Abstract
Cancer cells are often hypersensitive to the targeting of transcriptional regulators, which may reflect the deregulated gene expression programs that underlie malignant transformation. One of the most prominent transcriptional vulnerabilities in human cancer to emerge in recent years is the bromodomain and extraterminal (BET) family of proteins, which are coactivators that link acetylated transcription factors and histones to the activation of RNA polymerase II. Despite unclear mechanisms underlying the gene specificity of BET protein function, small molecules targeting these regulators preferentially suppress the transcription of cancer-promoting genes. As a consequence, BET inhibitors elicit anticancer activity in numerous malignant contexts at doses that can be tolerated by normal tissues, a finding supported by animal studies and by phase I clinical trials in human cancer patients. In this review, we will discuss the remarkable, and often perplexing, therapeutic effects of BET bromodomain inhibition in cancer.
Small molecules that target BET proteins preferentially suppress transcription of cancer-promoting genes and induce anticancer responses in vivo. The mechanisms that underlie this gene specificity are just beginning to be revealed.
In eukaryotic cells, sequence-specific DNA-binding transcription factors (TFs) activate their target genes by recruiting multisubunit coactivator complexes, which use diverse biochemical mechanisms to activate RNA polymerase II (Pol II). One important class of coactivators possess lysine acetyltransferase (KAT) activity, which transfers acetyl groups from acetyl-coenzyme A to the epsilon amino group of lysine residues of various substrate proteins. Many KAT enzymes (e.g., p300/CBP) have permissive substrate specificity and will acetylate unstructured, lysine-rich peptides found on TFs, histones, and various other components of the transcription apparatus (Dancy and Cole 2015). The pervasiveness of TF–KAT interactions in transcriptional regulation leads to a global partitioning of eukaryotic genomes into hyperacetylated and hypoacetylated domains, which strongly correlate with active and inactive cis-regulatory regulatory elements, respectively (Wang et al. 2008).
One mechanism by which acetylation influences transcription is by neutralizing the positive charge of lysine side chains to disrupt electrostatic interactions (e.g., between histones and DNA), which can lead to chromatin decompaction (Roth et al. 2001). In an alternative mechanism, lysine side-chain acetylation of many transcriptional regulators will create docking sites for proteins possessing acetyllysine binding/reader domains. In this setting, acetyllysine serves a vital function in the assembly of the transcriptional apparatus at enhancer and promoter elements. The most well established acetyllysine reader is the bromodomain, which is present on 46 different proteins encoded in the human genome (Dhalluin et al. 1999; Filippakopoulos et al. 2012). A bromodomain is composed of a left-handed bundle of four α helices, with interhelical loops forming a hydrophobic binding pocket that engages in acetyllysine recognition (Dhalluin et al. 1999). Studies spanning nearly two decades have implicated bromodomain-containing coactivator proteins as integral components of TF-mediated gene regulation by linking lysine acetylation to downstream effects on chromatin structure and transcription (Sanchez and Zhou 2009). Moreover, the functional diversity and emerging “drugability” of bromodomain modules with small molecules has motivated a widespread interest in this class of proteins as therapeutic targets (Filippakopoulos and Knapp 2014).
BET PROTEIN FAMILY OF TRANSCRIPTIONAL COACTIVATORS
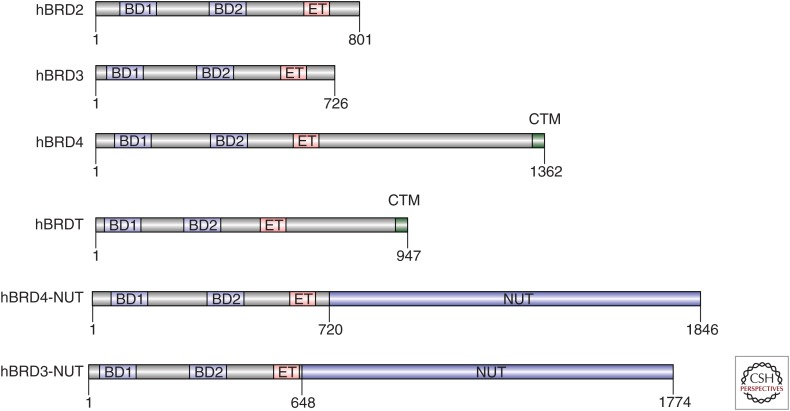
The mammalian BET (bromodomain and extraterminal domain-containing) protein family consists of four members, including the ubiquitously expressed BRD2, BRD3, BRD4, and the germ-cell-specific BRDT (Fig. 1) (Wu and Chiang 2007; Shi and Vakoc 2014). All four BET proteins have two conserved bromodomains that preferentially bind to multiacetylated peptides (Fig. 1) (Dey et al. 2003; Moriniere et al. 2009; Gamsjaeger et al. 2011; Filippakopoulos et al. 2012). The preferred ligand of the first bromodomain (BD1) is KacXXKac, with the intervening X amino acids having small side chains (e.g., glycine or alanine), whereas the second bromodomain (BD2) is more permissive to binding multiacetylated peptides in diverse sequence contexts (Dey et al. 2003; Moriniere et al. 2009; Gamsjaeger et al. 2011; Filippakopoulos et al. 2012). The acetylated tails of core histones H3 and H4 and acetylated regions of TFs are the most well-validated binding partners of BET bromodomains, which are generated as consequence of TF-mediated KAT recruitment (Dey et al. 2003; Lamonica et al. 2011; Shi et al. 2014; Roe et al. 2015). ChIP-seq studies in several cell types have shown that BRD4 localizes preferentially to the nucleosome-free site occupied by TFs at enhancers and promoters, which is consistent with acetylated TFs being important recruiters of BET proteins (Roe et al. 2015; Stonestrom et al. 2015). It is likely that a multitude of acetylated peptides contribute to BET protein recruitment to a particular DNA element, with different acetylated peptides being relevant at different cis elements. However, it is challenging to pinpoint the complete repertoire of acetylated peptides responsible for recruiting BET proteins to chromatin.
Figure 1.
Domain structure of the bromodomain and extraterminal (BET) protein family and BET–NUT fusion proteins. Each BET protein contains two bromodomains (BD1 and BD2) and an extraterminal (ET) domain. BRD4 and BRDT have an additional carboxy-terminal motif (CTM). BRD3/BRD4–NUT fusion proteins found in NUT midline carcinoma (NMC) patients fuse the amino terminus of BRD4 (or BRD3) with almost the entire NUT protein.
In addition to two bromodomains, all four BET proteins possess a conserved extraterminal (ET) domain that performs an effector role in transcriptional activation and in chromatin remodeling. The ET domain interacts with several different cofactors, including the demethylase protein JMJD6, the methyltransferase/adaptor protein NSD3, and the chromatin remodeling ATPases CHD4 and BRG1 (Rahman et al. 2011; Shen et al. 2015). The ET domain also interacts with the virally encoded proteins, such as murine leukemia virus (MLV) integrase and latency-associated nuclear antigen (LANA) peptide of Kaposi’s sarcoma-associated herpesvirus (KSHV) (Hellert et al. 2013; Crowe et al. 2016). All of these interactions are mediated by a hydrophobic groove on the ET domain that recognizes a consensus motif of alternating lysine and hydrophobic residues found on several of the above-mentioned cofactors (e.g., an IKLKI motif on NSD3 and a LKIKL motif on CHD4) (Hellert et al. 2013; Shen et al. 2015; Crowe et al. 2016; Zhang et al. 2016). The available evidence indicates that BRD4 relies on a unique subset of these ET-interacting proteins for transcriptional activation in particular cell types. In HEK293T cells, BRD4 uses JMJD6 as its ET-domain effector, which will demethylate histones and noncoding RNA to promote enhancer-mediated gene activation (Liu et al. 2013). In acute myeloid leukemia (AML) cells, the ET domain of BRD4 activates transcription by interacting with NSD3, which functions as a scaffold to recruit the chromatin-remodeling enzyme CHD8 (Shen et al. 2015). This apparent context-specificity of ET domain function is not well understood at present.
BRD4 and BRDT possess a unique carboxy-terminal motif (CTM), which binds to the serine/threonine kinase P-TEFb as an additional mechanism of gene activation (Bisgrove et al. 2007; Krueger et al. 2010). P-TEFb is a heterodimer of the kinase Cdk9 and a K, T1, or T2-type cyclin, which together can phosphorylate the serine 2 position of the Pol II carboxy-terminal domain (CTD), as well as serine and threonine residues on the pausing factors DSIF and NELF (Krueger et al. 2010; Jonkers and Lis 2015). Thus, BRD4-mediated P-TEFb recruitment will drive a variety of local phosphorylation events to bypass the paused state of Pol II and promote transcription elongation. At the biochemical level, BRD4 uses multiple mechanisms to regulate P-TEFb activity. Using purified proteins, the interaction with BRD4 is sufficient to stimulate P-TEFb kinase activity (Itzen et al. 2014). In cells, this BRD4 interaction is competitive with the interaction of P-TEFb with HEXIM1/7SK RNA, which are inhibitors of its kinase activity (Jang et al. 2005; Yang et al. 2005). BRD4 also contributes to the localization of P-TEFb to hyperacetylated enhancers and promoters across the genome, thus guiding P-TEFb to it relevant substrates near TF-bound sites (Jang et al. 2005; Yang et al. 2005). Another key player in the functional linkage between BRD4 and P-TEFb is the Mediator complex, which is a 30 subunit coactivator complex that physically associates with BRD4 and with P-TEFb (Jiang et al. 1998; Jang et al. 2005; Donner et al. 2010; Allen and Taatjes 2015). Although the precise interaction surface between BRD4 and Mediator has yet to be mapped, the MED23 subunit has been implicated in this interaction (Wang et al. 2013). BRD4 and Mediator stabilize each other’s occupancy at specific sites across the genome, and these two machineries cooperate in recruiting P-TEFb (Jang et al. 2005; Donner et al. 2010; Bhagwat et al. 2016). It should be noted that the CTM region is found on BRD4 (and BRDT), but not on BRD2 and BRD3, which may explain why BRD4 performs a broader nonredundant role in transcriptional activation than the other BET proteins. For example, genetic inactivation of BRD4 will lead to slow growth phenotypes in essentially all mammalian cell lines, whereas BRD2 and BRD3 lead to only subtle phenotypes when targeted (Vakoc CR, unpubl.). Only a few cellular contexts have been identified in which redundancy exists among the BET proteins, as was recently shown for BRD2 and BRD3 in hematopoietic cells (Stonestrom et al. 2015).
Recent studies have found that BRD4 protein possesses intrinsic kinase and KAT activity in in vitro assays (Devaiah et al. 2012, 2016). Similar to P-TEFb, BRD4 can directly phosphorylate the CTD of Pol II at the serine 2 position and can acetylate multiple residues on histone H3 and H4, including H3K112 found on the globular region of the nucleosome (Devaiah et al. 2012, 2016). Earlier work had also identified an intrinsic kinase activity in purified BRD2 and in FSH, the Drosophila melanogaster ortholog of BRD4 (Denis and Green 1996; Chang et al. 2007). The presence of these activities in biochemical assays is difficult to reconcile with the lack of an obvious kinase or KAT domain in the BRD4 polypeptide, and hence these activities should be considered provisional at present and await further validation to confirm their importance in vivo.
SMALL-MOLECULE INHIBITORS OF BET BROMODOMAINS
The above description of BET proteins suggests a general role of these regulators in transcriptional control, particularly because acetylated TFs and histones are found at all active promoters and enhancers in the genomes. It is only recently that the attention of the field has turned toward identifying biological processes that are disproportionately BET protein-dependent. This avenue of research was invigorated by two studies published in 2010 describing the first selective small-molecule inhibitors of BET bromodomains (Filippakopoulos et al. 2010; Nicodeme et al. 2010). The potency, specificity, and in vivo activity of these molecules in modulating BET proteins has allowed numerous studies in a myriad of animal models of disease. This work has exposed a remarkable gene specificity of transcriptional effects of BET inhibition that underpins a broad interest in BET proteins as therapeutic targets.
The first class of BET bromodomain inhibitors, which are a series of thienotriazolodiazepines, were originally filed as patents by the Mitsubishi Tanabe Pharmaceutical Corporation (Adachi et al. 2006; Miyoshi et al. 2010, 2013). The compounds belong to the diazepine family and are analogs of benzodiazepine, which has been used extensively in the clinic as psychoactive drug (Smith et al. 2014). Thereafter, the Bradner laboratory and researchers at GlaxoSmithKline independently published the highly potent and selective BET bromodomain inhibitors JQ1 (a thienotriazolodiazepine) and iBET (a benzodiazepine), respectively (Filippakopoulos et al. 2010; Nicodeme et al. 2010). Of note, both JQ1 and iBET are pan-BET bromodomain inhibitors, which do not discriminate between the two bromodomains within the same BET protein, nor among the four BET family members (Filippakopoulos et al. 2010; Nicodeme et al. 2010). Because the tandem bromodomains within the same BET protein have distinct functions and binding affinities toward acetylated peptides, the lack of specificity of JQ1 and iBET limits the potential use of these chemical probes to study the roles of individual BET proteins. Nonetheless, the paninhibitory activity of these compounds may contribute to their high potency in modulating biological processes in vivo. Because BRD4 tends to be the dominant transcriptional regulator within the BET protein family in somatic cell types, most studies have linked the transcriptional effects of BET inhibitors to BRD4 inhibition, with BRD2 and BRD3 contributing to a lesser degree.
Chemists have continued to optimize these compounds with the aim to improve the selectivity among the BET protein family and enhance drug potency and in vivo pharmacodynamics, in an effort to make these compounds suitable for clinical investigation. This includes multiple pan inhibitors, some of which have entered clinical trials (see below), BD1 selective inhibitors, such as MS-436, Olinone, and BI-2536, as well as the BD2 selective inhibitors RVX-208 and RVX-297 (Steegmaier et al. 2007; Park et al. 2013; Picaud et al. 2013; Zhang et al. 2013; McLure et al. 2014; Kharenko et al. 2016). As expected, inhibition of individual BET bromodomains will lead to different transcriptional and phenotypic outcomes. For example, the BD1-specific inhibitor Olineone induces differentiation of mouse primary oligodendrocytes, whereas pan BET bromodomain inhibitors have the opposite effect (Gacias et al. 2014). The BD2-selective inhibitor RVX-208 was originally identified in a cell-based chemical screen to enhance the production of apoA-1, and only recently was discovered to target BET proteins after completing phase III clinical trials for treatment of atherosclerosis (Bailey et al. 2010; Nicholls et al. 2011, 2012; Picaud et al. 2013; McLure et al. 2014). Interestingly, RVX-208 causes a milder effect on the transcriptome of cells when compared with inhibitors that bind to both BET bromodomains (Picaud et al. 2013).
All of the compounds described above bind to BET bromodomains in a competitive manner with acetyllysine to displace BET-containing protein complexes from chromatin. A more recent innovation in BET inhibitor design has been to conjugate JQ1 with chemical moieties that promote recruitment of E3 ubiquitin ligases, which leads to polyubiquitylation of BET proteins and proteasome-dependent degradation, a strategy known as proteolysis targeting chimera (PROTAC) (Lu et al. 2015; Winter et al. 2015; Zengerle et al. 2015). This new generation of inhibitors (known as dBET1, ARV-825, or MZ1) leads to more potent suppression of BET proteins in cells, and may provide an additional strategy for therapeutic targeting.
Recent studies have shown that many clinical-stage kinase inhibitors, including the CDK inhibitor dinaciclib, the JAK2 inhibitor TG101209, and the PLK1 inhibitor BI-2536, potently inhibit BET bromodomains as an unintended off-target effect (Martin et al. 2013; Ciceri et al. 2014; Dittmann et al. 2014; Ember et al. 2014). These findings raise the possibility that the off-target effect on BET proteins might contribute to the therapeutic effect of these kinase inhibitors. Moreover, these findings reveal an opportunity for the rational design of drugs that simultaneously target specific kinases and BET proteins to augment anticancer activity or prevent drug resistance (Ciceri et al. 2014).
TARGETING THE BRD4–NUT FUSION ONCOPROTEIN IN NUT MIDLINE CARCINOMA
The first malignant context in which BET proteins were proposed as therapeutic targets is in a rare cancer called NUT midline carcinoma (NMC), which is an aggressive subtype of squamous cell carcinoma with a median survival of only 6.7 months (French et al. 2003; French 2010). Most cases of NMC possess a chromosomal translocation that generates a fusion of the amino terminus of BRD4 (or less commonly BRD3 or NSD3) to the carboxyl terminus of NUT, which is normally only expressed in testes (French et al. 2007, 2014; French 2010). The resulting BRD4–NUT oncoprotein retains the two bromodomains and the ET domain fused to a region of NUT that binds to p300, a protein with KAT activity (French et al. 2003, 2007; Reynoird et al. 2010; Wang and You 2015). It is interesting to note that BRD4–NUT (and presumably BRD3–NUT) requires an interaction with NSD3 via its ET domain for its oncogenic function, whereas the NSD3–NUT fusion requires its BRD4-binding motif for its oncogenic function (French et al. 2014). This highlights a remarkable convergence of molecular functions among BRD3–, BRD4–, and NSD3–NUT fusion proteins, and implies that each of these proteins function through similar multisubunit complexes to regulate transcription.
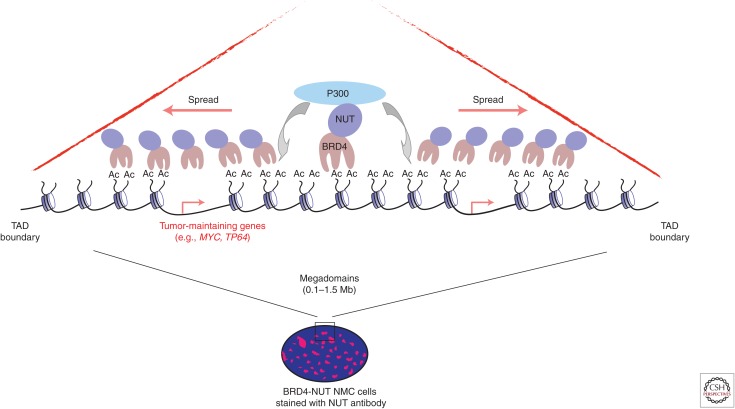
The tumorigenic properties of BRD4–NUT stems from the coupling of its bromodomains with the p300 binding site on NUT, which leads to a positive feedback loop that generates hyperacetylation-driven nuclear foci (Fig. 2) (French et al. 2007, 2014; Yan et al. 2011; Grayson et al. 2014; Alekseyenko et al. 2015; Wang and You 2015). At a genomic level, this positive feedback loop of acetylation and bromodomain-mediated binding generates large contiguous “megadomains” of active chromatin, which are enriched for BRD4–NUT, p300, and histone hyperacetylation, which span >1 Mb of the genome (Alekseyenko et al. 2015). The BRD4–NUT-induced active chromatin domain seems to propagate unfettered until it encounters the edge of a toplogical domain defined by CTCF/cohesion (Alekseyenko et al. 2015). Active chromatin domains of this size are not observed in normal mammalian chromatin, highlighting a unique chromatin-based mechanism of transformation used by BRD4–NUT. Despite the enormity of these megadomains, only a specific program of genes becomes aberrantly expressed by BRD4–NUT function, which includes MYC and the epithelial fate determinant TP63 (Alekseyenko et al. 2015).
Figure 2.
Oncogenic mechanism of the BRD4–NUT fusion protein. In NUT midline carcinoma (NMC) cells, BRD4–NUT forms 80 to 100 large nuclear foci, due to the aggregation of BRD4–NUT on the chromatin. On a molecular level, BRD4–NUT occupies a stretch of highly acetylated chromatin, spanning 0.1–1.5 Mb, termed “mega-domains.” The NUT portion of BRD4–NUT recruits p300, which is a histone acetyltransferases that drives further increases in local histone acetylation, thus forming a feed forward loop. The size and spreading of megadomains tends to be restricted to specific topologically associated domains (TADs). Megadomains can occur over genes that are important for the survival of NMC cells, like MYC and TP63.
Because the bromodomains of BRD4–NUT are essential to its oncogenic function, NMC provides a clear rationale for evaluating the therapeutic activity of BET bromodomain inhibitors. Exposing cultured NMC cell lines to JQ1 leads to a rapid eviction of BRD4–NUT from chromatin, followed by a rapid suppression of its direct target genes, such as MYC, and the induction of terminal squamous cell differentiation (Filippakopoulos et al. 2010; Yan et al. 2011; Grayson et al. 2014; Alekseyenko et al. 2015). Moreover, JQ1 treatment of mice bearing subcutaneous NMC patient-derived xenograft leads to a pronounced inhibition of tumor growth in vivo, with minimal toxicity to normal tissues (Filippakopoulos et al. 2010). This remarkable study provided the first evidence for BET inhibition having a therapeutic index in treating cancer, and provided rationale for subsequent clinical studies of BET inhibition in NMC patients (see below).
A WIDESPREAD SENSITIVITY OF CANCER CELLS TO BET BROMODOMAIN INHIBITION
Given the rarity of NMC, a key question arises as to whether malignancies that lack BRD4 rearrangements would also be sensitive to BET inhibitors. Because the original description of NMC sensitivity to BET inhibition, numerous studies have shown that BRD4 is a non-oncogene dependency in several forms of cancer. Two studies published in 2011 implicated BRD4 as a vulnerability in the MLL-rearranged subtype of AML (Dawson et al. 2011; Zuber et al. 2011). One used shRNA screening to reveal that AML cells were hypersensitive to genetic knockdown of BRD4, whereas the other study used a proteomic approach to link BRD4 with MLL-fusion cofactors (Dawson et al. 2011; Zuber et al. 2011). These studies, as well as others using multiple myeloma and lymphoma models, showed that BET inhibitors show therapeutic effects in diverse genetic contexts of hematological malignancy, with effects comparable to observations in NMC models (Dawson et al. 2011; Delmore et al. 2011; Mertz et al. 2011; Zuber et al. 2011). Although BET inhibitors show broad efficacy in the blood malignancies, the precise pattern of gene expression changes incurred by BET inhibitor treatment is remarkably heterogeneous among different cancer cell lines. In addition, some cancer cell types will terminally differentiate in response to JQ1/iBET exposure, whereas others undergo apoptosis (Dawson et al. 2011; Delmore et al. 2011; Mertz et al. 2011; Zuber et al. 2011). Although there is generally a lack of consistent gene-expression alterations following BET inhibitor treatment in these malignancies, the well-established oncogenes MYC, BCL2, and CDK6 are often suppressed by these drugs, whereas housekeeping genes tend to be unaffected (Shi and Vakoc 2014).
Over the past 5 years, the efficacy of BET inhibitors has been shown in numerous preclinical solid tumor models, including tumors of the prostate, breast, colon, intestine, pancreas, liver, and brain (Sahai et al. 2016). Large-scale profiling studies in human cancer cell lines have suggested that a specific subset of these different tumors harbor exceptional sensitivity to BET inhibitors (Rathert et al. 2015). However, it has been challenging to identify biomarkers that predict hypersensitivity to BET inhibition that might guide patient enrollment into clinical trials. Nonetheless, we can now appreciate that sensitivity to BET inhibition is pervasive across different malignancies.
Although the broad anticancer activity of BET inhibitors is remarkable, it should be emphasized that normal cell types are also affected in unique ways by these agents. This leads to pleiotropic phenotypes in the normal tissues of mice, and presumably in humans. Some of these effects may present therapeutic opportunities in areas outside of oncology. For example, iBET will attenuate cytokine production in innate immune cells, which can allow iBET-treated mice to survive septic shock (Nicodeme et al. 2010). BET inhibitors will also suppress the pathological remodeling of cardiomyoctyes in response to pressure overload (Anand et al. 2013). It has also been proposed that BET inhibitors might be used as a male contraceptive, owing to the reversible impairment in spermatogenesis consequent to BRDT inhibition in the testes (Matzuk et al. 2012; Berkovits and Wolgemuth 2013). BET inhibition will reactivate latent HIV infection, which might prove useful to eliminate viral reservoirs in infected patients (Banerjee et al. 2012; Zhang et al. 2012; Zhu et al. 2012; Li et al. 2013). However, it remains to be determined whether these indications for BET inhibition will be translated into human clinical investigation.
There are also more concerning effects of BET inhibition, such as an impairment in memory formation and an autism-like syndrome in the central nervous system and a worsening of viral/bacterial infections consequent to immunosuppression (Marazzi et al. 2012; Korb et al. 2015; Sullivan et al. 2015). Transgenic Brd4 shRNA mice, in which BRD4 levels are reduced conditionally in adult tissues, show stem cell depletion in the small intestine and hyperplasia of epidermal tissues (Bolden et al. 2014). The latter phenotype may reflect the emerging role for BRD4 in tumor suppression, and hence BET inhibition might be expected to worsen certain malignancies (Alsarraj and Hunter 2012; Fernandez et al. 2014; Tasdemir et al. 2016). Notably, the side effects of BRD4 knockdown in vivo are known to be reversible after restoring BET protein function (Matzuk et al. 2012; Bolden et al. 2014; Nagarajan et al. 2014).
PHASE I CLINICAL STUDIES OF BET INHIBITORS IN HUMAN CANCER PATIENTS
The efficacy of BET inhibitors in preclinical cancer models provided the rationale for a multitude of ongoing human clinical trials, which includes patients with hematological malignancies, BRD4–NUT expressing NMC, and various solid tumors. A summary of these ongoing trials can be found in Table 1. Although we cannot determine at the present time the ultimate impact BET inhibitors will have in oncology, there a few key observations than have been made from the initial administration of these agents to human patients thus far. Importantly, the toxicities in humans have been determined for three clinical BET inhibitors (OTX015, TEN-010, and CPI-0610) (Abramson et al. 2015; Shapiro et al. 2015; Amorim et al. 2016; Berthon et al. 2016; Stathis et al. 2016). In one set of phase 1 trials in hematological cancers, OTX015 was administered orally once or twice a day for 21-day cycles. At the higher doses (120–160 mg/d), OTX015 resulted in a substantial, yet reversible thrombocytopenia (drop in platelet counts), severe gastrointestinal events, and fatigue (Amorim et al. 2016; Berthon et al. 2016). The recommended dose for leukemia and lymphoma patients was identified in this study as 80 mg/d given in repetitive cycles of 14 days on followed by 7 days off (Amorim et al. 2016; Berthon et al. 2016). At different doses in these trials, evidence was reported of disease reduction in five out of 37 acute leukemia patients and five out of 17 diffuse large B cell lymphoma patients, whereas no responses were observed in any of the 12 multiple myeloma patients treated (Amorim et al. 2016; Berthon et al. 2016). Consistent with observations in mouse models, OTX015 was found to cause the terminal differentiation of myeloid leukemia cells, as indicated by an increase in peripheral neutrophil counts during treatment (Dawson et al. 2011; Zuber et al. 2011; Berthon et al. 2016). Unfortunately, a specific genetic mutation in leukemia patients has yet to be identified that correlates with responses to OTX015. This highlights the formidable challenge of identifying a predictive biomarker to guide patient enrollment in future studies. In NMC patients, rapid responses to OTX015 have also been identified, in association with tumor regression (Shapiro et al. 2015; Stathis et al. 2016). Although these findings are encouraging and will motivate further phase II evaluation, many of the patients that initially responded later relapsed several months after initiating treatment (Stathis et al. 2016). This indicates the importance of studying mechanisms of resistance and the potential of combining BET inhibitors with other agents to provide more durable responses. Taken together, these initial studies have generated sufficient enthusiasm within the pharmaceutical industry to justify a continuing of phase II clinical investigation.
Table 1.
Clinical trials of BET bromdomain inhibitors
| Compound | Sponsor | NCT identifier | Conditions | Clinical phase |
|---|---|---|---|---|
| ABBV-075 | AbbVie | NCT02391480 | Advanced cancer; breast cancer; non-small-cell lung cancer (NSCLC); acute myeloid leukemia (AML); multiple myeloma | Phase I (recruiting) |
| BAY 1238097 | Bayer | NCT02369029 | Neoplasms | Phase I (terminated) |
| BI 894999 | Boehringer Ingelheim | NCT02516553 | Neoplasms | Phase I (recruiting) |
| BMS-986158 | Bristol-Myers Squibb | NCT02419417 | Multiple indications cancer | Phase I/IIa (recruiting) |
| NCT01949883 | Lymphoma | Phase I (recruiting) | ||
| CPI-0610 | Constellation Pharmaceuticals | NCT02157636 | Multiple myeloma | Phase I (recruiting) |
| NCT02158858 | Leukemia, myelocytic, acute; myelodysplastic syndrome (MDS); meylodusplastic/myeloproliferative neoplasm, unclassifiable; meylofibrosis | Phase I (recruiting) | ||
| FT-1101 | Forma Therapeutics | NCT02543879 | AML; acute myelogenous leukemia; myelodysplastic syndrome | Phase I (recruiting) |
| GS-5829 | Gilead Sciences | NCT02607228 | Metastatic castration-resistant prostate cancer (CRPC) (as a single agent or in combination with enzalutamide) | Phase I (recruiting) |
| GSK2820151 | GlaxoSmithKline | NCT02630251 | Cancer | Phase I (not yet open for recruiting) |
| GSK525762/I-BET762 | GlaxoSmithKline | NCT01587703 | Carcinoma, midline | Phase I (recruiting) |
| NCT01943851 | Cancer | Phase I (recruiting) | ||
| INCB054329 | Incyte Corporation | NCT02431260 | Advanced cancer | Phase I/II (recruiting) |
| NCT02698189 | AML including AML de novo and AML secondary to MDSs; diffuse large B-cell lymphoma (DLBCL) | Phase I (recruiting) | ||
| MK-8628 | Merck Sharp & Dohme Corp. | NCT02698176 | NUT-midline carcinoma (NMC); triple-negative breast cancer (TNBC); NSCLC; CRPC | Phase I (recruiting) |
| N-methyl-2-pyrrolidone | Peter MacCallum Cancer Centre, Australia | NCT02468687 | Multiple myeloma | Phase I (recruiting) |
| NCT01713582 | AML; DLBCL; acute lymphoblastic leukemia; multiple myeloma | Phase I (active, not recruiting) | ||
| OTX015/MK-8628 | OncoEthix GmbH/Merck | NCT02259114 | NMC; TNBC; NSCLC with rearranged ALK gene/fusion protein or KRAS mutation; CRPC; pancreatic ductal adenocarcinoma |
Phase I (active, not recruiting) |
| NCT02296476 | Glioblastoma multiforme | Phase I (terminated) | ||
| RVX-208/ RVX 000222 | Resverlogix Corp. | NCT02586155 | Diabetes mellitus, type 2; coronary artery disease; cardiovascular diseases | Phase III (recruiting) |
| TEN 010 | Tensha Therapeutics/Roche | NCT01987362 | Solid tumors; advanced solid tumors | Phase I (recruiting) |
| NCT02308761 | MDSs; AML | Phase I (recruiting) | ||
| ZEN003694 | Zenith Epigenetics | NCT02711956 | Metastatic CRPC (in combination with enzalutamide) | Phase I (not yet open for recruiting) |
| NCT02705469 | Metastatic CRPC | Phase I (recruiting) |
See clinicaltrials.gov.
WHY ARE CANCER GENES HYPERSENSITIVE TO BET INHIBITION?
When considering the basic molecular function of BRD4 described above, it is difficult to understand why chemical inhibition of BET proteins would lead to preferential impairment to cancer cells versus nontransformed cell types. Transcriptome-level studies have revealed that BET inhibitors suppress hundreds of genes in each cell type (Anand et al. 2013; Chapuy et al. 2013; Lovén et al. 2013; Asangani et al. 2014). The identity of BET-dependent genes varies dramatically from cell type to cell type, which poses a major challenge in proposing a unified mechanism to explain the anticancer effects of BET inhibitors. At present, our understanding of these effects is limited to correlative observations that these compounds will preferentially suppress expression of cancer-promoting genes versus that of housekeeping genes (Chapuy et al. 2013; Lovén et al. 2013; Nagarajan et al. 2014; Roe et al. 2015; Bhagwat et al. 2016; Henssen et al. 2016). An alternative summation of the available evidence is that JQ1 preferentially suppresses “highly regulated” genes, that is, genes that are dynamically expressed in response to exogenous stimuli or those genes that are expressed in a lineage-specific manner, and hence are influenced by numerous trans- and cis-acting regulators. Indeed, many growth/cancer-promoting genes (e.g., MYC and BCL2) fall into this broad category (Lovén et al. 2013; Asangani et al. 2014; Roe et al. 2015; Shu et al. 2016). Such a model would also explain why JQ1 suppresses cytokine genes in immune cells and the immediate-early genes in cardiomyocytes and neurons (Nicodeme et al. 2010; Anand et al. 2013; Brown et al. 2014; Korb et al. 2015; Toniolo et al. 2015). It is important to note that the effects of JQ1 on transcription have been shown to be reversible, that is, withdrawing JQ1 leads to a rapid restoration of the preexisting transcription level (Mertz et al. 2011). This is likely to account for why normal tissues are able to recover in the setting of 14-day-on, 7-day-off BET inhibition treatment-cycles in humans (Amorim et al. 2016; Berthon et al. 2016). Cancer cells are perhaps less able to recover following BET inhibitor treatment, owing to their “addiction” to oncogenes like MYC (Arvanitis and Felsher 2006). Nevertheless, the specific pattern of genes that are suppressed by BET inhibition in each cell type is clearly of central importance to the therapeutic efficacy of these agents. Below, we describe studies that have defined molecular mechanisms that underlie the gene-specific transcriptional effects of BET bromodomain inhibition.
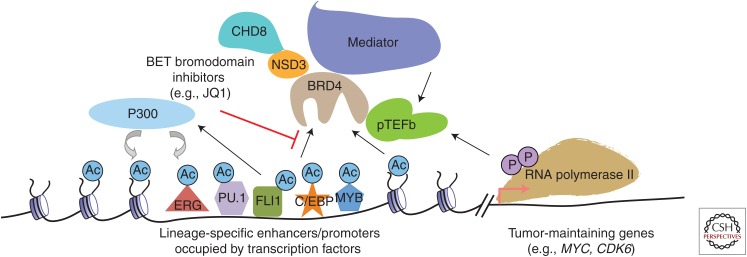
One explanation for the context-specific effects of BET inhibition is that each cell type expresses a different complement of acetylated TFs that bind to BET bromodomains. In AML cells, for example, it has been found that BRD4 binding across the genome is highly correlated with a set of hematopoietic lineage TFs (ERG, FLI1, PU.1, MYB, C/EBPα, and C/EBPβ), which can each physically associate BRD4 in either a bromodomain-dependent or independent manner (Roe et al. 2015). In JQ1-treated leukemia cells, the downstream target genes of this set of TFs are rapidly suppressed, whereas TF occupancy on DNA remains unaffected (Fig. 3) (Roe et al. 2015). Moreover, ectopic expression of these hematopoietic TFs in fibroblasts can recapitulate the JQ1 transcriptional response seen in leukemia cells, indicated that these TFs are sufficient to specify the effects of BET inhibition (Roe et al. 2015). An expanding body of literature is continuing to link the anticancer effects of BET inhibition to the functional suppression key TFs—the androgen receptor in prostate cancer (Asangani et al. 2014), the estrogen receptor and TWIST in breast cancer (Nagarajan et al. 2014; Shi et al. 2014), NF-κB in lymphoma and lung cancers (Asangani et al. 2014; Nagarajan et al. 2014; Zou et al. 2014; Gao et al. 2015). The broad suppression of TF function in normal cell types is also likely to be related to the on-target toxicities of BET inhibitors seen in human patients. As an example, the demonstrated interaction between the diacetylated TF GATA-1 and BET proteins provides a potential explanation for the thrombocytopenia observed in BET inhibitor-treated patients (Gamsjaeger et al. 2011; Lamonica et al. 2011; Stonestrom et al. 2015).
Figure 3.
The BRD4 pathway in acute myeloid leukemia (AML) cells. In AML cells, BRD4 is recruited to lineage-specific enhancers and promoters by acetylated histones and transcription factors (TFs), which are acetylated by p300. BRD4 will then recruit several proteins to regions through direct physical interaction, including NSD3/CHD8, the Mediator complex, and p-TEFb, which promotes transcriptional activation. BET bromodomain inhibitors will release BRD4 from chromatin along with its cofactors to suppress transcription. Because many BRD4-occupied enhancers are located at distal upstream or downstream sites, several of these activities are occurring at a distance from the target gene promoter.
It has been observed that many of the genes that are sensitive to BET inhibition have an unusually large repertoire of enhancer elements in their vicinity, which might be interpreted as another indicator of genes that are “highly regulated.” Indeed, genes with a large number of enhancers tend to encode lineage-specific and growth-regulatory factors (Hnisz et al. 2013; Whyte et al. 2013). Such enhancers have been given numerous labels (superenhancers, locus control regions, stretch enhancers) and often exist in clusters and harbor high levels of BRD4 enrichment (Li et al. 2002; Chapuy et al. 2013; Lovén et al. 2013; Parker et al. 2013). Using genomic approaches applied to several different cell types, it has been observed that genes with “superenhancers” nearby tend to be more suppressed by JQ1 than randomly chosen expressed genes (Chapuy et al. 2013; Lovén et al. 2013; Peeters et al. 2015). More recent evidence indicates that only a minority of superenhancers are in fact targeted by JQ1, as indicated by measurements of Mediator eviction following BET inhibitor exposure (Bhagwat et al. 2016).
From these studies, it is clear that specific cis-elements in the genome are more suppressed by BET inhibition than others, and this contributes to the biased effects of JQ1 on certain genes. The mechanistic basis underlying these heterogeneous effects is still not understood. One possibility is that enhancer-binding proteins like BRD4 have variable on–off rates at each cis-regulatory element, and that perhaps BET inhibitors will preferentially evict BRD4-containing proteins complexes from chromatin at sites that are more dynamic. Taken together, the context-specific consequences of BET inhibition can be attributed, at least in part, to the specific complement of TF-bound cis-elements (enhancers and promoters) that are suppressed by these small molecules.
MECHANISMS OF RESISTANCE TO BET INHIBITION
All cancer monotherapies are limited by the emergence of drug-resistant cell populations, with the ongoing clinical trials indicating that BET inhibitors are not an exception. Hence, an important area of ongoing investigation has been to define mechanisms of resistance to BET bromodomain inhibition. Several recent studies have shown that acquired resistance to BET inhibition is associated with nongenetic mechanisms, in association with a global alteration of gene expression that compensates for the effects of BET inhibition (Tang et al. 2014; Fong et al. 2015; Rathert et al. 2015; Shu et al. 2016). In MLL-fusion AML, this compensatory change in gene expression is linked to activation of the WNT signaling pathway, which can restore MYC expression despite chemical blockade of BRD4 (Fong et al. 2015; Rathert et al. 2015). In breast cancer cells, JQ1 resistance is associated with an elevated level of BRD4 phosphorylation, which in turn will bind more tightly to the Mediator complex to achieve bromodomain-independent recruitment to chromatin (Shu et al. 2016). Across a panel of heterogeneous cell lines, it has been found that the sensitivity to BET inhibitors can be correlated with the preexisting expression level of genes that encode apoptosis regulators (Conery et al. 2016). For example, leukemia cell lines with high BCL2 expression and low expression of BCL2L1 (also called BCL-xl) or BAD are generally correlated with higher sensitivity to BET inhibition (Conery et al. 2016). Consistent with this observation, acquired resistance to BET inhibition can be linked to an increase in the expression of BCL2L1 by gaining super enhancers upstream of the BCL2L1 gene (Shu et al. 2016). What is notable across these studies is that resistance to BET inhibition is not associated with BRD2/BRD3/BRD4 mutations, but instead is associated with selection for a rare (presumably preexisting) cell population harboring a pattern of gene expression that bypasses BET inhibition (Fong et al. 2015; Rathert et al. 2015; Conery et al. 2016).
A clear objective for future clinical investigation is to identify drugs that synergize with BET inhibitors in causing anticancer effects, but without having a synergistic increase in toxicity. One promising area of drug combinations is to use BET inhibition as a means to eliminate drug resistance to other targeted agents. In T-cell leukemia driven by activating NOTCH1 mutations, drug-tolerant cells are able to survive NOTCH pathway inhibition (Knoechel et al. 2014). These resistant T-ALL cells are more sensitive to BET bromodomain inhibition than the parental population, thus providing a rationale to combine NOTCH and BET-targeting agents in this disease (Knoechel et al. 2014). A similar scenario is found in breast cancer driven by PI3 kinase mutations, in which sensitivity to PI3K inhibitors is attenuated by feedback pathways that bypass PI3K through the activation of tyrosine kinase pathways (Stratikopoulos et al. 2015). Remarkably, these bypass pathways can be suppressed transcriptionally via BET bromodomain inhibition. Hence, combinations of BET and PI3K inhibitors are a promising therapeutic approach in breast cancer. Resistance to estrogen receptor modulation, Sonic hedgehog, and androgen receptor blockade can also be overcome by BET bromodomain inhibition, thus providing numerous opportunities to explore drug combinations in the clinic (Asangani et al. 2014; Nagarajan et al. 2014; Tang et al. 2014; Shu et al. 2016).
CONCLUDING REMARKS
There is a long history of treating cancer patients with agents that disrupt fundamental cellular processes (e.g., antimetabolites and DNA alkylating agents), which can cause more severe cell death responses in cancer cells than in normal tissues. A similar description could be applied to BET bromodomain inhibitors, which target a set of transcriptional coactivators to disrupt an important hub for numerous TF pathways. As we have outlined in this review, there is clearly specificity in the transcriptional effects of BET inhibition, but only to a degree. Hence, these agents can cause detrimental effects to cancer cells in association with tolerable pleiotropic biological effects in normal tissues. The basic and preclinical research performed in this field has provided a roadmap for the implementation of BET inhibition in the clinic. In our view, the success of BET inhibition in clinical studies will rest squarely on our ability to find predictive biomarkers of therapeutic response and the most effective drug combinations for achieving durable disease remissions.
ACKNOWLEDGMENTS
C.R.V. is supported by the Leukemia and Lymphoma Society, the Burroughs-Wellcome Fund, the Pershing Square Sohn Cancer Research Alliance, the Starr Cancer Consortium, and the National Institutes of Health/National Cancer Institute (NIH/NCI) Grant RO1 CA174793.
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Abramson JS, Blum KA, Flinn IW, Gutierrez M, Goy A, Maris M, Cooper M, Meara M, Borger D, Mertz J, et al. 2015. BET inhibitor CPI-0610 is well tolerated and induces responses in diffuse large B-cell lymphoma and follicular lymphoma: Preliminary analysis of an ongoing phase 1 study. Blood 126: 1491. [Google Scholar]
- Adachi K, Hikawa H, Hamada M, Endoh JI, Ishibuchi S, Fujie N, Tanaka M, Sugahara K, Oshita K, Murata M. 2006. Thienotriazolodiazepine compound and a medicinal use thereof. WPO Patent No. WO2006129623A1.
- Alekseyenko AA, Walsh EM, Wang X, Grayson AR, Hsi PT, Kharchenko PV, Kuroda MI, French CA. 2015. The oncogenic BRD4–NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev 29: 1507–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Taatjes DJ. 2015. The mediator complex: A central integrator of transcription. Nat Rev Mol Cell Biol 16: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsarraj J, Hunter KW. 2012. Bromodomain-containing protein 4: A dynamic regulator of breast cancer metastasis through modulation of the extracellular matrix. Int J Breast Cancer 2012: 670632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K, et al. 2016. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 3: e196–e204. [DOI] [PubMed] [Google Scholar]
- Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. 2013. BET bromodomains mediate transcriptional pause release in heart failure. Cell 154: 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis C, Felsher DW. 2006. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol 16: 313–317. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. 2014. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D, Jahagirdar R, Gordon A, Hafiane A, Campbell S, Chatur S, Wagner GS, Hansen HC, Chiacchia FS, Johansson J, et al. 2010. RVX-208: A small molecule that increases apolipoprotein A-I and high-density lipoprotein cholesterol in vitro and in vivo. J Am Coll Cardiol 55: 2580–2589. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. 2012. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol 92: 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Wolgemuth DJ. 2013. The role of the double bromodomain-containing BET genes during mammalian spermatogenesis. Curr Top Dev Biol 102: 293–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, et al. 2016. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol 3: e186–e195. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Roe JS, Mok BA, Hohmann AF, Shi J, Vakoc CR. 2016. BET bromodomain inhibition releases the Mediator complex from select cis-regulatory elements. Cell Rep 15: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. 2007. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci 104: 13690–13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden JE, Tasdemir N, Dow Lukas E, van Es Johan H, Wilkinson John E, Zhao Z, Clevers H, Lowe Scott W. 2014. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep 8: 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, Bair S, Newton G, Lichtman A, Kung A, et al. 2014. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell 56: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YL, King B, Lin SC, Kennison JA, Huang DH. 2007. A double-bromodomain protein, FSH-S, activates the homeotic gene ultrabithorax through a critical promoter-proximal region. Mol Cell Biol 27: 5486–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown Michael R, Lin Charles Y, Monti S, Roemer Margaretha GM, Qi J, Rahl Peter B, Sun Heather H, Yeda Kelly T, Doench John G, et al. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciceri P, Müller S, O’Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, Lasater EA, Pallares G, Picaud S, Wells C, et al. 2014. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol 10: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conery AR, Centore RC, Spillane KL, Follmer NE, Bommi-Reddy A, Hatton C, Bryant BM, Greninger P, Amzallag A, Benes CH, et al. 2016. Preclinical anticancer efficacy of BET bromodomain inhibitors is determined by the apoptotic response. Cancer Res 76: 1313–1319. [DOI] [PubMed] [Google Scholar]
- Crowe BL, Larue RC, Yuan C, Hess S, Kvaratskhelia M, Foster MP. 2016. Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction. Proc Natl Acad Sci 113: 2086–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancy BM, Cole PA. 2015. Protein lysine acetylation by p300/CBP. Chem Rev 115: 2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittman A, Giotopoulos G, Bantscheff M, Chan W-I, Robson SC, Chung Cw, Hopf C, Savitski MM, et al. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV, Green MR. 1996. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev 10: 261–271. [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, Singer DS. 2012. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci 109: 6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, Singer DS. 2016. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 23: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci 100: 8758–8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM, Zhou MM. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496. [DOI] [PubMed] [Google Scholar]
- Dittmann A, Werner T, Chung CW, Savitski MM, Fälth Savitski M, Grandi P, Hopf C, Lindon M, Neubauer G, Prinjha RK, et al. 2014. The commonly used PI3-kinase probe LY294002 is an inhibitor of BET bromodomains. ACS Chem Biol 9: 495–502. [DOI] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 17: 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ember SWJ, Zhu JY, Olesen SH, Martin MP, Becker A, Berndt N, Georg GI, Schönbrunn E. 2014. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem Biol 9: 1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P, Scaffidi P, Markert E, Lee JH, Rane S, Misteli T. 2014. Transformation resistance in a premature aging disorder identifies a tumor-protective function of BRD4. Cell Rep 9: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. 2014. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13: 337–356. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, et al. 2012. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149: 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong CY, Gilan O, Lam EYN, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, et al. 2015. BET inhibitor resistance emerges from leukaemia stem cells. Nature 525: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA. 2010. NUT midline carcinoma. Cancer Genet Cytogenet 203: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. 2003. BRD4–NUT fusion oncogene: A novel mechanism in aggressive carcinoma. Cancer Res 63: 304. [PubMed] [Google Scholar]
- French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. 2007. BRD–NUT oncoproteins: A family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 27: 2237–2242. [DOI] [PubMed] [Google Scholar]
- French CA, Rahman S, Walsh EM, Kühnle S, Grayson AR, Lemieux ME, Grunfeld N, Rubin BP, Antonescu CR, Zhang S, et al. 2014. NSD3–NUT fusion oncoprotein in NUT midline carcinoma: Implications for a novel oncogenic mechanism. Cancer Discov 4: 928–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, et al. 2014. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem Biol 21: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. 2011. Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3. Mol Cell Biol 31: 2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yang Y, Wang Z, Gao X, Zheng B. 2015. BRAD4 plays a critical role in germinal center response by regulating Bcl-6 and NF-κB activation. Cell Immunol 294: 1–8. [DOI] [PubMed] [Google Scholar]
- Grayson AR, Walsh EM, Cameron MJ, Godec J, Ashworth T, Ambrose JM, Aserlind AB, Wang H, Evan GI, Kluk MJ, et al. 2014. MYC, a downstream target of BRD–NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene 33: 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellert J, Weidner-Glunde M, Krausze J, Richter U, Adler H, Fedorov R, Pietrek M, Rückert J, Ritter C, Schulz TF, et al. 2013. A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine γ-herpesvirus LANA proteins. PLoS Pathog 9: e1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssen A, Althoff K, Odersky A, Beckers A, Koche R, Speleman F, Schäfers S, Bell E, Nortmeyer M, Westermann F, et al. 2016. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clin Cancer Res 22: 2470–2481. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke H, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen F, Greifenberg AK, Bösken CA, Geyer M. 2014. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res 42: 7577–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci 95: 8538–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. 2015. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharenko OA, Gesner EM, Patel RG, Norek K, White A, Fontano E, Suto RK, Young PR, McLure KG, Hansen HC. 2016. RVX-297—A novel BD2 selective inhibitor of BET bromodomains. Biochem Biophys Res Commun 477: 62–67. [DOI] [PubMed] [Google Scholar]
- Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, Gillespie SM, Fernandez D, Ku M, Wang H, et al. 2014. An epigenetic mechanism of resistance to targeted therapy in T-cell acute lymphoblastic leukemia. Nat Genet 46: 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD. 2015. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci 18: 1464–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger BJ, Varzavand K, Cooper JJ, Price DH. 2010. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes conformational change in 7SK. PLoS ONE 5: e12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, et al. 2011. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci 108: E159–E168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. 2002. Locus control regions. Blood 100: 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Guo J, Wu Y, Zhou Q. 2013. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res 41: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal A K, Rosenfeld M G. 2013. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155: 1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K, Hines J, Winkler J D, Crew Andrew P, Coleman K, et al. 2015. Hijacking the E3 ubiquitin ligase Cereblon to efficiently target BRD4. Chem Biol 22: 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JSY, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. 2012. Suppression of the antiviral response by an influenza histone mimic. Nature 483: 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Olesen SH, Georg GI, Schönbrunn E. 2013. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem Biol 8: 2360–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, et al. 2012. Small-molecule inhibition of BRDT for male contraception. Cell 150: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLure KG, Gesner EM, Tsujikawa L, Kharenko OA, Attwell S, Campeau E, Wasiak S, Stein A, White A, Fontano E, et al. 2014. RVX-208, an inducer of ApoA-I in humans, is a BET bromodomain antagonist. PLoS ONE 8: e83190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ. 2011. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci 108: 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi S, Ooike S, Iwata K, Hikawa H, Sugahara K. 2010. Antitumor agent. U.S. Patent 20100286127 A1.
- Miyoshi S, Ooike S, Iwata K, Hikawa H, Sugahara K. 2013. Antitumor agent. U.S. Patent 8476260 B2.
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461: 664–668. [DOI] [PubMed] [Google Scholar]
- Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, Taipaleenmäki H, Ben-Batalla I, Scheller M, Loges S, et al. 2014. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep 8: 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls SJ, Gordon A, Johansson J, Wolski K, Ballantyne CM, Kastelein JJP, Taylor A, Borgman M, Nissen SE. 2011. Efficacy and safety of a novel oral inducer of apolipoprotein A-I synthesis in statin-treated patients with stable coronary artery disease: A randomized controlled trial. J Am College Cardiol 57: 1111–1119. [DOI] [PubMed] [Google Scholar]
- Nicholls SJ, Gordon A, Johannson J, Ballantyne CM, Barter PJ, Brewer HB, Kastelein JJP, Wong NC, Borgman MRN, Nissen SE. 2012. ApoA-I induction as a potential cardioprotective strategy: Rationale for the SUSTAIN and ASSURE studies. Cardiovasc Drugs Ther 26: 181–187. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung Cw, Chandwani R, Marazzi I, Wilson P, Coste H, et al. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468: 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SR, Speranza G, Piekarz R, Wright JJ, Kinders RJ, Wang L, Pfister T, Trepel JB, Lee MJ, Alarcon S, et al. 2013. A multi-histology trial of fostamatinib in patients with advanced colorectal, non-small cell lung, head and neck, thyroid, and renal cell carcinomas, and pheochromocytomas. Cancer Chemother Pharmacol 71: 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Program NCS, et al. 2013. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci 110: 17921–19726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters JGC, Vervoort SJ, Tan Sander C, Mijnheer G, de Roock S, Vastert SJ, Nieuwenhuis EES, van Wijk F, Prakken BJ, Creyghton MP, et al. 2015. Inhibition of super-enhancer activity in autoinflammatory site-derived T cells reduces disease-associated gene expression. Cell Rep 12: 1986–1996. [DOI] [PubMed] [Google Scholar]
- Picaud S, Wells C, Felletar I, Brotherton D, Martin S, Savitsky P, Diez-Dacal B, Philpott M, Bountra C, Lingard H, et al. 2013. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci 110: 19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31: 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, et al. 2015. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525: 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, Caron C, Kimura H, Rousseaux S, Cole PA, et al. 2010. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J 29: 2943–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. 2015. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 58: 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. 2001. Histone acetyltransferases. Annu Rev Biochem 70: 81–120. [DOI] [PubMed] [Google Scholar]
- Sahai V, Redig AJ, Collier KA, Eckerdt FD, Munshi HG. 2016. Targeting bet bromodomain proteins in solid tumors. Oncotarget 10.18632/oncotarget.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. 2009. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Dev 12: 659–665. [PMC free article] [PubMed] [Google Scholar]
- Shapiro GI, Dowlati A, LoRusso PM, Eder JP, Anderson A, Do KT, Kagey MH, Sirard C, Bradner JE, Landau SB. 2015. Clinically efficacy of the BET bromodomain inhibitor TEN-010 in an open-label substudy with patients with documented NUT-midline carcinoma (NMC). AACR Annual Meeting, Abstract A49 Philadelphia, PA, April 18–22. [Google Scholar]
- Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. 2015. NSD3-Short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 60: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Vakoc CR. 2014. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 54: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, et al. 2014. Disrupting the interaction of BRD4 with di-acetylated twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25: 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Jin Huh S, Liang Y, Ryan J, et al. 2016. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529: 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SG, Sanchez R, Zhou MM. 2014. Privileged diazepine compounds and their emergence as bromodomain inhibitors. Chem Biol 21: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de La Motte Rouge T, Uro-Coste E, de Braud F, Pelosi G, French CA. 2016. Clinical response of carcinomas harboring the BRD4–NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov 6: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krššák M, Gürtler U, Garin-Chesa P, Lieb S, Quant J, et al. 2007. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol 17: 316–322. [DOI] [PubMed] [Google Scholar]
- Stonestrom AJ, Hsu SC, Jahn KS, Huang P, Keller CA, Giardine BM, Kadauke S, Campbell AE, Evans P, Hardison RC, et al. 2015. Functions of BET proteins in erythroid gene expression. Blood 125: 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratikopoulos E, Dendy M, Szabolcs M, Khaykin Alan J, Lefebvre C, Zhou MM, Parsons R. 2015. Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell 27: 837–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JM, Badimon A, Schaefer U, Ayata P, Gray J, Chung CW, von Schimmelmann M, Zhang F, Garton N, Smithers N, et al. 2015. Autism-like syndrome is induced by pharmacological suppression of BET proteins in young mice. J Exp Med 212: 1771–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N, et al. 2014. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 20: 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir N, Banito A, Roe JS, Alonso-Curbelo D, Camiolo M, Tschaharganeh DF, Huang C-H, Aksoy O, Bolden JE, Chen CC, et al. 2016. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov 6: 612–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toniolo PA, Liu S, Yeh JE, Moraes-Vieira PM, Walker SR, Vafaizadeh V, Barbuto JAM, Frank DA. 2015. Inhibiting STAT5 by the BET bromodomain inhibitor JQ1 disrupts human dendritic cell maturation. J Immunol 194: 3180–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, You J. 2015. Mechanistic analysis of the role of bromodomain-containing protein 4 (BRD4) in BRD4–NUT oncoprotein-induced transcriptional activation. J Biol Chem 290: 2744–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yao X, Huang Y, Hu X, Liu R, Hou D, Chen R, Wang G. 2013. Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription 4: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and Mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. 2015. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348: 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282: 13141–13145. [DOI] [PubMed] [Google Scholar]
- Yan J, Diaz J, Jiao J, Wang R, You J. 2011. Perturbation of BRD4 protein function by BRD4–NUT protein abrogates cellular differentiation in NUT midline carcinoma. J Biol Chem 286: 27663–27675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545. [DOI] [PubMed] [Google Scholar]
- Zengerle M, Chan KH, Ciulli A. 2015. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol 10: 1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, Rusinova E, Gerona-Nevarro G, Moshkina N, Joshua J, et al. 2012. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem 287: 28840–28851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Plotnikov AN, Rusinova E, Shen T, Morohashi K, Joshua J, Zeng L, Mujtaba S, Ohlmeyer M, Zhou MM. 2013. Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J Med Chem 56: 9251–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zeng L, Shen C, Ju Y, Konuma T, Zhao C, Vakoc Christopher R, Zhou MM. 2016. Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure 24: 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Gaiha GD, Sinu JP, Pertel T, Chin CR, Gao G, Qu H, Walker BD, Elledge SJ, Brass AL. 2012. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep 2: 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, Nair S, Chen LF. 2014. Brd4 maintains constitutively active NF-κB in cancer cells by binding to acetylated RelA. Oncogene 33: 2395–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]