Abstract
Plants are attractive platforms for synthetic biology and metabolic engineering. Plants’ modular and plastic body plans, capacity for photosynthesis, extensive secondary metabolism, and agronomic systems for large-scale production make them ideal targets for genetic reprogramming. However, efforts in this area have been constrained by slow growth, long life cycles, the requirement for specialized facilities, a paucity of efficient tools for genetic manipulation, and the complexity of multicellularity. There is a need for better experimental and theoretical frameworks to understand the way genetic networks, cellular populations, and tissue-wide physical processes interact at different scales. We highlight new approaches to the DNA-based manipulation of plants and the use of advanced quantitative imaging techniques in simple plant models such as Marchantia polymorpha. These offer the prospects of improved understanding of plant dynamics and new approaches to rational engineering of plant traits.
New approaches to the DNA-based manipulation of plants (e.g., CRISPR-Cas9) and the use of simple plant models (e.g., Marchantia) are helping us to better understand and engineer plants.
The development of new technologies for the production of larger and improved quantities of goods from less feedstock has defined human innovation for thousands of years, especially in food production. To increase agricultural productivity, plant breeders have been selecting for advantageous traits in crops since 9000–10,000 BC (Zohary et al. 2012). One notable success is the selective breeding of modern maize from teosinte (Beadle 1939; Doebley 2004), in which a handful of genetic differences caused substantial changes in ear morphology, kernel number, and crop yield.
Technologies such as mutagenesis and introgressive hybridization, developed in the early to mid-20th century, are commonly used to increase genetic diversity in breeding populations of food crops, which show lower allelic variation compared with wild populations. Selective breeding still plays a major role in the production of new varieties but the emergence of modern plant biotechnology has led to a more targeted approach to increasing crop yields. Traits of agricultural importance successfully introduced to plants using recombinant DNA technology include herbicide resistance (Comai et al. 1985), drought resistance (Kumar et al. 2014), pest resistance (Bates et al. 2005), pathogen resistance (Brunner et al. 2011; Horvath et al. 2012; Jones et al. 2014), abiotic stress resistance (Jaglo-Ottosen et al. 1998), enhanced photosynthetic capacity (Ku et al. 2001), improved nitrogen use efficiency (Yanagisawa et al. 2004), and added nutritional value (Ye et al. 2000). However, although plants hold a unique promise for bioproduction at the gigatonne scale, efforts in genetic engineering of plants are lagging compared with microbial systems (Antunes et al. 2009, 2011; Liu et al. 2011, 2013; Koschmann et al. 2012; Wend et al. 2013; Zurcher et al. 2013; Fethe et al. 2014; Müller et al. 2014).
To exploit the unused potential of plants in expressing complex traits, we require (1) efficient tools and methods for genetic engineering, (2) simpler multicellular plant chassis that are amenable to rapid, high-throughput analysis, and (3) control over the biosynthesis, transport, and storage of metabolites in specialized cells within complex plant tissues. In the following, we highlight progress made in these three key areas of concern for the future development of plant synthetic biology.
NUCLEAR TRANSFORMATION IN PLANTS
Implementation of synthetic biology in plants calls for efficient methods for genetic manipulation. The most widely used technique for transformation of most plant species relies on the native capacity of virulent strains of Agrobacterium tumefaciens to infect plant tissue and to transfer a segment of its DNA (T-DNA) to the host cell (reviewed in Gelvin 2003). Because this method was adapted to allow delivery of transgenes, it has been used extensively: thousands of transformants can be obtained in a single experiment (Meyerowitz 1989; Ishizaki et al. 2008), providing the throughput required for forward genetic screens. The random nature of this transformation method has been used to mine the underlying biology of plants by mutational T-DNA insertion as well as gene trap methods (reviewed in Springer 2000). Enhancer traps have been used to indicate the presence of nearby endogenous enhancer elements driving an orthogonal transcriptional activator such as GAL4-VP16. T-DNA insertion in the vicinity of an enhancer allows temporal and/or tissue-specific expression of the activator, leading to restricted reporter gene expression according to the endogenous enhancer (Johnson et al. 2005; Laplaze et al. 2005; Gardner et al. 2009).
In addition to stable nuclear transformation mediated by Agrobacterium, transient methods have been developed for quantification of gene expression (Kapila et al. 1997) and bioproduction. For instance, Nicotiana benthamiana leaves can be infiltrated with Agrobacterium at multiple regions of a single leaf and screened for gene expression a few days after infection. This allows relatively fast characterization of libraries of genetic components in a common genetic background (Sparkes et al. 2006; Engler et al. 2014; Brückner et al. 2015). Transient leaf agroinfiltration in Nicotiana has been adopted as an efficient method for the optimization of metabolic pathways, such as artemisinin (van Herpen et al. 2010) and triterpene biosynthesis pathways (reviewed in Thimmappa et al. 2014), and the bioproduction of vaccines (D’Aoust et al. 2008, Mardanova et al. 2015). Large investments have been made in both the private and the public sectors to scale up production for vaccines against viruses like Ebola (ZMapp, Mapp Biopharmaceuticals) and influenza (Medicago). Protoplasts, single cells derived from tissues by digestion of cell walls, can also be transformed transiently. Protoplast transformation has been performed by electroporation (Fromm et al. 1985; Ou-Lee et al. 1986; Hauptmann et al. 1987; Negrutiu et al. 1987; Nishiguchi et al. 1987; Jones et al. 1989) or incubation in a PEG solution (Krens et al. 1982; Potrykus et al. 1985), and is established as one of the preferred methods for studying signaling pathways (reviewed in Sheen 2001). Furthermore, high-throughput protoplast transformation has been used to perform quantitative characterization of large libraries of genetic elements in Arabidopsis thaliana (Arabidopsis) and Sorghum bicolor (sorghum) (Schaumberg et al. 2016).
Precise methods for genomic inspection and reverse genetics screens have also been implemented in plants: for instance, CRISPR-Cas9 genome editing has been applied in a number of plant model systems including Arabidopsis, Nicotiana spp., Solanum lycopersicum (tomato), Oryza sativa (rice), Triticum aestivum (wheat), Zea mays (maize), Citrus sinensis (orange), and Marchantia polymorpha (Marchantia), at varying efficiencies (reviewed in Bortesi and Fischer 2015). Notably, CRISPR-Cas9 genome editing has been successfully used to promote homologous recombination and to simultaneously target multiple loci in Arabidopsis and Nicotiana (Li et al. 2014), as well as to induce targeted deletions in Nicotiana and rice (Lowder et al. 2015). Such methods are enabling unprecedented efficiency of construct delivery, genome refactoring, and chassis engineering in plants.
A COMMON SYNTAX FOR SYNTHETIC PLANT GENES
Increasingly, synthetic biologists rely on the use of modular DNA components to implement genetic circuits, along with a facile chassis for prototyping and troubleshooting, and tools for predicting behavior from mechanistic models. The implementation of assembly standards that allow parts, even those from multiple manufacturers, to be assembled together has underpinned invention in engineering disciplines and the generation of libraries of well-characterized standardized components is at the core of the synthetic biology paradigm.
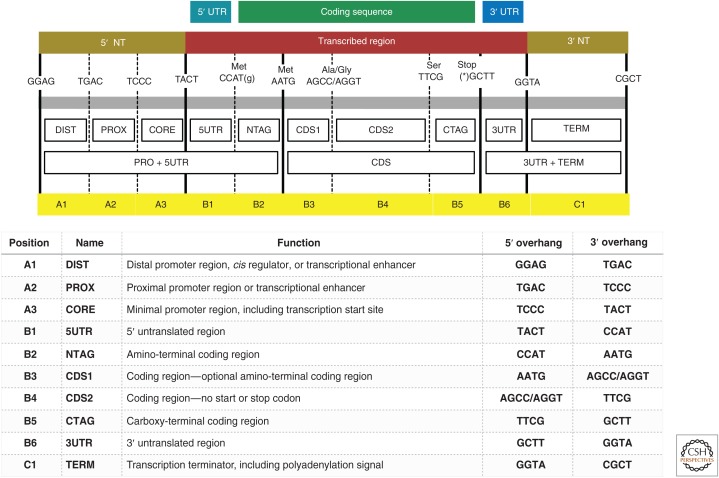
The BioBrick assembly standard was the first widely adopted biological standard (Knight 2003; Shetty et al. 2008) and large number of parts, primarily for engineering prokaryotes, have been submitted to the Registry of Standard Biological Parts (Endy 2005). Eukaryotic organisms such as plants, however, require a different genetic syntax for the assembly of genes and pathways. Additionally, plasmids used for plant transformation often require specific features, such as the ability to replicate in a shuttle chassis such as A. tumefaciens. Recently, several alternative DNA assembly technologies have gained prominence because they offer the advantage of allowing the assembly of multiple parts in a single reaction (Engler et al. 2008; Gibson et al. 2009; Quan and Tian 2009; Li and Elledge 2012; De Kok et al. 2014). Although overlap-dependent methods such as Gibson assembly are powerful, they require custom oligonucleotides and amplification of even well characterized standard parts for each new assembly (Ellis et al. 2009; Liu et al. 2013; Patron 2014) unless standardized overlaps are used (Torella et al. 2013; Casini et al. 2014). The application of Type IIS restriction enzymes for assembling standard parts, known widely as “Golden Gate Cloning,” has become widely used as an alternative approach because parts can be exchanged and assembled cheaply, easily, and in an automatable way without proprietary tools and reagents (Engler et al. 2009; Sarrion-Perdigones et al. 2011; Werner et al. 2012). Many commonly used sequences have been adapted for Type IIS assembly by various plant research laboratories (Sarrion-Perdigones et al. 2011; Weber et al. 2011; Emami et al. 2013; Lampropoulos et al. 2013; Binder et al. 2014; Engler et al. 2014; Vafaee et al. 2014), and a common syntax to enable the exchange of interoperable DNA parts for plants has recently been agreed by a large section of the plant research community (see Fig. 1; Patron et al. 2015).
Figure 1.
Summary of the Phytobrick syntax for standardized plant DNA part composition and assembly. The Phytobrick syntax is a consolidation of Golden Gate, MoClo, and Golden Braid standards (Patron et al. 2015) and defines 12 fusion sites that divide eukaryotic genes into 10 basic functional units (A1–C1). The domains are listed with a brief description of their encoded function. Phytobrick parts can comprise the region between an adjacent pair of fusion sites or span many sites, and consist of portion(s) of a gene cloned into a plasmid flanked by a convergent pair of BsaI Type IIS restriction endonuclease recognition sequences. Parts can be assembled into complete transcriptional units in a one-pot, one-step digestion–ligation reaction provided compatible overhangs are produced on digestion and the acceptor plasmid has divergent BsaI recognition sites as well as a unique bacterial selection cassette.
This common syntax describes 12 fusion sites to enable the facile assembly of eukaryotic transcriptional units and establishes standards that allow standard parts to be exchanged and reused without adaptation. This agreement will aid in establishing registries of genetic components for plants and thus facilitate the design of genetic circuits. The availability of well-characterized parts is the first stage for establishing a higher-order abstraction to implement more complex functions in plants. Combining extensive collections of genetic elements together with high-throughput quantification methods will provide robust measurements to inform mathematical models that describe the statistical normalization methods required for developing parts and devices that will work as expected in complex multicellular organisms.
CONTROL OF TRANSGENE EXPRESSION IN PLANTS
Control of transgene expression can be implemented at multiple levels in plants and other eukaryotes. Regulation can be exerted at the transcriptional, posttranscriptional, and translational levels by both endogenous and synthetic orthogonal systems. Among proven orthogonal systems for modulation of gene expression are transactivation systems like the aforementioned GAL4 activator, a tTA tetracycline inducible system based on the Escherichia coli tetR repressor (Weinmann et al. 1994), and an IPTG-inducible pOp/LhG4 system based on the E. coli lac operon (Moore et al. 1998). The various inducible expression systems implemented in plants to date include the dexamethasone responsive rat glucocorticoid (GR) ligand-binding domain (Aoyama and Chua 1997; Craft et al. 2005; Samalova et al. 2005), ethanol/acetaldehyde-inducible ALCR transcription factor and alcA promoter (Caddick et al. 1998; Salter et al. 1998; Roslan et al. 2001), DNA-binding domain of the lexA oestrogen receptor (Bruce et al. 2000; Zuo et al. 2000), copper-inducible ace1 promoter (Mett et al. 1993), and ecdysone receptor (EcR) ligand binding-domain inducible by insecticide methoxyfenozide (Martinez et al. 1999; Padidam et al. 2003; Koo et al. 2004). Other useful circuits comprise the Cre-Lox recombinase system that can be used for induction of transcription (Hoff et al. 2001) or transgene excision (Chakraborti et al. 2008), and a catalytically inactive version of Cas9 fused to regulatory domains. The latter has been shown to perform effective transcriptional activation and repression of an endogenous gene in Nicotiana (Piatek et al. 2015) and Arabidopsis (Lowder et al. 2015).
Posttranscriptional control over gene expression in plants has been enabled by a variety of RNA-based technologies. Artificial microRNAs (amiRNAs), for example, exploit the endogenous silencing machinery to repress gene expression. The extent of amiRNA-mediated repression can be tuned by altering their expression level (Yu and Pilot 2014). Systems for optimizing the specificity and the efficiency of the amiRNAs (Li et al. 2013) as well as for obtaining inducible multigene silencing (Goh et al. 2012) have also been implemented. Another approach to posttranscriptional control of gene expression in plants is based on riboswitches. Riboswitches are ligand-binding nucleic acid aptamers that are able to modulate gene expression by sequestering or exposing regulatory regions in transcripts via alternative nucleotide base-pairing (Anthony et al. 2012). The thiamine pyrophosphate riboswitch is an endogenous example in plants: It was identified in the 3′ end of the untranslated region of the Arabidopsis thiC gene, and exerts control by affecting pre-mRNA splicing of the transcript (Sudarsan et al. 2003; Wachter et al. 2007). Methods have been developed to tune the sensitivities of riboswitches for their cognate ligands (Beisel and Smolke 2009), and to expand their chemical diversity (reviewed in Link and Breaker 2009).
Technologies for nuclear transformation and control over gene expression have been combined to implement synthetic genetic logic systems in plants. Examples include a light-inducible gene expression system activated by red light and switched off by far-red light (Müller et al. 2014) as well as a range of in vivo biosensors to monitor levels of cytokinin (Müller and Sheen 2008; Zurcher et al. 2013), auxin (Wend et al. 2013), plant pathogens (Liu et al. 2011; Fethe et al. 2014), and elicitors (Koschmann et al. 2012). These are examples of synthetic circuits that have been constructed in plants using methods for DNA manipulation, transgene regulation, transformation, and analysis to yield basic systems for chassis engineering and genetic network design in plant synthetic biology.
METABOLIC ENGINEERING IN CELLULAR COMPARTMENTS
Although plant biotechnology has traditionally relied on the cumulative introduction of genes followed by selection (Bates et al. 2005), new technologies promise to streamline this process: methods such as genome-editing (reviewed in Raitskin and Patron 2016; and in Schiml and Puchta 2016), construction of synthetic eukaryotic chromosomes (Annaluru et al. 2014), or the application of operon-like gene clusters (Field et al. 2011; Nützmann and Osbourn 2014) enable unprecedented control over metabolic pathways.
Plants are ideally suited for the implementation of cooperative biosynthetic processes and dedicated storage compartments. One example of a highly beneficial metabolic interaction between different plant cell types is the cooperation of bundle sheath and mesophyll, which markedly enhances the efficiency of photosynthesis in C4 plants (Hibberd and Covshoff 2010). Another prominent instance of multicellular compartmentalization strategy is naturally observed in glandular trichomes of the wormwood Artemisia annua, which are specialized for production and storage of the antimalarial compound artemisinin (Graham et al. 2010). The potential of glandular tissues in various plant species for metabolic engineering of alkaloid, terpenoid, and fatty acid metabolism for applications in biotechnology has been reviewed in more detail elsewhere (McCaskill and Croteau 1999; Schilmiller et al. 2008).
Marchantia AS A BASAL MODEL CHASSIS FOR PLANT SYNTHETIC BIOLOGY
One of the major challenges in harnessing the potential of plant systems lies in our limited understanding of complex processes in cellular development. Morphogenetic processes in plants are driven by genetic programs, which shape the collective behavior of cohorts of cells. Vice versa, groups of cells display self-organizing properties, which affect the expression of individual genes. Plant metabolic processes take place in dedicated tissue compartments and are subject to multiscale feedback regulation. Thus, there is a need to develop suitable platforms for elucidating and engineering patterning processes defining the anatomy of plant tissues.
Ideally, such a platform requires a model system that serves as a link between microbes and higher plants. Genetic engineering of crop plants still remains a cumbersome undertaking owing to slow life cycles: Obtaining mature plants from seeds takes ∼4 months for tomato (Kimura and Sinha 2008), 5 months for maize (Green and Phillips 1975), and 7 months for wheat (McHughen 1983). Furthermore, polyploidy (Vaughan et al. 2007) and high levels of genetic redundancy in higher plants (Dean et al. 1999) require mutagenesis of several loci to dissect gene function. Arabidopsis became popular as a model organism for molecular studies in the 1980s mostly because of its relative simplicity among angiosperm plants (Somerville and Koornneef 2002). Features of Arabidopsis include its phylogenetic relevance, a comparatively rapid life cycle of 2–3 months, small stature, abundant seed production, and relative ease of transformation (Flavell 2009). Although easier to work with than many crop species, Arabidopsis still shows notable levels of genetic redundancy (Briggs et al. 2006), and crosses to obtain plants homozygous for integrated transgenes (generally integrated hemizygously) are laborious (Wijnker et al. 2012).
Promising alternative candidate species of lower complexity can be found in the group of bryophytes, descendants of the earliest terrestrial plants (Goffinet and Shaw 2009). The best studied of these are the moss Physcomitrella patens and the thalloid liverwort M. polymorpha. Although both species show ample promise for bioengineering, Marchantia offers several unique advantages including a shorter life cycle, the production of gemmae as accessible and robust vegetative propagules (Shimamura 2012), a more streamlined genome (Sasaki et al. 2007; Zobell et al. 2010), and a simple spore-based transformation method for genetic manipulation.
In particular, Marchantia represents an excellent model for studying aspects of plant development and morphology. Marchantia forms simple, sheet-like tissues that can be characterized in terms of its distinct surfaces (Heberlein 1929): The lower surface shows root-like cells, called rhizoids, which are responsible primarily for the uptake of water and organic nutrients as well as the anchoring of the plant body to the substrate. The body of the thallus features scattered differentiated cells called oil bodies, which have been shown to play a role in isoprenoid metabolism (Suire et al. 2000). Finally, the upper surface is composed of primitive modular complexes for photosynthesis, each of which contains a permanently open pore for gas exchange. The most striking features of the upper surface of the thallus arguably are conical splash cups. Within them, gemmae originate from single cells and are attractive specimens for engineering morphology, given their robustness, plasticity, and accessibility (Vötching 1885; Fitting 1935). In contrast to most developing organs in higher plants, gemmae are not buried within maternal or support tissues. Furthermore, gemmae retain their viability for approximately one year if stored at 4°C in agar, and are capable of vigorous germination on soil as well as artificial and sterile media (Miller 1964).
The morphological simplicity of Marchantia is matched by highly streamlined genetics. The sequences of the Y-chromosome, mitochondrial, and plastid genomes are available (Ohyama et al. 1986; Oda et al. 1992; Yamato et al. 2007), whereas an assembled nuclear genome sequence is expected to be published shortly, and can be found in draft form online (marchantia.info). The size of the nuclear genome has been estimated at 280 Mb by flow cytometry, and ∼20,000 protein-coding genes have been predicted. In comparison to the 32,670 genes present in Arabidopsis and the 35,938 genes in P. patens (Yamato and Kohchi 2012), lower levels of gene redundancy appear to be present in Marchantia. For instance, a study of receptor-like kinases (RLK) concluded that the Arabidopsis genome contains 52 different RLK-families comprised of >600 RLK genes, whereas in Marchantia 26 RLK-families comprised of only 29 RLK genes were found (Sasaki et al. 2007). Another example is illustrated by MIKC* MADS box transcription factors: P. patens has been shown to include 11 members of this gene family (Rensing et al. 2008) whereas Marchantia only encodes one of these.
Marchantia possesses features that qualify it as a plant model chassis for quick and easy genetic engineering. Like moss, the life cycle of Marchantia is characterized by a dominant haploid gametophyte phase (O’Hanlon 1926). This allows the immediate expression of mutant genotypes in the phenotype. Through propagation of gemmae, haploid isogenic plants are obtained. Because gemma development originates from a single cell, transgenic gemmae do not display chimeric gene expression patterns. Further, during the gametophytic phase, it is possible to determine the sex of haploid plants by means of a PCR-based assay (Okada et al. 2000), allowing easy handling and management of female and male lines. To prepare crossing, the sexual phase of the life cycle can be induced by exposure to far-red light (Nakazato et al. 1999), and fertilization is performed by transferring sperm from mature antheridiophores to archegonia. Following fertilization and zygote formation, the life cycle continues in its diploid phase, leading to formation of spores packaged within yellow sporangia. Each cross can produce approximately 7 million spores that can be stored at −80°C for several years. Spore development takes place as soon the spores find a suitable substrate for germination. Unlike seeds of higher plants, spores are unicellular and do not possess a testa, allowing direct visualization of all early processes in development. During spore germination, spores undergo asymmetric cell division, initiate photosynthesis after differentiation of plastids, and begin to develop differentiated tissues within only a few days (see Fig. 2). Following germination, new plants grow vigorously on solid (Takenaka et al. 2000) or liquid media (Ishizaki et al. 2008) in sterile culture, and robustly regenerate from vegetative gemmae (Miller 1964), protoplasts (Ono et al. 1979), or tissue cuttings (Kubota et al. 2013). Rapid photoautotrophic growth of Marchantia cultures has been reported with a 1.76-day doubling time for culture mass, a faster rate than Arabidopsis (Katoh et al. 1979). The high regenerative capacity of liverworts is independent of externally supplied hormones (Vötching 1885), and well exceeds the reported regeneration efficiency of Arabidopsis (Valvekens et al. 1992) or wheat (McHughen 1983). The life cycle of Marchantia takes ∼2 months to complete (Shimamura 2012), which is similar to Arabidopsis and faster than in P. patens (∼3 months [Cove 2005]).
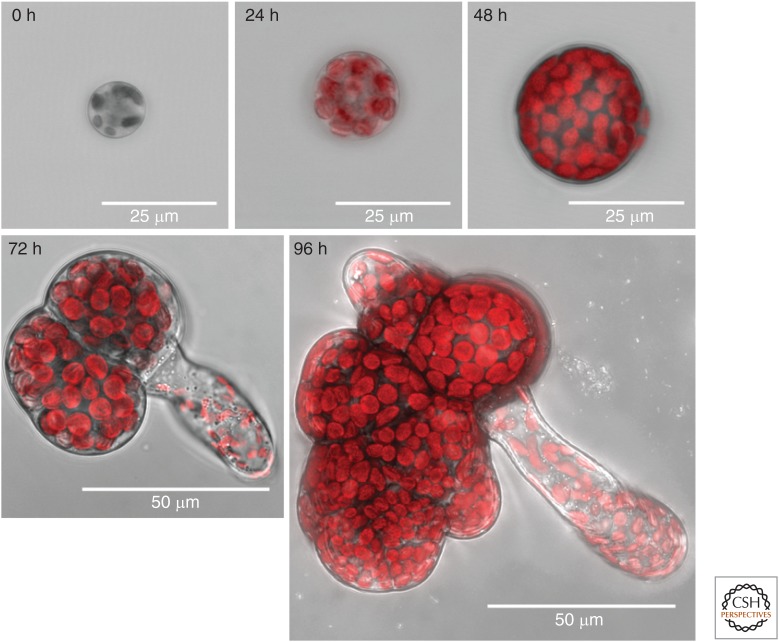
Figure 2.
Spore germination. Marchantia spores were germinated on a nutrient agar surface. The spores were examined under a 63×, NA 1.2 objective at 0, 24, 48, 72, and 96 h after germination using a Leica SP5 confocal laser scanning microscope. A 488-nm laser was used to collect transmission images (grayscale channel), and these were overlaid with images of chlorophyll fluorescence (488 nm excitation, 680–700 nm emission, red channel). Z-series of images (2 µm apart) were collected and merged to provide views of the developing sporelings at different stages of growth. Scale bars are indicated in each image.
Marchantia is one of few plant species for which chloroplast transformation protocols have been confirmed by at least two independent studies (Bock 2015). Stable transplastomic lines of Marchantia encoding dual spectinomycin and streptomycin resistance were generated through particle bombardment of sporelings (Chiyoda et al. 2007). This approach was recently extended to the expression of a fluorescent reporter gene from the Marchantia plastome (Boehm et al. 2016), which allows characterization of plastid promoters in this model system (see Fig. 3). A notable feature of the Marchantia plastome is the apparent absence of mechanisms for RNA editing (Ohyama et al. 2009), which may substantially simplify rational engineering of this organellar genome. Although simpler organisms may lack some of the complexity of angiosperms, the basal plant model Marchantia can enhance our still fragmented understanding of fundamental aspects of gene regulation, expression, and function in plant metabolism and development as well as provide a more rapid and tractable testbed for synthetic biology.
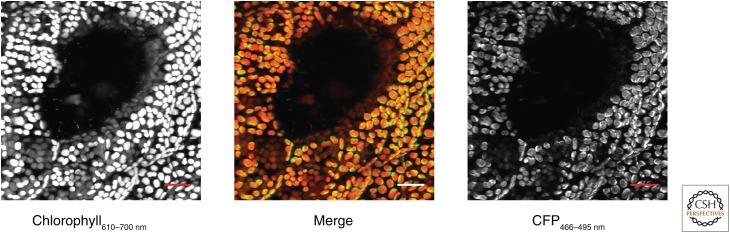
Figure 3.
Transgenic chloroplasts of Marchantia polymorpha expressing the cyan fluorescent protein mTurquoise2 under control of the tobacco psbA promoter. Mature thallus of Marchantia was mounted in water under a coverslip, and examined under a 63×, NA 1.2 objective using a Leica SP5 confocal laser-scanning microscope. Cells surrounding an air pore were imaged using 458-nm laser excitation. Emission wavelengths were collected for chloroplast autofluorescence (610–700 nm, left) and cyan fluorescent protein (CFP) fluorescence (466–495 nm, right), with merged images in red and green channels, respectively, shown center. Scale bar, 20 µm.
GENETIC MANIPULATION OF CHLOROPLASTS
In bridging the gap between microbes and plants as platforms for synthetic biology, a particularly attractive chassis is the chloroplast. Sharing its evolutionary heritage with cyanobacteria, this organelle possesses prokaryote-like regulation of gene expression (Mayfield et al. 1995), and is the major biosynthetic compartment in plants. The high biosynthetic capacity of chloroplasts has been used for production of a number of biofuel enzymes (Verma et al. 2010) as well as resistance proteins, antibodies, biopharmaceutical proteins, and vaccine antigens (Wani et al. 2010). Accumulation of large amounts of target protein has been achieved in chloroplasts, exceeding 70% of the plant’s total soluble protein (Oey et al. 2009). No gene silencing has been observed in chloroplasts despite such high accumulation of foreign transcripts (169 times higher than in nuclear transgenic plants, Lee et al. 2003) or foreign protein (46% of total leaf protein [De Cosa et al. 2001]). Metabolic engineering for the production of bioplastic monomers (Bohmert-Tatarev et al. 2011) and compounds of nutritional relevance (Craig et al. 2008; Hasunuma et al. 2008; Apel and Bock 2009) has also been applied to chloroplasts. The high biosynthetic capacity of the chloroplast is closely linked to the polyploid nature of the system: At 10–100 chloroplasts per cell, and 10–1000 genomes per chloroplast (Bendich 1987), stable integration of a transgene into the chloroplast genome enables a substantial amplification in transgene copy number. The high ploidy of the plastome can entail the need for several rounds of selective regeneration following transgene introduction to segregate transplastomic organelles and establish homoplasmy (Maliga 2004). However, homoplasmy has also been achieved immediately after primary selection (Chiyoda et al. 2014).
The process of transgene integration into the plastome following delivery into the chloroplast by means of particle bombardment (Svab et al. 1990) or PEG-mediated transformation (Golds et al. 1993) is mediated by homologous recombination. This mechanism has enabled the introduction of point mutations in a site-specific manner (Przibilla et al. 1991). As a consequence, the risk of inconsistent gene expression by the positional effect in T-DNA-mediated nuclear transformation is alleviated in the chloroplast environment (Daniell et al. 2002). The high efficiency of homologous recombination in the chloroplast (Blowers et al. 1989) also allows the simultaneous introduction of modifications to several sites of the chloroplast genome by means of cotransformation (Kindle et al. 1991), encouraging the implementation of phenotypic traits based on multiple foreign genes. Multiple genes may be conveniently organized in operon-like polycistronic units (Hasunuma et al. 2008), which can be processed into more efficiently translated monocistronic transcripts by the incorporation of intercistronic expression elements (Lu et al. 2013). Previous studies have also highlighted the activity of chloroplast promoters in bacteria (Brixey et al. 1997) and of bacterial promoters in the chloroplast (Newell et al. 2003). Another bacterial gene expression element, the widely used lac repressor from E. coli, has been adapted for IPTG-inducible chloroplast transgene expression (Mühlbauer and Koop 2005). Alternatively, chloroplast-based translational regulation of transgene expression can be implemented via riboswitches under control of the externally applied ligand theopylline (Verhounig et al. 2010; Emadpour et al. 2015).
Large DNA fragments exceeding 50 kb have been successfully incorporated into the plastome (Adachi et al. 2007). In fact, on the basis of recent advances in DNA synthesis and assembly technology, synthesis of entire tailor-made plastid genomes has become feasible (Scharff and Bock 2014). Today, metabolic engineering in microbial systems is predominantly driven by generation of large numbers of circuit variants followed by selection of a desired phenotype. Translation of this approach into plants has not been widely adopted primarily because of the requirement for generation and analysis of a sufficient number of transgenic events in the context of long generation times and polyploidy of established model species. Compared with random integration in the nuclear genome by transgenesis or interbreeding (Karunanandaa et al. 2005), integration in the chloroplast genome is achieved by homologous recombination and thus DNA is inserted at a chosen location. This allows a far smaller number of events to be compared as they are free from position effects as well as from epigenetic gene silencing, which is only encountered in the nuclear genome (Lu et al. 2013). Another notable advantage of transplastomic technology over nuclear transformation lies in maternal inheritance of plastid DNA, which greatly reduces the risk of unwanted transgene transmission via pollen and markedly increases the level of containment for field release of transgenic plants.
MODELS FOR MULTICELLULAR GROWTH AND INTEGRATED TECHNIQUES FOR QUANTITATIVE MICROSCOPY AND IMAGE ANALYSIS
Specialized structures in plants carry out a number of highly efficient processes for photosynthesis, secondary metabolite production, and compound storage. These structures are generated during morphogenesis through the concerted behavior of cell populations during development. Our ability to modify plant form is currently limited by our understanding and control of multicellular processes during morphogenesis. Increasing evidence from mutagenesis studies in angiosperms has suggested that morphogenesis is shaped by a combination of processes embodied in classical organismal (Green 1980) and cellular (Beemster et al. 2003) theories: the “neo”-cell theory proposes that feedback and interplay between cellular and organism-wide processes cooperate in the patterning of tissues (Tsukaya 2003). Evidence indicates that the polarity of cell division and elongation is regulated by the plant cell cytoskeleton and interaction with local cell wall determinants, wall strain, or geometry (Fig. 4). Subsequent genetic interactions mediated by protein or phytohormone traffic between cells can provide positional signals to trigger gene expression, cell proliferation, differentiation, and organ formation (Bohn-Courseau 2010). Consequent cellular growth generates physical strains that are instantaneously transmitted across tissues and constrain growth. The physical constraints on cell size and shape regulate timing and orientation of individual cell divisions and expansion, and therefore guide morphogenesis. This results in hierarchical, highly parallel, and feedback-regulated systems in which the DNA program and phenotype are not directly related, but part of an emergent self-organizing system. DNA-based reprogramming of plants can predictably alter individual cell behaviors, but the grand challenge of plant synthetic biology is to understand and predict how altered cell logic can propagate through large cell populations, modify tissue physics and local patterns of cellular growth, and produce defined morphologies. Effective multiscale modeling of cellular growth will be a prerequisite for predictable engineering of plant morphogenesis.
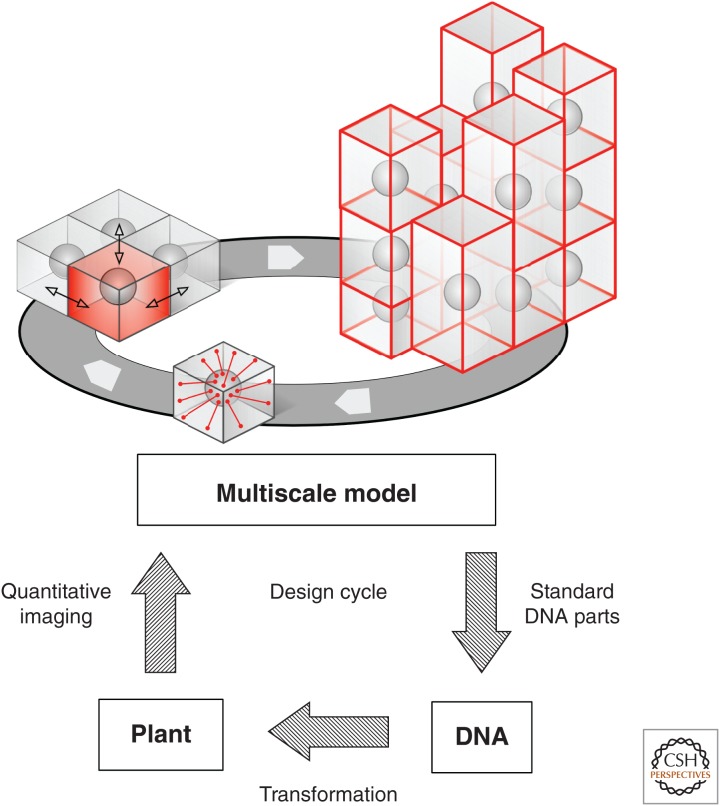
Figure 4.
Multiscale model of plant growth for engineering synthetic botanical forms. To model and predict the form of reprogrammed plants, integrated, multiscale models for plant growth are required. These software models need to capture (1) the interaction between cytoskeletal elements and local cell wall determinants, strain or geometry regulating the polarity of cell division and elongation, (2) genetic interactions between neighboring cells that can trigger gene expression and cell proliferation and differentiation, and (3) cellular growth that results in physical strains that are transmitted across tissues and constrain cell growth, because physical constraints on cell size and shape regulate timing and orientation of individual cell divisions and guide morphogenesis. Multiscale models provide an essential tool for engineering multicellular systems. Standardized DNA parts facilitate assembly of DNA circuits that may be introduced into plant systems by transformation, and the performance of DNA-based circuits can be measured using quantitative imaging techniques. Although a genetic circuit may regulate or alter the behavior of an individual cell in an easily predictable fashion, the consequences of altered cell interactions, propagation of changes across large cell populations, changes in tissue-wide physical and chemical interactions, and feedback on the properties of individual cells are difficult to predict. However, this type of system, characterized by cross talk and emergent properties, can be captured accurately by multiscale models. The models form an essential part of any design–build–test cycle for DNA-based engineering of plants.
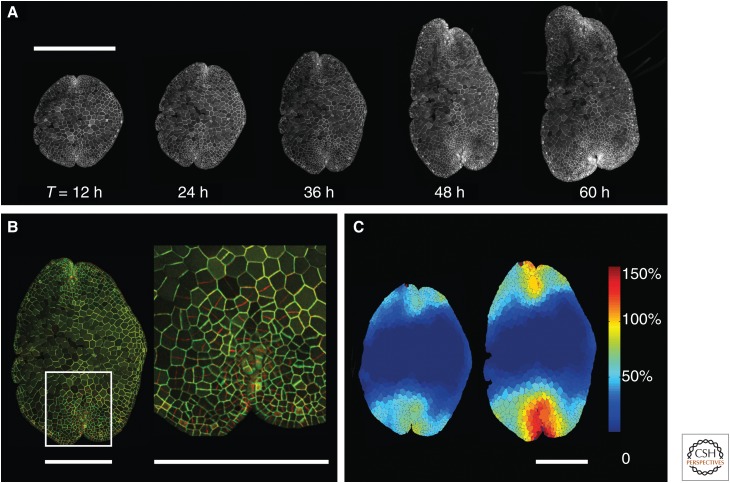
The past decade has seen the emergence of increasingly effective models for plant growth in which the software describes the genetic, cellular, and biophysical properties of growing tissues (Jönsson and Krupinski 2010; De Vos et al. 2012; Prusinkiewicz and Runions 2012). Although these models have been developed primarily to provide insight into plant developmental processes, they allow the integration of genetic, cellular, and whole-tissue properties, in a way that facilitates DNA-based reprogramming of large-scale plant growth. To complement these software tools, synthetic biologists require an experimental platform that allows simultaneous measurements of gene expression, growth, and division of individual cells at the scale of the entire organism. Sporelings and gemmae, the reproductive propagules of Marchantia, are well suited for such purpose. Their prostrate morphology and exposed mode of development facilitate the application of quantitative imaging techniques (Fig. 5): all cells in the surface layer are accessible for imaging to quantify gene expression based on in planta cytometry (Federici et al. 2012). Robust protocols to map the dynamics of cell expansion, division, and differentiation on the surface of the gemma during the early stages of gemma development have been developed (N Purswani, unpubl.).
Figure 5.
Marchantia gemmae as testbeds for quantitative parameterization of plant growth. (A) A transgenic line of Marchantia was generated that expressed a green fluorescent protein localized to the plasma membrane. A gemma from this transgenic line was transferred to a nutrient agar surface and examined after 12, 24, 36, 48, and 60 h of growth using a Leica SP5 confocal laser scanning microscope. Z-series of optical sections were collected for the same gemma, and maximum intensity projections are shown for each point during growth. Scale bar, 500 µm. (B) The 24-h (green channel) and 36-h (red channel) images from the time course were matched using warp-registration image-processing techniques and overlaid. A white box is positioned over one of the apical notches, and this corresponds to the enlarged view shown in the inset. The frequency and orientation of apex-localized cell divisions can be directly visualized in a single gemma. Scale bar, 200 µm. (C) The plasma membrane-localized marker allows accurate segmentation of cell geometry during growth of living plants. Quantitative parameters such as cell expansion rate can be mapped across a single gemma. Measurements of percent clonal sector expansion per 12 h are shown as a color map. Scale bar, 200 µm.
This combination of advanced imaging techniques and in silico models has proven useful for examining and predicting laws for cell division and tissue growth in multicellular algae (Dupuy et al. 2010; Besson and Dumais 2011). In an engineering context, these methods can form an essential part of a design–build–test cycle for reprogramming plant growth (see Fig. 4). Prototype designs can be assembled from modular DNA parts and transformed into plants, in which the properties of a synthetic gene circuit and impact on plant growth can be measured by quantitative microscopy. Multiscale software models can allow evaluation and even prediction of emergent processes. The simplicity, experimental accessibility, and morphological plasticity of Marchantia sporelings and gemmae (Vötching 1885; Fitting 1935), ease of quantitative measurements, and growing sophistication of software models for cellular growth provide a pathway to the rational design of morphogenetic programs in plants.
CONCLUDING REMARKS
Marchantia provides an experimental model to help bridge the gap between the relative simplicity of microbes and the complexity of higher plants. In particular, Marchantia is well-suited for prototyping genetic circuits before their application in a higher plant species of interest because of its amenability to high-throughput transformation and screening of large numbers of circuit variants: As an integral part of its rapid life cycle, Marchantia produces millions of spores, which can be transformed within a week using available protocols (Ishizaki et al. 2008; Tsuboyama and Kodama 2014). Owing to the haploid-dominant nature of Marchantia, phenotypic effects in transgenic plants can be immediately screened without the need for repeated selective regeneration to obtain homozygous lines. Screening can be performed by means of existing quantitative microscopy methods (Federici et al. 2012), and image processing and parameters can be extrapolated for physico-genetic modeling of multicellular plant tissues (Dupuy et al. 2010) to refine the next iteration of the design cycle. In the future, this process will benefit not only from the improvement of current techniques for DNA assembly, plant transformation, quantitative microscopy, and computational modeling, but especially from the availability of the Marchantia nuclear genome. This sequence information will guide the discovery of cell-type- and developmental stage–specific marker genes. The identification of tissue and cell-type-specific promoters in Marchantia has particular merit not only for developmental studies but also for metabolic engineering. Notably, compounds derived from plant secondary metabolism still play a dominant role in the discovery of new biopharmaceuticals (Newman and Cragg 2016), despite the advances of modern chemistry. In this light, the ultimate challenge for biological engineers will not be manipulating individual genes, but exercising control over the collective behavior of metabolic pathways and cohorts of cells and refactoring a multicellular organism’s body plan to generate specialized structures and organs for manufacturing and storage of compounds of interest. For example, applications of spatially controlled gene expression in Marchantia may include targeted manipulation of carbon fixation in assimilatory filaments (Goffinet and Shaw 2009) or production of biofuel components in oil cells (Suire et al. 2000). Control circuits and synthetic pathways proven in this context may serve as valuable tools for some of the largest contemporary challenges in plant metabolic engineering, such as the refactoring of nitrogen fixation pathways in plants (Rogers and Oldroyd 2014) or the introduction of C4 photosynthesis into C3 crops (Leegood 2013).
ACKNOWLEDGMENTS
The authors acknowledge support by the Gates Cambridge Trust (to C.R.B.), Becas Chile and the Cambridge Trust (to B.P.), and the Biotechnology and Biological Sciences Research Council and Engineering and Physical Sciences Research Council (OpenPlant Grant No. BB/L014130/1) to N.P. and J.H.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Adachi T, Takase H, Tomizawa K. 2007. Introduction of a 50 kbp DNA fragment into the plastid genome. Biosci Biotechnol Biochem 71: 2266–2273. [DOI] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, et al. 2014. Total synthesis of a functional designer eukaryotic chromosome. Science 344: 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony PC, Perez CF, Garcia-Garcia C, Block SM. 2012. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci 109: 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes MS, Morey KJ, Tewari-Singh N, Bowen TA, Smith JJ, Webb CT, Hellinga HW, Medford JI. 2009. Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol Syst Biol 5: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes MS, Morey KJ, Smith JJ, Albrecht KD, Bowen TA, Zdunek JK, Troupe JF, Cuneo MJ, Webb CT, Hellinga HW, et al. 2011. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLoS ONE 6: e16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Apel W, Bock R. 2009. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol 151: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates SL, Zhao JZ, Roush RT, Shelton AM. 2005. Insect resistance management in GM crops: Past, present and future. Nat Biotechnol 23: 57–62. [DOI] [PubMed] [Google Scholar]
- Beadle G. 1939. Teosinte and the origin of maize. J Hered 30: 245–247. [Google Scholar]
- Beemster GTS, Fiorani F, Inzé D. 2003. Cell cycle: The key to plant growth control? Trends Plant Sci 8: 154–158. [DOI] [PubMed] [Google Scholar]
- Beisel CL, Smolke CD. 2009. Design Principles for riboswitch function. PLoS Comput Biol 5: e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich AJ. 1987. Why do chloroplasts and mitochondria contain so many copies of their genome? BioEssays 6: 279–282. [DOI] [PubMed] [Google Scholar]
- Besson S, Dumais J. 2011. Universal rule for the symmetric division of plant cells. Proc Natl Acad Sci 108: 6294–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder A, Lambert J, Morbitzer R, Popp C, Ott T, Lahaye T, Parniske M. 2014. A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS ONE 9: e88218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers AD, Bogorad L, Shark KB, Sanford JC. 1989. Studies on Chlamydomonas chloroplast transformation: Foreign DNA can be stably maintained in the chromosome. Plant Cell 1: 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. 2015. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu Rev Plant Biol 66: 211–241. [DOI] [PubMed] [Google Scholar]
- Boehm CR, Ueda M, Nishimura Y, Shikanai T, Haseloff J. 2016. A cyan fluorescent reporter expressed from the chloroplast genome of Marchantia polymorpha. Plant Cell Physiol 57: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert-Tatarev K, McAvoy S, Daughtry S, Peoples OP, Snell KD. 2011. High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol 155: 1690–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn-Courseau I. 2010. Auxin: A major regulator of organogenesis. C R Biol 333: 290–296. [DOI] [PubMed] [Google Scholar]
- Bortesi L, Fischer R. 2015. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33: 41–52. [DOI] [PubMed] [Google Scholar]
- Briggs G, Osmont K, Shindo C, Sibout R, Hardtke C. 2006. Unequal genetic redundancies in Arabidopsis—A neglected phenomenon? Trends Plant Sci 11: 492–498. [DOI] [PubMed] [Google Scholar]
- Brixey PJ, Guda C, Daniell H. 1997. The chloroplast psbA promoter is more efficient in Escherichia coli than the T7 promoter for hyperexpression of a foreign protein. Biotechnol Lett 19: 395–400. [Google Scholar]
- Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B. 2000. Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K, Schäfer P, Weber E, Grützner R, Marillonnet S, Tissier A. 2015. A library of synthetic transcription activator-like effector-activated promoters for coordinated orthogonal gene expression in plants. Plant J 82: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner S, Hurni S, Herren G, Kalinina O, von Burg S, Zeller SL, Schmid B, Winzeler M, Keller B. 2011. Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol J 9: 897–910. [DOI] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson L, Krause KP, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB. 1998. An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol 16: 177–180. [DOI] [PubMed] [Google Scholar]
- Casini A, MacDonald JT, de Jonghe J, Christodoulou G, Freemont PS, Baldwin GS, Ellis T. 2014. One-pot DNA construction for synthetic biology: The modular overlap-directed assembly with linkers (MODAL) strategy. Nucleic Acids Res 42: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti D, Sarkar A, Mondal HA, Schuermann D, Hohn B, Sarmah BK, Das S. 2008. Cre/lox system to develop selectable marker free transgenic tobacco plants conferring resistance against sap sucking homopteran insect. Plant Cell Rep 27: 1623–1633. [DOI] [PubMed] [Google Scholar]
- Chiyoda S, Linley PJ, Yamato KT, Fukuzawa H, Yokota A, Kohchi T. 2007. Simple and efficient plastid transformation system for the liverwort Marchantia polymorpha L. suspension-culture cells. Transgenic Res 16: 41–49. [DOI] [PubMed] [Google Scholar]
- Chiyoda S, Yamato KT, Kohchi T. 2014. Plastid transformation of sporelings and suspension-cultured cells from the liverwort Marchantia polymorpha L. Methods Mol Biol 1132: 439–447. [DOI] [PubMed] [Google Scholar]
- Comai L, Facciotti D, Hiatt WR, Thompson G, Rose RE, Stalker DM. 1985. Expression in plants of a mutant aroA gene from Salmonella typhimurium confers tolerance to glyphosate. Nature 317: 741–744. [Google Scholar]
- Cove D. 2005. The moss Physcomitrella patens. Annu Rev Genet 39: 339–358. [DOI] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I. 2005. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J 41: 899–918. [DOI] [PubMed] [Google Scholar]
- Craig W, Lenzi P, Scotti N, De Palma M, Saggese P, Carbone V, McGrath Curran N, Magee AM, Medgyesy P, Kavanagh TA, et al. 2008. Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res 17: 769–782. [DOI] [PubMed] [Google Scholar]
- Daniell H, Khan MS, Allison L. 2002. Milestones in chloroplast genetic engineering: An environmentally friendly era in biotechnology. Trends Plant Sci 7: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aoust MA, Lavoie PO, Couture MMJ, Trépanier S, Guay JM, Dargis M, Mongrand S, Landry N, Ward BJ, Vézina LP. 2008. Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol J 6: 930–940. [DOI] [PubMed] [Google Scholar]
- Dean RE, Dahlberg JA, Hopkins MS, Mitchell SE, Kresovich S. 1999. Genetic redundancy and diversity among “Orange” accessions in the U.S. National Sorghum Collection as assessed with simple sequence repeat (SSR) markers. Crop Sci 39: 1215–1221. [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. 2001. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19: 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kok S, Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, et al. 2014. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol 3: 97–106. [DOI] [PubMed] [Google Scholar]
- De Vos D, Dzhurakhalov A, Draelants D, Bogaerts I, Kalve S, Prinsen E, Vissenberg K, Vanroose W, Broeckhove J, Beemster GTS. 2012. Towards mechanistic models of plant organ growth. J Exp Bot 63: 3325–3337. [DOI] [PubMed] [Google Scholar]
- Doebley J. 2004. The genetics of maize evolution. Annu Rev Genet 38: 37–59. [DOI] [PubMed] [Google Scholar]
- Dupuy L, Mackenzie J, Haseloff J. 2010. Coordination of plant cell division and expansion in a simple morphogenetic system. Proc Natl Acad Sci 107: 2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Wang X, Collins JJ. 2009. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol 27: 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emadpour M, Karcher D, Bock R. 2015. Boosting riboswitch efficiency by RNA amplification. Nucleic Acids Res 43: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami S, Yee MC, Dinneny JR. 2013. A robust family of Golden Gate Agrobacterium vectors for plant synthetic biology. Front Plant Sci 4: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy D. 2005. Foundations for engineering biology. Nature 438: 449–453. [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. 2009. Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JDG, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Federici F, Dupuy L, Laplaze L, Heisler M, Haseloff J. 2012. Integrated genetic and computation methods for in planta cytometry. Nat Methods 9: 483–485. [DOI] [PubMed] [Google Scholar]
- Fethe MH, Liu W, Burris JN, Millwood RJ, Mazarei M, Rudis MR, Yeaman DG, Dubosquielle M, Stewart CN. 2014. The performance of pathogenic bacterial phytosensing transgenic tobacco in the field. Plant Biotechnol J 12: 755–764. [DOI] [PubMed] [Google Scholar]
- Field B, Fiston-Lavier AS, Kemen A, Geisler K, Quesneville H, Osbourn AE. 2011. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci 108: 16116–16121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitting H. 1935. Untersuchungen über die induktion der dorsiventralität bei den keimenden brutkörpern Marchantia und Lunularia. Jahrb Wiss Bot 82: 333. [Google Scholar]
- Flavell R. 2009. Role of model plant species. Methods Mol Biol 513: 1–18. [DOI] [PubMed] [Google Scholar]
- Fromm M, Taylor LP, Walbot V. 1985. Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc Natl Acad Sci 82: 5824–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Baker AJ, Assie JM, Poethig RS, Haseloff JP, Webb AA. 2009. GAL4 GFP enhancer trap lines for analysis of stomatal guard cell development and gene expression. J Exp Bot 60: 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. 2003. Agrobacterium-mediated plant transformation: The biology behind the “Gene-Jockeying” Tool. Microbiol Mol Biol Rev 67: 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goffinet B, Shaw AJ. 2009. Bryophyte biology, 2nd ed Cambridge University Press, Cambridge. [Google Scholar]
- Goh HH, Sloan J, Dorca-Fornell C, Fleming A. 2012. Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol 159: 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golds T, Maliga P, Koop HU. 1993. Stable plastid transformation in PEG-treated protoplasts of Nicotiana tabacum. Nat Biotechnol 11: 95–97. [Google Scholar]
- Graham IA, Besser K, Blumer S, Branigan CA, Czechowski T, Elias L, Guterman I, Harvey D, Isaac PG, Khan AM, et al. 2010. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327: 328–331. [DOI] [PubMed] [Google Scholar]
- Green PB. 1980. Organogenesis—A biophysical view. Annu Rev Plant Physiol 31: 51–82. [Google Scholar]
- Green CE, Phillips RL. 1975. PPlant regeneration from tissue cultures of maize. Crop Sci 15: 417–421. [Google Scholar]
- Hasunuma T, Miyazawa SI, Yoshimura S, Shinzaki Y, Tomizawa KI, Shindo K, Choi SK, Misawa N, Miyake C. 2008. Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55: 857–868. [DOI] [PubMed] [Google Scholar]
- Hauptmann RM, Ozias-Akins P, Vasil V, Tabaeizadeh Z, Rogers SG, Horsch RB, Vasil IK, Fraley RT. 1987. Transient expression of electroporated DNA in monocotyledonous and dicotyledonous species. Plant Cell Rep 6: 265–270. [DOI] [PubMed] [Google Scholar]
- Heberlein EA. 1929. Morphological studies on a new species of Marchantia. Bot Gaz 88: 417–429. [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol 61: 181–207. [DOI] [PubMed] [Google Scholar]
- Hoff T, Schnorr KM, Mundy J. 2001. A recombinase-mediated transcriptional induction system in transgenic plants. Plant Mol Biol 45: 41–49. [DOI] [PubMed] [Google Scholar]
- Horvath DM, Stall RE, Jones JB, Pauly MH, Vallad GE, Dahlbeck D, Staskawicz BJ, Scott JW. 2012. Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS ONE 7: e42036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Chiyoda S, Yamato KT, Kohchi T. 2008. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol 49: 1084–1091. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. 1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106. [DOI] [PubMed] [Google Scholar]
- Johnson AAT, Hibberd JM, Gay C, Essah PA, Haseloff J, Tester M, Guiderdoni E. 2005. Spatial control of transgene expression in rice (Oryza sativa L.) using the GAL4 enhancer trapping system. Plant J 41: 779–789. [DOI] [PubMed] [Google Scholar]
- Jones H, Ooms G, Jones MGK. 1989. Transient gene expression in electroporated Solanum protoplasts. Plant Mol Biol 13: 503–511. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Witek K, Verweij W, Jupe F, Cooke D, Dorling S, Tomlinson L, Smoker M, Perkins S, Foster S. 2014. Elevating crop disease resistance with cloned genes. Philos Trans R Soc B Biol Sci 369: 20130087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson H, Krupinski P. 2010. Modeling plant growth and pattern formation. Curr Opin Plant Biol 13: 5–11. [DOI] [PubMed] [Google Scholar]
- Kapila J, De Rycke R, Van Montagu M, Angenon G. 1997. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci 122: 101–108. [Google Scholar]
- Karunanandaa B, Qi Q, Hao M, Baszis SR, Jensen PK, Wong YHH, Jiang J, Venkatramesh M, Gruys KJ, Moshiri F, et al. 2005. Metabolically engineered oilseed crops with enhanced seed tocopherol. Metab Eng 7: 384–400. [DOI] [PubMed] [Google Scholar]
- Katoh K, Ohta Y, Hirose Y, Iwamura T. 1979. Photoautotrophic growth of Marchantia polymorpha L. cells in suspension culture. Planta 144: 509–510. [DOI] [PubMed] [Google Scholar]
- Kimura S, Sinha N. 2008. How to grow tomatoes. CSH Protoc 2008: pdb.prot5081. [DOI] [PubMed] [Google Scholar]
- Kindle KL, Richards KL, Stern DB. 1991. Engineering the chloroplast genome: Techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc Natl Acad Sci 88: 1721–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight T. 2003. Idempotent vector design for standard assembly of biobricks. MIT Artificial Intelligence Laboratory; MIT Synthetic Biology Working Group.
- Koo JC, Asurmendi S, Bick J, Woodford-Thomas T, Beachy RN. 2004. Ecdysone agonist-inducible expression of a coat protein gene from tobacco mosaic virus confers viral resistance in transgenic Arabidopsis. Plant J 37: 439–448. [DOI] [PubMed] [Google Scholar]
- Koschmann J, Machens F, Becker M, Niemeyer J, Schulze J, Bulow L, Stahl DJ, Hehl R. 2012. Integration of bioinformatics and synthetic promoters leads to the discovery of novel elicitor-responsive cis-regulatory sequences in Arabidopsis. Plant Physiol 160: 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krens FA, Molendijk L, Wullems GJ, Schilperoort RA. 1982. In vitro transformation of plant protoplasts with Ti-plasmid DNA. Nature 296: 72–74. [Google Scholar]
- Ku MS, Cho D, Li X, Jiao DM, Pinto M, Miyao M, Matsuoka M. 2001. Introduction of genes encoding C4 photosynthesis enzymes into rice plants: Physiological consequences. Novartis Found Symp 236: 100–116. [DOI] [PubMed] [Google Scholar]
- Kubota A, Ishizaki K, Hosaka M, Kohchi T. 2013. Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating Thalli. Biosci Biotechnol Biochem 77: 167–172. [DOI] [PubMed] [Google Scholar]
- Kumar A, Dixit S, Ram T, Yadaw RB, Mishra KK, Mandal NP. 2014. Breeding high-yielding drought-tolerant rice: Genetic variations and conventional and molecular approaches. J Exp Bot 65: 6265–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J. 2013. GreenGate––A novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martinière A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J. 2005. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56: 2433–2442. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H. 2003. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed 11: 1–13. [Google Scholar]
- Leegood RC. 2013. Strategies for engineering C4 photosynthesis. J Plant Physiol 170: 378–388. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. 2012. SLIC: A method for sequence- and ligation-independent cloning. Methods Mol Biol 852: 51–59. [DOI] [PubMed] [Google Scholar]
- Li JF, Chung HS, Niu Y, Bush J, McCormack M, Sheen J. 2013. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25: 1507–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Zhang D, Sheen J. 2014. Cas9-based genome editing in Arabidopsis and tobacco. Methods Enzymol 546: 459–472. [DOI] [PubMed] [Google Scholar]
- Link KH, Breaker RR. 2009. Engineering ligand-responsive gene-control elements: Lessons learned from natural riboswitches. Gene Ther 16: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Mazarei M, Rudis MR, Fethe MH, Stewart CN. 2011. Rapid in vivo analysis of synthetic promoters for plant pathogen phytosensing. BMC Biotechnol 11: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Yuan JS, Stewart CN Jr. 2013. Advanced genetic tools for plant biotechnology. Nat Rev Genet 14: 781–793. [DOI] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. 2015. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169: 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Rijzaani H, Karcher D, Ruf S, Bock R. 2013. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc Natl Acad Sci 110: E623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. 2004. Plastid transformation in higher plants. Annu Rev Plant Biol 55: 289–313. [DOI] [PubMed] [Google Scholar]
- Mardanova ES, Kotlyarov RY, Kuprianov VV, Stepanova LA, Tsybalova LM, Lomonosoff GP, Ravin NV. 2015. Rapid high-yield expression of a candidate influenza vaccine based on the ectodomain of M2 protein linked to flagellin in plants using viral vectors. BMC Biotechnol 15: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Sparks C, Hart CA, Thompson J, Jepson I. 1999. Ecdysone agonist inducible transcription in transgenic tobacco plants. Plant J 19: 97–106. [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Yohn CB, Cohen A, Danon A. 1995. Regulation of chloroplast gene expression. Annu Rev Plant Physiol Plant Mol Biol 46: 147–166. [Google Scholar]
- McCaskill D, Croteau R. 1999. Strategies for bioengineering the development and metabolism of glandular tissues in plants. Nat Biotechnol 17: 31–36. [DOI] [PubMed] [Google Scholar]
- McHughen A. 1983. Rapid regeneration of wheat in vitro. Ann Bot 51: 851–853. [Google Scholar]
- Mett VL, Lochhead LP, Reynolds PH. 1993. Copper-controllable gene expression system for whole plants. Proc Natl Acad Sci 90: 4567–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM. 1989. Arabidopsis, a useful weed. Cell 56: 263–269. [DOI] [PubMed] [Google Scholar]
- Miller MW. 1964. A technique for isolating and culturing gemmae of Marchantia polymorpha L. under aseptic conditions. Bryologist 67: 317–320. [Google Scholar]
- Moore I, Galweiler L, Grosskopf D, Schell J, Palme K. 1998. A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci 95: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer SK, Koop H-U. 2005. External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J 43: 941–946. [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Siegel D, Rodriguez Jahnke F, Gerrer K, Wend S, Decker EL, Reski R, Weber W, Zurbriggen MD. 2014. A red light-controlled synthetic gene expression switch for plant systems. Mol BioSyst 10: 1679–1688. [DOI] [PubMed] [Google Scholar]
- Nakazato T, Kadota A, Wada M. 1999. Photoinduction of spore germination in Marchantia polymorpha L. is mediated by photosynthesis. Plant Cell Physiol 40: 1014–1020. [Google Scholar]
- Negrutiu I, Shillito R, Potrykus I, Biasini G, Sala F. 1987. Hybrid genes in the analysis of transformation conditions. Plant Mol Biol 8: 363–373. [DOI] [PubMed] [Google Scholar]
- Newell CA, Birch-Machin I, Hibberd JM, Gray JC. 2003. Expression of green fluorescent protein from bacterial and plastid promoters in tobacco chloroplasts. Transgenic Res 12: 631–634. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. 2016. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79: 629–661. [DOI] [PubMed] [Google Scholar]
- Nishiguchi M, Sato T, Motoyoshi F. 1987. An improved method for electroporation in plant protoplasts: Infection of tobacco protoplasts by tobacco mosaic virus particles. Plant Cell Rep 6: 90–93. [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Osbourn A. 2014. Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 26: 91–99. [DOI] [PubMed] [Google Scholar]
- Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, et al. 1992. Complete nucleotide sequence of the mitochondrial DNA from a liverwort, Marchantia polymorpha. Plant Mol Biol Report 10: 105–163. [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. 2009. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57: 436–445. [DOI] [PubMed] [Google Scholar]
- O’Hanlon ME. 1926. Germination of spores and early stages in development of gametophyte of Marchantia polymorpha. Bot Gaz 82: 215–222. [Google Scholar]
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, et al. 1986. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322: 572–574. [Google Scholar]
- Ohyama K, Takemura M, Oda K, Fukuzawa H, Kohchi T, Nakayama S, Ishizaki K, Fujisawa M, Yamato K. 2009. Gene content, organization and molecular evolution of plant organellar genomes and sex chromosomes: Insights from the case of the liverwort Marchantia polymorpha. Proc Jpn Acad Ser B Phys Biol Sci 85: 108–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Fujisawa M, Sone T, Nakayama S, Nishiyama R, Takenaka M, Yamaoka S, Sakaida M, Kono K, Takahama M, et al. 2000. Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. Plant J 24: 421–428. [DOI] [PubMed] [Google Scholar]
- Ono K, Ohyama K, Gamborg OL. 1979. Regeneration of the liverwort Marchantia polymorpha L. from protoplasts isolated from cell suspension culture. Plant Sci Lett 14: 225–229. [Google Scholar]
- Ou-Lee TM, Turgeon R, Wu R. 1986. Expression of a foreign gene linked to either a plant-virus or a Drosophila promoter, after electroporation of protoplasts of rice, wheat, and sorghum. Proc Natl Acad Sci 83: 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M, Gore M, Lu DL, Smirnova O. 2003. Chemical-inducible, ecdysone receptor-based gene expression system for plants. Transgenic Res 12: 101–109. [DOI] [PubMed] [Google Scholar]
- Patron NJ. 2014. DNA assembly for plant biology: Techniques and tools. Curr Opin Plant Biol 19: 14–19. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Orzaez D, Marillonnet S, Warzecha H, Matthewman C, Youles M, Raitskin O, Leveau A, Farré G, Rogers C, et al. 2015. Standards for plant synthetic biology: A common syntax for exchange of DNA parts. New Phytol 208: 13–19. [DOI] [PubMed] [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. 2015. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13: 578–589. [DOI] [PubMed] [Google Scholar]
- Potrykus I, Paszkowski J, Saul MW, Petruska J, Shillito RD. 1985. Molecular and general genetics of a hybrid foreign gene introduced into tobacco by direct gene transfer. MGG Mol Gen Genet 199: 169–177. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Runions A. 2012. Computational models of plant development and form. New Phytol 193: 549–569. [DOI] [PubMed] [Google Scholar]
- Przibilla E, Heiss S, Johanningmeier U, Trebst A. 1991. Site-specific mutagenesis of the D1 subunit of photosystem II in wild-type Chlamydomonas. Plant Cell 3: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Tian J. 2009. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4: e6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitskin O, Patron NJ. 2016. Multi-gene engineering in plants with RNA-guided Cas9 nuclease. Curr Opin Biotechnol 37: 69–75 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y, et al. 2008. The physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. [DOI] [PubMed] [Google Scholar]
- Rogers C, Oldroyd GED. 2014. Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot 65: 1939–1946. [DOI] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, White MRH, Croft KP, Robson F, Coupland G, Doonan J, Laufs P, Tomsett AB, et al. 2001. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J 28: 225–235. [DOI] [PubMed] [Google Scholar]
- Salter MG, Paine JA, Riddell K V, Jepson I, Greenland AJ, Caddick MX, Tomsett AB. 1998. Characterisation of the ethanol-inducible alc gene expression system for transgenic plants. Plant J 16: 127–132. [Google Scholar]
- Samalova M, Brzobohaty B, Moore I. 2005. pOp6/LhGR: A stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. Plant J 41: 919–935. [DOI] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Falconi EE, Zandalinas SI, Juárez P, Fernández-del-Carmen A, Granell A, Orzaez D. 2011. GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 6: e21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki G, Katoh K, Hirose N, Suga H, Kuma K, Miyata T, Su ZH. 2007. Multiple receptor-like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergii and Nitella axillaris: Extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene 401: 135–144. [DOI] [PubMed] [Google Scholar]
- Scharff LB, Bock R. 2014. Synthetic biology in plastids. Plant J 78: 783–798. [DOI] [PubMed] [Google Scholar]
- Schaumberg KA, Antunes MS, Kassaw TK, Xu W, Zalewski CS, Medford JI, Prasad A. 2016. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat Methods 13: 94–100. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Last RL, Pichersky E. 2008. Harnessing plant trichome biochemistry for the production of useful compounds. Plant J 54: 702–711. [DOI] [PubMed] [Google Scholar]
- Schiml S, Puchta H. 2016. Revolutionizing plant biology: Multiple ways of genome engineering by CRISPR/Cas. Plant Methods 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. 2001. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF. 2008. Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura M. 2012. An introduction to the taxonomy and morphology of Marchantia polymorpha. BSJ-Review 3: 84–113. [Google Scholar]
- Somerville C, Koornneef M. 2002. A fortunate choice: The history of Arabidopsis as a model plant. Nat Rev Genet 3: 883–889. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Springer PS. 2000. Gene traps: Tools for plant development and genomics. Plant Cell 12: 1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suire C, Bouvier F, Backhaus RA, Bégu D, Bonneu M, Camara B. 2000. Cellular localization of isoprenoid biosynthetic enzymes in Marchantia polymorpha. Uncovering a new role of oil bodies. Plant Physiol 124: 971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. 1990. Stable transformation of plastids in higher plants. Proc Natl Acad Sci 87: 8526–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Yamaoka S, Hanajiri T, Shimizu-Ueda Y, Yamato KT, Fukuzawa H, Ohyama K. 2000. Direct transformation and plant regeneration of the haploid liverwort Marchantia polymorpha L. Transgenic Res 9: 179–185. [DOI] [PubMed] [Google Scholar]
- Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A. 2014. Triterpene biosynthesis in plants. Annu Rev Plant Biol 65: 225–257. [DOI] [PubMed] [Google Scholar]
- Torella JP, Boehm CR, Lienert F, Chen JH, Way JC, Silver PA. 2013. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic Acids Res 42: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama S, Kodama Y. 2014. AgarTrap: A simplified Agrobacterium-mediated transformation method for sporelings of the liverwort Marchantia polymorpha L. Plant Cell Physiol 55: 229–236. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2003. Organ shape and size: A lesson from studies of leaf morphogenesis. Curr Opin Plant Biol 6: 57–62. [DOI] [PubMed] [Google Scholar]
- Vafaee Y, Staniek A, Mancheno-Solano M, Warzecha H. 2014. A modular cloning toolbox for the generation of chloroplast transformation vectors. PLoS ONE 9: e110222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Lijsebettens M, Van Montagu M. 1992. Arabidopsis regeneration and transformation (Root Explant System). In Plant tissue culture manual, pp. 1–17. Springer, Boston. [Google Scholar]
- van Herpen TWJM, Cankar K, Nogueira M, Bosch D, Bouwmeester HJ, Beekwilder J. 2010. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE 5: e14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA, Balazs E, Heslop-Harrison JS. 2007. From crop domestication to super-domestication. Ann Bot 100: 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R. 2010. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci 107: 6204–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Kanagaraj A, Jin S, Singh ND, Kolattukudy PE, Daniell H. 2010. Chloroplast-derived enzyme cocktails hydrolyse lignocellulosic biomass and release fermentable sugars. Plant Biotechnol J 8: 332–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vötching H. 1885. Über die regeneration der Marchantiaceen. Jahrbuecher fur Wissenschaftliche Bot 16: 367–414. [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. 2007. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19: 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani S, Haider N, Kumar HB, Singh N. 2010. Plant plastid engineering. Curr Genomics 11: 500–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. 2011. a modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann P, Gossen M, Hillen W, Bujard H, Gatz C. 1994. A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J 5: 559–569. [DOI] [PubMed] [Google Scholar]
- Wend S, Bosco CD, Kämpf MM, Ren F, Palme K, Weber W, Dovzhenko A, Zurbriggen MD. 2013. A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci Rep 3: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Engler C, Weber E, Gruetzner R, Marillonnet S. 2012. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs 3: 38–43. [DOI] [PubMed] [Google Scholar]
- Wijnker E, van Dun K, de Snoo CB, Lelivelt CLC, Keurentjes JJB, Naharudin NS, Ravi M, Chan SWL, de Jong H, Dirks R. 2012. Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat Genet 44: 467–470. [DOI] [PubMed] [Google Scholar]
- Yamato KT, Kohchi T. 2012. The “sneak” preview of the Marchantia polymorpha genome. BSJ Rev 3: 71–83. [Google Scholar]
- Yamato KT, Ishizaki K, Fujisawa M, Okada S, Nakayama S, Fujishita M, Bando H, Yodoya K, Hayashi K, Bando T, et al. 2007. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc Natl Acad Sci 104: 6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. 2004. Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci 101: 7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I. 2000. Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305. [DOI] [PubMed] [Google Scholar]
- Yu S, Pilot G. 2014. Testing the efficiency of plant artificial microRNAs by transient expression in Nicotiana benthamiana reveals additional action at the translational level. Front Plant Sci 5: 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell O, Faigl W, Saedler H, Munster T. 2010. MIKC* MADS-Box Proteins: Conserved regulators of the gametophytic generation of land plants. Mol Biol Evol 27: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Zohary D, Hopf M, Weiss E. 2012. Domestication of plants in the old world: The origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean basin. Oxford University Press, New York. [Google Scholar]
- Zuo J, Niu QW, Chua NH. 2000. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273. [DOI] [PubMed] [Google Scholar]
- Zurcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Muller B. 2013. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol 161: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]