Abstract
RNA editing, a widespread post-transcriptional mechanism, has emerged as a new player in cancer biology. Recent studies have reported key roles for individual miRNA editing events, but a comprehensive picture of miRNA editing in human cancers remains largely unexplored. Here, we systematically characterized the miRNA editing profiles of 8595 samples across 20 cancer types from miRNA sequencing data of The Cancer Genome Atlas and identified 19 adenosine-to-inosine (A-to-I) RNA editing hotspots. We independently validated 15 of them by perturbation experiments in several cancer cell lines. These miRNA editing events show extensive correlations with key clinical variables (e.g., tumor subtype, disease stage, and patient survival time) and other molecular drivers. Focusing on the RNA editing hotspot in miR-200b, a key tumor metastasis suppressor, we found that the miR-200b editing level correlates with patient prognosis opposite to the pattern observed for the wild-type miR-200b expression. We further experimentally showed that, in contrast to wild-type miRNA, the edited miR-200b can promote cell invasion and migration through its impaired ability to inhibit ZEB1/ZEB2 and acquired concomitant ability to repress new targets, including LIFR, a well-characterized metastasis suppressor. Our study highlights the importance of miRNA editing in gene regulation and suggests its potential as a biomarker for cancer prognosis and therapy.
RNA editing is a widespread post-transcriptional modification mechanism that confers specific nucleic changes at the RNA level but does not affect the corresponding DNA sequence (Keegan et al. 2001; Bass 2002). The most prevalent type of RNA editing in humans is the conversion of adenosine to inosine (A to I), which is catalyzed by adenosine deaminase acting on RNA (ADAR) enzymes. Computational analyses on RNA sequencing data have detected more than 2 million RNA editing sites in the human transcriptome that potentially affect a number of protein-coding genes (Ramaswami et al. 2012; Bazak et al. 2014; Ramaswami and Li 2014). Several studies have characterized the critical role of individual missense A-to-I RNA editing events in cancer development (Chen et al. 2013; Galeano et al. 2013; Han et al. 2014). More recently, we and other groups have systematically characterized the RNA-editing genomic landscape in various cancer types using mRNA-seq data from The Cancer Genome Atlas (TCGA) (Fumagalli et al. 2015; Han et al. 2015; Paz-Yaacov et al. 2015). These studies revealed a large number of dysregulated A-to-I RNA editing events in tumor samples relative to normal samples, many of which show clinically relevant patterns and suggested that, like “driver” somatic mutations, RNA editing events alter the growth of cancer cells and also selectively change drug sensitivity in cell lines consistent with therapeutic relevance in patients.
In addition to protein-coding genes, RNA editing can occur in noncoding genes, such as lncRNAs and microRNAs (miRNAs) (Gong et al. 2014, 2017). miRNAs are ∼22-nt small RNAs that function in post-transcriptional gene regulation and participate in various cellular pathways and pathological processes, including cancer (Bartel 2004; Garzon et al. 2009). By base-pairing with their target mRNAs, miRNAs can result in mRNA degradation or protein translational repression (Valencia-Sanchez et al. 2006; Bartel 2009), and most human coding genes are conserved target genes of miRNAs (Friedman et al. 2009; Ha and Kim 2014). RNA editing in miRNAs has the potential to regulate the processing of precursor miRNAs into mature miRNAs (Yang et al. 2006; Kawahara et al. 2007a). Moreover, because miRNA regulation requires a perfect base-pairing of the seed region (2–8 position) of miRNA, a single-nucleotide change can markedly alter miRNA target recognition (Kawahara et al. 2007b; Negi et al. 2015). Intriguingly, several miRNA editing events appear to be critical in cancer. For example, in glioma, a low editing level of miR-376a increases the migration ability of tumor cells and is associated with more aggressive tumors. These effects are mediated by the ability of the edited miRNA to target AMFR combined with an inability to inhibit the original target RAP2A (Choudhury et al. 2012). In melanoma, edited miR-455 has been proposed to suppress tumor growth and metastasis by up-regulating tumor suppressor CPEB1 (Shoshan et al. 2015).
With the availability of large-scale small RNA sequencing data, several groups have characterized miRNA editing events in a more systematic way (Landgraf et al. 2007; Morin et al. 2008; Ebhardt et al. 2009; Eisenberg et al. 2010). However, because of the short read length and high similarities among miRNAs, high-quality bioinformatic tools for miRNA editing calling were not being developed until very recently (Alon et al. 2012, 2015; Gong et al. 2014). As a result, systematic analyses of RNA editing events in miRNAs have been very limited and highly biased toward specific diseases such as glioma (Alon et al. 2012; Tomaselli et al. 2015). Here, we performed a systematic analysis of miRNA editing events based on high-throughput TCGA small RNA sequencing data of 20 different cancer types.
Results
Systematic detection of RNA editing hotspots in miRNAs across cancer types
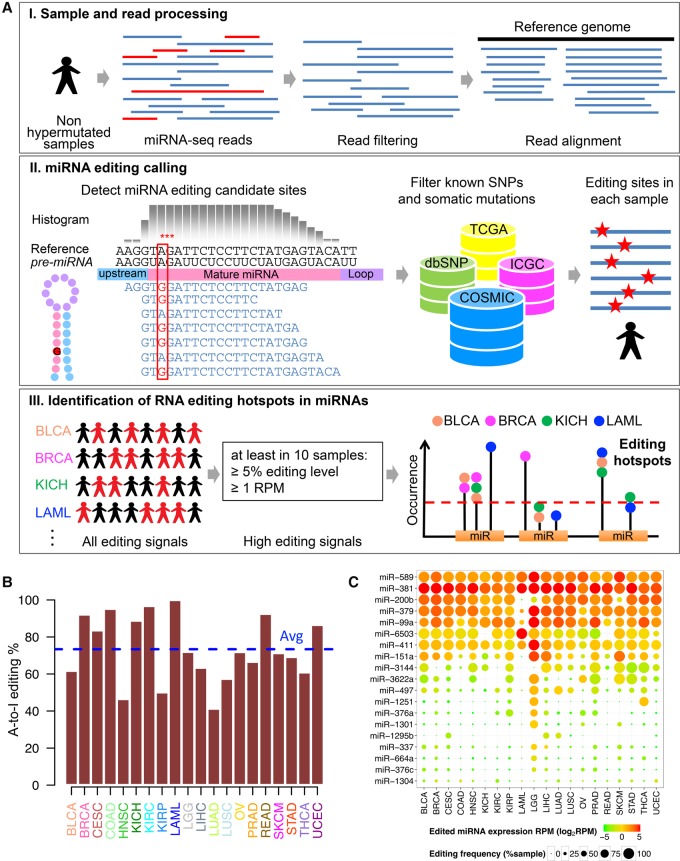
To comprehensively characterize the profiles of miRNA editing events in human cancers, we developed a computational pipeline that is similar to a previous analysis of miRNA editing in the human brain (Alon et al. 2012), but with some key improvements, and applied it to TCGA small RNA sequencing data (Fig. 1A; Methods; The Cancer Genome Atlas Research Network et al. 2013). First, to reduce false positives in detecting RNA editing events, we filtered cancer samples with an ultra-high mutation rate for each cancer type and low-quality reads for each sample. In total, we analyzed 8595 TCGA samples (including 7961 cancer samples and 634 noncancer samples) from 20 major cancer types (Supplemental Table S1). On average, we obtained 4.76 million mapped reads per cancer sample and 7.50 million mapped reads per normal sample. Second, based on the read alignments of each individual sample, we detected RNA editing candidates in mature miRNAs and then filtered the sites with known SNPs and somatic mutations to remove noise due to potential variation at the DNA level. Third, we focused on 28,816 high-confidence RNA editing events (editing level ≥ 5% and edited miRNA expression amount ≥ 1 RPM [reads per million reads aligned to miRNAs]) and identified the “miRNA editing hotspots” (recurrent in ≥10 tumor samples) for each cancer type (Supplemental Table S2). Across the cancer types surveyed, on average, 73% of the identified RNA editing events at these hotspots are A-to-I modifications, consistent with the notion that A-to-I editing is the most prevalent type in humans (Fig. 1B). Further, we compiled a list of miRNA editing sites from the literature and found that 89% of the identified A-to-I editing events have been reported in at least one previous study. These results suggest that the miRNA editing hotspot candidates we identified are of both high sensitivity and specificity. In total, we identified 19 unique A-to-I RNA editing hotspots across the 20 cancer types (a subset of 15 editing hotspots was identified if a 5% recurrence rate within a cancer type was used) (Table 1; Supplemental Table S3). To ensure the quality of these RNA editing hotspots, we further ruled out the possibility of any unreported SNPs and somatic mutations or cross-mapping (Methods). Among these editing hotspots, 18 (95%) are located at the position 1–8 of mature miRNAs, 12 (63%) have been reported in previous studies (Blow et al. 2006; Kawahara et al. 2007a,b, 2008; Alon et al. 2012; Choudhury et al. 2012; Gong et al. 2014; Warnefors et al. 2014; Saiselet et al. 2015; Tomaselli et al. 2015; Nishikura 2016), and two editing sites (miR-151a and miR-376a) have been functionally characterized (Kawahara et al. 2007a; Choudhury et al. 2012). To further characterize these miRNA editing hotspots, we calculated their editing frequency (defined as the fraction of the tumor samples with detectable editing signals), the edited miRNA expression amount (defined as the RPM), and the editing level across cancer types. These miRNA editing hotspots can be separated into two distinct groups: One group has extensive strong RNA editing signals in almost all cancer types surveyed, while the other group shows cancer-specific patterns, with high editing signals only in one or a few cancer types (Fig. 1C). Among the cancer types, low-grade glioma (LGG) shows the most intensive editing signal with the largest number of miRNA editing hotspots being identified, which is consistent with the high expression level of ADAR in brain tissues.
Figure 1.
Overview of bioinformatic pipeline and RNA editing profiles in miRNAs across cancer types. (A) Schematic of bioinformatic pipeline. For each cancer type, sample and read processing, miRNA editing calling (filtering DNA variants using various resources), and identification of miRNA editing hotspots. (B) Proportions of A-to-I RNA editing events among all RNA editing events observed at hotspots in different cancer types; average proportion across 20 cancer types is 73.4%. (C) Heat map of miRNA editing hotspots. Edited miRNA expression amounts (log2RPM) are in color; editing frequencies (% of samples with editing signals) are indicated by circle size.
Table 1.
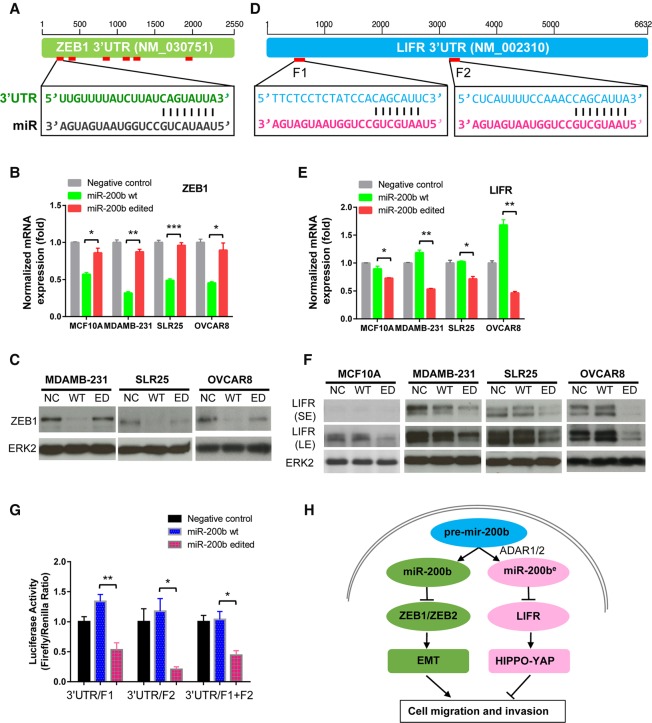
Summary of the 19 A-to-I RNA editing hotspots identified in miRNAs in TCGA data analysis
Validation of A-to-I miRNA editing hotpots
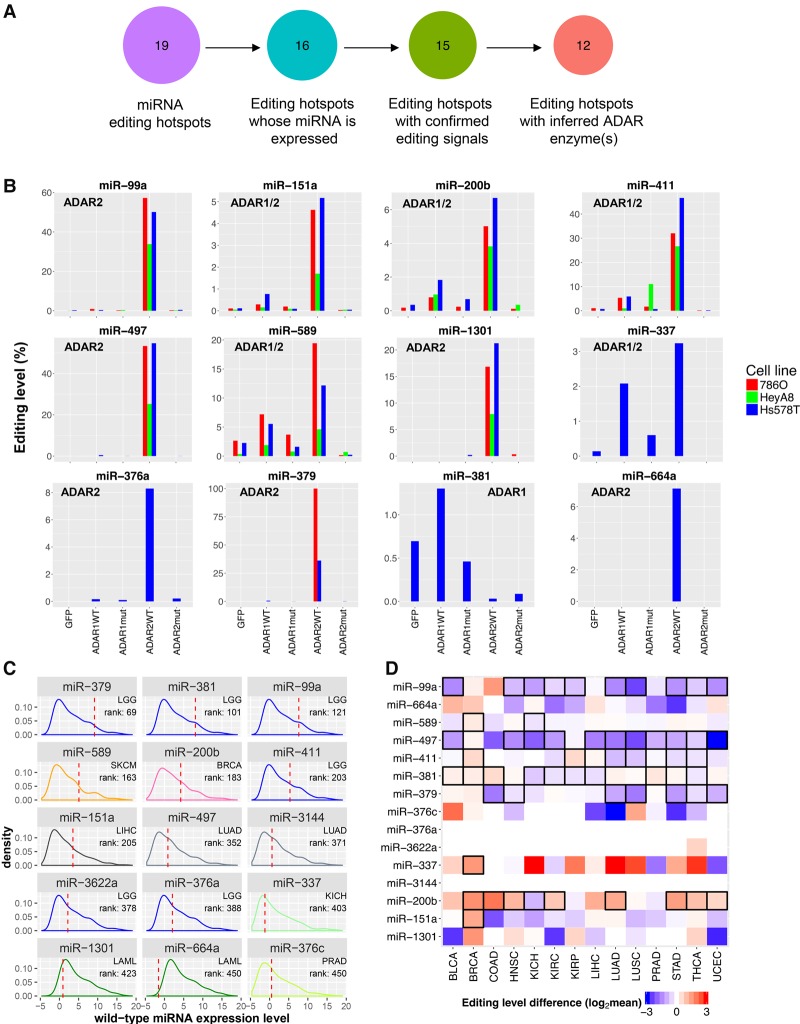
To independently validate these editing hotspots, we performed miRNA sequencing in three cancer cell lines (786O, HeyA8, and Hs578T) with overexpression of an ADAR enzyme or its mutant (Supplemental Fig. S1; Methods). Among the 19 miRNAs containing an editing hotspot, 16 of them (84.2%) were expressed in at least one cell line surveyed. Among the 16 miRNA editing hotspots, 15 (93.8%) showed detectable editing signals, and for 12 of them, we further inferred the specific ADAR enzymes underlying the editing event, because (1) the editing level increased dramatically given the overexpression of the wild-type (WT) ADAR enzyme, and (2) there were no significant changes in the editing level given the overexpression of the inactive form of ADAR enzyme (Fig. 2A,B). These results are also consistent with the patterns observed in an ADAR2-perturbed experiment (Supplemental Table S4; Tomaselli et al. 2015).
Figure 2.
Validation and molecular profiles of cancer miRNA editing hotspots. (A) Numbers of miRNA editing hotspots in the analysis of ADAR-perturbation experiments in 786O, HeyA8, and Hs578T cell lines. (B) The editing level changes after transfection of wild-type ADAR enzymes (ADAR WT), and inactive ADAR enzymes (ADAR mut). Inferred ADAR determinant(s) of each miRNA editing hotspot are shown in each miRNA subpanel. (C) For each edited miRNA, the highest rank of the edited miRNA expression amount (upper-quartile value across samples in a cancer type) relative to the WT miRNA expression amounts (median value) across cancer types is shown. (D) A heat map showing editing-level differences of 19 miRNA editing hotspots between cancerous and matched normal samples (two-sided paired Wilcoxon rank test, n ≥ 5). Red indicates overediting and blue indicates underediting in cancer samples; boxes highlight significant differences (FDR < 0.05).
We next focused on these validated miRNA editing hotspots for further analyses. The editing level of these hotspots in edited samples shows great variation, ranging from 80% to a few percent (Supplemental Fig. S2), and their correlation with the WT miRNA expression level varies, depending on both specific editing sites and tumor context (Supplemental Fig. S3). To assess in a biologically relevant context, we calculated the ranks of the expression amounts of edited miRNAs relative to the WT miRNAs in different cancer types and found that almost all the edited miRNAs have an expression amount comparable to that of the top 400 WT miRNAs in a cancer type (Fig. 2C). Although the total number of human miRNAs is under debate, it is commonly believed that at least 300–400 highly expressed miRNAs are evolutionarily conserved and play important biological roles (Liang and Li 2009; Chiang et al. 2010). Thus, these edited miRNAs have the potential to functionally regulate target genes, at least in some cancers.
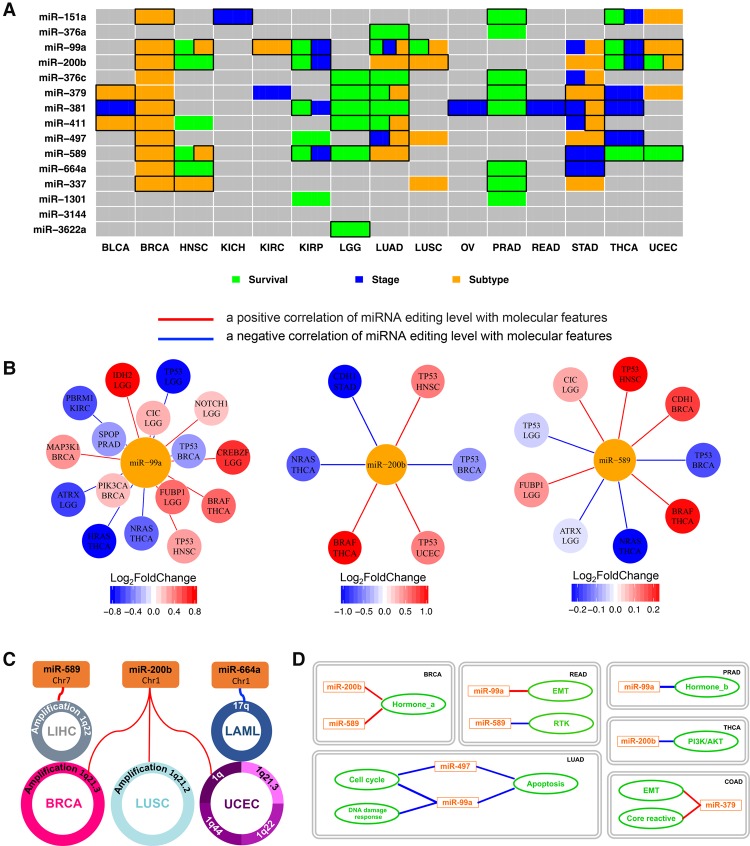
We examined the editing levels in 14 cancer types with available normal miRNA sequencing data (at least five tumor and normal matched pairs) (Fig. 2D). We found that nine of the 15 editing hotspots show a significant editing difference between tumor and normal tissue samples (paired Wilcoxon rank test, false discovery rate [FDR] < 0.05), suggesting potential biomedical relevance. As the cell composition of tumor and normal samples can be very different (e.g., the fraction of epithelial cells), the observed differential RNA editing activities at these hotspots may not be directly related to tumorigenesis. To further explore clinical relevance of the miRNA editing hotspots, we examined the correlations of their editing levels with tumor subtype, disease stage, and patient survival time within each cancer type. We found that 14 out of the 15 editing sites show significant patterns across 15 different cancer types (FDR < 0.2) (Fig. 3A), and many of the editing sites, such as miR-151a, miR-99a, miR-200b, miR-376c, miR-381, miR-411, and miR-664a, are significant in multiple cancer types. Moreover, these observations are robust to potential confounding effects of tumor purity (Methods). Since tumor subtype, disease stage, and the patient's survival time are key clinical parameters, the nonrandom patterns of miRNA editing in regard to these variables provides further support for miRNA editing as biomarkers of tumor behavior.
Figure 3.
Correlation of miRNA editing hotspots with clinical features, molecular drivers, and signaling pathways. (A) A heat map showing the clinical relevance of miRNA editing hotspots. Significant correlations with tumor subtype (orange, two-sided Wilcoxon or Kruskal-Wallis test), disease stage (blue, two-sided Wilcoxon or Kruskal-Wallis test), and patient survival times (green, univariate Cox proportional hazards model or log-rank test for median-based two-group comparison) (FDR < 0.2); after considering tumor purity, boxes highlight remaining significant correlations. (B) Significant correlations between the miRNA editing levels and significantly mutated genes (the gene nodes are colored according to the fold change between the mutated samples and the WT sample groups; two-sided Wilcoxon test, FDR < 0.05). The specific cancer types showing the significant correlations are listed below gene names. (C) Correlations between the miRNA editing levels and frequent SCNAs (Spearman rank correlation, |RS| > 0.5 and FDR < 0.05). (D) Correlations between the miRNA editing levels and signaling pathway scores (derived from TCGA protein expression data; Spearman rank correlation, |RS| > 0.3 and FDR < 0.05). Red lines indicate positive correlations, while blue lines are negative correlations.
Correlations of miRNA editing with molecular drivers and signaling pathways
To gain the mechanistic insights of these miRNA editing hotspots, we examined the correlations of the miRNA editing events with other molecular drivers and signaling pathways. First, we focused on mutated driver genes (i.e., significantly mutated genes) and performed a differential analysis of miRNA editing level between the sample groups with and without a mutation of a specific driver gene. In total, we identified 60 statistically significant miRNA-gene pairs (FDR < 0.05) between 12 miRNA editing hotspots and 20 significantly mutated genes across 10 individual cancer types (Fig. 3B; Supplemental Fig. S4A). Editing hotspots in miR-99a, miR-589, and miR-200b demonstrate the largest numbers of significant associations. For example, miR-200b editing shows correlations with TP53 in breast, head and neck, and endometrial cancers (BRCA, HNSC, and UCEC), with BRAF and NRAS in thyroid cancer (THCA), and with CDH1 in gastric cancer (STAD), while miR-99a editing shows correlations with TP53 in BRCA and HNSC, with PIK3CA and MAP3K1 in BRCA, and with HRAS and NRAS in THCA. Second, we assessed the relationships of miRNA editing with significant somatic copy number alterations (SCNAs). In total, we detected 10 significant SCNA-miRNA associations between four miRNA editing hotspots and 10 SCNAs in five cancer types (|RS| > 0.5) (Fig. 3C; Supplemental Fig. S4B). Interestingly, Chr1q21.3, which hosts ADAR1, shows positive associations with the miR-200b editing level in different cancer types, supporting ADAR1 as one key enzyme mediator for this editing event. Third, we examined the correlations of miRNA editing with signaling pathways based on TCGA reverse-phase protein expression data (Methods). In total, we detected 13 significant correlations between six edited miRNAs and nine signaling pathways across six cancer types (|RS| ≥ 0.3, FDR < 0.05) (Fig. 3D). Several edited miRNAs, such as miR-99a and miR-200b, show correlation with distinct pathways in different cancer types.
Functional effects of RNA editing in miR-200b on cell migration and invasion
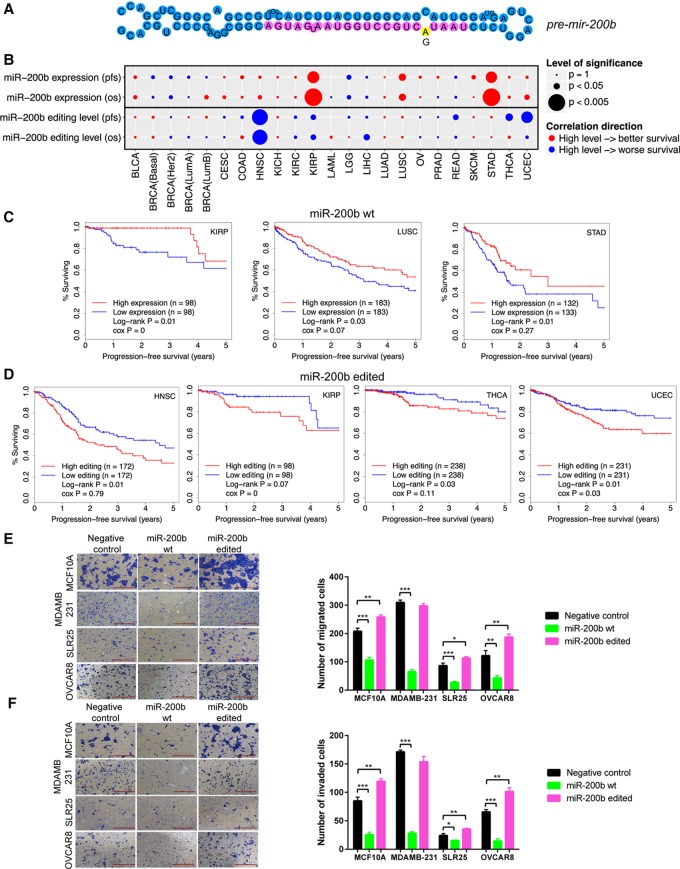
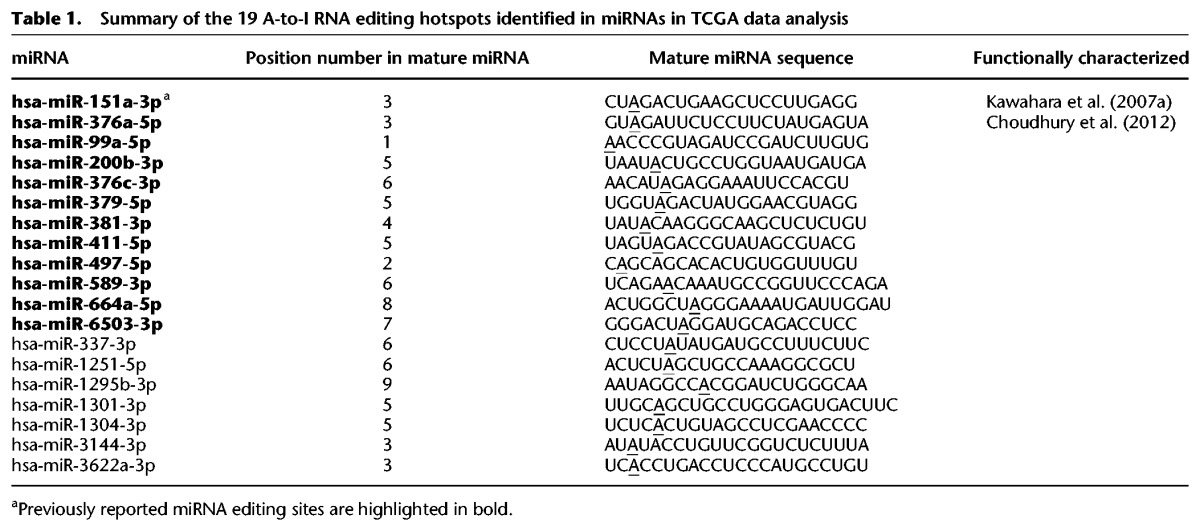
To deeply investigate the functional effect of miRNA editing, we focused on the RNA editing event in miR-200b (the fifth position in the mature miR) for experimental investigation, which to our knowledge has not previously been functionally characterized (Fig. 4A). We chose this target for several reasons. First, miR-200b is a highly expressed key member in the miR-200 family that plays an essential role in cancer metastasis by inhibiting epithelial mesenchymal transition (EMT) (Gregory et al. 2008; Park et al. 2008). Currently, miR-200 is under intensive clinical investigation as a promising cancer therapeutic target (Feng et al. 2014). Second, RNA editing in miR-200b is quite pervasive in various cancer types studied in terms of both editing frequency and edited miRNA expression (Fig. 1C). Further, it shows an overediting pattern in tumor samples related to the matched normal samples across eight cancer types (Fig. 2C). Third, miR-200b editing shows extensive correlations with molecular drivers (such as TP53 mutations) and key pathways in various cancers (Fig. 3). Last but not least, one striking observation about miR-200b editing is that the WT miR-200b expression and miR-200b editing level show opposite correlations with patient survival (Fig. 4B–D). Consistent with its established role as a tumor suppressor, high miR-200b expression is associated with better clinical outcomes across cancer types (Fig. 4C; Hu et al. 2009; Gravgaard et al. 2012; Kurashige et al. 2012). In contrast, patients with a high miR-200b editing level tend to have worse survival, including HNSC, KIRP, THCA, and UCEC (Fig. 4D). For ADAR1 and ADAR2, their mRNA expression levels show varied patterns with patient survival times across cancer types and do not behave similarly to miR-200b or edited miR-200b (Supplemental Fig. S5). These intriguing observations suggest a critical but distinct role of edited miR-200b in multiple cancer types.
Figure 4.
Effects of RNA editing in miR-200b on cell migration and invasion, and correlation with clinical outcomes. (A) Cartoon of A-to-I editing in stem–loop structure of pre-mir-200b. (B) Summary of correlations of WT miR-200b expression and miR-200 editing level with patient survival times across cancer types. Circle size represents statistical significance; color represents direction. In general, high expression of miR-200b is associated with better patient survival; high editing level in miR-200b is associated with worse patient survival. (C) Kaplan-Meier plots of patients grouped by miR-200b expression in individual cancer types. (D) Kaplan-Meier plots of patients grouped by editing level in miR-200b in individual cancer types. (E,F) Effects of miR-200b mimics on (E) migration and (F) invasion in MCF10A, MDAMB-231, SLR25, and OVCAR8 cells (n = 2 or 3). Two-sided t-test was used to assess the difference. Error bars denote ±SEM; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. Scale bar length is 500 µm.
We first examined the effects of miRNA editing on cell invasion and migration in MCF10A (normal mammary epithelial cells), MDAMB-231 (breast cancer), SLR25 (renal cancer), and OVCAR8 (ovarian cancer) cell lines using “mimics” of WT and edited miR-200b. Upon the mimic transfection, we measured the miRNA expression amount using qRT-PCR and found that the overexpressed amount was generally compatible with the range observed in TCGA patient samples (Supplemental Fig. S6). Given the same amount of the transfected mimics, WT miR-200b substantially decreased cell migration and invasion compared to control mimics (t-test, P < 0.05) (Fig. 4E,F), which is in good agreement with findings from previous studies (Bendoraite et al. 2010; Li et al. 2010; Shinozaki et al. 2010). In contrast, edited miR-200b significantly increased cell migration and invasion in MCF10A, SLR25, and OVCAR8 (t-test, P < 0.05) (Fig. 4E,F) but not in MDAMD-231, a cell line with very high motility (although we observed increases in some repeats). In addition, we performed cell viability assays to examine the impact on cell proliferation and survival and found that, compared with control mimics, WT miR-200b decreased cell viability, but edited miR-200b led to an even more dramatic decrease (t-test, P < 0.05) (Supplemental Fig. S7). This observation further confirmed that the elevated migration and invasiveness resulting from edited miR-200b is not due to an increase in cell proliferation. Compatible with the effects of the edited miR-200b, enhanced migration is frequently associated with decreased proliferation with a requirement for cells to return to an epithelial phenotype (MET) to enter proliferation (Evdokimova et al. 2009; May et al. 2011). Taken together, the functional assays and TCGA patient cohort survival analyses suggest that the single-nucleotide RNA editing event in miR-200b may switch its function from suppressing tumor metastasis to promoting metastasis.
Redirected target genes of edited miR-200b
We next sought to elucidate the molecular mechanisms underlying the “switch” of the phenotypic effects of miR-200b editing and reasoned that the effects may be achieved via two distinct mechanisms: (1) a loss of function through the impaired ability to inhibit the original targets of WT miR-200b; and (2) a gain of function by inhibiting new target genes. To systematically test this hypothesis in an unbiased manner, we performed RNA sequencing experiments on MCF10A, MDAMB-231, and SLR25 cells and identified genes that were significantly down-regulated (paired t-test, P < 2 × 10−5) upon transfection of WT or edited miR-200b mimics (relative to the control mimics). We then searched for sequence motifs that were overrepresented in the 3′ UTRs of these two groups of down-regulated genes. For WT miR-200b, the consensus motif we identified was CAGUAUU, the exact sequence that is complementary to the seed region of miR-200b, position 2–8 (Fig. 5A); for edited miR-200b, the consensus motif identified was CAGCAUU, the exact sequence that is complementary to the seed region of edited miR-200b (Fig. 5B). These results indicate that the editing of miR-200b redirects the miRNA to a completely new set of target genes by altering the seed sequence complementarity. To further pinpoint key altered targeting relationships, we integrated the sequence motif information and the gene expression data and predicted 68 high-confidence targets of WT miR-200b and 47 targets of edited miR-200b, with three genes overlapping between the two sets (Fig. 5C; Methods).
Figure 5.
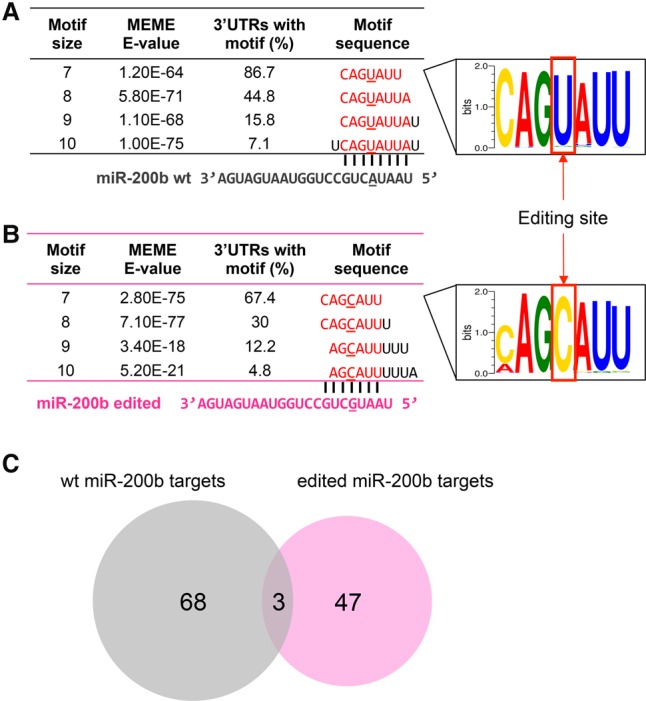
RNA editing in miR-200b redirects the target genes. (A) Sequence motif identified in 3′ UTRs of down-regulated genes upon transfection with WT miR-200b mimics (vs. negative control), corresponding to seed match of WT miR-200b. (B) Sequence motif identified in 3′ UTRs of down-regulated genes upon transfection with edited miR-200b mimics (vs. negative control), corresponding to seed match of edited miR-200b. (C) High-confidence predicted target genes of WT miR-200b and edited miR-200b by integrating gene expression and sequence motif data.
Among the predicted targets of WT miR-200b, ZEB1 and ZEB2 are established miR-200b targets and master EMT regulators (Fig. 6A; Supplemental Fig. S8), and they showed significant down-regulation upon transfection with WT miR-200b mimics compared to transfection with edited miR-200b mimics. Among the predicted targets of edited miR-200b, we focused on those genes for which the 3′ UTRs do not contain potential seed matches to the WT miR-200b and which showed specific down-regulation upon transfection with edited miR-200b compared to transfection with WT miR-200b mimics (Supplemental Table S5). The top candidate, leukemia inhibitory factor receptor (LIFR), is of particular interest. LIFR is a previously identified breast cancer metastasis suppressor that functions through altering the Hippo-YAP pathway (Chen et al. 2012). Down-regulation of LIFR has been observed in a variety of human cancers, and in vitro experiments have shown that silencing LIFR expression strongly promotes cell migration, invasion, and metastasis; conversely, restoring LIFR in highly malignant tumor cells effectively suppresses metastasis (Chen et al. 2012). Clinically, high expression of LIFR correlates with better patient survival across cancer types (Supplemental Fig. S9). Therefore, we focused on these three genes for further characterization. A 3′ UTR sequence survey shows that ZEB1 and ZEB2, respectively, have six and five binding sites for the WT miR-200b seed region (Fig. 6A; Supplemental Fig. S8; Supplemental Table S6). Given similar amounts of transfected WT and edited miR-200b mimics, we observed substantial inhibition of ZEB1 and ZEB2 expression for WT miR-200b but no similar changes for edited miR-200b by qRT-PCR (Fig. 6B; Supplemental Fig. S8). This trend was similar in three cell lines, and the suppression effects on ZEB1 were also verified by Western blot results at the protein level in MDAMB-231, SLR25, and OVCAR8 (Fig. 6C). For the novel target LIFR, which has two predicted binding sites (Fig. 6D; Supplemental Table S6), edited miR-200b dramatically reduced the expression of LIFR at both mRNA and protein expression levels in all four lines (Fig. 6E,F). In contrast, WT miR-200b had little effect on LIFR expression (Fig. 6E,F). We verified this new targeting relationship using luciferase reporter assays that contained the predicted target sites (Fig. 6G). Further, there was a significant negative correlation between edited miR-200b and LIFR mRNA expression across cancer types (RS = −0.19, P = 0) (Supplemental Fig. S10). These results suggest that A-to-I editing in miR-200b creates a neomorphic miRNA molecule that loses the ability to repress the original targets, such as ZEB1, but acquires the ability to target a new set of genes, such as LIFR, thereby affecting different downstream pathways and reversing functional outcomes.
Figure 6.
Molecular mechanisms of WT and edited miR-200b in cancer cells. (A) 3′ UTR representation of WT miR-200b target gene ZEB1. (B) qRT-PCR of ZEB1 upon 24-h transfection with WT miR-200b and edited miR-200b mimics in MCF10A, MDAMB-231, SLR25, and OVCAR8 cells. (C) Western blots of ZEB1 upon 48-h transfection with WT miR-200b and edited miR-200b mimics in MDAMB-231, SLR25, and OVCAR8 cells. (D) 3′ UTR representation of edited miR-200b target, LIFR. (E) qRT-PCR of LIFR upon 24-h transfection with WT miR-200b and edited miR-200b mimics in MCF10A, MDAMB-231, SLR25, and OVCAR8 cells. (F) Western blots of LIFR upon 48-h transfection with WT miR-200b and edited miR-200b mimics in MCF10A, MDAMB-231, SLR25, and OVCAR8 cells. Blots with short-time (SE) and long-time exposure (LE) are shown. (G) Luciferase reporter assays that contain two predicted binding sites of edited miR-200b (F1 and F2) in LIFR. In B, E, and G, two-sided t-test was used to assess the difference, n = 2 or 3, and error bars denote ±SEM; (*) P < 0.05. (**) P < 0.01, (***) P < 0.001. (H) Proposed mechanistic model in which WT miR-200b inhibits key EMT regulators ZEB1 and ZEB2, thereby suppressing cell migration and invasion, whereas edited miR-200b (catalyzed by both ADAR1 and ADAR2) inhibits a new target LIFR, a well-characterized metastasis suppressor, thereby promoting cell migration and invasion.
Taken together, we put forward a mechanistic model to explain the functional consequences of miR-200b editing (Fig. 6H). Generated from premature pre-mir-200b, WT mature miR-200b regulates the EMT pathway through inhibition of ZEB1 and ZEB2. In contrast, modified by the ADAR1/2 enzyme, edited miR-200b binds to the LIFR 3′ UTR based on sequence complementarity and inhibits its expression at both mRNA and protein levels. Subsequently, a decreased level of LIFR deactivates the Hippo phosphorylation cascade that leads to elevated cell migration, invasion, and metastasis. Therefore, this RNA editing event changes the fate of miR-200b from a tumor suppressor to an “oncogene” that promotes the metastatic process.
Discussion
Here, we performed a systematic analysis of miRNA editing patterns in >8000 samples of 20 different cancer types, which is the largest data set assessed for such an analysis. We observed a clear enrichment of A-to-I editing signals across cancer types, consistent with the notion that A-to-I editing is the dominant RNA editing type in humans. Our study thus provides a comprehensive view of RNA editing in miRNAs, a key class of regulatory genes, across a broad range of cancer types.
To reduce the noise in identifying miRNA editing events from highly heterogeneous tumor samples, we implemented a previous computational pipeline (Alon et al. 2012) with several key improvements. First, we filtered out those hypermutated samples since they contain large numbers of DNA mutations. Second, we removed all possible SNPs or mutations by compiling several databases such as dbSNP, COSMIC, ICGC, and TCGA. These efforts further reduce the possibility that the inferred editing events come from variants at the DNA level. The vast majority of RNA editing events identified are canonical A-to-I or C-to-U RNA editing, but there are still considerable numbers of other editing types. Since miRNAs are short, often duplicate in families, contain repeats, and undergo extensive post-transcriptional modifications, these “noncanonical” RNA editing events likely result from the prevalent cross-mapping issue of miRNA sequencing reads.
Applying relatively stringent criteria, we focused on A-to-I miRNA editing hotspots for detailed analysis because these recurrent epigenetic modifications represent the most promising “driver” candidates for mechanistic evaluation and for potential clinical utility. As a tradeoff, we may have missed some low-recurrence, low-level functionally relevant RNA editing events. Among the 19 miRNA editing hotspots, we performed cell line perturbation experiments to validate their editing signals and infer the responsible ADAR enzyme(s). It should be noted that the ectopic ADAR1/2 expression and concomitant measurement of editing in an miRNA in cell lines does not necessarily identify the editing enzyme underlying the editing event in vivo. Further in vivo experiments are important to confirm these ADAR1/2 associations. Intriguingly, the levels of the high-confidence editing hotspots we identified show extensive associations with clinical outcomes and established tumor subtype. Further, there are correlations between edited miRNAs and mutated driver genes, frequent SCNAs, and key signaling pathways. These nonrandom patterns do not necessarily indicate causal relationships but suggest that the miRNA editing hotspot events may coordinate with other molecular drivers to define the complexity of cancer phenotypes. Collectively, our analysis provides a rationale for further exploring miRNA editing-based diagnosis, prognosis, and possibly therapeutic strategies.
Because miRNA function strongly depends on its sequence complementarity with target genes, RNA editing in a mature miRNA not only can decrease the “effective” amount of WT miRNA but also have the potential to generate another new miRNA that functions as a neomorph by inhibiting a completely different set of targets. Here, we focused on an RNA editing event in miR-200b, a key regulator of EMT and cancer metastasis. Although its editing level is low, the absolute expression amount of edited miRNA could be significant because of the extremely high level of WT miR-200b. Indeed, in most cancer types surveyed (12 out of 20), the up-quantile expression amount of edited miR-200b ranks among the top 220 WT miRNAs (median). Our in vitro functional assays suggest that a single-nucleotide change in its seed region may switch the role of this miRNA in cancer metastasis. This functional change is due, at least in part, to an impaired ability to inhibit ZEB1/ZEB2 and the acquired ability to target LIFR, thereby conferring a net effect of increasing motility and invasion. The expression amount of this edited miRNA is comparable to that of miR-125a, a known regulator of LIFR (Nandy et al. 2015), in a considerable fraction of patient samples of various cancer types. One limitation of our study is that we mainly relied on the miRNA mimic-mediated overexpression experiments in cancer cell lines, which might not represent the real tumor context. Future efforts, especially in vivo experiments, should be undertaken to confirm the functional consequences of edited miR-200b and investigate its clinical utility.
Methods
Characterization of miRNA editing profiles
We downloaded miRNA sequencing BAM files of 8164 patient tumor samples and 643 related normal tissue samples (as available) for 20 cancer types from the UCSC Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). We identified 203 of the cancer samples with more than 1000 somatic mutations in the exome as hypermutated samples and excluded these samples to reduce false positives in the downstream analysis. All BAM files were converted back to FASTQ files, and the sequencing reads were trimmed by 2 nt at the 3′ end. We filtered the reads that (1) were shorter than 15 bp or longer than 28 bp, or (2) contained more than three low-quality positions (quality score cutoff, 20). We then remapped the reads to the human reference genome (hg19) using Bowtie (Langmead 2010), allowing at most one mismatch per read, the best alignment, and no cross-mapping. We focused on the reads aligned to the genomic regions of known pre-miRNA in miRBase (release 21) (Kozomara and Griffiths-Jones 2014) for mismatch calling based on the binomial test as described in the literature (Alon et al. 2012). Using the cutoff of the Bonferroni-corrected P-value of 0.05 and the mismatch base quality score ≥ 30, we detected miRNA editing candidates in each sample. As these candidates may result from variation at the DNA level, we removed all annotated SNPs and somatic mutations from dbSNP (build 142), COSMIC, ICGC, and TCGA mutation data. To identify miRNA editing events of potential significance, we focused on high-confidence editing events with (1) the editing level ≥ 5% (the editing level was defined as the proportion of the mapped reads containing the edited nucleotide relative to the total mapped reads at the given position) and (2) the edited miRNA amount ≥ 1 RPM (mapped to miRNA), and identified the RNA editing hotspots (defined as those that occurred in ≥10 tumor samples in any cancer type). To further exclude unreported SNPs and somatic mutations, we assessed the ICGC whole-genome sequencing data and TCGA whole-exome sequencing data at the identified miRNA editing hotspots and removed sites with potential DNA mutational signals. This analysis yielded 20 unique miRNA editing sites with strong A-to-I editing signals. The editing site in miR-4510 was removed due to its extremely high editing level (e.g., 100%) in multiple cancer types, which is unlikely to be real RNA editing. The scripts used for miRNA editing calling were previously published in Alon et al. (2012) and obtained from http://www.tau.ac.il/∼elieis/miR_editing/. We modified the scripts to fit miRBase 21 and these are available in Synapse (https://www.synapse.org/, syn6020318).
Profile analysis of miRNA editing hotspots
For the samples with sufficient coverage (≥10×), we calculated the editing frequency for each miRNA editing hotspot as the proportion of cancer samples with detectable editing signals (edited reads ≥ 1) among the total number of cancer samples in a cancer type. The edited miRNA expression amount was defined as RPM. To assess the significance of the edited miRNA expression amount, we first calculated the median expression values (RPM) of all expressed WT mature miRNAs (out of 2588) across samples in each cancer type. Then, for samples with detectable editing signals, we measured the upper-quartile expression value of the validated 15 edited miRNAs and ranked them against the median expression distribution of WT miRNAs in each cancer type. Finally, we reported the cancer type with the highest rank for each miRNA editing hotspot. To compare miRNA editing patterns between cancer and normal samples, we considered only the cancer types with ≥5 matched cancer and normal sample pairs (with sufficient coverage) for the sites of interest of miRNA editing. We used a paired Wilcoxon test to compare the editing level between tumor and normal samples and detected a statistically significant difference at FDR < 0.05. We obtained TCGA mRNA expression data from the TCGA data portal (https://tcgadata.nci.nih.gov/tcga/) and used the Spearman rank correlation test to quantify correlations of the miRNA editing level with the mRNA expression level of ADAR1, ADAR2, and ADAR3. A significant correlation was detected as RS ≥ 0.5 and FDR < 0.05. To verify the correlations of ADAR enzymes with the RNA editing levels observed in TCGA patient samples, we obtained miRNA sequencing data of two glioblastoma cell lines U87 control (NCBI SRA SRX098102), U87 ADAR2 overexpression (SRX098101), U118 control (SRX098107), U118 ADAR2 overexpression (SRX735409), U118 inactive ADAR2 overexpression (SRX735410), and U118 silencing ADAR2 (SRX764455) (Alon et al. 2012; Tomaselli et al. 2015). All samples were analyzed using our miRNA-editing calling pipeline. For six editing hotspots showing a significantly strong correlation with ADAR2 in LGG patient samples, we compared the editing level across all ADAR2-perturbed cell line samples.
Clinical relevance analysis of miRNA editing hotspots
We obtained clinical information, including tumor subtypes, disease stage, and patient overall and progression-free survival times from TCGA marker papers and the TCGA data portal. For each cancer type, we used the Wilcoxon test or Kruskal-Wallis nonparametric ANOVA to detect differential editing levels among cancer subtypes or disease stages and considered FDR < 0.2 as statistically significant. For the survival analysis, we capped the survival data at 5 yr to reduce the effect of non-disease-related deaths. We used two methods to test whether the editing level of an miRNA was correlated with patient survival time in each cancer type. We used (1) the univariate Cox proportional hazards model to assess the correlation with the patient's progression-free survival time (Cox 1992), considering FDR < 0.2 as statistically significant; and (2) the log-rank test to compare patient survival curves between the high- and low-editing level groups (separated by the median), considering FDR < 0.2 as statistically significant. To assess the potential confounding effects of tumor purity, we obtained tumor purity data from Synapse (syn1710466). We then repeated the above analyses by including tumor purity as a covariate in the ANOVA.
Correlations of miRNA editing hotspots with molecular drivers and pathways
We obtained TCGA somatic mutation data from Firehose (http://gdac.broadinstitute.org/, analysis 2014-04-16). For each cancer type, we used the top 20 significantly mutated genes in our analysis. We applied the Wilcoxon test to assess the editing level at the miRNA editing hotspots between mutated versus WT groups, considering FDR < 0.05 as statistically significant. We obtained significant SCNAs (FDR < 0.25) from Firehose (analysis 2014-04-16). For both significantly altered focal and arm-level features, we used the Spearman rank correlation to measure associations of miRNA editing level with SCNAs, considering FDR < 0.05 and |RS| > 0.5 as significant hits. To investigate correlation of the miRNA editing level with signaling pathways, we first integrated the normalized protein expression data (Z-score) from the TCGA reverse-phase protein array platform into 11 core cellular pathways (Akbani et al. 2014) and then quantified the correlations of the editing level with all core pathways. We considered FDR < 0.05 and |RS| > 0.3 as statistically significant. To assess the effect of tumor purity, we repeated the above analyses with tumor purity as a covariate in ANOVA or a linear model.
Cell culture and transfection with miRNA mimics
The MCF10A and MDAMB-231 cell lines were purchased from the American Type Culture Collection (ATCC) and were cultured under the conditions specified by ATCC. SLR25, OVCAR8, 786O, HeyA8, and Hs578T cells were purchased from the MD Anderson Characterized Cell Line Core Facility. All the cell lines were confirmed by short tandem repeat (STR) analysis, and mycoplasma testing was found to be negative. MCF10A cells were maintained in complete DMEM/F12 (Invitrogen) full medium with 5% horse serum (Invitrogen), 20 ng/mL EGF (Peprotech), 10 µg/mL insulin (Sigma), 100 ng/mL Cholera Toxin (Sigma), and 0.5 mg/mL hydrocortisone. MDAMB-231, SLR25, OVCAR8, 786O, HeyA8, and Hs578T cells were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum. MiR-200b mimics were from Sigma-Aldrich including MISSION miRNA Negative Control 1 (HMC0002), and MISSION microRNA Mimic hsa-miR-200b-3p (HMI0352). The sense sequence of hsa-miR-200b-3p edited mimics is [AmC6F] UCAUCAUUACCAGGCAGCAUUUAdTdT, and the antisense sequence is UAAUGCUGCCUGGUAAUGAUGA. Cells were transfected with 50 nM of the indicated miRNA mimics using Lipofectamine RNAiMAX Reagent (ThermoFisher Scientific). Cells were plated for proliferation, migration, and invasion assays 24 h after transfection.
miRNA sequencing-based ADAR perturbation experiments and generation of stable cell lines
The mutant open reading frames (ORFs) corresponding to the RNA editing sites in mutant ADAR1 and ADAR2 were made by site-directed mutagenesis and confirmed by Sanger sequencing as previously described (Han et al. 2015). ADAR1-E912A and ADAR2-E396A contain an E-to-A amino acid change that abolishes ADAR editase activity (Macbeth et al. 2005). Virus was produced by transfecting HEK293PA cells with the GFP control vectors, pHAGE-V5-puromycin expression vectors (carrying ADAR1-WT or ADAR1-E912A; ADAR2-WT or ADAR2-E396A), and the Lentiviral Packaging Mix (psPAX2 and pMD2.G). 786O, HeyA8, and Hs578T cells were transduced by the virus followed by selection with puromycin (Hs578T 1 µg/mL, HeyA8 1 µg/mL, and 786O 3 µg/mL), and after 7 d of antibiotic selection, expression of the constructs was verified by Western blots. To validate the miRNA-editing sites, we chose 786O, HeyA8, and Hs578T for perturbation studies. Small RNA of post-transfection was subjected to miRNA sequencing and analyzed using the exact same pipeline used for TCGA samples. Editing level of each miRNA editing hotspot was then calculated and compared. For the 19 miRNAs containing an RNA editing hotspot, 16 of them were expressed with at least three RNA-seq reads in any cell line under any condition surveyed. In these expressed miRNAs, 15 miRNA editing hotspots showed detectable editing signals (at least one edited read in any cell line under any condition). Among them, 12 editing sites had sufficient coverage to observe editing level changes (edited reads > 1, and editing level given the overexpression of a WT ADAR enzyme was higher than 1% in any of the three cell lines). For qualified miRNAs, the cell lines with good coverage and editing signals (edited reads > 1 in the WT ADAR enzyme overexpression conditions) were used to infer ADAR determinants.
RNA isolation and quantitative real-time RT-PCR
To measure the miRNA expression, total RNA from cultured cells, with efficient recovery of small RNAs, was isolated using the mirVana miRNA Isolation kit with phenol (Ambion). Detection of the mature form of miRNAs was performed using the TaqMan MicroRNA Reverse Transcription kit and TaqMan Universal Master Mix II (no UNG), according to the manufacturer's instructions (ThermoFisher). The U6 small nuclear RNA was used as an internal control. The hsa-miR-200b TaqMan MicroRNA Assay (Catalog #4427975, Assay ID: 002251) and U6 snRNA TaqMan microRNA Control Assay (Catalog # 4427975, Assay ID: 001973) were purchased from ThermoFisher. To examine the effects on the potential miRNA target genes, total RNA was isolated using the RNeasy Plus Mini kit (Qiagen). RNAs were transcribed into cDNAs using the High-Capacity cDNA Reverse Transcription kit (Life technologies). Expression levels were normalized to beta actin. Reactions were done in duplicate using TaqMan Fast Universal PCR Master Mix (2×), no AmpErase UNG (Life Technologies). The primers from ThermoFisher were as follows: LIFR primer (Hs01123581_m1), ZEB1 primer (Hs00232783_m1), and ZEB2 primer (Hs00207691_m1). qRT-PCR was performed by Applied Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems). The relative expression was calculated by the 2(–DDCt) method.
Generation and analysis of cell line mRNA sequencing data
MCF10A, MDAMB-231, and SLR25 cells transfected with WT and edited miR-200b mimics were subjected to mRNA sequencing (the sequencing platform was HiSeq 4000 and the paired end reads were 2 × 76 bp) at the MD Anderson Sequencing and Microarray Core Facility. We mapped FASTQ raw reads and performed gene differential expression analysis using TopHat2 alignment with default parameters (Kim et al. 2013) and HTSeq-count with mode “union” (Anders et al. 2015), followed by EdgeR (Robinson et al. 2010). We identified a set of potential target genes of WT miR-200b by comparing the expression of the paired samples of WT miR-200b and control mimics and a set of potential target genes of edited miR-200b by comparing the expression of the paired samples of edited miR-200b and control mimics. Differentially expressed genes with FDR < 2 × 10−5 in the comparison were used in subsequent analysis. The 3′ UTRs of the two gene sets were then extracted on the basis of RefSeq gene annotation, and the sequence motifs were discovered by first using RepeatMasker (http://repeatmasker.org) to mask repeats, and then MEME (the “zoops” model) with the motif length ranging from 7 to 10 nt (Bailey and Elkan 1994). The sequence logos were generated using WebLogo (http://weblogo.threeplusone.com). We identified high-confidence predicted targets of edited miR-200b by two additional criteria: (1) The 3′ UTR contains more than one seed match, and (2) the differential fold change is >1.5.
Immunoblotting
Whole-cell lysates for Western blotting were extracted with RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease, and phosphatase inhibitor cocktail). Protein concentrations were determined using bicinchoninic acid (Pierce) assays according to the manufacturer's instructions. Cell lysates (30 µg) were loaded onto 8% or 12% SDS-PAGE and transferred to a polyvinylidene fluoride membrane, and protein expression was depicted with an enhanced chemiluminescence Western blot detection kit (Amersham Biosciences). The following antibodies were used: LIFR (1:500, Santa Cruz Biotechnology, sc-659), ZEB1 (1:500, Novus Biologicals, NBP1-05987), and ERK2 (1:2000, Santa Cruz Biotechnology, sc-154), ADAR1 (1:1000, Santa Cruz Biotechnology, sc-271854), ADAR2 (1:1000, Genetex, GTX114237), and GAPDH (1:3000, Santa Cruz Biotechnology, sc-25778).
Cell viability assay
The MCF10A, MDAMB-231, and SLR25 cell lines were seeded into 96-well plates and transfected with miRNA mimics. Twenty-four hours after transfection, CellTiter-Glo 2.0 (Promega) was added to assess cell viability according to the manufacturer's instructions at 1, 2, 4, and 6 d. The significance of the differences was analyzed with Student's t-test, and P < 0.05 was considered statistically significant.
In vitro migration and invasion assay
For transwell migration assays, 2.5 × 104 to 1 × 105 cells were plated in the top chamber with a noncoated membrane (Corning BioCoat Control Insert; 8.0 µm; 24-well; 24/CS 354578). For invasion assays, 2.5 × 104 to 1 × 105 cells were plated in the top chamber with Matrigel-coated membrane (Corning BioCoat Matrigel Invasion Chamber; 24-well; 24/CS 354483). In both assays, cells were plated in medium without serum or growth factors, and medium supplemented with growth factors (for MCF10A) or serum (for MDAMB-231, SLR25, and OVCAR8) was used as a chemoattractant in the lower chamber. The cells were incubated for 18 h or 30 h and cells that did not migrate or invade through the pores were removed with a cotton swab. Cells on the lower surface of the membrane were fixed with ethanol, and then stained with Coomassie brilliant blue and counted under 10 different low-power (100×) microscopic fields.
Luciferase reporter assay
The human LIFR 3′ UTR fragments (F1 fragment: 484 bp, F2 fragment: 450 bp, F1+F2 fragment: 3289 bp) were PCR-amplified from genomic DNA and cloned into the pMIR-REPORT luciferase construct using the following cloning primers: F1 fragment forward, 5′-GATTGGGAGCTCTTTGTGATCAAGCCAAAG-3′; F1 fragment reverse, 5′-CCGGCACGCGTCTATCACAGCAAGAAAACATGATTATGATTTGGGC-3′; F2 fragment forward, 5′-CCGGCGAGCTCGCATAGATTAGCACATGTATGCAATCTAGGTCATC-3′; F2 fragment reverse, 5′-CCGGCACGCGTCCCCTAGTCTCCTTCACTAAATTACAAATAATTACCC-3′.
MDAMB-231 cells of 50% confluence in 24-well plates were transfected using Lipofectamine 3000 reagent (Invitrogen). The pMIR-REPORT firefly luciferase reporter gene construct (20 ng), 2 ng of the pRL-TK Renilla luciferase construct (for normalization), and 50 nM miRNA mimics were cotransfected per well. Cell extracts were prepared 48 h after transfection, and the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Data access
The RNA sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP075557.
Supplementary Material
Acknowledgments
We acknowledge contributions from the TCGA Research Network and TCGA Pan-Cancer Analysis Working Group. This study was supported by the National Institutes of Health, National Cancer Institute (CA168394, CA098258, and U24CA143883 to G.B.M.; R01CA175486 and U24CA209851 to H.L.; and CCSG grant CA016672); the R. Lee Clark Fellow Award from The Jeanne F. Shelby Scholarship Fund; a grant from the Cancer Prevention and Research Institute of Texas (RP140462 to H.L.); and the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine (to H.L.). We thank the MD Anderson high-performance computing core facility for computing and LeeAnn Chastain for editorial assistance.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.219741.116.
References
- Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al. 2014. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun 5: 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Mor E, Vigneault F, Church GM, Locatelli F, Galeano F, Gallo A, Shomron N, Eisenberg E. 2012. Systematic identification of edited microRNAs in the human brain. Genome Res 22: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Erew M, Eisenberg E. 2015. DREAM: a webserver for the identification of editing sites in mature miRNAs using deep sequencing data. Bioinformatics 31: 2568–2570. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intellt Syst Mol Biol 2: 28–36. [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O'Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, et al. 2010. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol Oncol 116: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. 2006. RNA editing of human microRNAs. Genome Biol 7: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, et al. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, et al. 2012. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med 18: 1511–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Li Y, Lin CH, Chan THM, Chow RKK, Song YY, Liu M, Yuan YF, Fu L, Kong KL, et al. 2013. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 19: 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, Wang S. 2012. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest 122: 4059–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. 1992. Regression models and life-tables. In Breakthroughs in statistics: methodology and distribution (ed. Kotz S, Johnson NL), Vol. II, pp. 527–541. Springer, New York. [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. 2009. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res 37: 2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Li JB, Levanon EY. 2010. Sequence based identification of RNA editing sites. RNA Biol 7: 248–252. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Sorensen PH. 2009. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle 8: 2901–2906. [DOI] [PubMed] [Google Scholar]
- Feng X, Wang Z, Fillmore R, Xi Y. 2014. MiR-200, a new star miRNA in human cancer. Cancer Lett 344: 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli D, Gacquer D, Rothe F, Lefort A, Libert F, Brown D, Kheddoumi N, Shlien A, Konopka T, Salgado R, et al. 2015. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep 13: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano F, Rossetti C, Tomaselli S, Cifaldi L, Lezzerini M, Pezzullo M, Boldrini R, Massimi L, Di Rocco CM, Locatelli F, et al. 2013. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 32: 998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. 2009. MicroRNAs in cancer. Annu Rev Med 60: 167–179. [DOI] [PubMed] [Google Scholar]
- Gong J, Wu Y, Zhang X, Liao Y, Sibanda VL, Liu W, Guo AY. 2014. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. RNA Biol 11: 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Liu C, Liu W, Xiang Y, Diao L, Guo AY, Han L. 2017. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res 45: D79–D84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravgaard KH, Lyng MB, Laenkholm AV, Sokilde R, Nielsen BS, Litman T, Ditzel HJ. 2012. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat 134: 207–217. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Bio 10: 593–601. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Han SW, Kim HP, Shin JY, Jeong EG, Lee WC, Kim KY, Park SY, Lee DW, Won JK, Jeong SY, et al. 2014. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med 211: 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HM, Eterovic AK, Yuan Y, et al. 2015. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell 28: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. 2009. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol 114: 457–464. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. 2007a. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8: 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. 2007b. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. 2008. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O'Connell MA. 2001. The many roles of an RNA editor. Nat Rev Genet 2: 869–878. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S, Baba Y, Baba H. 2012. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol 19: S656–S664. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11 17 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, Borges M, Goggins M. 2010. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res 70: 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li WH. 2009. Lowly expressed human microRNA genes evolve rapidly. Mol Biol Evol 26: 1195–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May CD, Sphyris N, Evans KW, Werden SJ, Guo W, Mani SA. 2011. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res 13: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy SB, Arumugam A, Subramani R, Pedroza D, Hernandez K, Saltzstein E, Lakshmanaswamy R. 2015. MicroRNA-125a influences breast cancer stem cells by targeting leukemia inhibitory factor receptor which regulates the Hippo signaling pathway. Oncotarget 6: 17366–17378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi V, Paul D, Das S, Bajpai P, Singh S, Mukhopadhyay A, Agrawal A, Ghosh B. 2015. Altered expression and editing of miRNA-100 regulates iTreg differentiation. Nucleic Acids Res 43: 8057–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. 2016. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 17: 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N, Bazak L, Buchumenski I, Porath HT, Danan-Gotthold M, Knisbacher BA, Eisenberg E, Levanon EY. 2015. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep 13: 267–276. [DOI] [PubMed] [Google Scholar]
- Ramaswami G, Li JB. 2014. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res 42: D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. 2012. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods 9: 579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiselet M, Gacquer D, Spinette A, Craciun L, Decaussin-Petrucci M, Andry G, Detours V, Maenhaut C. 2015. New global analysis of the microRNA transcriptome of primary tumors and lymph node metastases of papillary thyroid cancer. BMC Genomics 16: 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, Uozaki H, Takada K, Fukayama M. 2010. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res 70: 4719–4727. [DOI] [PubMed] [Google Scholar]
- Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al. 2015. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol 17: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaselli S, Galeano F, Alon S, Raho S, Galardi S, Polito VA, Presutti C, Vincenti S, Eisenberg E, Locatelli F, et al. 2015. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol 16: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524. [DOI] [PubMed] [Google Scholar]
- Warnefors M, Liechti A, Halbert J, Valloton D, Kaessmann H. 2014. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol 15: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.