triple-negative breast cancer (TNBC) is associated with a higher recurrence rate and a less favorable prognosis than receptor-positive breast cancer. About 10% of younger patients with TNBC have a germline BRCA1 risk variant for contralateral breast cancer and ovarian cancer (Robertson et al. 2012). Breast cancer patients with a familial history of cancer or who are young are increasingly being offered genetic counseling and testing for germline risk variants in BRCA1, BRCA2, TP53, and other genes. Patients with germline BRCA risk variants are then offered bilateral mastectomy. Patients without germline TP53 risk variants may be offered adjuvant radiotherapy, whereas germline TP53 risk variant carriers may be at an increased risk for radiation induced secondary cancers. In addition to germline testing, breast cancer tissue testing is increasingly being proposed to match approved cancer drugs with the patient's individual somatic mutations or gene expression profile (Vasan et al. 2014; Meißner et al. 2015).
After the initial diagnosis, the now deceased TNBC patient was tested for BRCA1, BRCA2, and TP53 germline risk variants when she underwent standard treatments. After segmentectomy, relapse, bilateral mastectomy, and completing the last line of standard treatments, the patient decided to undergo additional experimental off-label therapy. Her treatments, which included bortezomib, eribulin, and pembrolizumab, are summarized in Figure 1. Whole-exome sequencing was performed on peripheral blood (PB), primary tumor (PT), and a metastasis (MK), after standard treatments failed. Whole-exome sequencing was chosen to allow the analysis of ∼90% of all genomic protein coding regions, giving a much more complete scientific and potential clinical data yield than gene panel testing. Two years later, RNA and whole-exome sequencing were performed on a fine needle aspirate of a metastasis (MF) taken at the final stage of the disease. Detailed somatic mutation analysis was performed but interpretation could not be completed until postmortem. The mutational signatures (Fig. 2) were associated with defective DNA mismatch repair, which nowadays would probably trigger immunotherapy (Le et al. 2015).
Figure 1.
Patient history. Time line in months summarizing surgical procedures, diagnoses, drug treatments, adverse events, and molecular findings. Previously reported associations with gene variants/mutations are shown in red, and associations with gene expression are shown in green. Time line colors and boxes are yellow for germline, red for ancestral tumor cell, blue for mv2 (see Fig. 3), brown for primary tumor, lilac for first metasis (MK), gray for final metasis (MF), and orange for unsampled terminal tumor clones. Gene names are in italics, up-regulation is shown by an arrow, PSM members PSMB1 and PSMB5 of the proteasome family. tox, toxicity; mets, metastases; p53mut, mutated p53.
Figure 2.
Somatic signatures. Pie charts showing the weights of each Catalogue of Somatic Mutations in Cancer (COSMIC) somatic signature assigned for the samples primary tumor (A), MK (B), and MF (C). Signature 1: aging (i.e., the result of an endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine). Signature 3: found in breast, ovarian, and pancreatic cancers. It is associated with failure of DNA double-strand break repair by homologous recombination. It is strongly associated with germline and somatic BRCA1 and BRCA2 mutations in breast, pancreatic, and ovarian cancers. In pancreatic cancer, responders to platinum therapy usually exhibit Signature 3 mutations. Signature 5: found in all cancer types and most cancer samples. Etiology is unknown. Signature 6: associated with defective DNA mismatch repair and found in microsatellite unstable tumors. Signature 7: likely due to ultraviolet light exposure. Signature 12: usually contributes a small percentage (<20%) of the mutations observed in a liver cancer sample. Signature 15: associated with defective DNA mismatch repair. Signature 26: believed to be associated with defective DNA mismatch repair. Signature 29: has been observed only in gingivo-buccal oral squamous cell carcinoma, pattern of C>A mutations due to tobacco chewing.
The mutations furthermore allowed us to infer tumor evolution and an ancestral tumor cell (Fig. 3), which may have been of interest for treatment decisions, for diagnostic surveillance, and for the patient's own interest in understanding her disease. We analyzed germline pharmacogenomic variants in the exome data and found mutations associated with adverse drug toxicities from which the patient suffered. However, PharmGKB evidence levels for many of these drug–variant combinations are currently low. In summary, our aims were to find out (a) whether gene expression or somatic mutations may explain the efficacy of bortezomib in this patient, or her bone and soft tissue metastases, which were not detected early using CT and bone scintigraphy imaging, and (b) whether certain variants may explain her adverse reactions to anthracyclines, capecitabine, and cisplatin. Finally, we also discuss hypothetical treatment options arising from the completed analysis results.
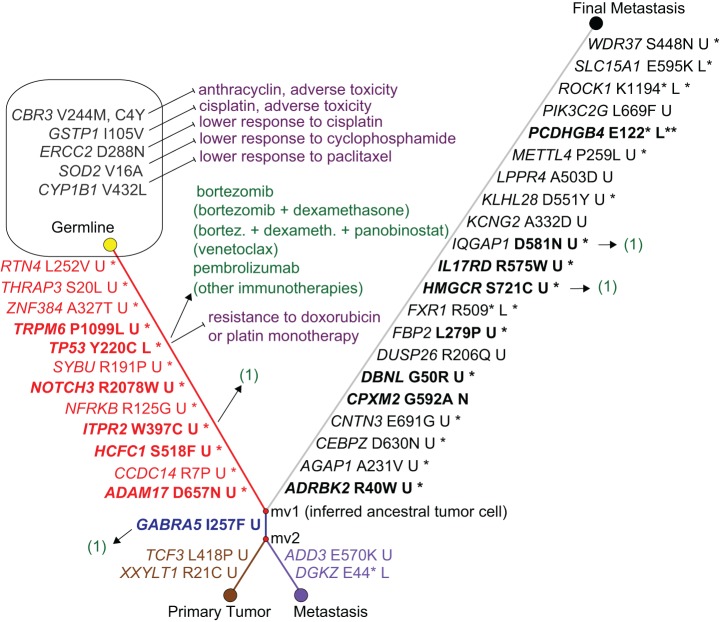
Figure 3.
Tumor evolution phylogeny, ancestral tumor cell and mutation-matched drug associations. The phylogenetic network shows that all three tumor nodes branch from the inferred ancestral tumor cell node mv1, which is characterized by 12 somatically mutated genes including TP53, NOTCH3, and ADAM17. The gene names on the links are the differences between a pair of nodes, harboring a rare/novel mutation predicted or known as disease-causing. Bold names indicate genes previously reported for breast cancer/TNBC. Amino acid changes are shown in Human Genome Variation Society (HGVS) nomenclature, followed by functional consequences: (U) unknown; (L) loss of function; (N) unknown but possibly neutral. A star indicates that the mutated allele was detected in the RNA-seq of the final metastasis, and two stars indicate nonsense-mediated decay of the stopgain-mutated allele. TP53 harbors a known pathogenic mutation that is expressed. The primary tumor and first metastasis each harbor 3 additional somatically mutated genes including shared GABRA5, and the final metastasis shows 21 additional somatically mutated genes. Potentially favorable drugs are associated to the mutated genes with an arrow, and potentially unfavorable drugs with a “stop” arrow. Drugs that were administrated to the patient are not in parentheses; hypothetical drugs that target the known TP53 mutation with at least some human in vivo evidence that were not administered are in parentheses. (1) Stands for hypothetical drugs that are discussed in the Discussion. The patient's polymorphisms in the shown germline genes are associated with adverse toxicity and lower response.
RESULTS
Clinical Presentation
Primary Tumor: Diagnosis and Treatment
A 55-yr-old female of central European ancestry presented with breast cancer of the left mamma. Figure 1 summarizes her patient history, course of treatments, adverse events, and associated molecular findings. She had a previous history of pollinosis and of surgeries for appendicitis, tonsillitis, a cyst at the left thyroid gland, and varicosis. Her primary tumor was a poorly differentiated invasive ductal breast carcinoma in the left lower quadrant with distinct lymphangiosis carcinomatosa. Immunohistochemical (IHC) staining of tumor tissue for the estrogen, progesterone, and Her2 receptors was negative. In a blood test, no pathogenic germline risk variants were detected in BRCA1, BRCA2, or TP53. Surgical treatment consisted of segmentectomy of the left mamma after sonographic marking, sentinel node biopsy, and axillary lymphonodectomy. Surgery was followed by dose escalating dose-dense sequential adjuvant chemotherapy (Citron et al. 2003; Citron 2008) with 4× concurrent epirubicin and cyclophosphamide (EC) every 2 wk followed by paclitaxel, a microtubule-targeting agent, every 2 wk. In months 5 and 6, postoperative radiotherapy was performed on the left breast with a dose of 50.4 Gy and a boost to 61.2 Gy onto the tumor region in the lower left quadrant. Radiotherapy was furthermore applied to the ipsilateral supraclavicular and axillary lymph drainage region with a dose of 50.4 Gy.
First Recurrence: Diagnosis and Treatment
In month 17, no signs of metastases were visible using computed tomography (CT) imaging. In month 18, we diagnosed a local recurrence on the left side, a secondary tumor in the operation scar region, and another tumor at 3 o'clock. Mastectomy was performed on the left breast and also—at the patient's decision—on the contralateral breast. Axillary dissection by the surgeon and histology by the pathologist showed histologically poorly differentiated invasive ductal mamma carcinoma and associated ductal carcinoma in situ (12 mm) with a high core malignancy score. Bone scintigraphy showed no signs of skeletal metastases. From months 18 to 20, three cycles of chemotherapy with cisplatin and ifosfamide were applied and then stopped because of complications (hyponatremia, genital abscess, superinfected mamma) and grade 3 adverse events (protracted nausea/emesis, hematological toxicity, polyneuropathy). From months 23 to 39, adjuvant therapy with bortezomib was applied in 14-d intervals.
Second Recurrence: Diagnosis
In month 36 the first clinical signs of local recurrence occurred. In month 39, an extensive local recurrence was diagnosed with diffuse lymphangiosis carcinomatosa and soft tissue infiltration in the left thoracic aperture, the left thoracic wall, and the shoulder girdle muscles, with infiltration into the truncus inferior of the left plexus brachialis. The patient suffered from paresis of the left shoulder. Multifocal locoregional soft tissue metastases were found prepectorally on the right side, mediastinally, along the left arteria thoracica interna, paraaortic in the abdomen, and interaortocavic immediately below the diaphragm. Bone metastases were seen in the marrow of the right proximal femur. Magnetic resonance imaging (MRI) indicated the possibility of pulmonal metastasis, and differential diagnosis came up with lymphonodular metastasis.
Palliative Chemotherapy
Palliative chemotherapy was applied using a combination of capecitabine and bevacizumab (antibody targeting VEGF) from months 39 to 43 with partial response and grade 2 and 3 hand–foot syndrome as subjectively tolerable adverse events. General system-wide and local progression occurred very suddenly in month 43, and the therapy was changed to vinorelbin monotherapy, a microtubule targeting agent. In month 46, CA15-3 increased considerably and local progression was seen, therefore the therapy was switched to gemcitabine. In month 47, rapid progression, pain, and swelling occurred. A skin metastasis biopsy was taken for IHC and next-generation sequencing analyses, and the therapy was changed to eribulin, a microtubule-targeting agent, leading to a clear response. In the course of month 48, progression occurred. Therefore, in month 49, therapy was changed to a combination of eribulin with pegylated doxorubicin, with grade 3–4 stomatitis as adverse events. Hospitalization then became necessary. Although differential diagnosis was not clear at this point, as to whether adverse events or tumor progression were the cause for the patient's deterioration, the doses of eribulin and pegylated doxorubicin were reduced. Pembrolizumab then was finally applied according to the patients’ wish. The patient died 6 d later as a result of cancer progression at 51 mo after first diagnosis.
Genomic Analysis
Next-Generation Sequencing
Sequencing statistics for whole-exome sequencing (WES) and RNA sequencing of the final metastasis sample are listed in Table 1. For samples PB, PT, and MK, a variant call format (VCF) file was available only, hence sequencing statistics were not available to us.
Table 1.
Next-generation sequencing statistics for sample MF (WES and RNA-seq)
| MF WES | RNA-seq 50 ng | RNA-seq 100 ng | |
|---|---|---|---|
| Total reads | 41,389,563 | 110,576,133 | 81,360,029 |
| Mapped reads | 99.15% | 97.42% | 97.64% |
| Duplicated reads | 39.3% | 70.8% | 70.8% |
| On target ratea | 77.6% | 93.2% | 91.7% |
| Mean coveragea | 122× | Not applicable | Not applicable |
MF, final metastasis; WES, whole-exome sequencing.
aMapping statistics are based on TruSeq Exome regions and computed using Qualimap without the duplicated reads.
Somatic Signatures
Mutational processes can be characterized by unique combinations of mutation types in the form of mutational signatures. We analyzed somatic mutations from the primary tumor sample and the two metastatic samples for mutational signatures based on a set of 30 signatures identified and hosted at Catalogue of Somatic Mutations in Cancer (COSMIC). We detected a total of 9/30 signatures (Signatures 1, 3, 5, 6, 7, 12, 15, 26, 29) among the three samples. Signatures 1, 3, and 12 were detected in all three samples. PT and MK shared signatures 1, 3, 7, 12, and 15, with signature 26 in addition being present in PT. Compared with PT and MK, MF did not have signatures 7, 15, and 26, but signatures 5, 6, and 29 were present (Fig. 2).
Pharmacogenomic Germline Variants
Relevant germline variants were detected in CBR3, CYBA, CYP1B1, ERCC2, GSTP1, and SOD2, which are associated with toxicity or response to anthracyclines, cyclophosphamide, and cisplatin. These variants and the evidence levels as curated by PharmGKB (Whirl-Carrillo et al. 2012) are shown in Table 2.
Table 2.
Pharmacogenomic germline variants associated with toxicity and/or effectiveness
| Gene | Genomic coordinates (hg19) | HGVS cDNA | HGVS protein | Variant type | PharmGKB evidence level | dbSNP | Genotype | Comments |
|---|---|---|---|---|---|---|---|---|
| CBR3 | Chr 21: 37518706 | NM_001236: c.730G>A | p.V244M | Missense | 2B | rs1056892 | het | Higher doxorubicin cardiotox. |
| CBR3 | Chr 21: 37507501 | NM_001236: c.11G>A | p.C4Y | Missense | 3 | rs8133052 | het | Doxorub. neutropenia, cardiotox. |
| CYBA | Chr 16: 88713236 | NM_000101: c.214T>C | p.Y72H | Missense | 3 | rs4673 | hom | Lower acute doxorub. cardiotox. |
| CYP1B1 | Chr 2: 38298203 | NM_000104: c.1294G>C | p.V432L | Missense | 3 | rs1056836 | het | Less effective dose-intense paclitaxel |
| DPYD | Chr 1: 98348885 | NM_000110: c.85C>T | p.R29C | Missense | 3 | rs1801265 | hom | Decreased 5-FU tox risk |
| ERCC2 | Chr 19: 45867259 | NM_001130867: c.862G>A | p.D288N | Missense | 3 | rs1799793 | hom | Lower cisplatin response |
| GSTP1 | Chr 11: 67352689 | NM_000852: c.313A>G | p.I105V | Missense | 2A | rs1695 | het | Increased cisplatin tox risk |
| SOD2 | Chr 6: 160113872 | NM_000636: c.47T>C | p.V16A | Missense | 2B | rs4880 | het | Less effective cyclophosphamide, lower tox |
PharmGKB evidence levels: 2A, moderate evidence and variant in Very Important Pharmacogene; 2B, moderate evidence; 3, unreplicated or conflicting evidence.
HGVS, Human Genome Variation Society; dbSNP, Database for Short Genetic Variations; het, heterozygous; hom, homozygous.
Tumor Evolution Phylogeny and Ancestral Tumor Cell Clone
We constructed a phylogenetic network from somatic mutations in the primary tumor, the first metastasis, and the final metastasis, using the peripheral blood sample as the germline node in the network. The somatic mutations that we focused on are shown in Figure 3. No homozygous mutation was seen among these somatic mutations. All included somatic mutations were either unknown or occur with a low allele frequency in the general population. Six of these mutations are known to occur in cancer patients (i.e., the TP53, HCFC1, ADD3, IL17RD, PIK3C2G, ROCK1 mutations) and all remaining mutations were predicted to be damaging by two prediction tools, MutationTaster (Schwarz et al. 2014) and Combined Annotation-Dependent Depletion (CADD) (Kircher et al. 2014). The network shows that all three tumor samples evolved from a common precursor tumor clone with 12 somatic mutations shared in all three tumor samples (Fig. 3). Of note, this precursor clone includes somatic missense mutations in TP53, HCFC1, ADAM17, and ITPR2, which are detailed in Table 3. The primary tumor additionally includes missense mutations in GABRA (shared with the first metastasis, Table 3), CHAD, and others (Table 3). The first metastasis (month 18) includes missense mutations in DKGZ and ADD3 (Table 3), and the final metastasis (month 47) harbors 18 additional missense mutations and also three stopgain mutations, in FXR1, PCDHGB4, and ROCK1 (Table 3).
Table 3.
Somatic rare pathogenic or predicted damaging mutations, which are discussed in the Results or Discussion
| Gene | Genomic coordinates (hg19) | HGVS cDNA | HGVS protein | Variant type | Predicted effect | dbSNP, COSMIC | Genotype | Comments |
|---|---|---|---|---|---|---|---|---|
| Shared mutations in all tumor samples | ||||||||
| ADAM17 | Chr 2: 9633900 | NM_003183: c.1969G>A | p.D657N | Missense | D/34 | het | RNA. TNBC, BrCa progression | |
| CCDC14 | Chr 3: 123680145 | NM_022757: c.20G>C | p.R7P | Missense | D/16 | het | RNA. Centriole duplication | |
| HCFC1 | Chr X: 153224834 | NM_005334: c.1553C>T | p.S518F | Missense | D/25 | het | RNA. BrCa driver, BRCA1 pathway | |
| ITPR2 | Chr 12: 26835564 | NM_002223: c.1191G>T | p.W397C | Missense | D/21 | het | RNA. BrCa | |
| NFRKB | Chr 11: 129758492 | NM_006165: c.373C>G | p.R125G | Missense | D/17 | het | RNA. Candidate oncogene | |
| NOTCH3 | Chr 19: 15272207 | NM_000435: c.6232C>T | p.R2078W | Missense | D/17 | het | Neural development, BrCa | |
| RTN4 | Chr 2: 55254481 | NM_020532: c.754C>G | p.L252V | Missense | D/15 | het | Cell adhesion, migration, metastasis, and apoptosis | |
| SYBU | Chr 8: 110588198 | NM_001099746: c.572G>C | p.R191P | Missense | D/21 | het | RNA. Microtubule and neural associated | |
| THRAP3 | Chr 1: 36748223 | NM_005119: c.59C>T | p.S20L | Missense | D/28 | het | RNA. Cancer growth, damage response | |
| TP53 | Chr 17: 7578190 | NM_000546: c.659A>G | p.Y220C | Missense | D/22 | rs121912666 COSM10758 | het | RNA. Pathogenic. Germline in Li–Fraumeni, somatic |
| TRPM6 | Chr 9: 77390891 | NM_001177310: c.3296C>T | p.P1099L | Missense | D/21 | het | BrCa, magnesium homeostasis | |
| ZNF384 | Chr 12: 6781631 | NM_001135734: c.979G>A | p.A327T | Missense | D/21 | het | RNA. Fusions with TAF15 or EP300 in ALL | |
| Shared mutations in primary tumor and first metastasis | ||||||||
| GABRA5 | Chr 15: 27185116 | NM_000810: c.769A>T | p.I257F | Missense | D/23 | het | GABA inhibits BrCa cell migration | |
| Private mutations in primary tumor | ||||||||
| TCF3 | Chr 19: 1619388 | NM_001136139: c.1253T>C | p.L418P | Missense | D/17 | het | Not a mutational driver in BrCa | |
| XXYLT1 | Chr 3: 194991727 | NM_152531: c.61C>T | p.R21C | Missense | D/24 | het | NOTCH pathway | |
| Private mutations in first metastasis | ||||||||
| ADD3 | Chr 10: 111890220 | NM_001121: c.1708G>A | p.E570K | Missense | D/33 | COSM914716 | het | Endometrial carcinoma |
| DGKZ | Chr 11: 46354955 | NM_201532: c.130G>T | p.E44X | Stopgain | D/37 | het | Induces cell-cycle arrest, is target of p53 | |
| Private mutations in final metastasis | ||||||||
| FXR1 | Chr 3: 180688068 | NM_001013438: c.1525C>T | p.R509X | Stopgain | A/36 | het | RNA. Activation of cyclin-dependent kinase inhibitors, p53 | |
| HMGCR | Chr 5: 74655244 | NM_001130996: c.2161A>T | p.S721C | Missense | D/24 | het | RNA. BrCa, dereg. lipogenesis ass. with mut. p53 | |
| IL17RD | Chr 3: 57132008 | NM_017563: c.1723C>T | p.R575W | Missense | D/16 | COSM249014 | het | RNA. BrCa. epithelial to mesenchymal transformation, migration/invas. |
| IQGAP1 | Chr 15: 90999512 | NM_003870: c.1741G>A | p.D581N | Missense | D/20 | het | RNA. BrCa. Cell invasiveness | |
| PCDHGB4 | Chr 5: 140767815 | NM_003736: c.364G>T | p.E122X | Stopgain | A/34 | het | Nonsense-mediated decay. BrCa. | |
| PIK3C2G | Chr 12: 18552594 | NM_004570: c.2005C>T | p.L669F | Missense | D/19 | rs61754413 | het | Gene implicated in cancer development |
| ROCK1 | Chr 18: 18535139 | NM_005406: c.3580A>T | p.K1194X | Stopgain | A/50 | het | RNA. Malignant ascites in gastric cancer | |
| SLC15A1 | Chr 13: 99339879 | NM_005073: c.1783G>A | p.E595K | Missense | D/35 | het | RNA. Overexpressed in some cancers. Substrate transport. | |
HGVS, Human Genome Variation Society; dbSNP, Database for Short Genetic Variations; COSMIC, Catalogue of Somatic Mutations in Cancer; RNA, somatic mutation also detected in RNA-seq of final metastasis; TNBC, gene associated with triple-negative breast cancer; BrCa, gene associated with breast cancer; ALL, gene associated with acute lymphoblastic leukemia; het, heterozygous; hom, homozygous. Predicted effect from MutationTaster is disease-causing (D) or disease-causing-automatic (A), and from Combined Annotation-Dependent Depletion (CADD) is the Phred-like C score.
Recurrent Somatic Nonsynonymous Mutations in All Three Tumor Tissue Samples
The somatic TP53 mutation p.Y220C (or p.Y61C, p.Y88C, or p.Y181C, depending on transcript variant; Database for Short Genetic Variations [dbSNP] ID rs121912666) (Table 3), was heterozygous in each tumor sample, with an allele frequency of ∼0.5 (between 0.36 and 0.58). It is a known somatic mutation in COSMIC in at least 37 breast cancer tissue samples (at time of writing), and also in ovary, esophagus, upper aerodigestive tract, and lung cancers. In ClinVar, the mutation has not previously been reported as a somatic tumor tissue mutation but has been reported as a pathogenic germline Li–Fraumeni syndrome variant. The mutation p.Y220C is localized in the core domain of p53 and by introducing a surface crevice it leads to accelerated denaturing of the protein at body temperature (Boeckler et al. 2008). The HCFC1 p.S518F mutation has been reported in a melanoma patient (Hugo et al. 2016) (patient 13). The remaining recurrent somatic mutations in all three tumor samples are not yet reported, to our knowledge. For SYBU the G>A and G>T mutations have been reported in COSMIC but not the patient's c.572G>C mutation. All somatic mutations were clearly detected in the RNA-seq data of the final metastasis, except for NOTCH3, TRPM6, and RTN4, with only one mutated RNA read each. For NOTCH3 and TRPM6, the wild-type expression was halved compared with the in-house healthy controls.
Recurrent Somatic Nonsynonymous Mutation in Primary Tumor and First Metastasis
The GABRA5 mutation (Table 3) has not been reported to our knowledge and its functional consequences are unknown.
Private Somatic Nonsynonymous Mutations in Primary Tumor
The mutations in TCF3 and XXYLT1 (Table 3) are novel to our knowledge.
Private Somatic Nonsynonymous Mutations in First Metastasis
The stop mutation DGKZ p.E44* (Table 3) is novel to our knowledge. The missense mutation ADD3 p.E570K is reported in COSMIC to be seen in three The Cancer Genome Atlas (TCGA) endometrial carcinoma samples (http://cancer.sanger.ac.uk/cosmic/mutation/overview?id=914716). ADD3 p.E570K is predicted as tolerated by Alamut Visual (AlignGVD class C0, SIFT tolerated) but as disease-causing by MutationTaster and CADD.
Private Somatic Nonsynonymous Mutations in Final Metastasis
Of the 21 private mutations, all three stopgain mutations (ROCK1 p.K1194*, FXR1 p.R509*, PCDHGB4 p.E122*) are expressed on the RNA level and are predicted to be the most damaging (Table 3). The ROCK1 p.K1194* mutation lies in the first exon, knocking out all protein-binding regions of this allele, and has been reported in a lung cancer cell line (Sosonkina et al. 2014). The PCDHGB4 p.E122* mutation also lies in the first exon, within the cadherin protein domain. The mutation in FXR1 lies in exon 16 near the protein-binding region. Thirteen of the 18 private missense mutations were found expressed on the RNA level: ADRBK2, AGAP1, CEBPZ, CNTN3, DBNL, FBP2, HMGCR, IL17RD, IQGAP1, KLHL28, METTL4, SLC15A1, and WDR37 (Supplemental Table 1). IL17RD p.R575W has been reported as a somatic mutation in pancreatic cancer (Wu et al. 2011). The SLC15A1 (aka HPEPT1) p.E595K mutation has been reported as a loss-of-function mutation (Xu et al. 2009). The following genes or their somatically mutated alleles were not detected as expressed: CPXM2, DUSP26, KCNG2, LPPR4, PIK3C2G, and PCDHGB4. CPXM2 p.G50R was imputed as a germline variant for a noncancer patient (A Kainz, pers. comm.).
Differential Gene Expression
We analyzed differential gene expression by comparing the patient's expression data (two replicates) from the final metastasis sample against a set of 11 normal breast tissue controls. Of note, 9810/19,301 genes (51%) were significantly differentially expressed between the patient's tumor samples and the controls (Supplemental Table 2). A total of 4591 genes were up-regulated and 5219 genes were down-regulated. Of these genes, 4851 had a greater than twofold (log2) and 1536 a greater than fourfold (log2) difference in expression level. The 10 most up- and down-regulated genes are presented in Supplemental Table 3. Among the differentially expressed genes, 410 could be assigned as “breast cancer–relevant” based on a list of 433 breast cancer–relevant genes generated using GLAD4U webservice. The 10 most up- and down-regulated breast cancer relevant genes as defined by this method are presented in Supplemental Table 4. KEGG Pathway enrichment analysis on the differentially expressed, breast cancer–relevant genes revealed 31 KEGG pathways showing significant enrichment with those genes (Supplemental Table 5).
Drug–Gene Interaction
Querying the Drug–Gene Interaction Database (DGIdb; http://dgidb.genome.wustl.edu/) (Griffith et al. 2013) for drug–gene interactions using differential up-regulated genes (log2 FC > 1) revealed a candidate list of 60 compounds across 16 genes (Table 4). With respect to the applied anthracyclines (epirubicin, doxorubicin), microtubule inhibitors (paclitaxel, vinorelbine, and eribulin), and bortezomib: TOP2A, members of the microtubule gene family, and members of the proteasome gene family and were up-regulated 29-fold, 3.6-fold, and 5.6-fold, respectively (Supplemental Table 2).
Table 4.
Overexpressed breast cancer relevant genes associated with clinical compounds
| Symbol | Entrez | Log2FC | Clinically actionable compound(s) |
|---|---|---|---|
| RAD51 | 5888 | 5 | Amuvatinib |
| TOP2A | 7153 | 4.9 | Daunorubicin, Doxorubicin, Epirubicin, Idarubicin, Dexrazoxane, Lucanthone, Amsacrine, Mitoxantrone, Teniposide |
| AURKB | 9212 | 4.5 | Danusertib, Tozasertib, AT9283, GSK1070916, PF-03814735, Barasertib, SNS-314, ML8237 |
| AURKA | 6790 | 4.3 | PF-03814735, Alisertib, SNS-314, AT9283, ENMD-2076, Danusertib, Tozasertib, ML8237 |
| CHEK1 | 1111 | 3.6 | Rabusertib, SCH900776, AZD7762 |
| CHEK2 | 11200 | 2.0 | AZD7762 |
| NOTCH1 | 4851 | 2.0 | RO4929097, γ-Secretase |
| PARP1 | 142 | 1.9 | Veliparib, Olaparib, Rucaparib |
| PIK3R2 | 5296 | 1.9 | Apitolisib, Pilaralisib, GSK2636771, Pictilisib, PI-103, Gedatolisib, SF1126, Sophoretin, Omipalisib |
| RRM1 | 6240 | 1.8 | Fludarabine, Gemcitabine, Hydroxyurea, Cladribine |
| SYK | 6850 | 1.7 | Fostamatinib |
| CDK4 | 1019 | 1.6 | Palbociclib, Abemaciclib, Ribociclib, Alvocidib |
| AKT1 | 207 | 1.5 | MK2206, Ipatasertib, Perifosine, AZD5363 |
| MAP2K1 | 5604 | 1.4 | TAK-733, RO4987655, Pimasertib, Refametinib, AZD8330, Trametinib, Selumetinib, GDC-0623, Cobimetinib |
| ALK | 238 | 1.3 | Brigatinib, Crizotinib, Ganetespib, Ensartinib, ASP3026 |
| ERBB3 | 2065 | 1.1 | AV-203, GE-huMab-HER3, Patritumab, Seribantumab, LMJ716, REGN1400, H4B-121Ab, MM-111, Duligotuzumab, Istiratumab, GSK2849330, KTN3379, Osimertinib |
Sources: MyCancerGenomeClinicalTrial, TALC, DrugBank, MyCancerGenome, CancerCommons (aggregated by DGIdb)—initial results returned from DGIdb have been manually revised and curated.
Bold indicates Federal Drug Administration (FDA)-approved drugs (on-label as well as off-label); all other drugs are preclinical or in clinical trials. Names ending in ib (small molecule inhibitor) or ab (antibody) indicate gene (or mutation) specificity.
Log2FC, log2 of fold change.
Immunohistochemical Staining
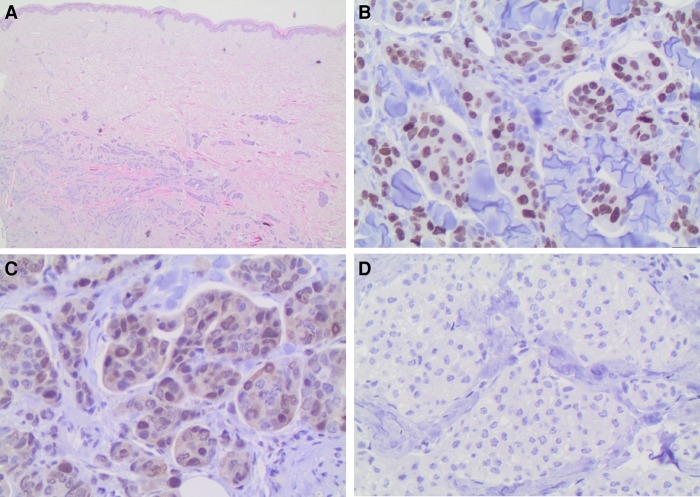
To validate the RNA-seq overexpression findings for sample MF in the absence of matched normal tissue from the same patient, we used antibody staining on the same tissue sample. We used antibodies that were available and properly established in our pathology laboratories. Figure 4 shows that CDK4 transcript up-regulation (threefold) and MKI67 up-regulation (13-fold) is clearly validated by IHC. ALK staining was negative, although RNA-seq analysis revealed a 2.4-fold up-regulation of transcript levels. Androgen receptor staining was weakly positive in 20% of lesional cells although RNA-seq indicated strong down-regulation in the patient's metastasis sample compared with the control samples.
Figure 4.
CDK4-immunohistochemical staining of final skin metastasis. Histopathological findings. (A) Islands of tumor cells are identified within the dermis (hematoxylin and eosin, 100×). (B) High Ki-67 proliferation index marks the nuclei of many neoplastic cells. This independently validates the 13-fold RNA-seq overexpression that was averaged over the final metastasis tissue sample (400×). (C) Tumor cell nuclei stain weakly but distinctly for CDK4, independently validating the threefold averaged RNA-seq overexpression (400×). (D) Isotype negative control with no detectable staining (400×).
DISCUSSION
In this case study, we aimed to identify potential targeted drugs using an exome- and transcriptome-based molecular profile of the patient's tumor and metastases. We also examined whether the molecular profile may explain the response or nonresponse to the administered drugs, the adverse events to some of the drugs, and the high aggressiveness seen in this individual TNBC case. To rapidly inform of potential targeted drugs, we scanned the existing whole-exome results for novel and known drug-associated somatic tumor tissue mutations in only those genes that are listed in https://www.mycancergenome.org/ and some commercial cancer panels, as well as in TP53, BRCA1, BRCA2. This first scan only found a single mutation—that is, the heterozygous damaging somatic mutation in TP53 in both tumor samples but not the germline sample. Our later in-depth analyses confirmed the TP53 mutation to be drug- and disease-relevant in the context of the complete set of somatic mutations in the tumor samples. To inform of potential further targeted treatments (Table 4), we also sequenced the whole transcriptome from the final metastasis sample, scanned for overexpressed genes by comparing the patient's sample with a set of healthy breast tissue controls, and validated the overexpressed genes on the protein level using available antibody stains. Metastatic tumor nodes accumulate a significant number of mutations that should be considered for treatment and that were not present in the primary tumor. For this reason, we analyzed the complete whole-exome data of the primary tumor and the two metastatic samples, classified each tumor sample using the COSMIC somatic signatures, and performed extensive database and literature research for the identified somatic tumor tissue mutations that were predicted as damaging. Because more than one tumor sample was available, we were able to see the shared mutations in each tumor sample that could be addressed with the same treatment, which serves to narrow down the lengthy list of potential treatments.
Somatic Signatures
Somatic signatures from the Sanger Center's COSMIC website (http://cancer.sanger.ac.uk/cosmic/signatures) allow a quick classification based on lists of somatic tumor tissue mutations that may otherwise be difficult to interpret, and may provide a first answer for patients who ask how and why their tumor started. Figure 2 shows that the earlier tumors PT and MK are associated with aging and ultraviolet light exposure (∼25%–30% Signature 1 and ∼10% Signature 7) and failure of DNA damage repair (∼20%–25% Signature 3—BRCA1/2-related, and ∼15%–25% Signature 15). For PT, there is also ∼10% Signature 26, which is associated with defective DNA mismatch repair. The causes of Signature 12 (∼10%–20%) are unknown, probably because it is usually seen in liver metastases, which may arise from a wide range of primary tumors. For MF the main signature is DNA damage repair failure (∼40% Signature 3 and ∼15% Signature 6), aging (∼10% Signature 1), and relatively uninformative remaining signature contributions (∼10% unknown, ∼10% Signature 29—gingivo-buccal oral squamous cell carcinoma, ∼10% Signature 12—liver cancer, ∼10% Signature 5—found in most cancers with unknown etiology).
Tumor Evolution Phylogeny
The concept of applying phylogenetic methods to analyze tumor evolution is not new (Shibata 2012; Murtaza et al. 2015). Having established the tumor evolution phylogeny, it is tempting to estimate the timespan for the ancestral tumor cell to evolve into the primary tumor. This may give an indication of the time window for early detection of the expanding tumor cell population. A stringent estimation method including error bounds is given in Saillard et al. (2000), which assumes a star-like phylogeny and a linear mutation rate. In cancer cells, the mutation rate depends on the number of mutations and the genes that are mutated (Loeb and Loeb 2000). Our patient's tumor evolution phylogeny is nearly star-like, but with only three tumor samples it is too sparse for applying the stringent estimation method. Therefore, we compared the discovery times and branch lengths of the final metastasis and the primary tumor with each other, and taking the mutation rate to be identical for both branches, we estimated that there was a time window of possibly only 6 mo for earlier detection of the primary tumor in our patient.
Somatic Mutations in the Inferred Ancestral Tumor Cell and Detected in All Three Tumor Samples
TP53 p.Y220C
The heterozygous pathogenic somatic tumor tissue mutation rs121912666 in TP53 is probably not the single driver mutation, because it is reported in ClinVar as a pathogenic germline variant for Li–Fraumeni cases, meaning that affected patients can live disease free for years or decades. The 12 predicted damaging somatic tumor tissue mutations in the trunk of our phylogenetic tumor evolution tree (Fig. 3) also strongly suggest that the viable ancestral tumor cell required multiple damaging mutations that may include several or all of these 12 mutations or the nine mutations detected to be highly expressed in the RNA-seq. However, p53 is a key apoptosis protein. On the functional level, this specific p.Y220C mutation does not affect the binding regions but leads to protein destabilization and much more rapid denaturing than wild-type p53 (Boeckler et al. 2008). On the frequency level, the p.Y220C mutation is reported as the ninth most frequent p53 mutation, affecting approximately 75,000 new cancer patients worldwide every year (Boeckler et al. 2008). Specifically, the p.Y220C mutation (aka p.Y88C) has been reported as a heterozygous somatic driver mutation in a Chinese breast cancer patient (Song et al. 2015). In our patient, the mutated TP53 allele was expressed in 90% of all transcripts in the final metastasis that we biopsied. The p53-mediated apoptosis pathway may therefore have been impaired significantly in her tumor cells.
It can hence be argued that the success of 11 mo of progression-free survival under bortezomib (proteasome inhibitor) experimental treatment was due to p53-independent apoptosis. Bortezomib was selected by the treating clinician as off-label therapy because of a published role of proteasome inhibitors in TNBC as determined by short interfering RNA (siRNA) screens (Petrocca et al. 2013). The course of disease suggests but does not prove transient therapeutic activity. Bortezomib has previously been reported to up-regulate CASP3, CASP8, and CASP9 (Saha et al. 2010), and down-regulate BCL2 (Goktas et al. 2010). Significant up-regulation of some proteasome transcripts underlined this possible activity of bortezomib in the TNBC patient. The TP53 mutation also suggested use of BCL2 inhibition (venetoclax); however, the RNA-seq data were inconclusive because of low coverage in BCL2. Impaired p53 has been associated with doxorubicin resistance (Dunkern et al. 2003). To overcome the resistance, an in vitro experiment successfully combined doxorubicin with a PARP inhibitor (Muñoz-Gámez et al. 2005). APR-246 (aka PRIMA-1), the first drug in clinical trials that directly targets mutant p53 is currently in a Phase 1b/II clinical trial (PiSSARO, ClinicalTrials.gov ID NCT02098343). APR-246 showed strong synergies with cisplatin in p.Y220C-mutated cancer cell lines (Mohell et al. 2015), and also with olaparib in p53-mutated non-small-cell lung cancer cell lines (Deben et al. 2016). Further compounds such as PK7088 that specifically target the p.Y220C induced crevice are in preclinical development (Joerger and Fersht 2010; Liu et al. 2013).
ADAM17 p.D657N and NOTCH3 p.R2078W
The enzyme encoded by ADAM17, also known as tumor necrosis factor-α-converting enzyme (TACE) sheds the extracellular domain of various receptors such as heparin-binding EGF, transforming growth factor-α (TGFA), amphiregulin (AREG), neuregulin (NRG), epiregulin (EREG), and β-cellulin (BTC) (Meng et al. 2016) from the cell membrane leading to an activation of downstream signaling pathways with significance in triple-negative breast cancer cells (Caiazza et al. 2015). NOTCH3 is a reported breast cancer driver gene where inactivating mutations or deletions in the PEST domain activate the Notch pathway (Wang et al. 2015). For breast cancer, the known somatic mutations form a cluster in the PEST region of our patient's p.R2078G mutation (https://www.intogen.org/search?gene=NOTCH3&cancer=BRCA). NOTCH3 expression was nearly undetectable in our RNA-seq data of the tumor tissue. At the Avera Cancer Institute, we see ADAM17 and Notch aberrations quite often in TNBC (B Leyland-Jones, pers. comm.). γ-Secretase inhibitors are under development for this pathway (Olsauskas-Kuprys et al. 2013; Wang et al. 2015).
The Remaining Shared Somatic Mutations and Their Potential Co-Driver Roles
The HCFC1 gene is reported as a major downstream effector of BRCA1-associated protein 1 (Kamburov et al. 2015) and as a breast cancer driver gene (The Cancer Genome Atlas Network 2012). The HCFC1 mutation ties in with somatic signature 3, which is reported for BRCA1- or BRCA2-associated failure of DNA double-strand break repair. ITPR2 is associated with estradiol-induced breast cancer; ITPR2 inhibitors include caffeine and the experimental compounds 2-APB and xestospongin C (XeC) (Szatkowski et al. 2010). NFRKB modulates NFKB1 and is being evaluated as a candidate oncogene (Bueno et al. 2010). THRAP3 has been associated with prostate cancer growth (Ino et al. 2016) and DNA damage response (Beli et al. 2012). ZNF384 fusions with TAF15 or EP300 are associated with acute lymphoblastic leukemia (Ping et al. 2015; Kim et al. 2016). Together, these seven genes and their expressed mutated alleles appear to be a plausible driver mutation combination required for a viable ancestral tumor cell. The SYBU mutation also appears to be relevant because its expression is detected in MF, despite being so far known only to be expressed in nerve cells. SYBU encodes syntabulin, which is a microtubule-associated protein that mediates transport of vesicles to neuronal processes (Su et al. 2004; Ying et al. 2012). TRPM6 is involved in magnesium homeostasis and epithelial magnesium transport, and somatic mutations clustering in the same region have been reported in Chinese breast cancer patients (Zhang et al. 2015). The role of magnesium in cancer development is much debated (Trapani et al. 2016), and it seems to vary depending on stage such as carcinogenesis, primary tumor growth, and metastasis formation (V Trapani, pers. comm.). RTN4 is associated with cell adhesion, migration, metastasis, and apoptosis in various cancers (Chi et al. 2015; Xue et al. 2015). Mutated alleles of TRPM6 and RTN4 were nearly undetectable in our RNA-seq data of the tumor tissue.
Perhaps Actionable Somatic Nonsynonymous Mutations That Do Not Appear in All Three Tumor Samples
We identified 26 mutations that were predicted damaging and that were not shared in all three tumor samples. The therapeutical and functional consequences of these mutations are currently not known, therefore any inferences are speculative. However, it is striking that nearly all of the mutated genes are known to be associated with TNBC, breast cancer, other cancers, invasion, metastasis, proliferation, relapse, and so on. A full discussion of all 26 genes is beyond the scope of this section, but interested readers can refer to the Supplemental Material for a short comment on the relevance of each gene. We now briefly discuss potential targets found to be mutated in the patient, for which inexpensive repurposed drugs and an expensive leukemia treatment could be considered.
Primary Tumor and First Metastasis
Regarding mutated GABRA5 in the primary tumor (PT) and the first metastasis (MK), the wild-type gene encodes a receptor for its ligand GABA, which inhibits norepinephrine-induced migration of breast and colon carcinoma cells (Entschladen et al. 2004). Lorazepam, an approved GABA agonist often used to treat anxiety disorders in cancer patients, may have a potential use as an anti-metastatic drug (Entschladen et al. 2004). The nonspecific β-blocker propranolol inhibits norepinephrine-induced migration of breast cancer cells and increases the cytotoxicity of natural killer cells, reducing distant metastases and improving 10-yr survival significantly from 70% to 90% (Powe et al. 2010). In nonrandomized comparisons of treated versus untreated ovarian cancer cohorts—characterized by mutated p53 in most patients—propranolol cohorts showed increased median overall survival (Watkins et al. 2015).
Primary Tumor
Regarding mutated TCF3 in the primary tumor, the wild-type gene encodes a key transcription factor for lymphopoiesis (Fischer et al. 2015), and a successful therapy protocol exists for the TCF3-PBX1 acute lymphoblastic leukemia subtype.
Final Metastasis
Regarding the mutated genes HMGCR and IQGAP1 in the final metastasis (MF): HMGCR encodes the rate-limiting enzyme in the mevalonite pathway. Pharmacologic inhibition of HMGCR with statins is reported to inhibit breast cancer cell growth in vitro (Pampalakis et al. 2015), reduce the risk and/or severity of breast cancer (Clendening et al. 2010), exhibit antimetastatic and antitumorigenic effects in ovarian cancer (Stine et al. 2016), and show anticancer effects in gastric cells (Chushi et al. 2016). IQGAP1 was associated with breast cancer cell invasion (Alemayehu et al. 2013), and its protein was reported as one of two binding targets of disulfuram (an anti–alcohol addiction drug), as a suggested TNBC combination treatment with doxorubicin (Alemayehu et al. 2013).
Whole-Transcriptome Sequencing
Whole-transcriptome sequencing of tumor RNA is of immense value to inform of overexpressed target genes and fusions in addition to the targets that can be identified by DNA-based mutation analysis alone. Furthermore, the functional effect of a tumor mutation can sometimes be tracked by looking at the expression of genes downstream from the mutated gene. For example, a FLT3 mutation of “unknown significance” in combination with overexpression of the downstream RNA transcripts for RAS, RAF, and MEK would be hypothesized as an activating FLT3 mutation; therapeutic consequences would be consideration of FLT3 inhibitors quizartinib or sorafinib, or downstream MEK inhibitors trametinib or cobimetinib. (The FLT3 mutation was a low-confidence mutation in this case study, which turned out to be an artefact.) Lastly, mutations that are not seen in RNA may be of secondary interest as probably being passenger mutations, whereas mutations detected in DNA can be considered validated if also seen in RNA.
In this case study we compared the RNA-seq data from the final metastasis sample against in-house healthy controls to obtain gene expression profiles. We saw an up-regulation of breast cancer relevant genes, many of which have been reported to be associated with proliferation, tumor progression, poor prognosis, or resistance to therapy. A small selection will be discussed here, and the interested reader is referred to Supplemental Data for a discussion of further genes. The significance of the TP53 mutation that was detected in the exomes ties in with the expression-based gene set enrichment analysis that ranks the p53 signaling pathway on rank 2 (Supplemental Table 5). On the single-gene level, overexpression of survivin (BIRC5) supports the anti-apoptosis theme. Survivin overexpression has been shown to inhibit apoptosis, promote mitosis, and to facilitate angiogenesis (Lv et al. 2010). It has further been linked to drug/radiation resistance (Lv et al. 2010) as well as being associated with poor prognosis (Tanaka et al. 2000; Span et al. 2004). Survivin has been established as a specific cancer cell target for preclinical immunotherapeutics (Bertino et al. 2013). Overexpression of homeodomain transcription factor HOXB13 has been shown to correlate with tamoxifen resistance in ER-positive breast cancer (Shah et al. 2013). HOXB13 is used as a biomarker within the breast cancer index (BCI) (Sgroi et al. 2013; Sgroi and Brufsky 2016). Cyclin E1 (CCNE1) is a well characterized oncogene. Overexpression in breast cancer is associated with proliferation and poor prognosis (Caldon and Musgrove 2010). At the Avera Cancer Institute, we frequently observe aberrations in CCNE1 in TNBC and ovarian tumors (B Leyland-Jones, pers. comm.). CDK2 inhibitors are in clinical trials to target CCNE1 amplification. CCNE1 overexpression is in agreement with the highly significant cell-cycle pathway that the gene set enrichment analysis placed on rank 3 (Supplemental Table 5). The RAD51 overexpression ties in with the highly significant homologous recombination pathway in the gene set enrichment analysis (rank 6) and with the Somatic Signature 3 (Fig. 1, Homologous recombination failure in breast and other cancers). At the Avera Cancer Institute, we would consider a poly ADP ribose polymerase (PARP) inhibitor or a platinum-based therapy. RAD51 overexpression was also reported to increase during breast cancer progression and metastasis and to promote the development of distant metastasis in TNBC (Wiegmans et al. 2014).
Down-regulation of the progesterone receptor (PGR) (Purdie et al. 2014) and of GSTM1 (S Willis, unpubl.) are prognostically bad (Supplemental Table 4). Topoisomerase II α (TOP2A) overexpression, independent from amplification, is highly associated with cell proliferation and aggressive tumor subtypes (Romero et al. 2011). TOP2A was up-regulated 29-fold, providing an expression-based rationale for therapeutic attempts with epirubicin and doxorubicin, but in the patient's case neither drug worked and she suffered from grade 3–4 stomatitis. The tubulin β chain (TUBB) expression was up-regulated 3.3-fold. The patient received paclitaxel, vinorelbin, and eribulin, which target tubulin β chain according to DrugBank (www.drugbank.ca). Although paclitaxel was applied within the Citron protocol, and vinorelbin and eribulin appeared to halt progression temporarily, their efficacy was disappointing compared with a Belgian trial (Aftimos et al. 2016) and not obvious compared with bortezomib. PSMB5 and PSMB1 were up-regulated 5.6-fold and threefold, respectively. These members of the proteasome gene family are inhibited by bortezomib (www.drugbank.ca), showing that not necessarily the highest up-regulated genes need be prioritized for a relatively long period of progression-free survival. A typical combination therapy to enhance the response to bortezomib in multiple myeloma is to add dexamethasone or to combine bortezomib, dexamethasone, and the histone deacetylase (HDAC) inhibitor panobinostat (which would have addressed the up to sevenfold up-regulated HDAC gene family members).
Hypothetical Surveillance of Treatment Response Using cfDNA Panels
In the terminal stage, the standard blood biomarker CA15-3 was stable, but skin metastases showed a completely different picture of rapid disease progression. The longitudinal detection at different treatment time points of somatic mutations in cell-free DNA (cfDNA) from blood plasma was first introduced on a large scale in the United States in 2014. In Germany, these longitudinal tests are not reimbursed by the public health insurers and are little known. The surveillance of the TP53 mutation, which would have been detected by any current cfDNA gene panel, possibly could have been more accurate than CA15-3 (Dawson et al. 2013; Heidary et al. 2014; Olsson et al. 2015). To gain information on actionable mutations, a broad cfDNA panel is needed. cfDNA next-generation sequencing (NGS) tests may pay for themselves by guiding drug choice and saving costs by detecting the lack of drug response at an early stage of treatment.
Conclusion
This was a first pilot for our ongoing multicenter collaboration to synergize our efforts and investigate methods to improve patient testing and the selection of therapies. This case study demonstrates that molecular profiling, if performed fast and early in the course of disease, can inform on potential targets and drugs, with the proviso that it alone cannot always reliably predict whether a drug will work: specifically, the patient's very high TOP2A expression level was an unsuitable biomarker for the selection of the anthracyclines that the patient received. Likewise, the RRM2 and TYMS overexpression do not appear suitable biomarkers for the indication of gemcitabine. However, the moderate TUBB overexpression tied in with the moderate and short-lived successes of vinorelbin and eribulin. The TYMS overexpression tied in with the moderate capecitabine response. The molecular case for possible bortezomib activity was plausible on the TP53 mutation level and the proteasome overexpression level. However, facing the multitude of genetic information and their potential link to pharmacologically actionable target patterns, from a clinical perspective it will be a major task to build relevance-based algorithms for interventions. Furthermore, every NGS-guided choice of targeted drugs has to be prospectively compared with clinical choice of therapy regimen delineated from several generations of controlled trials to place them correctly into our therapeutic armamentarium. In view of the tremendous amount of information that can be extracted from NGS and needs to be researched, validated, and summarized, timely whole-exome and transcriptome diagnostics should be considered at an early point in time, when the first line of treatment begins and when healthy reference tissue is still available. We suggest that mutated TP53 does not need to be seen as unrelated to treatments or as a desolate prognosis, but that many p53-independent treatments already exist that may be considered.
METHODS
DNA library preparation and exome capture were executed using the Kapa Hyper Plus Library Prep Kit and the SeqCap EZ MedExome Target Enrichment Kit (Roche). Paired-end whole-exome sequencing was performed on the Illumina NextSeq 500 platform, with 150-bp reads. Alignment was done with Burrows–Wheeler alignment (BWA)-MEM against the human reference hg19 (Li and Durbin 2009). DNA alignments were processed according to GATK best practices steps (McKenna et al. 2010). GATK HaplotypeCaller version 3.3-0 and VarScan version 2.3.9 (Koboldt et al. 2012) were used to call variants. ANNOVAR was used to annotate the union of the two call sets (Wang et al. 2010). The phylogenetic network was constructed with Network 5.0.0.0 (Fluxus). Somatic signatures in the primary tumor and the metastatic samples were inferred using the deconstructSigs method (Rosenthal et al. 2016).
RNA library preparation and targeted capture of libraries with exome baits were performed with the Illumina TruSeq RNA Access kit on two RNA aliquots as technical replicates. Paired-end RNA sequencing was performed on the Illumina NextSeq 500 platform with 76-bp reads. Mapping of RNA reads against the human reference hg19 and generation of gene counts were done using STAR (Dobin et al. 2013). RNA-seq count data were processed and differential expression analysis was performed using the DESeq2 Bioconductor package (Anders and Huber 2010), comparing the two patient tumor samples against a set of 11 in-house generated breast tissue controls. KEGG gene set enrichment analysis on differentially expressed and breast cancer relevant genes was performed using the g:ProfileR Bioconductor package (https://cran.r-project.org/web/packages/g.ProfileR/index.html). Drug–gene interactions for up-regulated genes were retrieved from the DGIdb database (Griffith et al. 2013).
A detailed description of samples, NGS, and data analysis methods is available in the Supplemental Methods.
ADDITIONAL INFORMATION
Data Deposition and Access
Somatic variants were reported to COSMIC under COSP identifier COSP42774 (Supplemental Table 6). Raw sequence data were submitted to the NCBI Sequence Read Archive under SRA BioProject ID PRJNA358250, BioSample IDs SAMN06168104, SAMN06168105, and SAMN06168106.
Ethics Statement
The patient provided written and verbal consent for the research, publication, and sharing of her data. This case study was approved by the ethics board of the University Hospital of Schleswig-Holstein under reference number D 433/17.
Acknowledgments
We thank the patient for her medical data, samples, and cooperation. We thank Brandon Young, Amanda Andrews, Cayce Conolly, Dorina Ölsner, and Melanie Vollstedt for technical assistance and Peter Forster, Go Ito, Alexander Kainz, Klaus Pantel, Pradip De, and Valentina Trappani for valuable discussions.
Author Contributions
T.M. and M.F. designed research, analyzed the data, and wrote the manuscript. C.M.-T. organized sequencing of peripheral blood, primary tumor, and first metastasis. A.M. performed data analysis and helped to write the manuscript. E.W. and T.G. performed immunohistochemical staining and analysis. C.W. and B.L.-J. helped to interpret results and write the manuscript, especially the discussion of hypothetical treatment options. N.A., P.R., and A.F. helped to write the manuscript. M.S. relayed medical information from the patient, helped to design research, organize the study, analyze the data, interpret results, and write the manuscript. W.E.B., S.W., and A.K. treated the patient and helped to write the manuscript. All authors read and approved the final manuscript.
Funding
W.E.B.’s laboratory is supported by Deutsche Forschungsgemeinschaft (DFG EXC 1003 cells in motion cluster of excellence). The Institute of Clinical Molecular Biology is supported by the German Federal Ministry of Education and Research (BMBF) as part of the e:Med framework (“sysINFLAME”, grant 01ZX1306), the Cluster of Excellence “Inflammation at Interfaces” (ExC 306), SysMedIBD EU FP7 under grant agreement no. 305564 and DEEP IHEC (TP2.3 and 5.2, BMBF).
Competing Interest Statement
M.F. cofounded Fluxus Technology Ltd (free Network 5.0.0.0 software).
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Aftimos P, Polastro L, Ameye L, Jungels C, Vakili J, Paesmans M, van den Eerenbeemt J, Buttice A, Gombos A, de Valeriola D, et al. 2016. Results of the Belgian expanded access program of eribulin in the treatment of metastatic breast cancer closely mirror those of the pivotal phase III trial. Eur J Cancer 60: 117–124. [DOI] [PubMed] [Google Scholar]
- Alemayehu M, Dragan M, Pape C, Siddiqui I, Sacks DB, Di Guglielmo GM, Babwah AV, Bhattacharya M. 2013. β-Arrestin2 regulates lysophosphatidic acid-induced human breast tumor cell migration and invasion via Rap1 and IQGAP1. PLoS One 8: e56174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. 2012. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 46: 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino P, Panigada M, Soprana E, Bianchi V, Bertilaccio S, Sanvito F, Rose AH, Yang H, Gaudino G, Hoffmann PR, et al. 2013. Fowlpox-based survivin vaccination for malignant mesothelioma therapy. Int J Cancer 133: 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. 2008. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci 105: 10360–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno R, De Rienzo A, Dong L, Gordon GJ, Hercus CF, Richards WG, Jensen RV, Anwar A, Maulik G, Chirieac LR, et al. 2010. Second generation sequencing of the mesothelioma tumor genome. PLoS One 5: e10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza F, McGowan PM, Mullooly M, Murray A, Synnott N, O'Donovan N, Flanagan L, Tape CJ, Murphy G, Crown J, et al. 2015. Targeting ADAM-17 with an inhibitory monoclonal antibody has antitumour effects in triple-negative breast cancer cells. Br J Cancer 112: 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon CE, Musgrove EA. 2010. Distinct and redundant functions of cyclin E1 and cyclin E2 in development and cancer. Cell Div 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C, Liu N, Yue L, Qi W-W, Xu L-L, Qiu W-S. 2015. RTN4/Nogo is an independent prognostic marker for gastric cancer: preliminary results. Eur Rev Med Pharmacol Sci 19: 241–246. [PubMed] [Google Scholar]
- Chushi L, Wei W, Kangkang X, Yongzeng F, Ning X, Xiaolei C. 2016. HMGCR is up-regulated in gastric cancer and promotes the growth and migration of the cancer cells. Gene 587: 42–47. [DOI] [PubMed] [Google Scholar]
- Citron ML. 2008. Dose-dense chemotherapy: principles, clinical results and future perspectives. Breast Care (Basel) 3: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron ML, Berry DA, Cirrincione C, Hudis C, Winer EP, Gradishar WJ, Davidson NE, Martino S, Livingston R, Ingle JN, et al. 2003. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: first report of Intergroup Trial C9741/Cancer and Leukemia. J Clin Oncol 21: 1431–1439. [DOI] [PubMed] [Google Scholar]
- Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, Trentin GA, Martirosyan A, Hakem A, Hakem R, Jurisica I, et al. 2010. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci 107: 15051–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. 2013. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- Deben C, Lardon F, Wouters A, Op de Beeck K, Van den Bossche J, Jacobs J, Van Der Steen N, Peeters M, Rolfo C, Deschoolmeester V, et al. 2016. APR-246 (PRIMA-1MET) strongly synergizes with AZD2281 (olaparib) induced PARP inhibition to induce apoptosis in non-small cell lung cancer cell lines. Cancer Lett 375: 313–322. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkern TR, Wedemeyer I, Baumgärtner M, Fritz G, Kaina B. 2003. Resistance of p53 knockout cells to doxorubicin is related to reduced formation of DNA strand breaks rather than impaired apoptotic signaling. DNA Repair (Amst) 2: 49–60. [DOI] [PubMed] [Google Scholar]
- Entschladen F, Drell TL IV, Lang K, Joseph J, Zaenker KS. 2004. Tumour-cell migration, invasion, and metastasis: navigation by neurotransmitters. Lancet Oncol 5: 254–258. [DOI] [PubMed] [Google Scholar]
- Fischer U, Forster M, Rinaldi A, Risch T, Sungalee S, Warnatz H-J, Bornhauser B, Gombert M, Kratsch C, Stütz AM, et al. 2015. Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options. Nat Genet 47: 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goktas S, Baran Y, Ural AU, Yazici S, Aydur E, Basal S, Avcu F, Pekel A, Dirican B, Beyzadeoglu M. 2010. Proteasome inhibitor bortezomib increases radiation sensitivity in androgen independent human prostate cancer cells. Urology 75: 793–798. [DOI] [PubMed] [Google Scholar]
- Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB, Eldred JM, et al. 2013. DGIdb: mining the druggable genome. Nat Methods 10: 1209–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidary M, Auer M, Ulz P, Heitzer E, Petru E, Gasch C, Riethdorf S, Mauermann O, Lafer I, Pristauz G, et al. 2014. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res 16: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. 2016. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ino Y, Arakawa N, Ishiguro H, Uemura H, Kubota Y, Hirano H, Toda T. 2016. Phosphoproteome analysis demonstrates the potential role of THRAP3 phosphorylation in androgen-independent prostate cancer cell growth. Proteomics 16: 1069–1078. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. 2010. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol 2: a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, Lander ES, Getz G. 2015. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci 112: E5486–E5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim HS, Shin S, Lee ST, Choi JR. 2016. t(12;17)(p13;q12)/TAF15-ZNF384 rearrangement in acute lymphoblastic leukemia. Ann Lab Med 36: 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D, Zhang Q, Larson D, Shen D, McLellan M, Lin L, Miller C, Mardis E, Ding L, Wilson R. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. 2015. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wilcken R, Joerger AC, Chuckowree IS, Amin J, Spencer J, Fersht AR. 2013. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res 41: 6034–6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb KR, Loeb LA. 2000. Significance of multiple mutations in cancer. Carcinogenesis 21: 379–385. [DOI] [PubMed] [Google Scholar]
- Lv Y-G, Yu F, Yao Q, Chen J-H, Wang L. 2010. The role of survivin in diagnosis, prognosis and treatment of breast cancer. J Thorac Dis 2: 100–110. [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meißner T, Fisch KM, Gioia L, Su AI. 2015. OncoRep: an n-of-1 reporting tool to support genome-guided treatment for breast cancer patients using RNA-sequencing. BMC Med Genomics 8: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Hu B, Hossain MM, Chen G, Sun Y, Zhang X. 2016. ADAM17-siRNA inhibits MCF-7 breast cancer through EGFR-PI3K-AKT activation. Int J Oncol 49: 682–690. [DOI] [PubMed] [Google Scholar]
- Mohell N, Alfredsson J, Fransson Å, Uustalu M, Byström S, Gullbo J, Hallberg A, Bykov VJN, Björklund U, Wiman KG. 2015. APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis 6: e1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gámez JA, Martín-Oliva D, Aguilar-Quesada R, Cañuelo A, Nuñez MI, Valenzuela MT, Ruiz de Almodóvar JM, De Murcia G, Oliver FJ. 2005. PARP inhibition sensitizes p53-deficient breast cancer cells to doxorubicin-induced apoptosis. Biochem J 386: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M, Dawson S-J, Pogrebniak K, Rueda OM, Provenzano E, Grant J, Chin S-F, Tsui DWY, Marass F, Gale D, et al. 2015. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 6: 8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsauskas-Kuprys R, Zlobin A, Osipo C. 2013. γ-Secretase inhibitors of Notch signaling. Onco Targets Ther 6: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson E, Winter C, George A, Chen Y, Howlin J, Tang MH, Dahlgren M, Schulz R, Grabau D, van Westen D, et al. 2015. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 7: 1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalakis G, Politi AL, Papanastasiou A, Sotiropoulou G. 2015. Distinct cholesterogenic and lipidogenic gene expression patterns in ovarian cancer—a new pool of biomarkers. Genes Cancer 6: 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocca F, Altschuler G, Tan SM, Mendillo ML, Yan H, Jerry DJ, Kung AL, Hide W, Ince TA, Lieberman J. 2013. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell 24: 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping N, Qiu H, Wang Q, Dai H, Ruan C, Ehrentraut S, Drexler HG, MacLeod RAF, Chen S. 2015. Establishment and genetic characterization of a novel mixed-phenotype acute leukemia cell line with EP300-ZNF384 fusion. J Hematol Oncol 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powe DG, Voss MJ, Zänker KS, Habashy HO, Green AR, Ellis IO, Entschladen F. 2010. β-blocker drug therapy reduces secondary cancer formation in breast cancer and improves cancer specific survival. Oncotarget 1: 628–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie CA, Quinlan P, Jordan LB, Ashfield A, Ogston S, Dewar JA, Thompson AM. 2014. Progesterone receptor expression is an independent prognostic variable in early breast cancer: a population-based study. Br J Cancer 110: 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L, Hanson H, Seal S, Warren-Perry M, Hughes D, Howell I, Turnbull C, Houlston R, Shanley S, Butler S, et al. 2012. BRCA1 testing should be offered to individuals with triple-negative breast cancer diagnosed below 50 years. Br J Cancer 106: 1234–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero A, Martín M, Cheang MCU, López García-Asenjo JA, Oliva B, He X, de la Hoya M, García Sáenz JÁ, Arroyo Fernández M, Díaz Rubio E, et al. 2011. Assessment of Topoisomerase II α status in breast cancer by quantitative PCR, gene expression microarrays, immunohistochemistry, and fluorescence in situ hybridization. Am J Pathol 178: 1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. 2016. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha MN, Jiang H, Jayakar J, Reece D, Branch DR, Chang H. 2010. MDM2 antagonist nutlin plus proteasome inhibitor velcade combination displays a synergistic anti-myeloma activity. Cancer Biol Ther 9: 936–944. [DOI] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt HJ, Nørby S. 2000. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67: 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. 2014. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11: 361–362. [DOI] [PubMed] [Google Scholar]
- Sgroi DC, Brufsky A. 2016. Biomarkers for early-stage breast cancer: clinical utility for extended adjuvant treatment decisions. J Clin Oncol 34: 3941–3942. [DOI] [PubMed] [Google Scholar]
- Sgroi DC, Sestak I, Cuzick J, Zhang Y, Schnabel CA, Schroeder B, Erlander MG, Dunbier A, Sidhu K, Lopez-Knowles E, et al. 2013. Prediction of late distant recurrence in patients with oestrogen-receptor-positive breast cancer: A prospective comparison of the breast-cancer index (BCI) assay, 21-gene recurrence score, and IHC4 in the TransATAC study population. Lancet Oncol 14: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Jin K, Cruz LA, Park S, Sadik H, Cho S, Goswami CP, Nakshatri H, Gupta R, Chang HY, et al. 2013. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERα and inducing IL-6 expression. Cancer Res 73: 5449–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata D. 2012. Cancer. Heterogeneity and tumor history. Science 336: 304–305. [DOI] [PubMed] [Google Scholar]
- Song F, Li X, Song F, Zhao Y, Li H, Zheng H, Gao Z, Wang J, Zhang W, Chen K. 2015. Comparative genomic analysis reveals bilateral breast cancers are genetically independent. Oncotarget 6: 31820–31829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosonkina N, Hong S-K, Starenki D, Park J-I. 2014. Kinome sequencing reveals RET G691S polymorphism in human neuroendocrine lung cancer cell lines. Genes Genomics 36: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Span PN, Sweep FCGJ, Wiegerinck ETG, Tjan-Heijnen VCG, Manders P, Beex LVAM, De Kok JB. 2004. Survivin is an independent prognostic marker for risk stratification of breast cancer patients. Clin Chem 50: 1986–1993. [DOI] [PubMed] [Google Scholar]
- Stine JE, Guo H, Sheng X, Han X, Schointuch MN, Gilliam TP, Gehrig PA, Zhou C, Bae-Jump VL. 2016. The HMG-CoA reductase inhibitor, simvastatin, exhibits anti-metastatic and anti-tumorigenic effects in ovarian cancer. Oncotarget 7: 946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Cai Q, Gerwin C, Smith CL, Sheng Z-H. 2004. Syntabulin is a microtubule-associated protein implicated in syntaxin transport in neurons. Nat Cell Biol 6: 941–953. [DOI] [PubMed] [Google Scholar]
- Szatkowski C, Parys JB, Ouadid-Ahidouch H, Matifat F. 2010. Inositol 1,4,5-trisphosphate-induced Ca2+ signalling is involved in estradiol-induced breast cancer epithelial cell growth. Mol Cancer 9: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. 2000. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res 6: 127–134. [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapani V, Luongo F, Arduini D, Wolf FI. 2016. Magnesium modulates doxorubicin activity through drug lysosomal sequestration and trafficking. Chem Res Toxicol 29: 317–322. [DOI] [PubMed] [Google Scholar]
- Vasan N, Yelensky R, Wang K, Moulder S, Dzimitrowicz H, Avritscher R, Wang B, Wu Y, Cronin MT, Palmer G, et al. 2014. A targeted next-generation sequencing assay detects a high frequency of therapeutically targetable alterations in primary and metastatic breast cancers: implications for clinical practice. Oncologist 19: 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang Q, Li D, Ching K, Zhang C, Zheng X, Ozeck M, Shi S, Li X, Wang H, et al. 2015. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a γ-secretase inhibitor. Clin Cancer Res 21: 1487–1496. [DOI] [PubMed] [Google Scholar]
- Watkins JL, Thaker PH, Nick AM, Ramondetta LM, Kumar S, Urbauer DL, Matsuo K, Squires KC, Coleman RL, Lutgendorf SK, et al. 2015. Clinical impact of selective and nonselective β-blockers on survival in patients with ovarian cancer. Cancer 121: 3444–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. 2012. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92: 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmans AP, Al-Ejeh F, Chee N, Yap P-Y, Gorski JJ, Da Silva L, Bolderson E, Chenevix-Trench G, Anderson R, Simpson PT, et al. 2014. Rad51 supports triple negative breast cancer metastasis. Oncotarget 5: 3261–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, Eshleman JR, Goggins MG, Wolfgang CL, Canto MI, et al. 2011. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci 108: 21188–21193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Haworth IS, Kulkarni AA, Bolger MB, Davies DL. 2009. Mutagenesis and cysteine scanning of transmembrane domain 10 of the human dipeptide transporter. Pharm Res 26: 2358–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Wang Z, Chen J, Yang Z, Tang J. 2015. Knockdown of reticulon 4C by lentivirus inhibits human colorectal cancer cell growth. Mol Med Rep 12: 2063–2067. [DOI] [PubMed] [Google Scholar]
- Ying Y, Li L, Cao W, Yan D, Zeng Q, Kong X, Lu L, Yan M, Xu X, Qu J, et al. 2012. The microtubule associated protein syntabulin is required for glucose-stimulated and cAMP-potentiated insulin secretion. FEBS Lett 586: 3674–3680. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cai Q, Shu X-O, Gao Y-T, Li C, Zheng W, Long J. 2015. Whole-exome sequencing identifies novel somatic mutations in Chinese breast cancer patients. J Mol Genet Med 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.