Abstract
The 3D organization of the genome changes concomitantly with expression changes during hematopoiesis and immune activation. Studies have focused either on lamina-associated domains (LADs) or on topologically associated domains (TADs), defined by preferential local chromatin interactions, and chromosome compartments, defined as higher-order interactions between TADs sharing functionally similar states. However, few studies have investigated how these affect one another. To address this, we mapped LADs using Lamin B1–DamID during Jurkat T-cell activation, finding significant genome reorganization at the nuclear periphery dominated by release of loci frequently important for T-cell function. To assess how these changes at the nuclear periphery influence wider genome organization, our DamID data sets were contrasted with TADs and compartments. Features of specific repositioning events were then tested by fluorescence in situ hybridization during T-cell activation. First, considerable overlap between TADs and LADs was observed with the TAD repositioning as a unit. Second, A1 and A2 subcompartments are segregated in 3D space through differences in proximity to LADs along chromosomes. Third, genes and a putative enhancer in LADs that were released from the periphery during T-cell activation became preferentially associated with A2 subcompartments and were constrained to the relative proximity of the lamina. Thus, lamina associations influence internal nuclear organization, and changes in LADs during T-cell activation may provide an important additional mode of gene regulation.
The genome is organized within the three-dimensional interphase nucleus. Regulatory elements influence target genes hundreds of kilobase pairs to many megabase pairs distal by moving proximal through looping in three-dimensional space (Sanyal et al. 2012; Shen et al. 2012; Arner et al. 2015; Schoenfelder et al. 2015). Chromosomes are also organized along their length into discrete regions displaying preferential internal interactions detected by chromosome conformation capture (Hi-C) approaches, termed topologically associated domains (TADs) or contact domains (CDs) depending on the algorithm used to calculate them (Dixon et al. 2012; Hou et al. 2012; Nora et al. 2012; Sexton et al. 2012). As TADs and CDs likely represent orthogonal measures of the same structural units, we use the term TADs unless specifically referring to published data sets employing the CD approach. Topological constraints imposed by TADs favor looping interactions to cis-regulatory element:gene pairings within the same TAD (Lupianez et al. 2015; Franke et al. 2016; Hnisz et al. 2016). However, a higher level of genome organization also occurs where multiple TADs many megabase pairs distal on one chromosome or on different chromosomes can be adjacent and share a functional state (Lieberman-Aiden et al. 2009). Such associations form compartments, which segregate into two classes: A, transcriptionally active; B, transcriptionally repressed (Lieberman-Aiden et al. 2009). While TAD composition is largely invariant, compartment composition changes during differentiation and between cell types correlating with gene activity (Dixon et al. 2015; Fraser et al. 2015); however, how compartment interactions are controlled during differentiation or other cellular transitions is largely unexplored.
The nuclear periphery, in contrast, contributes radial genome organization. Genome regions proximal to the lamin intermediate filament polymer lining the inner nuclear membrane are called lamina-associated domains (LADs) (Guelen et al. 2008). However, while they are implicated in genome organization (Mewborn et al. 2010; Mattout et al. 2011; Solovei et al. 2013), it remains unclear if lamins themselves are absolutely required for tethering LADs to the periphery (Amendola and van Steensel 2015; Zheng et al. 2015). The nuclear periphery is generally repressive, with genes in LADs tending to contain silencing marks (Guelen et al. 2008). Constitutive LADs are defined as unchanging, but the number defined as facultative, changing with the cell cycle or between cell types, is constantly increasing (Peric-Hupkes et al. 2010; Kind et al. 2013; Meuleman et al. 2013; Chandra et al. 2015; Robson et al. 2016). The importance of facultative LADs is underscored by the fact that their disruption has profound negative consequences for tissue differentiation (Robson et al. 2016). Tellingly, such facultative LADs move both to and from the periphery during differentiation and often contain genes requiring fine-tuned regulation (Peric-Hupkes et al. 2010; Robson et al. 2016).
Though LADs correlate with the repressive B compartment and TADs (Dixon et al. 2012; Rao et al. 2014; Fraser et al. 2015), few studies have investigated how alterations to one can influence the other. Moreover, this correlation is imperfect and insufficient, for example, to predict TADs from LADs or vice versa. Several questions come to mind: Does a TAD as a functional unit move between the periphery and interior and thus perhaps encompass a facultative LAD? Can a new LAD that forms during differentiation influence the organization of TADs/compartments or vice versa?
Facultative LAD repositioning tends to associate with repression or activation of genes within them. In neurogenesis and myogenesis, 5%–15% of genes move from the nuclear periphery to the interior, while another 5%–10% move from the interior to the periphery (Peric-Hupkes et al. 2010; Robson et al. 2016). Though lacking LAD data sets, lymphocyte-specific gene-positioning changes are reported, such as IGH locus release from the periphery upon induction of V-D-J recombination (Kosak et al. 2002) and MAF locus repositioning to the periphery with its repression (Hewitt et al. 2004). Contrasting differentiation, lymphocyte activation is much more rapid and dynamic and involves significant genome restructuring (Drings and Sonnemann 1974). Compacted peripheral chromatin dissipates upon activation concomitant with large-scale gene activation (Pompidou et al. 1984). Our previous finding of changes in nuclear membrane protein composition during lymphocyte activation (Korfali et al. 2010) led us to investigate whether there are also specifically regulated gene exchanges at the nuclear envelope.
Results
Mapping gene expression and repositioning changes during T-cell activation
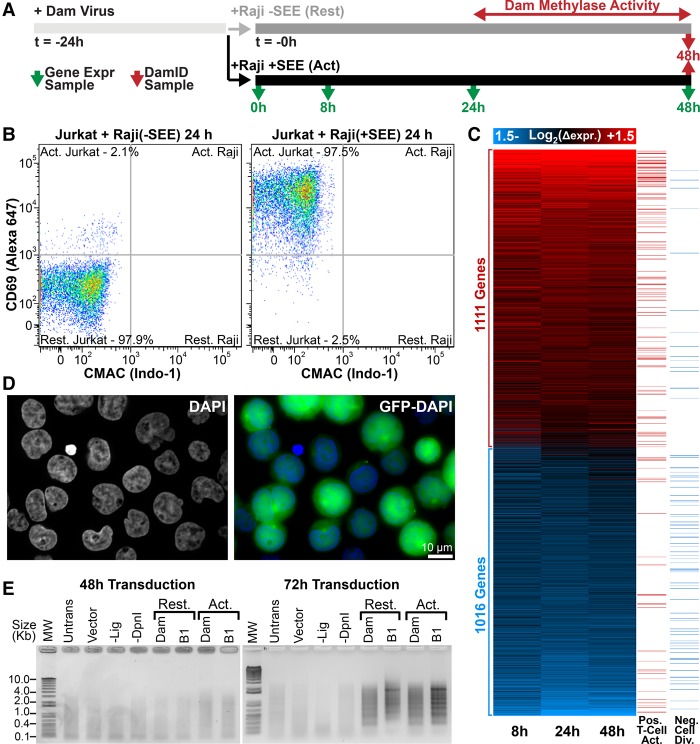
The extensive electron-dense peripheral heterochromatin of resting T cells dissipates during activation concomitantly with the induction of immunogenic genes (Hirschhorn et al. 1971; Pompidou et al. 1984; Manteifel et al. 1992; Rawlings et al. 2011). However, it is unclear whether this reorganization is whole-scale or whether there is also more specific reorganization for immune activation. We predicted that loss of peripheral heterochromatin would correlate with release of T-cell activation–associated genes from the periphery. Accordingly, we investigated coordinated gene expression and genome organization changes during T-cell activation using microarrays and DamID (Fig. 1). Jurkat cells were incubated with Raji B cells that had been preconjugated with staphylococcal enterotoxin E (SEE), resulting in the formation of antigen-independent immunological synapses (Fig. 1A; Gonzalez-Granado et al. 2014). This yielded >95% activation of Jurkat cells by fluorescence-activated cell sorting (FACS) (Fig. 1B). To determine associated gene expression changes, RNA was extracted at 0, 8, 24, and 48 h post SEE stimulation and analyzed by microarray. Upon activation, 1111 genes were up-regulated at least 1.4-fold at any of the three time points, and these were significantly enriched in Gene Ontology (GO) terms positively supporting T-cell activation and early effector function (Fig. 1C; Supplemental Fig. S1). Similarly, 1016 genes were repressed at least 1.4-fold that were enriched in GO terms inhibiting mitosis and cell division, presumably permitting accelerated proliferation of activated cells.
Figure 1.
Transcription and genome organizational changes during T-cell activation. (A) Transcriptome and DamID experimental strategy. (B) FACS plot of CD69 versus CMAC staining for Jurkat T cells exposed to Raji B cells lacking/presenting SEE antigen. Raji B cells were gated from CMAC staining to distinguish them from Jurkat T cells, revealing 97.5% Jurkat T-cell activation. (C) Heatmap of time-course gene expression changes with a 1.4-fold cutoff highlighting GO terms positively associated with T-cell activation or negatively associated with cell division in the right-most columns (GO terms in Supplemental Fig. S1). (D) Representative micrograph of GFP-encoding lentivirus-transduced Jurkat T cells indicating high efficiencies for parallel transduced DamID constructs. (E) PCR amplified Dam-methylated genomic DNA. Amplification is only observed in resting (Rest.) or activated (Act.) cells 72 h post transduction. T4 DNA ligase-null (-Lig) and DpnI-null (-DpnI) controls represent background amplification. DamID results are derived from a single experiment, while gene expression analysis was performed in triplicate.
The same cell populations were subjected to Lamin B1–DamID to globally map changes in genome-nuclear envelope associations. A GFP reporter confirmed efficient lentiviral transduction of Jurkat T cells (Fig. 1D), but DamID signal was not observed until ∼48 h post transduction (Fig. 1E). Hence, as no carry over DamID signal from the resting state would be observed, Jurkat T cells were transduced with DamID constructs 24 h before antigen presentation so that measured changes reflect genome organization between 24 and 48 h into activation (Fig. 1A). Bacterial deoxyadenosine methylase (Dam)–Lamin B1 fusions were expressed to uniquely methylate DNA proximal to the lamin polymer (Vogel et al. 2007). To control for local variation in chromatin accessibility, parallel experiments expressed Dam methylase alone (Dam-only). The methylated DNA was then isolated and identified by sequencing.
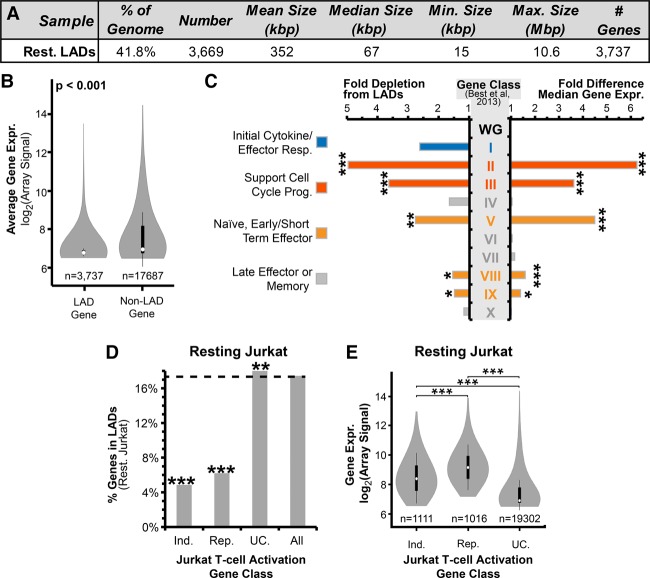
Log2(Lamin B1-Dam/Dam-only) ratios were generated and used to identify LADs in resting and activated Jurkat T cells based on a circular binary segmentation algorithm slightly modified from that previously described (see Methods) (Harr et al. 2015). LADs were mapped onto the hg19 assembly because hg19 was used for earlier data sets used for comparison, but all regions examined by fluorescence in situ hybridization (FISH; see below) were confirmed to be unaltered in the newer hg38 assembly. Interestingly, the magnitude of changes was similar between this rapid activation system and previous differentiation DamID studies (Peric-Hupkes et al. 2010; Robson et al. 2016), with 96% of LAD coverage shared between resting and activated cells and the remaining 4% roughly equally distributed between lost and newly formed LADs (Figs. 2A, 3A). The 18.6% of genes found within LADs were less transcriptionally active than the 81.3% of genes in non-LADs (Fig. 2B), consistent with the periphery being repressive (Pickersgill et al. 2006; Pindyurin et al. 2007). Similar gene expression differences between LADs and non-LADs were observed when comparing the Jurkat DamID data with published expression data from primary CD4+ cells (Kundaje et al. 2015; Supplemental Fig. S2A). Hence, the immortalized T-cell lymphoblastic leukemia Jurkat and naïve CD4+ T cells display a broad correspondence between their gene expression state and lamina association.
Figure 2.
Frequency of lamina association relates to gene expression and function. (A) Summary of resting Jurkat T-cell LAD parameters. (B) Violin plot comparing the microarray-determined signal between genes within LADs and non-LADs. (C) Fold differences in gene expression and frequency of lamina association in resting Jurkat T cells for 10 published categories of genes displaying distinct transcriptional behaviors during T-cell activation (Supplemental Fig. S2; Best et al. 2013). Fold differences were calculated relative to the whole genome (WG) in resting Jurkat T cells. Higher expressed genes tend to be depleted from LADs, such as categories associated with naïve cells, early effector function, and cell division. Lamina association correlated more with late effector/memory functional classes. (D) Percentage of genes in resting Jurkat LADs from different expression categories: genes induced (Ind.) or repressed (Rep.) at least 1.4-fold during T-cell activation, unchanged genes (UC.), and all genes. Both induced and repressed genes are three to four times less frequently found in LADs compared with unchanging genes. (E) Violin plot for expression levels of induced, repressed, and unchanging genes. Both induced and repressed categories are higher expressed than the unchanging genes in resting Jurkat T cells. (B,C,E) Significance of differential gene expression was determined by a Dunn test following a significant Kruskal-Wallis test. (C,D) Lamina-association frequency statistical significance was determined by Fisher's exact tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. See also Supplemental Figures S2 and S3 and Supplemental Table S1.
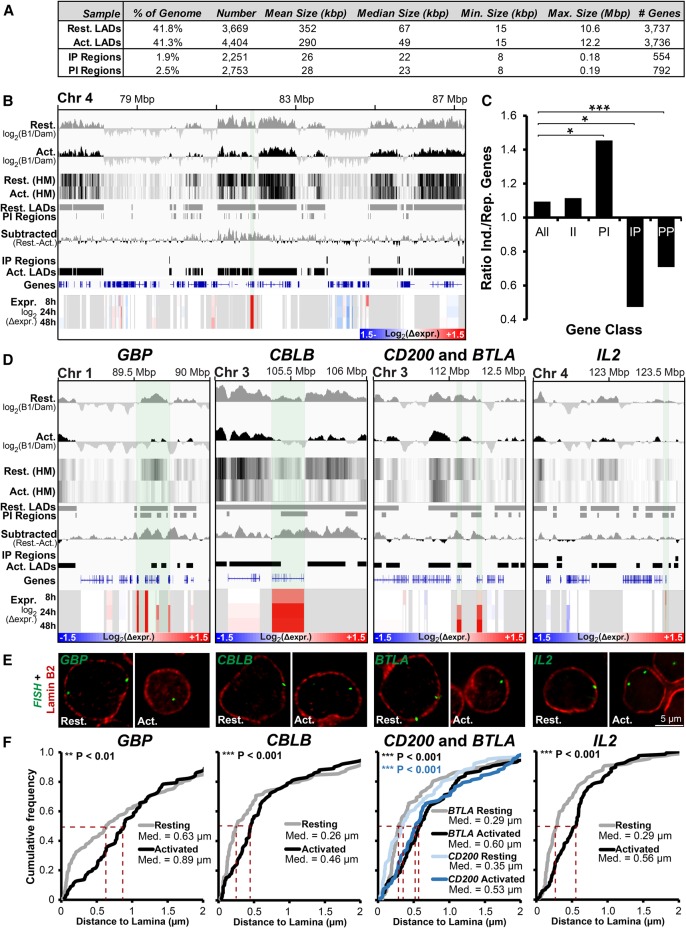
Figure 3.
Important genes are released from the periphery during T-cell activation. (A) Summary of LADs, IP (interior-to-periphery), and PI (periphery-to-interior) regions between resting and activated Jurkat T cells. (B) Genomic region surrounding ANTRX2 showing DamID signal bar plots and heatmaps (HM), LADs, IP and PI regions, subtracted resting-activated DamID signal, and expression changes in Jurkat T cells during activation from the resting state at 8, 24, and 48 h. The key for log2(Δexpr.) is given in the bottom right corner. ANTRX2 locus is highlighted by light shading. (C) Ratio of genes induced (Ind.) and repressed (Rep.) during T-cell activation for the whole genome (All), PI, IP, II (genes in the interior in both cell states), and PP (genes peripheral in both states) regions. Statistical significance was determined by Fisher's exact tests. (*) P < 0.05; (***) P < 0.001. (D) Genome browser views for the genomic region surrounding the GBP gene cluster and CBLB, CD200, BTLA, and IL2 loci covering the same parameters as in B. (E,F) Representative micrographs and quantification of gene positions in resting and activated cells relative to lamin B2. Median points are marked with dashed red lines. For quantification statistics, the position of loci in the activated sample was compared with the resting sample by KS tests. (**) P < 0.01; (***) P < 0.001. Distance measurements were performed on a minimum of 50 nuclei per sample. See also Supplemental Figure S4.
Gene expression and function reflects lamina-association frequency
On a genome-wide scale, lamina-association frequency relates to gene expression levels (Fig. 2B; Guelen et al. 2008). However, as LADs are enriched in developmentally regulated loci (Peric-Hupkes et al. 2010), we tested if this genome-wide relationship applied to functional gene classes. Ten classes of genes (Supplemental Fig. S2B) that displayed distinct transcriptional dynamics in a published CD8+ T-cell activation time-course (Best et al. 2013) were analyzed for lamina-association frequency compared with their expression in resting Jurkat T cells (Fig. 2C). This revealed that five gene classes involved in early stages of T-cell activation, such as cell cycle progression and early/short-term effector function, were significantly depleted from LADs relative to the whole genome while displaying increased gene expression. In contrast, four gene classes not needed until later in T-cell responses, such as late effector and memory T-cell function, were found in LADs as frequently as the whole genome and were mostly transcriptionally repressed. These relationships are largely maintained when instead considering published expression data from naïve CD4+ T cells (Supplemental Fig.S2C; Ni et al. 2016). However, two of the five gene classes depleted from LADs and expressed in resting Jurkat T cells—active cell division and short-term effector functions—were not expressed in naïve CD4+ T cells (Supplemental Fig. S2C). This likely reflects Jurkat T-cell origins as a constitutively dividing cancer cell line in contrast to the strong suppression of cell division found in primary resting naïve T cells. Also, one class associated with naïve memory precursor function displayed elevated transcriptional activity in the naïve CD4+ T cells, which was absent in resting Jurkat T cells.
All genes displaying induction and repression in Jurkat T-cell activation were similarly analyzed. As expected, genes becoming repressed during T-cell activation are active and depleted from LADs in resting cells, but surprisingly, many induced genes were similarly depleted from LADs (Fig. 2D). However, although displaying elevated expression upon Jurkat T-cell activation, the induced genes also displayed elevated basal expression in the resting state relative to the whole genome (Fig. 2E), and this was recapitulated with naïve CD4+ T cells (Supplemental Fig. S2D). Interestingly, a similar depletion from LADs of genes induced or repressed during myogenesis was also observed in undifferentiated myoblasts prior to the onset of myogenic differentiation (Supplemental Fig. S3A,B; Robson et al. 2016). Hence, LAD depletion invariably corresponds to increased gene expression for all gene classes tested. Moreover, such elevated basal expression may permit genes that are required in subsequent cell types or cell states, such as myotubes or activated T cells, to be maintained away from LADs.
Important T-cell genes are released from the periphery and induced during T-cell activation
We next sought to determine how gain or loss of LADs during T-cell activation correlates with gene expression changes. To ensure repositioning events to or from the periphery were identified with high confidence, we employed a stringent binary LAD or non-LAD definition. Whereas ∼40% of the genome was in LADs in both resting and activated Jurkat T cells, only 1.9% of the genome entered LADs in activated versus resting cells (interior-to-periphery regions [IP]) constituting 2251 regions containing 554 genes. The opposite tendency (periphery-to-interior [PI]) was observed for 2.5% of the genome in 2753 regions containing 792 genes (Fig. 3A). The size parameters of all LADs and differential IP/PI regions are summarized in Figure 3A. An example of log2(B1/Dam-only) signal intensity PI shifts is shown in Figure 3B. Consistent with peripheral heterochromatin dissipation, decreased associations with the nuclear periphery dominated over increased associations during T-cell activation.
Accordingly, released PI regions contained significantly more induced than repressed genes (Fig. 3C). In the opposite direction, IP regions favored repression (Fig. 3C). Loci remaining at the periphery (PP) similarly favored repression. Nonetheless, the majority of genes moving to or from the nuclear periphery displayed no change in expression (Supplemental Fig. S3D). The trend toward the periphery being repressive was also observed in a recent myogenic study (Robson et al. 2016), but the association between repositioning and altered gene expression was stronger than for Jurkat activation (Supplemental Fig. S3C–E). DamID and microarray data for all genes is in Supplemental Table S1.
The subset of genes undergoing both repositioning and expression changes tended to be important for T-cell regulation based on the aforementioned lymphocyte activation functional classes and GO terms. To confirm their repositioning inferred from DamID, five genes were directly tested by FISH: guanylate binding protein (GBP) gene cluster, interleukin 2 (IL2), Cbl proto-oncogene B (CBLB), CD200 molecule (CD200), and B- and T-lymphocyte associated (BTLA). These were chosen for their being induced throughout the Jurkat T-cell activation time-course (Fig. 3D, Expr.) and presence in PI regions (Fig. 3D, log2(B1/Dam) and PI Regions). GBP1 regulates the cytoskeleton during T-cell activation (Sharma et al. 2011), while IL2 is a potent inducer of T cells (Boyman and Sprent 2012). The other three genes regulate or attenuate immune responses, encoding, respectively, a ubiquitin ligase, receptor, and inhibitory ligand (Watanabe et al. 2003; Rygiel et al. 2009; Wallner et al. 2012). The absolute distance between the peak fluorescence intensity of the gene and costained Lamin B2 was measured in the mid-plane from deconvolved wide-field images and plotted as a cumulative frequency plot. FISH confirmed activation-induced release for all genes tested with differences statistically significant by Kolmogorov-Smirnov test (Fig. 3E,F). In contrast, an activated PP gene important for T-cell vascular endothelium associations, vascular cell adhesion molecule 1 (VCAM1), was not repositioned following T-cell activation (Supplemental Fig. S4A,B), confirming that repositioning from the periphery is not whole-scale. As an additional control, peripheral distance measurements for IL2RA (also known as CD25), encoding the high-affinity IL2 receptor subunit alpha that was in a non-LAD in both resting and activated cells, confirmed the DamID data (Supplemental Fig. S4C,D). The absence of a change in measured distance from the periphery for IL2RA also serves as a control that the increases measured for other loci represent bona fide gene repositioning rather than reflecting a global increase to nuclear volume.
Peripherally sequestered enhancers are released from the periphery upon their induction
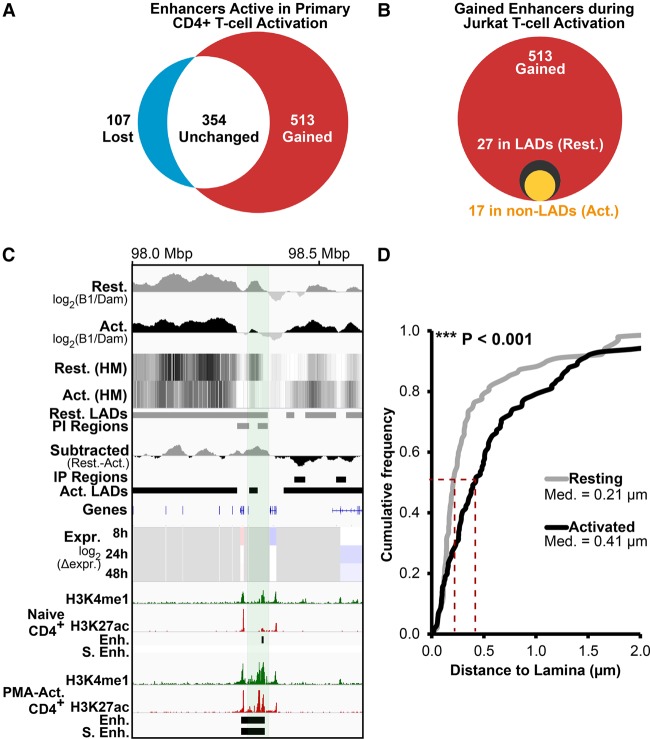
Enhancers, cis-regulatory elements that activate target genes by physical proximity, become transcribed prior to their functional activation (Arner et al. 2015). We predicted that enhancers in LADs would, like genes, reposition away from the periphery to be activated. To test this, a list of enhancer clusters/super-enhancers identified as functioning during human T-cell activation by H3K27ac accumulation (Hnisz et al. 2013) were compared to Jurkat activation LADs. Roughly half of enhancer clusters were induced only in activated T cells (Fig. 4A). Of these, only 27 occurred in LADs in the resting Jurkat T cells, but over half of these were released from the periphery in the activated Jurkat T cells (Fig. 4B).
Figure 4.
Enhancers are released from the periphery during T-cell activation. (A) Upon stimulation of naïve CD4+ T cells with PMA, 513 out of 974 putative enhancer clusters identified by Hnisz et al. (2013) are induced (red), while only 107 are silenced (blue). (B) Of the 513 induced enhancer clusters, 27 were in LADs in resting Jurkat T cells (black circle). Of these, 17 become released upon T-cell activation (yellow circle). (C) A 66-kbp Chromosome 3 region containing a putative enhancer induced during T-cell activation releases from the periphery (PI) in activated Jurkat T cells. In addition to Jurkat activation DamID and gene expression tracks, ChIP-seq tracks from primary naïve and PMA-activated CD4+ T cells for H3K4me1 and H3K27ac histone modifications marking active enhancers are shown (data from Bernstein et al. 2010). The H3K27ac peaks defined as enhancers (Enh.) and super enhancers (S. Enh.) by Hnisz et al. (2013) are also displayed as tracks. (D) Quantification of putative enhancer position in resting and activated Jurkat T cells relative to Lamin B2, confirming its activation-induced release. For quantification statistics, the position of loci in the activated sample was compared to the resting sample by KS tests. (***) P < 0.001. Distance measurements were performed on a minimum of 50 nuclei per sample.
FISH confirmed the PI repositioning upon activation of an enhancer cluster devoid of genes that accumulated H3K27ac and H3K4me1 upon activation of human CD4+ T cells (Fig. 4C,D; Bernstein et al. 2010). Hence, similar to genes, peripherally positioned enhancers are released concomitantly with their activation and acquisition of H3K27ac during T-cell activation.
TADs form the unit of gene repositioning
LADs are reported to significantly overlap with TADs, chromosome regions displaying preferential local interactions (Dixon et al. 2012; Fraser et al. 2015; Kind et al. 2015; Jabbari and Bernardi 2017). As TADs are largely invariant between cell types (Rao et al. 2014; Dixon et al. 2015; Fraser et al. 2015), we hypothesized that regions with altered peripheral localization would be delimited to single TADs rather than spanning multiple TADs.
To investigate this, we contrasted Jurkat activation DamID maps to the exact CD coordinates from a published GM12878 lymphoblastoid cell Hi-C data set (Rao et al. 2014). Global analysis of their CD and our LAD distributions revealed that both resting and activated Jurkat LADs are frequently entirely contained within CDs (Supplemental Fig. S5A). Five hundred randomly selected LADs were tested for their complete envelopment within CDs. As a control, the frequency of complete envelopment was measured for the same 500 LADs after random shuffling along their chromosomes with nonmappable regions (e.g., centromeres) excluded from the shuffling. This process was repeated over 1000 iterations to ensure all LADs in the genome were analyzed multiple times and frequencies of LAD envelopment within CDs plotted (Supplemental Fig. S5). All LADs occurred in their entirety within CDs significantly more frequently than for shuffled genome iterations. No statistically significant difference was observed between resting and activated LADs. We infer from this that a CD overlapping with a LAD repositions as a unit. A similar analysis using the GenometriCorr package, a tool that performs multiple analyses testing the spatial independence of domains along chromosomes (Favorov et al. 2012), confirmed this correspondence between CDs and LADs which displayed a uniform distance separation and an increased overlap frequency compared to chance (Supplemental Fig. S5B).
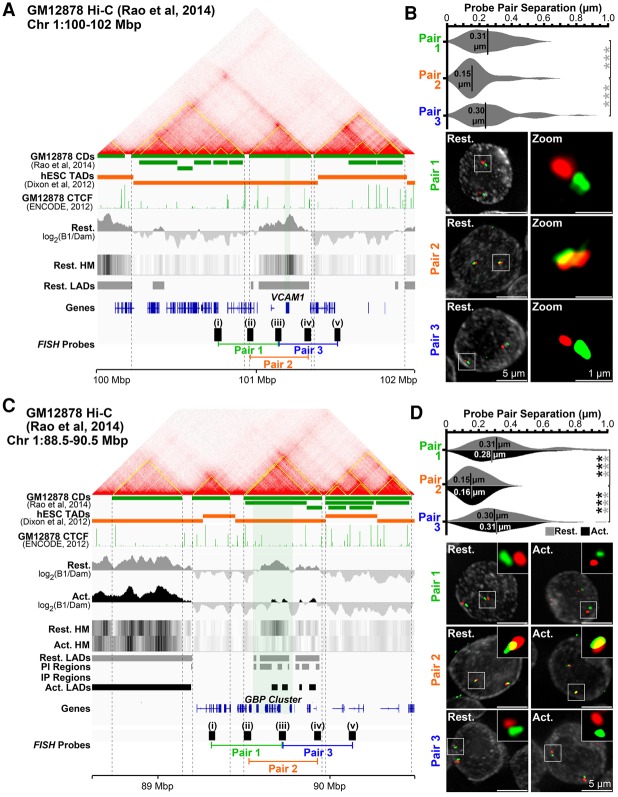
To determine if CD structure is similar in GM12878 and Jurkat cells, we performed pairwise FISH measurements of five fosmid probes separated by 149 kbp from each other in series over the VCAM1 locus (Fig. 5A). Probes i and v were in neighboring CDs upstream of and downstream from VCAM1. Probes ii and iv were within the VCAM1 CD boundary but close to the edges; probe iii, in the center. The distance between different equally spaced probe pairs, two crossing a CD boundary (pair 1: i+iii; pair 3: iii+v) and one within the same CD (pair 2: ii+iv), was determined in 2D from max projection deconvolved images. Supporting similar GM12878 and resting Jurkat T-cell CD structure, probes within the same CD were significantly closer (0.15 µm) than probes between adjacent CDs (∼0.30 µm) (Fig. 5B).
Figure 5.
The unit of repositioning is the CD. (A) Genomic region surrounding VCAM1 showing Hi-C data, associated GM12878 lymphoblastoid cell CDs (yellow lines on contact frequency heatmap and green track) (from Rao et al. 2014), and human embryonic stem cell (hESC) TADs (orange track) (from Dixon et al. 2012). Published ChIP-seq CTCF GM12878 cell data (The ENCODE Project Consortium 2012), DamID signals, LADs, and probes and probe pairings tested by FISH are also shown. (B) Quantification and representative max projection images measuring distance between pairs of ∼150-kbp spaced FISH probes. Pairs 1 (i-iii) and 3 (iii-v), containing probe pairings from different CDs, are separated significantly further than pair 2 (ii-iv), a pairing of probes from the same CD. (C) Genomic region view of the GBP gene cluster and probes tested. (D) Similar analysis as B for pairs of ∼175-kbp spaced FISH probes at the GBP locus in resting and activated Jurkat T cells. Probe pairs crossing CDs are similarly separated more than those within the same CD, and these separations remain unchanged during T-cell activation, indicating CD structure is maintained. Quantification statistics were determined by comparing the separation of pairs 1 and 3, with pair 2 in resting (gray) and activated (black) cells by KS tests. (***) P < 0.001. Distance measurements were performed on a minimum of 50 nuclei per sample. See also Supplemental Figure S5.
To test whether a CD is the unit of repositioning or whether its structure changes during lymphocyte activation-induced gene repositioning, FISH was performed on resting and activated Jurkat T cells using a similarly generated set of five probes (except 174 kbp apart) covering the GBP gene cluster (Fig. 5C). Again, the distance was significantly reduced for probe pairs within the same CD versus between adjacent CDs, and this organization was maintained in both resting and activated states (Fig. 5D). Hence, TADs form the unit of radial repositioning within which LADs are frequently subsumed.
Changing LADs correlate with specific chromosome compartments
Compartments represent interactions between regions of chromosomes sharing similar functional states (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Nora et al. 2012; Rao et al. 2014). Transcriptionally inactive B compartments can be further subdivided into B1, enriched in Polycomb H3K27me3, B2 and B3 correlating with LADs (Lieberman-Aiden et al. 2009; Gibcus and Dekker 2013; Rao et al. 2014), and B4 subcompartments (not considered as they are restricted to Chromosome 19). The A subcompartments are similar to one another in having active chromatin, but A2 displays increased H3K9me3 (Rao et al. 2014). As we now have mapped regions specifically changing at the periphery between resting and activated Jurkat T cells, we sought to determine if Jurkat LADs and IP/PI regions correlated with any particular GM12878 subcompartment.
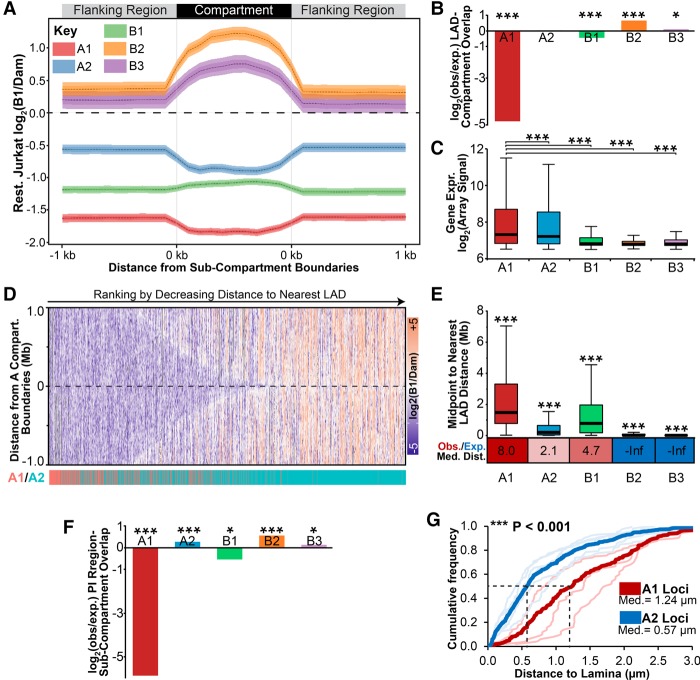
First, we extracted the Rao et al. (2014) subcompartments and separately the 1 kb upstream and downstream flanking DNA. As compartment size varies, individual compartment regions were plotted with lengths equalized. The average log2(Lamin B1/Dam-only) ratio from resting Jurkat T cells was then plotted over these size-normalized compartment regions and their 1-kb sequences (Fig. 6A). As anticipated, in resting Jurkat T cells, the B2 and B3 subcompartments exhibited a peak of log2(Lamin B1/Dam-only) signal relative to flanking regions, while the A1 and A2 subcompartments exhibited depletion. In contrast, Polycomb-associated B1 subcompartments exhibited only a modest increase in log2(Lamin B1/Dam-only) signal. Comparing these data to a genome with randomly shuffled lamina association, LADs are significantly enriched in B2 and B3 compartments and depleted in B1 and A1 subcompartments (Fig. 6B). Moreover, correlating with the degree of diminished lamina association, A1 subcompartments were the most highly expressed followed by the A2, B1, and B2/B3 subcompartments (Fig. 6C).
Figure 6.
Compartments are distributed nonrandomly with respect to LADs. (A) Plot displaying the average log2(Lamin B1/Dam-only) signal from resting Jurkat T cells across all size-normalized A1, A2, B1, B2, or B3 GM12878 subcompartment regions and 1 kbp of flanking DNA. Compartment data from Rao et al. (2014). (B) Fold enrichment analysis of LADs observed (obs) in each compartment compared with that expected in a genome with randomly shuffled LADs (exp). (C) Box plot displaying the expression distribution in resting Jurkat T cells for each GM12878 subcompartment. (D) Heatmap of DamID log2(laminB1/Dam) signal across ±1 Mbp surrounding A subcompartment boundaries. Regions are ranked to the right by decreasing distance to nearest LADs, and their identity as an A1 or A2 subcompartment region is highlighted below in red or blue, respectively. A2 domains are observed more frequently closer to LADs than are A1. (E) Box plot displaying the distribution of chromosomal distances between the midpoint of subcompartment regions and the nearest LAD in resting Jurkat T cells. Below is the ratio between observed (Obs.) median distance and that expected (Exp.) in a randomly LAD-shuffled genome. Values are color-coded to show increased (red) or decreased (blue) observed distance compared with that expected. (F) Fold enrichment analysis of PI regions observed in each compartment compared with that expected in a genome with randomly shuffled PI regions. (G) Quantification of average distance between A1 (thick red line) or A2 loci (thick blue line) and the lamina determined by FISH for activated Jurkat T cells where PI loci are detached from the lamina. Lighter lines designate individual A2 (GBP, NFKBIZ, CBLB, CD200, BTLA) and A1 (IL2RA, CCND2, TNF) loci. Distance measurements were performed on a minimum of 50 nuclei per sample. (B,F) Significance was determined by Fisher's exact tests; (C,E) significance was by Dunn test for multiple comparison testing after a significant Kruskal-Wallis test. (G) Significance of the difference between A1 and A2 distances to lamina was determined by KS tests. (*) P < 0.05; (***) P < 0.001. See also Supplemental Figure S6.
Interestingly, the absolute DamID signal was much higher in the regions flanking the A2 subcompartment than the regions flanking B1 and A1 subcompartments (Fig. 6A). This striking difference in flanking lamina association suggested that these compartments are spatially segregated with respect to the nuclear periphery. The log2(Lamin B1/Dam-only) signal within 1 Mbp of all A subcompartment region boundaries was plotted as a heatmap, revealing that A1 subcompartments tended to be further from LADs than A2 subcompartments (Fig. 6D). To quantitate this, we plotted the distances from the midpoint of subcompartment regions to the nearest LAD (Fig. 6E). This revealed, as expected, that B2 and B3 subcompartments correspond to LADs (median distance, 0 bp to the nearest LAD). The A2 subcompartments were next closest to LADs (median distance, 193 kbp), while B1 and A1 subcompartments were significantly further away (median distances, respectively, 0.767 and 1.468 Mbp). Supporting the specificity of these results, A1, A2, and B1 subcompartments all were further from LADs than expected in a genome with randomly shuffled LADs (Fig. 6E, Obs./Exp.). Similar statistically significant results were also obtained by GenometriCorr analysis (Supplemental Fig. S6A). Therefore, we propose, due to differences in their proximity to LADs along chromosomes, that A1 and A2 subcompartments represent radially segregated active chromatin with respect to the periphery.
It follows that a gene being released from the periphery during lymphocyte activation would be more likely to enter an A2 than an A1 subcompartment. Unfortunately, the GM12878 Hi-C data did not follow activation, nor was the activation state of their cells established. However, comparing the Jurkat activation DamID data with the GM12878 compartment maps revealed that many of the genes that were released from the periphery upon activation appeared in the A2 compartment. PI genes also displayed elevated expression in GM12878 cells relative to resting Jurkat T cells (Supplemental Fig. S6B). Therefore, we presume that the GM12878 lymphoblastoid cell data reflect a state more similar to that of an activated than resting T cell. Thus, the presence of PI genes in LADs in the resting Jurkat T cells likely indicates that they are in a B2/B3 subcompartment until T-cell activation, when they move to the A2 subcompartment. Indeed, we observe that PI regions are significantly enriched in the A2 subcompartment in GM12878 cells (Fig. 6F). Similar results were observed using GenometriCorr analysis, except that the B1 subcompartment was also enriched in PI regions (Supplemental Fig. S6C). Compellingly, the radial segregation of A1 and A2 subcompartments was directly confirmed for several loci tested (Fig. 6G).
Release of a predicted T-cell activation–specific enhancer from the nuclear periphery enables its association with similarly released genes in the interior
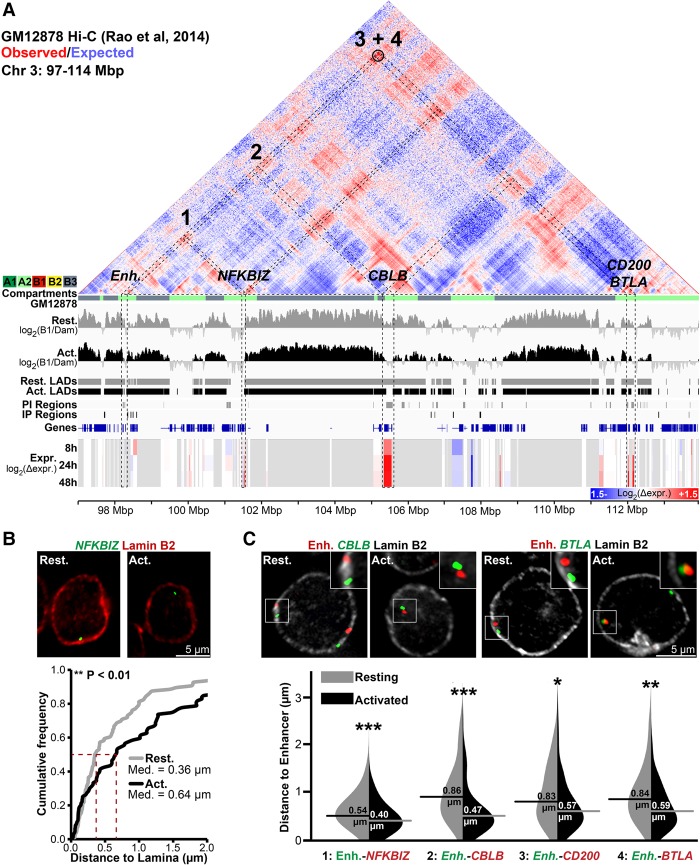
The Jurkat activation system further enabled direct testing of this repositioning between compartments by FISH. Loci released from different B compartment LADs to an A2 compartment during T-cell activation would be predicted to display increased proximity in stimulated T cells. Therefore, a 16-Mbp region of Chromosome 3 was selected where several Jurkat activation PI genes and a predicted PI enhancer corresponded to an A2 compartment identified in GM12878 cells (Fig. 7A). Several of these genes were tested in Figure 3 to confirm release from LADs during Jurkat T-cell activation (CBLB, BTLA, and CD200). We additionally tested NFKBIZ, which was induced during T-cell activation and in a GM12878 A2 subcompartment, but only proximal to and not overlapping a PI region. This locus was close to the periphery in resting cells and became more internal during activation, indicating that genomic context is important to consider in addition to direct lamina association itself (Fig. 7B).
Figure 7.
Genes released from LADs during T-cell activation associate in the interior in activated Jurkat T cells. (A) Genomic view of a 17-Mbp region of Chromosome 3 displaying GM12878 cell Hi-C interactions from published contact maps (Rao et al. 2014). Hi-C data are displayed as a heatmap of the ratio of observed versus expected contact frequencies in a distance-scaled model. GM12878 subcompartments (A1, A2, B1, B2, B3) are color-coded directly beneath the map. Tracks are displayed as in previous figures. The black circle represents a highly significant DNA–DNA contact detected by Rao et al. (2014). Several genes—NFKBIZ, CBLB, CD200, and BTLA—and a T-cell activation–specific enhancer (Enh.) display reduced lamina association during T-cell activation concomitant with transcriptional induction and are present in the GM12878 A2 subcompartment. (B) Representative micrographs and quantification NFKBIZ loci positions in resting and activated cells relative to Lamin B2. (C) Representative micrographs and violin plot quantification of distances between the enhancer and indicated genes in resting and activated Jurkat T cells. All enhancer-gene FISH probe combinations display reduced inter-probe distance following activation. For quantification statistics, relative distance measurements between loci or between loci and Lamin B2 were compared between the activated and resting cells by KS tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Distance measurements were performed on a minimum of 50 nuclei per sample. See also Supplemental Figure S7.
All these genes were compared to the PI locus containing the predicted enhancer described in Figure 4, C and D, that occurs in a GM12878 A2 subcompartment. FISH performed on combinations of the putative enhancer probe with the individual gene probes in resting and activated cells revealed that the distance between the putative enhancer and NFKBIZ, CBLB, CD200, and BTLA was much greater when the loci were at the periphery in resting cells than after their release upon activation (Fig. 7C). This is consistent with their being released from B2/B3 subcompartments in resting Jurkat T cells to associate in an A2 subcompartment. In contrast, the GBP cluster and VCAM1 are similarly spaced along Chromosome 1 (11 Mbp apart) and are both in LADs in resting cells; however, only GBP gets released upon Jurkat T-cell activation. Accordingly, no significant change in spatial distance was observed upon activation (Supplemental Fig. S7), and the loci display no GM12878 Hi-C interactions. Likewise, IL2 and IL2RA, located on separate chromosomes, did not exhibit any significant change in distance (Supplemental Fig. S7B,C). Thus, multiple loci released and activated following T-cell activation display increased spatial proximity once in the nuclear interior.
Lamina detachment and increase enhancer gene proximity also occurs in primary T-cell activation
The DamID was necessarily performed in the Jurkat model T-cell activation system due to poor transduction efficiencies of primary lymphocytes and the possibility of activation during lentiviral treatment. Thus, activation state differences for the small subset of activation gene classes (Fig. 2) necessitated validation of these findings in primary human lymphocytes.
Naïve CD4+ T cells were isolated from fresh human peripheral blood mononuclear cells by polychromatic FACS (Supplemental Fig. S8). Some were directly fixed for FISH, and some were activated with phorbol 12-myristate 13-acetate (PMA) for 24 h. PMA was chosen for activation to better match results to the published ChIP-seq study on naïve CD4+ T cells (Bernstein et al. 2010). The activated population was split for FISH and activation level determined by CD69+ FACS (78%) (Fig. 8A). The same probe pairs used in Figure 7 were used for FISH. In all cases, the predicted enhancer became closer to the gene locus upon activation, and this was significant for all except NFKBIZ (Fig. 8B). Also, as expected, the predicted enhancer, NFKBIZ, CBLB, CD200, and BTLA were all at the periphery in the resting primary lymphocytes, all moved away upon activation, and repositioning was highly significant (Fig. 8C). Thus, Jurkat T cells accurately reflect genome organization changes that occur upon human lymphocyte activation at least for the genes tested.
Figure 8.
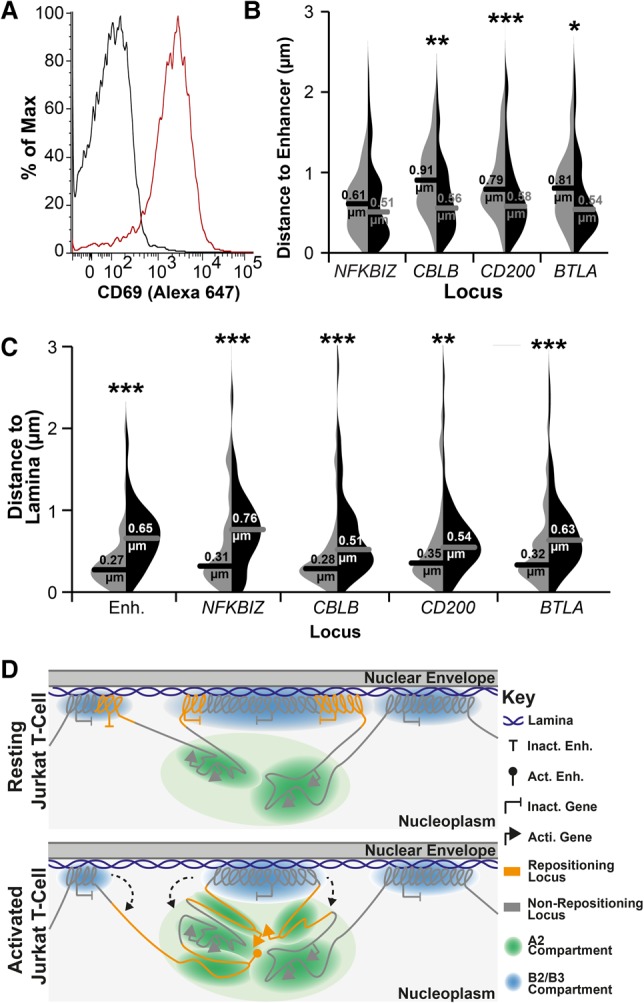
LAD release and association of genes in the nuclear interior also occurs in primary CD4+ T cells. (A) FACS plot of CD69 staining for naïve (resting; black line) and PMA-treated (activated; red line) CD4+ T cells, indicating 78% activation. For details of the naïve population, see also Supplemental Figure S8. (B) Violin plot quantification of distances between the predicted enhancer and indicated genes in naïve (light gray) and PMA-treated (black) cells. All loci displayed closer proximity to the predicted enhancer locus upon PMA activation of naïve CD4+ T cells. (C) Violin plot quantification of distances between indicated loci and Lamin B2 in naïve (light gray) and PMA-treated (black) cells. All loci displayed increased distance from the periphery upon PMA activation. For quantification statistics, relative distance measurements between loci or between loci and Lamin B2 were compared between the activated and resting cells by KS tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Distance measurements were performed on a minimum of 50 nuclei per sample. (D) Model of the relationship between LADs, compartments, and gene release. In resting T cells, B2 and B3 subcompartments exist as LADs, while A2 subcompartment regions remain near to the lamina due to their LAD proximity along the chromosome. In contrast, the A1 and B1 compartments, being further from LADs along chromosomes, are distributed more distally in the nuclear interior (Fig. 6). Following T-cell activation, regions of LADs containing necessary gene or enhancer loci are released from the periphery, where they associate in the A2 compartment proximal to the lamina in a transcriptionally active state.
Discussion
Genes subject to spatial regulation are important for T-cell activation
Nuclear envelope–regulated genes in a myogenesis study were often required early in differentiation but were inhibitory at later stages and so needed to be tightly regulated (Robson et al. 2016). Similarly, here, though first responder genes that initiate activation were absent from LADs, those subject to spatial regulation were important for subsequent T-cell function and regulation. For example, IL2 encodes Interleukin 2, which functions in propagating an immune response (Boyman and Sprent 2012), while the GBP1 product regulates the cytoskeleton to influence T-cell signaling (Sharma et al. 2011). If these activities were not tightly shut down in resting cells, they could potentially activate an immune response in the absence of infection. Others function in modulating the activation response. For example, induction of the nuclear IκB family member encoded by NFKBIZ is required for the subsequent expression of IL17 following T-cell stimulation (Okamoto et al. 2010). Similarly, receptor-ligand products of BTLA and CD200 and the CBLB-encoded ubiquitin ligase act to attenuate T-cell activation and thus fine-tune the immune response (Watanabe et al. 2003; Rygiel et al. 2009; Wallner et al. 2012). Consistent with these important regulatory activities, the dysregulation or loss of function of a number of these peripherally regulated gene products is associated with autoimmunity (Watanabe et al. 2003; Rosenblum et al. 2004; Sharma et al. 2011; Wallner et al. 2012; Johansen et al. 2015; Hussman et al. 2016). Thus, in all cases, the tighter regulation of these genes in resting lymphocytes from adding spatial control would benefit the organism. Similarly, regulating cis-regulatory elements such as enhancers is likely beneficial due to their ability to act on multiple target genes (Symmons and Spitz 2013). Importantly, the proximity of the A2 subcompartment into which they are released from the periphery would potentially enable regulation also in the opposite direction.
The relationship between gene activity and lamina association
Specific gene repositioning to and from the periphery occurs during lymphocyte activation, but what controls tethering or release remains unclear. We find that frequency of lamina association inversely relates to gene expression levels, consistent with previous studies (Guelen et al. 2008; Peric-Hupkes et al. 2010; Akhtar et al. 2013; Robson et al. 2016). It has been suggested that transcriptional activation drives gene release from the periphery (Tumbar et al. 1999; Tumbar and Belmont 2001; Chuang et al. 2006; Therizols et al. 2014), and the same could apply for enhancers that are frequently transcribed during induction (Arner et al. 2015). However, the relationship between gene position and expression is not absolute as we and others found the activity of most repositioning genes remains unaffected (Peric-Hupkes et al. 2010; Robson et al. 2016).
Depletion of repressive chromatin modifications enriched in LADs such as H3K9me2/3 results in disrupted lamina association (Zullo et al. 2012; Bian et al. 2013; Demmerle et al. 2013; Kind et al. 2013; Harr et al. 2015), suggesting that the marks not only provide for silencing but also themselves contribute to locus affinity for the periphery. At the same time, specific nuclear envelope proteins are important for peripheral tethering of endogenous developmentally repositioning loci (Zullo et al. 2012; Solovei et al. 2013; Robson et al. 2016). Several studies argue that physical recruitment of a locus alone is insufficient for silencing but works together with transcriptional regulators and other silencing factors at the periphery (Finlan et al. 2008; Robson et al. 2016). Thus, tethering to the nuclear envelope is a function of the chromatin state together with the milieu of nuclear envelope proteins.
Lamina association provides a compass for interpreting compartment information
By contrasting Hi-C data with LADs, we now place lamina association and gene release within the context of higher-order chromatin structure. Chromosome compartments are defined based on what is close to each other, but this does not reveal location within the nucleus. Previous studies comparing Hi-C and DamID data sets revealed a tendency for B compartments to be enriched in LADs (Lieberman-Aiden et al. 2009; Gibcus and Dekker 2013; Rao et al. 2014); however, no spatial discrimination of active compartments has been described. Here we found that A1 and A2 subcompartments segregate with their proximity to LADs, with A2 subcompartments closer to the periphery. Thus, the physical anchoring of distal regions in LADs can determine relative spatial positions for the wider genome. It follows that DamID differences between different cell types and differentiation stages could have wider consequences for the regulation of genes in the nuclear interior.
Our data suggests T-cell activation PI genes are switching from a LAD-associated B subcompartment to a LAD-proximal A2 subcompartment. The restriction in distance from the periphery for released loci could contribute to explaining a central question in the spatial genome organization field: how compartments are established and how particular genes find one another within the three-dimensional space of the nucleus. In the analysis here, once released, loci remained relatively close to the periphery, generally <0.6 µm. This is an important limitation because modeling studies indicate that loci 10 Mbp apart are unlikely to meet in a single cell cycle by diffusion alone (Dekker and Mirny 2016). In contrast, a 0.5- to 0.8-µm space could be sampled by a locus in 1 h, well within the rapid response timeframe for lymphocyte activation and the distances we measured for released loci. This “constrained release” could thus increase the likelihood of incorporation of a released locus into an A2 subcompartment (Fig. 8D) while keeping it close enough to the periphery to enable its sequestration again once T-cell activation has abated. Indeed, the activities of released genes in regulating T-cell function suggest such tight control may be necessary. A second functional consequence of constrained release may be to promote proximity between enhancers and distal target genes while at the same time sequestering them away from other inappropriate targets. Intriguingly, the putative enhancer described herein displays a long distance Hi-C looping contact with BTLA in GM12878 cells suggestive of such a functional association (Fig. 7A; Rao et al. 2014); however, it remains untested if this is functionally significant. Nonetheless, data presented here clearly show that LADs are intrinsically associated with gene regulation and have significant physical and, possibly, functional consequences for genes located in the nuclear interior.
Methods
Cell culture, transduction, and immune conjugation
Jurkat clone E6-1 T cells and Raji B cells were cultured in RPMI 1640 supplemented with 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin. VSV-G pseudotyped lentiviruses encoding DamID, GFP-tagged, or pLKO constructs were generated and transduced as described in the Supplemental Methods. Raji B cells were incubated 30 min at 1 million cells/mL in media containing 0.5 µg/mL SEE (Toxin Technologies, EP404), followed by pelleting at 1000 rpm for 5 min, two PBS washes, and resuspension in growth medium. Conjugated Raji cells were mixed 1:10 with Jurkat T cells to a total density of 1 million cells/mL for times indicated.
Isolation and FACS of peripheral blood mononuclear cells
Buffy coats supplied by the Scottish National Blood Transfusion Service were processed with the approval of the Committee for Governance of Blood and Tissue Samples for Nontherapeutic Use. Human peripheral blood mononuclear cells (PBMCs) were isolated using Histopaque 1077 (Sigma) per the manufacturer's instructions. CD3+ CD4+ CD62L+ CD25− CD44− CD69− naïve CD4+ T cells were then incubated with selected antibodies and isolated from PMBCs using a four-laser FACS Aria IIu flow cytometer (Beckton Dickinson). For a more detailed description of primary cell FACs isolation, see the Supplemental Methods.
FISH
PBS-washed Jurkat or primary CD4+ T cells were pelleted, spotted on polylysine-coated coverslips, and fixed with 4% paraformaldehyde (PFA) and FISH performed as described previously (Zuleger et al. 2013). A detailed description of the FISH method and probes or antibodies used can be found in the Supplemental Methods. Distance measurements were performed using Imaris with costaining with a Lamin B2 antibody before FISH, allowing the distance between genes to be determined using Imaris. A minimum of 50 nuclei were analyzed for each experiment.
Microarrays
RNA from resting and activated Jurkat T cells was extracted with TRI-reagent (Invitrogen). The RNA was converted to biotin labeled-cRNA using the TotalPrep RNA amplification kit (Ambion, AMIL1791) following the manufacturer's instructions. For each experiment, three biological replicates were hybridized to Illumina whole-genome gene expression arrays (HumanHT-12 v4 BeadChip). The data were quantile normalized using R (R Core Team 2010) and the Bioconductor package Limma (Smyth 2005). Differentially expressed transcripts with a log2 ratio above 0.5 in absolute value were selected and adjusted for a false-discovery rate of 5% (Benjamini and Hochberg 1995).
DamID
DamID was performed as previously described (Vogel et al. 2007) and is described in further detail in the Supplemental Methods. Briefly, Resting Jurkat T cells were transduced with Dam-Lamin B1-encoding or Dam-only–encoding lentiviruses over 24 h. Transduced cells were then incubated with Raji B cells ± SEE for a further 48 h, after which genomic DNA was sequenced (BGI). Sequencing reads were mapped to human hg19 genome and the log2(Lamin B1/Dam) value determined for all genomic DpnI fragments in resting and activated cells, obtaining 2.2- and 2.1-fold genome coverage, respectively. LADs were determined using a peak finder function in the Bioconductor package DNAcopy (http://bioconductor.org/packages/DNAcopy/). DamID analysis results are derived from a single experiment.
Hi-C data visualization
The displayed processed contact frequency maps are taken directly from Rao et al. (2014) and displayed using the Juicebox Hi-C viewer (Rao et al. 2014). A description of all external data sets used can be found in Supplemental Methods.
Data access
The Jurkat T-cell DamID and gene expression data sets from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE94972.
Supplementary Material
Acknowledgments
We thank Lior Pytowski and Anastasia Sokolidi for assistance with taking images, Shelagh Boyle for assistance with probe labeling, Dr. David Kelly for microscopy support, and Louise Evenden at the Wellcome Trust Clinical Research Facility for microarray analysis. M.I.R. was supported by a Wellcome Trust PhD Studentship, and funding for this work was provided by Wellcome Trust grants 095209 to E.C.S. and 092076 for the Centre for Cell Biology.
Author contributions: M.I.R. and E.C.S. conceived of the study and designed experiments; M.I.R., R.C., and A.S. performed experiments; M.I.R., J.I.d.l.H., and A.R.W.K. analyzed data; and M.I.R. and E.C.S. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.212308.116.
Freely available online through the Genome Research Open Access option.
References
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LF, van Lohuizen M, van Steensel B. 2013. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927. [DOI] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. 2015. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep 16: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drablos F, Lennartsson A, Ronnerblad M, Hrydziuszko O, Vitezic M, et al. 2015. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347: 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 57: 289–300. [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, Dustin ML, Goldrath AW, Immunological Genome Project Consortium. 2013. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat Immunol 14: 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Khanna N, Alvikas J, Belmont AS. 2013. β-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol 203: 767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyman O, Sprent J. 2012. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol 12: 180–190. [DOI] [PubMed] [Google Scholar]
- Chandra T, Ewels PA, Schoenfelder S, Furlan-Magaril M, Wingett SW, Kirschner K, Thuret JY, Andrews S, Fraser P, Reik W. 2015. Global reorganization of the nuclear landscape in senescent cells. Cell Rep 10: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831. [DOI] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmerle J, Koch AJ, Holaska JM. 2013. Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate expression and nuclear positions of MyoD, Myf5, and Pax7 genes during myogenesis. Chromosome Res 21: 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drings P, Sonnemann E. 1974. [Phytohemagglutinin-induced increase of euchromatin contents in human lymphocytes]. Res Exp Med 164: 63–76. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favorov A, Mularoni L, Cope LM, Medvedeva Y, Mironov AA, Makeev VJ, Wheelan SJ. 2012. Exploring massive, genome scale datasets with the GenometriCorr package. PLoS Comput Biol 8: e1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schopflin R, Kraft K, Kempfer R, Jerkovic I, Chan WL, et al. 2016. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265–269. [DOI] [PubMed] [Google Scholar]
- Fraser J, Ferrai C, Chiariello AM, Schueler M, Rito T, Laudanno G, Barbieri M, Moore BL, Kraemer DC, Aitken S, et al. 2015. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol 11: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Mol Cell 49: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Granado JM, Silvestre-Roig C, Rocha-Perugini V, Trigueros-Motos L, Cibrian D, Morlino G, Blanco-Berrocal M, Osorio FG, Freije JM, Lopez-Otin C, et al. 2014. Nuclear envelope lamin-A couples actin dynamics with immunological synapse architecture and T cell activation. Sci Signal 7: ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. 2015. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 208: 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. 2004. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur J Immunol 34: 3604–3613. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R, Decsy MI, Troll W. 1971. The effect of PHA stimulation of human peripheral blood lymphocytes upon cellular content of euchromatin and heterochromatin. Cell Immunol 2: 696–701. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351: 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. 2012. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell 48: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussman JP, Beecham AH, Schmidt M, Martin ER, McCauley JL, Vance JM, Haines JL, Pericak-Vance MA. 2016. GWAS analysis implicates NF-κB-mediated induction of inflammatory T cells in multiple sclerosis. Genes Immun 17: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari K, Bernardi G. 2017. An isochore framework underlies chromatin architecture. PLoS One 12: e0168023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen C, Mose M, Ommen P, Bertelsen T, Vinter H, Hailfinger S, Lorscheid S, Schulze-Osthoff K, Iversen L. 2015. IκBζ is a key driver in the development of psoriasis. Proc Natl Acad Sci 112: E5825–E5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfali N, Wilkie GS, Swanson SK, Srsen V, Batrakou DG, Fairley EA, Malik P, Zuleger N, Goncharevich A, de Las Heras J, et al. 2010. The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture. Mol Cell Proteomics 9: 2571–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162. [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteifel VM, Andreichuk TN, Karu TI. 1992. A comparative study of chromatin from lymphocyte nuclei upon activation of transcription by irradiation from an He-Ne-laser or phytohemagglutinin. Mol Biol (Mosk) 26: 1054–1062. [PubMed] [Google Scholar]
- Mattout A, Pike BL, Towbin BD, Bank EM, Gonzalez-Sandoval A, Stadler MB, Meister P, Gruenbaum Y, Gasser SM. 2011. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr Biol 21: 1603–1614. [DOI] [PubMed] [Google Scholar]
- Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. 2013. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res 23: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, Dellefave L, Pytel P, Selig S, Labno CM, et al. 2010. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One 5: e14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Yang W, Han M, Zhang Y, Shen T, Nie H, Zhou Z, Dai Y, Yang Y, Liu P, et al. 2016. Global intron retention mediated gene regulation during CD4+ T cell activation. Nucleic Acids Res 44: 6817–6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, Ohya K, Jetten AM, Akira S, Muta T, et al. 2010. IκBζ regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature 464: 1381–1385. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014. [DOI] [PubMed] [Google Scholar]
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaeva ES, Christophides GK, Kafatos FC, van Steensel B, Zhimulev IF. 2007. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci 120: 2344–2351. [DOI] [PubMed] [Google Scholar]
- Pompidou A, Rousset S, Mace B, Michel P, Esnous D, Renard N. 1984. Chromatin structure and nucleic acid synthesis in human lymphocyte activation by phytohemagglutinin. Exp Cell Res 150: 213–225. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Gatzka M, Thomas PG, Ihle JN. 2011. Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J 30: 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson MI, de Las Heras JI, Czapiewski R, Le Thanh P, Booth DG, Kelly DA, Webb S, Kerr AR, Schirmer EC. 2016. Tissue-specific gene repositioning by muscle nuclear membrane proteins enhances repression of critical developmental genes during myogenesis. Mol Cell 62: 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum MD, Olasz EB, Yancey KB, Woodliff JE, Lazarova Z, Gerber KA, Truitt RL. 2004. Expression of CD200 on epithelial cells of the murine hair follicle: a role in tissue-specific immune tolerance? J Invest Dermatol 123: 880–887. [DOI] [PubMed] [Google Scholar]
- Rygiel TP, Rijkers ES, de Ruiter T, Stolte EH, van der Valk M, Rimmelzwaan GF, Boon L, van Loon AM, Coenjaerts FE, Hoek RM, et al. 2009. Lack of CD200 enhances pathological T cell responses during influenza infection. J Immunol 183: 1990–1996. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, Javierre BM, Nagano T, Katsman Y, Sakthidevi M, Wingett SW, et al. 2015. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res 25: 582–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Sharma R, Fu SM, Ju ST. 2011. IL-2: a two-faced master regulator of autoimmunity. J Autoimmun 36: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In Bioinformatics and computational biology solutions using R and Bioconductor (ed. Gentleman R, et al. ), pp. 397–420. Springer, New York. [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. [DOI] [PubMed] [Google Scholar]
- Symmons O, Spitz F. 2013. From remote enhancers to gene regulation: charting the genome's regulatory landscapes. Philos Trans R Soc Lond B Biol Sci 368: 20120358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. 2014. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346: 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Belmont AS. 2001. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol 3: 134–139. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145: 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B. 2007. Detection of in vivo protein–DNA interactions using DamID in mammalian cells. Nat Protoc 2: 1467–1478. [DOI] [PubMed] [Google Scholar]
- Wallner S, Gruber T, Baier G, Wolf D. 2012. Releasing the brake: targeting Cbl-b to enhance lymphocyte effector functions. Clin Dev Immunol 2012: 692639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, et al. 2003. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol 4: 670–679. [DOI] [PubMed] [Google Scholar]
- Zheng X, Kim Y, Zheng Y. 2015. Identification of lamin B–regulated chromatin regions based on chromatin landscapes. Mol Biol Cell 26: 2685–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N, Boyle S, Kelly DA, de Las Heras JI, Lazou V, Korfali N, Batrakou DG, Randles KN, Morris GE, Harrison DJ, et al. 2013. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol 14: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. 2012. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 149: 1474–1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.