Abstract
Long noncoding RNAs (lncRNAs) play a conserved role in regulating gene expression, chromatin dynamics, and cell differentiation. They serve as a platform for RNA interference (RNAi)–mediated heterochromatin formation or DNA methylation in many eukaryotic organisms. We found in Schizosaccharomyces pombe that heterochromatin is lost at transcribed regions in the absence of RNA degradation. We show that heterochromatic RNAs are retained on chromatin, form DNA:RNA hybrids, and need to be degraded by the Ccr4-Not complex or RNAi to maintain heterochromatic silencing. The Ccr4-Not complex is localized to chromatin independently of H3K9me and degrades chromatin-associated transcripts, which is required for transcriptional silencing. Overexpression of heterochromatic RNA, but not euchromatic RNA, leads to chromatin localization and loss of silencing of a distant ade6 reporter in wild-type cells. Our results demonstrate that chromatin-bound RNAs disrupt heterochromatin organization and need to be degraded in a process of heterochromatin formation.
Regulation of genome expression is required for many cellular processes, including cell proliferation, differentiation, development, and viability. Heterochromatin, or silent chromatin, is essential to maintain genome stability and chromosome segregation. In fission yeast, heterochromatin is established by small RNA pathways that act at the chromatin level (Bond and Baulcombe 2014; Holoch and Moazed 2015). Small RNAs are loaded on Argonaute and interact with target RNAs through base-pairing interactions. The Argonaute-containing RNA-induced transcriptional silencing (RITS) complex is guided by the small RNAs to heterochromatic repeats. To amplify small RNAs, RITS recruits the RNA-dependent RNA Polymerase complex (RDRC), which synthesizes double-stranded RNA that is processed by Dicer into siRNAs. In addition, RITS recruits the H3K9 methyltransferase complex CLRC to chromatin, which leads to repressive histone 3 lysine 9 methylation (H3K9me), recruitment of HP1 proteins, and heterochromatin formation (Volpe et al. 2002; Verdel et al. 2004; Allshire and Ekwall 2015; Holoch and Moazed 2015; Martienssen and Moazed 2015; Zocco et al. 2016).
In fission yeast, constitutive heterochromatin is found at centromeres, subtelomeres, and the mating type (mat) locus (Allshire 1995; Cam et al. 2005). At centromeric repeats, RNAi is needed for the establishment and maintenance of heterochromatin. The establishment of centromeric heterochromatin is initiated by a class of single-stranded small RNAs called priRNAs (Halic and Moazed 2010), which are generated by Argonaute and the PARN nuclease Triman (Marasovic et al. 2013). In cells lacking components of the RNAi pathway, heterochromatin is not lost at the mat locus and subtelomeres. At those loci, heterochromatin maintenance is RNAi independent, but RNAi is required to establish heterochromatin (Kanoh et al. 2005; Hansen et al. 2006; Allshire and Ekwall 2015). At subtelomeres, the Shelterin complex binds telomeres and recruits the CLRC methyltransferase and the SHREC deacetylase complexes to maintain heterochromatin (Kanoh et al. 2005; Hansen et al. 2006; Sugiyama et al. 2007; Motamedi et al. 2008; Tadeo et al. 2013; Wang et al. 2016). At the mat locus, the cenH element shares homology with the centromeric repeats and is essential for RNAi-mediated heterochromatin establishment (Hall et al. 2002). Once established, heterochromatin can be maintained in an RNAi-independent way by the ATF/CREB transcription factors Atf1/Pcr1 (Jia et al. 2004).
We found that the Ccr4-Not complex and RNAi are required for degradation of heterochromatic RNAs, and this is essential for constitutive heterochromatin formation. Ccr4-Not is a conserved complex involved in RNA surveillance, transcription elongation, RNA export, and DNA repair (Collart and Panasenko 2012; Miller and Reese 2012; Wahle and Winkler 2013; Inada and Makino 2014). The Ccr4-Not complex is the primary deadenylase and is essential for downstream decapping and mRNA decay by Xrn1 (Schizosaccharomyces pombe: Exo2), a conserved 5′-3′ exoribonuclease (Collart and Panasenko 2012; Miller and Reese 2012). Caf1 and Ccr4 build the deadenylase module of Ccr4-Not, while Not1, Not2, Not4 (S. pombe: Mot2), and Not5 belong to the Not group of the complex. Caf1 bridges Ccr4 to Not1 and is essential for the deadenylation activity (Basquin et al. 2012).
In RNAi-mediated heterochromatin formation, heterochromatic transcripts serve as a template for siRNA generation, Argonaute targeting, and recruitment of the methyltransferase complex CLRC (Holoch and Moazed 2015). At the same time, heterochromatic transcripts are degraded by RNAi. A link between various RNA degradation machineries and H3K9me at meiotic genes was also recently reported (Hiriart et al. 2012; Zofall et al. 2012; Egan et al. 2014; Chalamcharla et al. 2015; Cotobal et al. 2015; Tucker et al. 2016). Several of those studies suggested a direct recruitment of the H3K9 methyltransferase by different RNA degradation machineries to deposit H3K9me. The aim of our work was to analyze if degradation of heterochromatic RNAs is required for functional heterochromatin organization.
Results
Caf1 and RNAi are required for heterochromatin formation
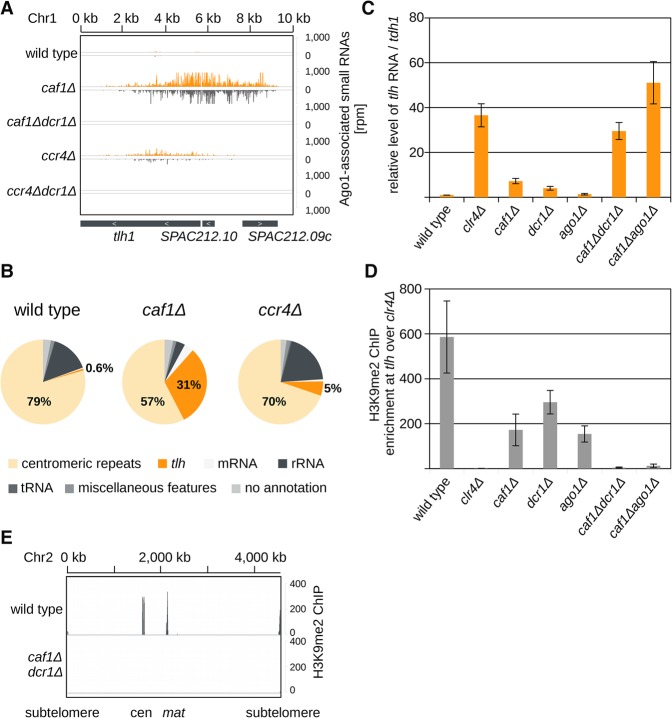
We sequenced Argonaute-bound small RNAs from cells having deletions of different RNA degradation pathways to determine if they might act redundantly with RNAi. In deletion of caf1, a deadenylase subunit of the Ccr4-Not complex, we observed a high amount of small RNAs generated from subtelomeric repeats (Fig. 1A,B). In wild-type cells, <1% of Argonaute-bound small RNAs map to the subtelomeric region, indicating that RNAi is not the major contributor to silencing at the subtelomeric repeats (Fig. 1B). On the contrary, in caf1Δ cells >30% of Argonaute-bound small RNAs map to the subtelomeric repeats (Fig. 1B). These small RNAs are Dcr1 dependent and show all features of siRNAs (Fig. 1A; Supplemental Fig. 1A,B). Subtelomeric siRNAs are generated from tlh1, SPAC212.10, and SPAC212.09c, covering a region from 0 to 9 kb on the left arm of Chromosome 1 (Fig. 1A) and the homologous regions on both arms of Chromosomes 1 and 2.
Figure 1.
Caf1 and RNAi are required for heterochromatin formation at transcribed regions. (A) Endogenously tagged Argonaute-bound small RNA reads in the indicated cells were plotted over the subtelomeric region. The location of genes is indicated as gray boxes below the small RNA peaks. Reads from + and − strands are depicted in orange and gray, respectively. Scale bars on the right denote small RNA read numbers normalized per one million reads. tlh is present at subtelomeres of both arms of Chromosomes 1 and 2 (Mandell et al. 2005; Hansen et al. 2006). (B) Classification of Argonaute-bound small RNAs from wild-type, caf1Δ, and ccr4Δ cells. Pie charts illustrate percentages for the individual small RNA classes relative to the total number of reads for each strain. Argonaute-bound subtelomeric siRNAs are increased by more than 50-fold in caf1Δ cells compared with the wild type. (C) Quantification of subtelomeric tlh transcripts in indicated strains by RT-qPCR. In caf1Δdcr1Δ and caf1Δago1Δ cells, silencing of subtelomeric repeats is lost. Error bars, SE of more than seven independent experiments. For caf1Δdcr1Δ and caf1Δago1Δ, experiments of two independent colonies were averaged, respectively. Reverse transcription was performed with specific primers; the wild type was set to one. (D) ChIP experiment showing that H3K9me2 is lost at subtelomeric tlh repeats in caf1Δdcr1Δ cells. Error bars, SE of at least four independent experiments. For caf1Δdcr1Δ and caf1Δago1Δ, data of two independent colonies were averaged, respectively. clr4Δ was set to one. (E) ChIP-seq showing that H3K9me2 is lost at all constitutive heterochromatic loci in caf1Δdcr1Δ cells. Scale bars on the right denote read numbers per million reads normalized to the TAS region (Chr 2: 4534 kb–4538 kb) where H3K9me2 is not lost in the mutant.
Similar to the subtelomeric region, higher amounts of siRNAs were generated at the mat locus in caf1Δ cells (Supplemental Fig. 1C). Centromeric siRNAs were generated near wild-type levels at dg and dh repeats but were strongly reduced at the IRC3 element in caf1Δ cells (Supplemental Fig. 1C; Halic and Moazed 2010). A similar siRNA pattern was detected in deletion of ccr4, the second deadenylase of the Ccr4-Not complex; however, lower amounts of siRNAs were generated from the subtelomeric and mat regions in ccr4Δ cells (Fig. 1A,B; Supplemental Fig. 1C). We did not observe a defect in length of Argonaute-bound small RNAs in caf1Δ or ccr4Δ cells, indicating that Caf1 and Ccr4, in contrast to Triman, are not directly processing small RNAs (Supplemental Fig. S1A,B; Marasovic et al. 2013). Our data suggest that the Caf1 and Ccr4 nucleases degrade subtelomeric, centromeric, and mat locus transcripts, and in their absence, RNAi acts as a redundant degradation mechanism.
We observed that centromeric dg and subtelomeric tlh transcripts were four- to fivefold up-regulated in caf1Δ cells, and silencing of a centromeric ade6 reporter was reduced (Fig. 1C; Supplemental Fig. 1D,E). These data show a partial loss of heterochromatic silencing at the subtelomeric and centromeric repeats in caf1Δ cells. Next, we generated caf1Δdcr1Δ and caf1Δago1Δ double mutants in several genetic backgrounds to remove both degradation pathways, RNAi and Ccr4-Not. While single deletions of caf1 and ago1/dcr1 had a small impact on RNA levels in the subtelomeric region, deletion of both pathways completely de-repressed subtelomeric transcripts (tlh1, SPAC212.10 and SPAC212.09c) to the level of deletion of the H3K9 methyltransferase Clr4 (Fig. 1C; Supplemental Fig. S1F,G). We performed chromatin immunoprecipitation (ChIP) with an antibody against H3K9me2 and observed loss of H3K9me2 at the subtelomeric, centromeric, and mat locus in caf1Δdcr1Δ and caf1Δago1Δ cells (Fig. 1D,E; Supplemental Fig. S2A,B). These results show that H3K9me2 and heterochromatic silencing cannot be maintained in these mutants at all constitutive heterochromatic loci.
Caf1 and Ccr4 activity is required for heterochromatin assembly at the tlh region
Next, we analyzed whether other subunits of the Ccr4-Not complex are required for subtelomeric silencing. In the double mutant of the second deadenylase Ccr4 and RNAi (ccr4Δdcr1Δ), subtelomeric transcripts were also accumulating, but H3K9me2 was not completely lost at the subtelomeric tlh region (Fig. 2A,B). not2Δdcr1Δ, rcd1Δdcr1Δ (RQCD1, Caf40), and caf16Δdcr1Δ cells showed no or little effect on tlh RNA levels; however, in mot2Δdcr1Δ cells, tlh transcripts were de-repressed (Fig. 2A; Supplemental Fig. S2C). H3K9me2 was also reduced at subtelomeric tlh and centromeric dg repeats in mot2Δdcr1Δ cells (Fig. 2B; Supplemental Fig. 2D). Our data indicate that the Ccr4-Not complex subunits Caf1, Ccr4, and also the E3 ubiquitin ligase Mot2 are involved in silencing and heterochromatin formation at constitutive heterochromatic loci. At the most distal region (toward centromeres), subtelomeric H3K9me2 was also lost in caf1Δ (Supplemental Fig. S2E), ccr4Δ, and mot2Δ cells (Cotobal et al. 2015).
Figure 2.
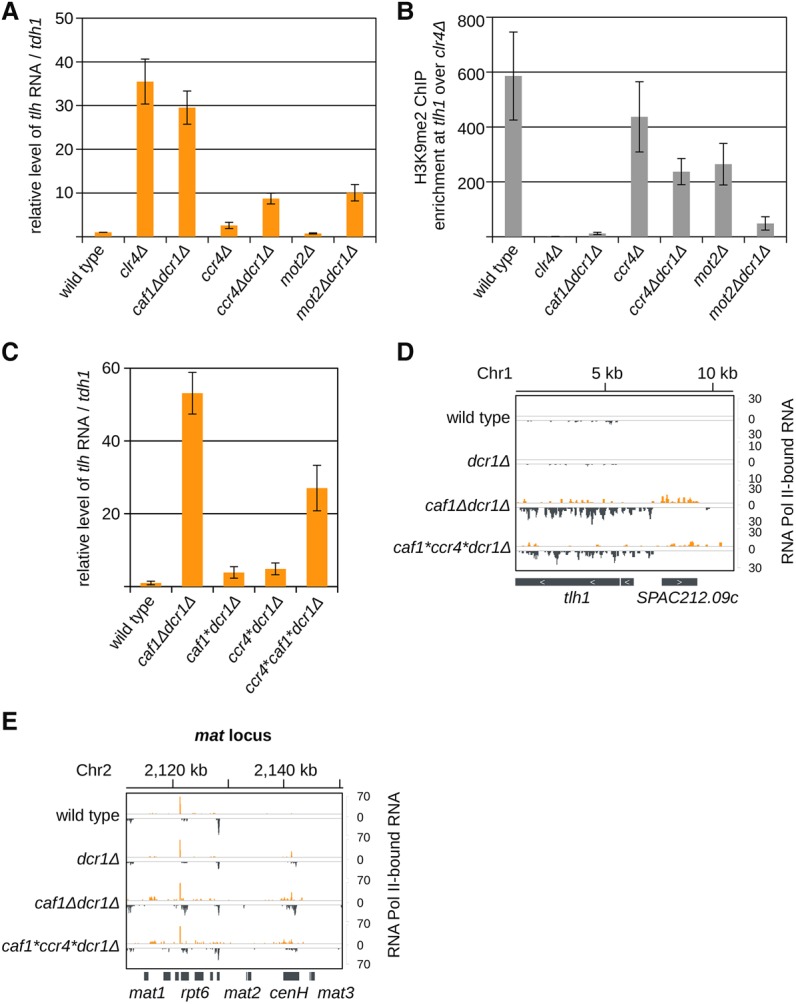
Caf1 and Ccr4 nuclease activity is required for heterochromatic silencing. (A) Quantification of subtelomeric tlh transcripts in indicated strains by RT-qPCR. In ccr4Δdcr1Δ and mot2Δdcr1Δ cells, silencing of subtelomeric repeats is defective, but not as much as in caf1Δdcr1Δ or clr4Δ cells. Error bars, SE of three or more independent experiments. Reverse transcription was performed with specific primers; the wild type was set to one. (B) ChIP experiment showing that H3K9me2 is lost at subtelomeric tlh repeats in mot2Δdcr1Δ cells but not in ccr4Δdcr1Δ. Error bars, SE of two (mot2Δ and mot2Δdcr1Δ) or more independent experiments. clr4Δ was set to one. (C) Quantification of subtelomeric tlh transcripts by RT-qPCR in the wild type or caf1Δdcr1Δ controls or in dcr1Δ strains expressing a Caf1* (Caf1D53AD243AD174A) or/and a Ccr4* (Ccr4H665A) activity mutant. Expression of Caf1* or Ccr4* silences tlh, but when both deadenylases are mutated, silencing of tlh is lost. Error bars, SE of three independent experiments. Reverse transcription was performed with specific primers; the wild type was set to one. (D,E) Sequencing reads of RNA Polymerase II (Pol II)–associated RNA in indicated cells are plotted over the subtelomeric tlh region (D) and over the mat locus (E). In caf1*ccr4*dcrΔ cells, transcription at etlh and SPAC212.09c and at the mat locus is increased compared with wild-type cells. Reads from + and − strands are depicted in orange and gray, respectively. Scale bars on the right denote RNA read numbers normalized to the total number of reads mapping to protein coding genes.
Of all subunits of the Ccr4-Not complex, deletion of the deadenylase Caf1 had the greatest loss of silencing of heterochromatic transcripts. We investigated if the nuclease activity of Caf1 was required for tlh silencing and heterochromatin assembly. We genomically integrated Caf1* (Caf1D53AD243AD174A) and Ccr4* (H665A) activity mutants into caf1Δdcr1Δ and dcr1Δ cells (Chen et al. 2002; Jonstrup et al. 2007). Introduction of the Caf1* or Ccr4* activity mutants showed only a minor increase in tlh RNA in caf1*dcr1Δ and ccr4*dcr1Δ cells (Fig. 2C), suggesting that both deadenylases, Caf1 and Ccr4, might act redundantly. Since Caf1 is required for Ccr4 recruitment to the Ccr4-Not complex, deletion of Caf1 eliminates the activity of both deadenylases (Basquin et al. 2012). Mutation of the active site of both Caf1 and Ccr4 resulted in a strong accumulation of subtelomeric transcripts and reduction in H3K9me2 in caf1*ccr4*dcr1Δ cells (Fig. 2C; Supplemental Fig. S3A). Sequencing of nascent RNA associated with RNA Polymerase II (Pol II) showed increased transcription in caf1*ccr4*dcr1Δ cells at subtelomeric, mat, and centromeric loci, similar to caf1Δdcr1Δ cells (Fig. 2D,E; Supplemental Fig. S3B). These results show that deadenlyation by Caf1 and Ccr4 is required for transcriptional silencing at all heterochromatic loci. We note that caf1*ccr4*dcr1Δ cells have a slightly weaker defect compared with caf1Δdcr1Δ cells, suggesting that Caf1 might recruit an additional factor contributing to RNA degradation or heterochromatin formation.
This suggested that other nucleases and RNA processing factors could also be involved in degradation of heterochromatic RNAs. In addition to caf1Δ, deletion of the 5′-3′ exonuclease Exo2 (Xrn1) showed strong accumulation of subtelomeric siRNAs (Supplemental Fig. S3C). Subtelomeric siRNAs were also increased in the deletion of the RNA processing factor mlo3 (Zhang et al. 2011). In the deletion of the exosome subunit rrp6, the TRAMP component cid14, the PAF complex subunit leo1 (Sadeghi et al. 2015), HP1 protein swi6, and in the dis3-53 mutant, we observed only a smaller accumulation of subtelomeric siRNAs (Supplemental Fig. S3C,D). There was little or no effect on siRNA generation at centromeric repeats in these mutants (Supplemental Fig. S3C). Our results indicate that the Ccr4-Not/Xrn1 pathway eliminates heterochromatic transcripts, and this is required for heterochromatic silencing.
Heterochromatin is lost at transcribed regions
The Shelterin complex recruits CLRC and SHREC to the telomeres, from where they spread to maintain subtelomeric heterochromatin (Kanoh et al. 2005; Sugiyama et al. 2007; Wang et al. 2016). In caf1Δdcr1Δ cells, Taz1, a DNA binding protein of the Shelterin complex, is still recruited to the telomeric repeats (Supplemental Fig. S4A). This indicates that the Shelterin complex could still recruit CLRC to maintain heterochromatin. In taz1Δ cells, we observe a small increase in subtelomeric siRNAs, and consistent with previous studies, in taz1Δdcr1Δ cells when both CLRC recruitment pathways are eliminated, tlh transcripts accumulate (Supplemental Fig. S4B,C,E; Kanoh et al. 2005; Hansen et al. 2006). In caf1Δtaz1Δ cells, we did not observe an additional loss of H3K9me2 at tlh compared with caf1Δ (Supplemental Fig. S4D).
Consistent with Taz1 localization (Supplemental Fig. S4A), H3K9me2 was not lost at the telomere-associated sequence (TAS) between telomeric repeats and tlh in caf1Δdcr1Δ cells (Fig. 3A). This indicates that the Shelterin complex can still deposit H3K9me at the telomeric borders, which can spread until the tlh locus. Heterochromatin, however, cannot spread over the transcribed tlh gene in caf1Δdcr1Δ cells (Supplemental Fig. S2E). These data show that Caf1 and RNAi are not essential for the maintenance and spreading of H3K9me from telomeric repeats until the tlh region.
Figure 3.
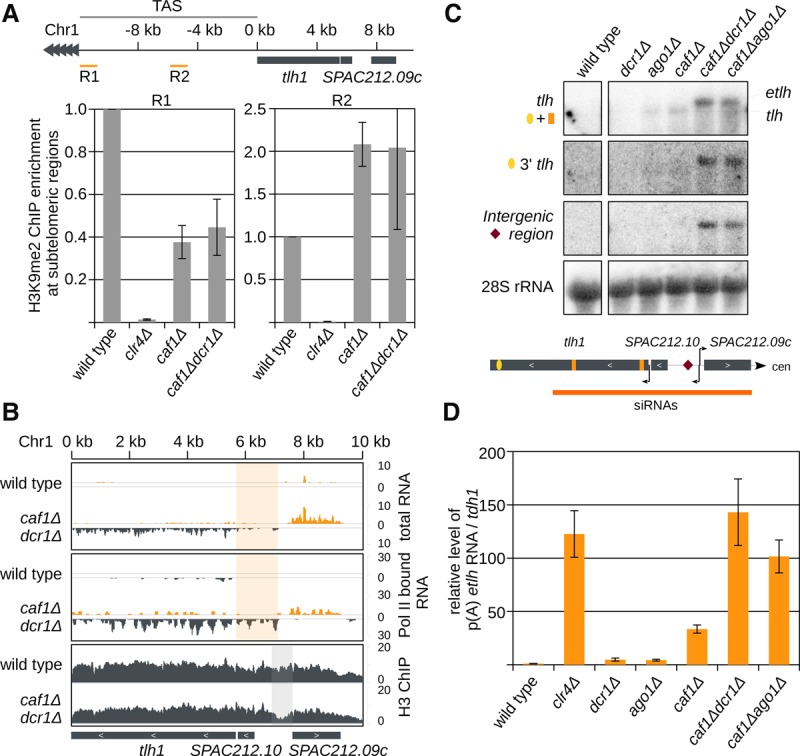
In caf1Δdcr1Δ cells, transcriptional silencing is lost. (A) ChIP experiment showing that in caf1Δdcr1Δ cells H3K9me2 is not lost at regions between telomeric repeats and the transcribed tlh gene. The S. pombe genome assembly is incomplete in the subtelomeric region, but additional insert clones from the telomere plasmid library with the sequence of the telomere-associated region (TAS) are available at www.pombase.com. Error bars, SE of three independent experiments. Region 1 (R1) is next to the telomere, and region 2 (R2) is ∼6 kb away from telomeric repeats. tlh is ∼10 kb upstream of the telomeric repeats. (B) Sequencing reads in indicated cells are plotted over the subtelomeric region. (Top) In caf1Δdcr1Δ cells, silencing in the subtelomeric region is lost. Reads between SPAC212.10 and SPAC212.09c can be detected. Reads from + and − strands are depicted in orange and gray, respectively. Scale bars on the right denote total RNA read numbers normalized to all reads mapping to protein coding genes. Highlighted in orange are reads that map upstream of tlh1. (Middle) RNA Pol II–associated RNA. Pol II starts transcription in caf1Δdcr1Δ cells upstream of the annotated tlh gene; in wild-type cells, it starts at the annotated transcription start. Reads from + and − strands are depicted in orange and gray, respectively. Scale bars on the right denote RNA read numbers normalized to total number of reads mapping to protein coding genes. Highlighted in orange are reads that map upstream of tlh1. (Bottom) H3 ChIP sequencing, showing a nucleosome-free region (highlighted in gray) upstream of the Pol II–associated RNA tlh reads. Scale bars on the right denote reads per million. (C) Northern blot showing accumulation of two distinct tlh products. (tlh) three probes mapping to the annotated tlh sequence; (3′ tlh) one probe mapping to the 3′ end of tlh; (Intergenic region) one probe hybridizing between SPAC212.10 and SPAC212.09c; and (28S rRNA) 28S rRNA probe as loading control. In caf1Δdcr1Δ and caf1Δago1Δ cells, a longer transcript (etlh) is accumulating. (D) Quantification of subtelomeric etlh transcripts in indicated strains by RT-qPCR reverse transcribed with oligo(dT). In caf1Δ cells, etlh silencing is strongly reduced. In caf1Δdcr1Δ and caf1Δago1Δ cells, silencing of the etlh transcript is lost. Error bars, SE of more than three independent experiments. Reverse transcription was performed with oligo(dT) primer, and qRT-PCR was performed with primers for etlh. The wild type was set to one.
Absence of RNA degradation leads to changes in chromatin organization
Small RNA sequencing data in caf1Δ cells show that siRNAs are generated from tlh, SPAC212.10, SPAC212.09c, and their intergenic regions (Fig. 1A). Sequencing of RNA Pol II–associated nascent RNA and also total RNA showed increased transcriptional activity in the intergenic region in caf1Δdcr1Δ cells but not in wild-type cells (Fig. 3B). This is consistent with the H3 ChIP-seq and nucleosome positioning data that show a nucleosome-free region between SPAC212.10 and SPAC212.09c in caf1Δdcr1Δ (Fig. 3B) and clr4Δ cells (Garcia et al. 2010). Northern blot analysis confirms that tlh RNA is longer in caf1Δdcr1Δ and caf1Δago1Δ cells than in ago1Δ or caf1Δ cells (Fig. 3C). tlh probes from the 3′ end as well as from the 5′ intergenic region hybridized to the longer transcript, indicating that in caf1Δdcr1Δ cells, transcripts start 2 kb upstream of the annotated tlh promoter and terminate at the tlh terminator (Fig. 3C). The extended tlh (etlh) transcript is polyadenylated and shows increased amounts in caf1Δ, but not in ago1Δ cells, and is completely de-repressed in caf1Δdcr1Δ and caf1Δago1Δ cells (Fig. 3D). The etlh transcript was also accumulating in clr4Δ and at lower levels in ccr4Δdcr1Δ cells (Fig. 3D; Supplemental Fig. S4F). These data indicate that small RNAs in caf1Δ cells are generated from one longer transcript and that, in the absence of RNA degradation, subtelomeric chromatin organization is changed.
Heterochromatic RNAs accumulate on chromatin and form DNA:RNA hybrids
Our data show that RNA degradation is required for heterochromatic silencing. This suggests that heterochromatic transcripts might accumulate on chromatin. We performed chromatin fractionation and also sequenced histone H3-bound RNA and observed that tlh RNA is enriched at chromatin in wild-type cells (Fig. 4A; Supplemental Fig. S5A,B). In caf1Δ and even more in caf1Δdcr1Δ cells, we observed increased levels of tlh transcripts in the chromatin fraction compared with wild-type cells (Fig. 4A; Supplemental Fig. S5A,B). Genome-wide comparison of histone H3-bound RNA and RNA Pol II–bound nascent RNA revealed that in wild-type cells, heterochromatic transcripts and lncRNA are more retained at chromatin than mRNAs relative to their transcription (Fig. 4B,C; Supplemental Figs. S5C, S6A). We found that transcripts from all constitutive heterochromatic loci accumulate on chromatin (Supplemental Fig. S6B). Our data suggest that heterochromatic transcripts and euchromatic lncRNA are processed less efficiently and are retained on chromatin post-transcriptionally.
Figure 4.
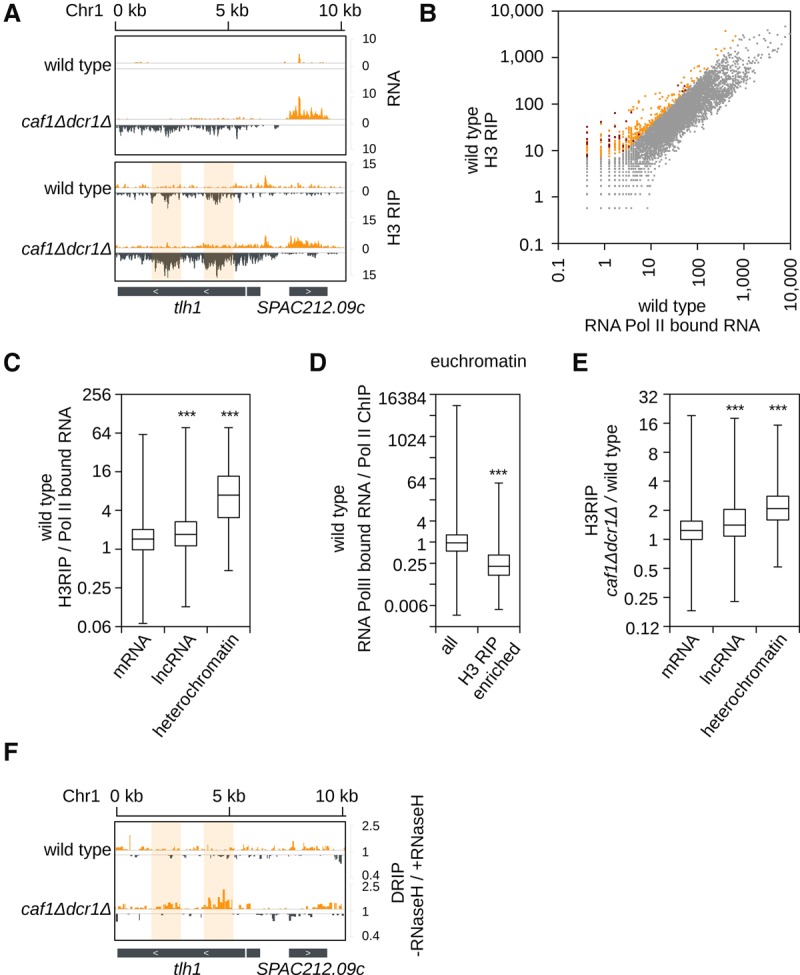
Heterochromatic transcripts accumulate on chromatin. (A) H3-RIP sequencing reads in indicated cells are plotted over the subtelomeric region. Total RNA sequencing reads on top as control. Reads from + and − strands are depicted in orange and gray, respectively. Scale bars on the right denote RNA read numbers per total number of reads mapping to protein coding genes. Compared with total RNA, tlh is enriched at the chromatin in wild-type and caf1Δdcr1Δ cells. Region where DNA:RNA hybrids are enriched is highlighted. Scale bars on the right denote RNA read numbers normalized to total number of reads mapping to protein coding genes. (B) Scatter plot showing H3-associated RNA relative to RNA Pol II–bound nascent RNA. A subset of genes is more enriched in H3 RIP than bound to RNA Pol II, indicating post-transcriptional retention on chromatin. Lowly expressed genes show stronger enrichment on chromatin. RNAs enriched on chromatin are shown in orange or red for heterochromatic RNA. (C) Box plot analysis of H3-associated RNA relative to nascent RNA bound to RNA Pol II in wild-type cells. (lncRNA) Euchromatic noncoding RNA with annotation SPNCRNA, n = 1354; (mRNA) all protein coding genes without genes in heterochromatic areas, n = 5014; (heterochromatic) genes which are located in constitutive heterochromatin areas, n = 62. Relative to their transcript levels, ncRNAs and especially heterochromatic RNA are more likely to be bound to chromatin than mRNA. (***) P < 0.001. (D) Box plot analysis of nascent RNA bound to RNA Pol II relative to RNA Pol II ChIP in wild-type cells. Genes that have RNA enriched on chromatin show reduced transcription compared with the quantity of RNA Pol II on the chromatin. (all) All mRNA and lncRNA, n = 6345; (chromatin enriched) euchromatic genes (mRNA and lncRNA) with greater than fourfold enrichment in H3-RIP over nascent RNA, n = 258. (***) P < 0.001. (E) Box plot analysis of H3-associated RNA in caf1Δdcr1Δ cells relative to wild-type cells. In caf1Δdcr1Δ cells, heterochromatic RNA and lncRNA are even more associated with chromatin than in wild-type cells. (mRNA) n = 5058; (lncRNA) n = 1467; (heterochromatin) n = 90. (***) P < 0.001. (F) DNA:RNA hybrid sequencing in wild-type and caf1Δdcr1Δ cells plotted over the subtelomeric region. DNA:RNA hybrids are formed with same sequences, which are also enriched in H3-RIP (highlighted). Scale bars on the right denote the ratio of “sample not treated” over “sample treated” with RNase H (−RNase H/+RNase H) before immunoprecipitation.
In euchromatin we observed that genes with chromatin-retained RNAs were less efficiently transcribed by RNA Pol II (Fig. 4D). The same amount of RNA Pol II synthesized less nascent RNAs at these genes than at genes with no chromatin-bound RNAs (Fig. 4D; Supplemental Fig. S6C). These data show that in wild-type cells, chromatin-bound RNAs reduce transcription at euchromatic genes. In caf1Δdcr1Δ cells, heterochromatic transcripts from all loci and euchromatic lncRNA associate even more with chromatin than in wild-type cells, suggesting that the Ccr4-Not complex is degrading these transcripts on chromatin (Fig. 4E; Supplemental Fig. S6D,E).
Consistent with a previous observation (Nakama et al. 2012), we found low levels of DNA:RNA hybrids at subtelomeres in wild-type cells (Fig. 4F). In caf1Δdcr1Δ cells, heterochromatin is lost, and heterochromatic transcripts show a higher amount of DNA:RNA hybrids than in wild-type cells (Fig. 4F). Our data indicate that in wild-type cells, RNAi and Ccr4-Not degrade heterochromatic RNAs on chromatin to prevent formation of DNA:RNA hybrids. These results also show that DNA:RNA hybrid formation is not heterochromatin dependent.
DNA:RNA hybrid formation was shown to induce H3S10 phosphorylation (H3S10P) (Castellano-Pozo et al. 2013), a chromatin mark that reduces HP1 binding to H3K9 methylated nucleosomes and interferes with heterochromatin formation and silencing (Fischle et al. 2005; Kloc et al. 2008). We determined H3S10P levels; however, we observed reduction of H3S10P in caf1Δdcr1Δ cells at tlh (Supplemental Fig. 6F). Consistent with our findings, an H3S10A mutant was previously shown to have reduced H3K9me2, indicating rather a codependence of these two histone marks (Kloc et al. 2008).
Caf1 eliminates heterochromatic RNA on chromatin
Our results show that heterochromatic transcripts are chromatin bound and suggest that Caf1 degrades them on chromatin. By using ChIP and ChIP-exo sequencing, we found that Caf1 is localized at the tlh and SPAC212.09c region in wild-type cells (Fig. 5A). This resembles the localization of Ccr4 and Not subunits that were found over open reading frames in Saccharomyces cerevisiae (Kruk et al. 2011; Venters et al. 2011). Caf1 localization on the chromatin was weak but detectable, indicating that Caf1 is not tightly bound to the chromatin. In clr4Δ cells, where heterochromatin is lost, even higher amounts of Caf1 were bound to the heterochromatic regions (Fig. 5B,C). In wild-type cells, Caf1 is bound to the chromatin in the same region where siRNAs were generated in caf1Δ cells (Fig. 5A; Supplemental Fig. S7A); however in clr4Δ cells, Caf1 was enriched over the larger subtelomeric region (Fig. 5B; Supplemental Fig. S7B). Our data show that the Ccr4-Not complex is associated with chromatin; however, the complex is not recruited by heterochromatin. This suggests that RNAs, which are more bound to chromatin in clr4Δ cells, recruit Caf1 to chromatin.
Figure 5.
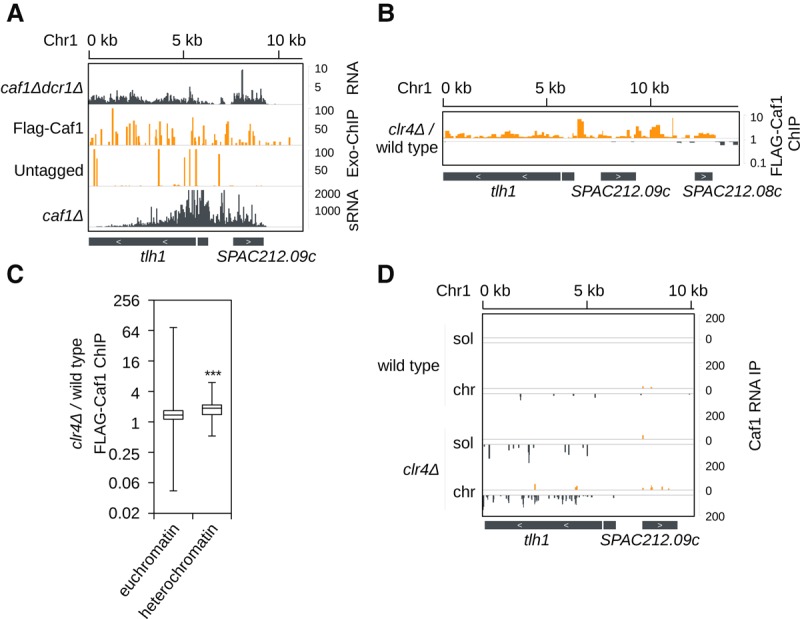
Heterochromatic transcripts are degraded on chromatin by the Ccr4-Not complex. (A) ChIP-exo (orange; middle) sequencing of endogenously tagged Flag-Caf1 in wild-type cells and untagged cells showing that Caf1 is enriched at subtelomeric tlh and SPAC212.09c genes. Caf1 localization overlaps with transcription (top; gray) and siRNA generation (bottom; gray). Scale bars on the right denote RNA read numbers normalized to total number of reads mapping to protein coding genes (RNA) or reads per million (Exo-ChIP and sRNA). (B) ChIP sequencing of endogenously tagged Flag-Caf1 in clr4Δ cells over wild-type cells. Caf1 is enriched at chromatin when clr4 is deleted. Scale bars on the right denote fold change of Flag-Caf1 ChIP in clr4Δ over wild-type cells. (C) Box plots of Flag-Caf1 ChIP showing that in clr4Δ cells, Caf1 is more localized to heterochromatic loci than in wild-type cells. (***) P < 0.001. (D) Sequencing of Flag-Caf1–bound RNA from soluble (sol) and chromatin (chr) fractions. tlh RNA associates more with Caf1 from the chromatin than the soluble fraction in both wild-type and in clr4Δ cells. Reads from + and − strands are depicted in orange and gray, respectively. Scale bars on the right denote RNA read numbers normalized to total number of reads mapping to protein coding genes.
We also performed Caf1 RNA-IP from chromatin and soluble fractions. In wild-type cells, chromatin-bound Caf1 associates with more tlh RNA than soluble Caf1 (Fig. 5D). This suggests that Caf1 degrades tlh RNA on chromatin. Other heterochromatic transcripts from subtelomeric and mat regions are degraded by Caf1 on chromatin as well (Supplemental Fig. S7C). At centromeric repeats we found that Caf1 does not degrade chromatin-associated RNAs in wild-type cells, which is consistent with RNAi being the primary degradation machinery at this locus (Supplemental Fig. S7C). In clr4Δ cells, chromatin-bound Caf1 associates with higher amounts of heterochromatic transcripts than soluble Caf1 (Fig. 5D; Supplemental Fig. S7C). This is consistent with increased Caf1 localization to heterochromatic DNA in clr4Δ cells and shows that Caf1 degrades heterochromatic RNA on chromatin (Fig. 5B–D). We also found that Caf1 binds Caf1 mRNA and, in this way, might regulate its own activity (Supplemental Fig. S7D). Our data suggest that chromatin-bound RNAs are degraded by Ccr4-Not on chromatin, while mRNAs that are exported are degraded in cytosol.
Expression of heterochromatic tlh RNA leads to loss of silencing
Our data suggest that chromatin-bound RNAs negatively regulate heterochromatin assembly. To determine if chromatin-bound RNAs cause the loss of heterochromatin and to exclude secondary effects of mutant backgrounds, we generated wild-type strains expressing the heterochromatic tlh transcript under the thiamine-repressible nmt1 promoter at a locus 5.5 kb upstream of tlh (Fig. 6A; Supplemental Fig. S8A). We split the tlh gene into two halves (5′tlh and 3′tlh) and inserted both halves into the genome at the place of SPNCRNA.70. As a control, we generated the same construct expressing the euchromatic LEU2 RNA.
Figure 6.
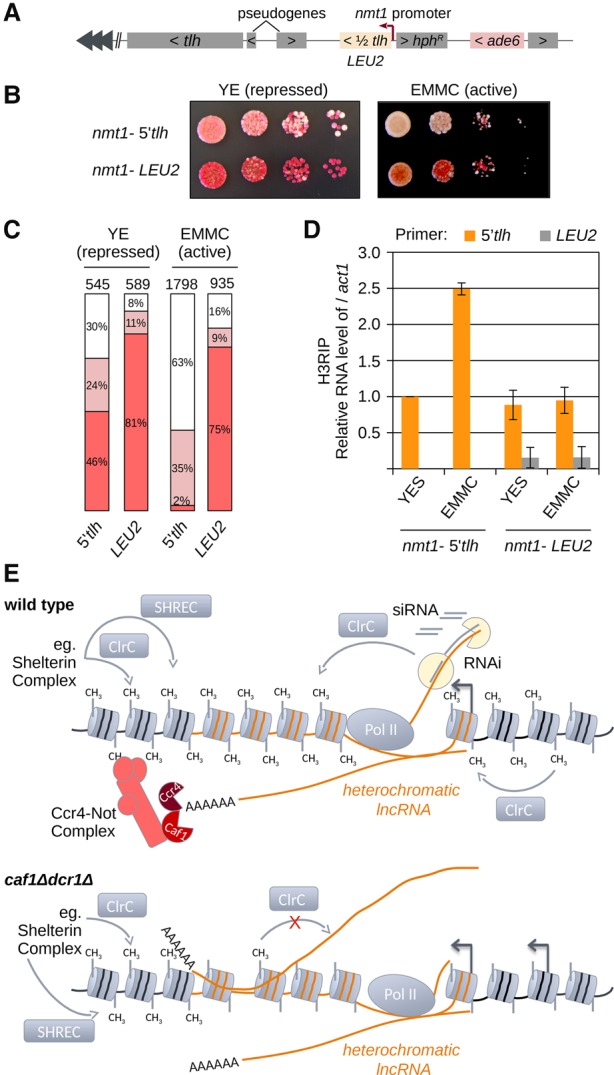
Accumulation of RNA on chromatin disrupts heterochromatin. (A) Scheme showing the constructs used. One half of tlh (5′tlh: first half; 3′tlh: second half) or LEU2 were inserted ∼5.5 kb upstream of tlh under the thiamine-inducible nmt1 promoter. The ade6 reporter gene was inserted 5 kb upstream of the inducible expression system. Cells that silence ade6 are red, while ade6-expressing cells are white when grown on low adenine plates. Heterochromatin maintenance of a silenced, red colony was analyzed for each strain on YE (low adenine, thiamine +) or EMMC low ade (10 mg/L adenine, thiamine −) plates. (B) Growth assay with ade6 reporter gene showing nmt1-5′tlh and nmt1-LEU2 cells. nmt1-5′tlh cells show strong loss of heterochromatic silencing when grown under inducible conditions. Expression of a control LEU2 gene did not lead to increased loss of heterochromatin when expression was induced. Cells were plated in 10-fold dilutions starting with 105 cells. (C) Quantification of different colored cells is shown for the indicated strains and expression, respectively. The total number of colonies counted is indicated above the bars. (D) Quantification of chromatin-associated RNA in indicated strains and conditions by H3 RIP RT-qPCR. In nmt1-5′tlh strains, 5′tlh RNA is enriched at chromatin in EMMC (induced) compared with YES medium (repressed). In nmt1-LEU2 strains, neither tlh nor LEU2 RNA is changed upon induction of LEU2 expression. (E) RNAi or Shelterin complex recruit the CLRC methyltransferase and SHREC deacetylase complexes to establish heterochromatin in wild-type cells. CLRC and SHREC spread into repeats to establish heterochromatin. The Ccr4-Not complex eliminates heterochromatic RNA and promotes spreading over the transcribed region. In the absence of the Ccr4-Not complex and RNAi, heterochromatic transcripts accumulate on the chromatin. This leads to loss of heterochromatin. At the transcribed loci, heterochromatin is lost in caf1Δdcr1Δ cells, indicating that at transcribed regions, RNA degradation is required for heterochromatin assembly.
We inserted an ade6 reporter gene 5 kb upstream of the tlh/LEU2-expressing constructs (Fig. 6A). When grown on low adenine medium, cells that silence the ade6 reporter gene are red, and cells that express ade6 are white. When grown on nmt1-repressive low adenine medium (YE), the cells expressing the LEU2 gene had mainly red colonies, indicating that the ade6 reporter is silenced (Fig. 6B,C; Supplemental Fig. S8B). Expression of both 5′tlh and 3′tlh constructs showed a higher percentage of white colonies even under promoter-repressive conditions (YE) (Fig. 6B,C; Supplemental Fig. S8B).
When plated on EMMC low adenine media, the nmt1 promoter is activated and RNA is transcribed at much higher levels with all constructs being expressed at similar levels (Supplemental Fig. S8C,D). Both 5′tlh- and 3′tlh-expressing cells had mainly white colonies on EMMC media, showing strong loss of heterochromatin (Fig. 6B,C; Supplemental Fig. S8E,F). On the contrary, the LEU2-expressing control cells were red, which indicates no loss of silencing on EMMC media (Fig. 6B,C). These data also show that the reporter gene is distant enough from the tlh/LEU-expressing locus, and it is not affected by transcription from that locus.
In both nmt1-5′tlh– and nmt1-3′tlh–expressing cells, we found an increase in chromatin retention of tlh RNA after induction of tlh expression (Fig. 6D; Supplemental Fig. S8G). Although expressed at the same level, LEU2 RNA shows very little enrichment on chromatin (Fig. 6D; Supplemental Fig. S8C). In the nmt1-LEU2 strain, we also do not observe a change in chromatin retention of endogenous tlh RNA (Fig. 6D). Our data show that only chromatin-bound tlh RNAs lead to loss of heterochromatin, while released LEU2 RNA did not have an effect on heterochromatic silencing. These results confirm that high amounts of chromatin-bound heterochromatic transcripts lead to loss of heterochromatin even in wild-type cells. This also excludes the possibility that chromatin association of heterochromatic RNAs is merely a consequence of increased transcription. Our data show that accumulation of RNA on chromatin leads to loss of heterochromatin and that degradation of chromatin-bound RNAs is essential for heterochromatin organization (Fig. 6E).
Discussion
Although RNA is required for heterochromatin formation, our data show that RNA needs to be degraded and chromatin-bound RNAs disrupt heterochromatin organization. We found that the Ccr4-Not complex eliminates heterochromatic transcripts on chromatin to maintain heterochromatin structure. The Ccr4-Not complex was initially described as a chromatin-associated complex involved in transcription (Miller and Reese 2012); however, its role on chromatin remained unclear. In fission yeast, the Ccr4-Not complex was recently shown also to be involved in deposition of H3K9 methylation at rDNA and meiotic genes (Cotobal et al. 2015; Sugiyama et al. 2016). Additionally, the Ccr4-Not complex was shown to play a role in silencing of subtelomeric transposons in Caenorhabditis elegans and Drosophila melanogaster (Fischer et al. 2013; Morgunova et al. 2015). Transposons are targeted by piRNAs in animal germline cells that establish heterochromatin in a similar way to siRNAs in fission yeast (Hirakata and Siomi 2016). The observations in C. elegans and D. melanogaster are analogous to our findings in S. pombe and imply a conserved role of the Ccr4-Not complex in degradation of chromatin-associated heterochromatic transcripts.
Our data suggest that degradation of heterochromatic transcripts by the Ccr4-Not complex and RNAi reduces the chance of DNA:RNA hybrid formation. It has been shown that chromatin retention of RNA and DNA:RNA hybrid formation establish RNAi-mediated heterochromatin (Sun et al. 2013; Skourti-Stathaki et al. 2014; Yu et al. 2014). Deletion of RNase H, however, has a defect in heterochromatin maintenance, indicating that an excess of DNA:RNA hybrids has a negative impact on heterochromatin assembly (Nakama et al. 2012). DNA:RNA hybrids were also shown to inhibit nucleosome formation (Dunn and Griffith 1980), which would interfere with chromatin organization. These data show that an increase in DNA:RNA hybrids will interfere with chromatin organization and heterochromatin assembly.
We show that the deadenylation activity of the Ccr4-Not complex is important for heterochromatin maintenance, indicating that degradation of polyadenylated RNAs occurs on chromatin post-transcriptionally. We observed, however, a slightly weaker defect in the activity mutants than in the knock-out strains, suggesting that the activity mutant can recruit other RNA processing enzymes, like Exo2 (Xrn1) (Collart and Panasenko 2012; Miller and Reese 2012). Another possibility is that other functions of the Ccr4-Not complex are impaired in the deletion mutants. Recently, the Ccr4-Not complex was shown to act as a transcription elongation factor suggested to reactivate arrested RNA Pol II (Kruk et al. 2011; Dutta et al. 2015). Dicer was also implicated in release of stalled RNA Pol II, which reduces DNA:RNA hybrid formation (Zaratiegui et al. 2011; Castel et al. 2014). These functions might also contribute to heterochromatin formation.
Our data show that chromatin-bound tlh RNA will recruit the Ccr4-Not complex to chromatin. In clr4Δ cells, we observe higher amounts of the Ccr4-Not complex on chromatin than in wild-type cells. This suggests that higher amounts of chromatin-bound RNAs recruit more Ccr4-Not to chromatin in these mutants. A recent study suggests that in mammalian cells, some lncRNAs might be degraded cotranscriptionally on chromatin, while others are degraded in the nucleoplasm (Schlackow et al. 2017). It remains to be analyzed why specific RNAs are degraded on chromatin and others in the soluble fraction. The Ccr4-Not complex is found in the cytosol and the nucleus (Collart 2016), and our data suggest that Ccr4-Not localization might be a result of RNA localization.
We also found that many euchromatic lncRNAs are retained at the chromatin in wild-type cells and even more in the absence of the Ccr4-Not complex. Although many euchromatic RNAs are retained at the chromatin, they do not recruit RNAi (Marasovic et al. 2013). Our data show that RNA localization on chromatin is not sufficient to initiate RNAi-mediated heterochromatin formation. This indicates that mutations in the Paf1 complex, which promote establishment of ectopic RNAi-dependent heterochromatin, interfere with chromatin functions of the Paf1 complex rather than RNA release from chromatin (Sadeghi et al. 2015; Verrier et al. 2015).
Many lncRNAs have been implicated in chromatin-related processes from yeast to human cells and are overexpressed in numerous human diseases and cancer (Sánchez and Huarte 2013). Several human lncRNAs were also suggested to be bound at chromatin (Böhmdorfer and Wierzbicki 2015; Yamashita et al. 2016). This indicates that the degradation of chromatin-associated transcripts might be a conserved mechanism to maintain chromatin structure.
Methods
Strain construction
All S. pombe strains used in this study are listed in the Supplemental Table S1. Strains were generated as previously described (Marasovic et al. 2013). The point mutations for Caf1D53AD243AD174A were chosen as previously described (Jonstrup et al. 2007). D50A corresponds to D53A according to a new PomBase annotation. The Ccr4 activity mutant, Ccr4H665A, was taken corresponding to the homologous Ccr4H818A mutant in S. cerevisiae (Chen et al. 2002). The tlh overexpression strains contain one-half of tlh1 (5′tlh: 1–2800 bp; 3′tlh: 2801–5664 bp) or LEU2 (from S. cerevisiae; from start codon to stop codon without 3′ UTR) under control of the nmt1 promoter and adh terminator with a downstream ade6 reporter. Cloning was done in a standard way with detailed description in the Supplemental Methods. Primers are listed in the Supplemental Table S2.
siRNA sequencing
Endogenously tagged 3×Flag-Ago1 was immunoprecipitated and Ago1-bound siRNA libraries were generated as previously described (Marasovic et al. 2013). A detailed description can be found in the Supplemental Methods.
Total RNA isolation, reverse transcription, and qRT-PCR
Total RNA isolation, reverse transcription, and quantitative real-time PCR (qRT-PCR) were performed according to standard procedures, described in detail in the Supplemental Methods. Primers are listed in the Supplemental Table S2.
Total RNA and poly(A) RNA sequencing
rRNA of 1 µg total RNA was degraded with Terminator nuclease (Epicentre) in buffer A for 2 h at 30°C. For poly(A) RNA sequencing, poly-adenylated RNA was extracted from total RNA with oligo d(T)25 magnetic beads (NEB). The RNA library was obtained using the NEBNext Ultra Directional RNA library prep kit for Illumina (NEB).
Northern Blot
Ten micrograms of total RNA was used for Northern Blot, which was performed in a standard way as described in the Supplemental Methods.
Growth assay and ade6 reporter spot assay
Tenfold serial dilutions of cultures with OD600 between 0.7 and 1.5 were made so that the highest density spot contained 105 cells. Cells were spotted on nonselective YES medium, not supplemented YE (low adenine) medium or EMMC low adenine (10 mg/L adenine, 226 mg/L leucine, 226 mg/uracil). The plates were incubated at 32°C for 2–3 d and imaged.
Chromatin immunoprecipitation
ChIP was performed as previously described (Marasovic et al. 2013) using 1.2 µg immobilized (Dynabeads Protein A, Thermo Scientific) antibody against dimethylated H3K9 (H3K9me2, Abcam AB1220), H3 (ab1791, Abcam), H3S10P (ab5176, Abcam), anti-Flag M2-magnetic beads (Sigma-Aldrich), or 5 µL agarose conjugated Pierce HA epitope tag antibody (no. 26181, Thermo Scientific). Oligonucleotides used for quantification are listed in the Supplemental Table S2. For sequencing, a ChIP-seq library was made using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB).
DRIP sequencing
DNA–RNA hybrid immunoprecipitation (DRIP) was performed as previously described (Wahba et al. 2016). Lysis was performed like in ChIP from a 50-mL pellet without crosslinking. For immunoprecipitation, the S9.6 antibody (Kerafast) was used. Sample treated with RNase H was compared with sample not treated with RNase H. For a detailed protocol, see the Supplemental Methods.
RNA IP/Pol II–bound nascent RNA/H3-bound RNA
RNA IP was performed like ChIP but without RNase A digestion, using anti-RNA Pol II CTD repeat YSPTSPS antibody [8WG16] (ab817, Abcam), anti-H3 antibody (ab1791, Abcam), or anti-Flag M2-magnetic beads (Sigma-Aldrich). After phenol-chloroform-isoamyl alcohol extraction, DNA was digested with DNase I (Thermo Scientific) for 2 h at 37°C. RNA was recovered with a second phenol-chloroform-isoamyl alcohol purification and ethanol precipitation. Either RNA was taken for making a sequencing library using the NEBNext Ultra Directional RNA library prep kit for Illumina (NEB) or it was reverse transcribed into cDNA with specific primers for subsequent qRT-PCR.
ChIP-exo sequencing
ChIP-exo was performed similarly as previously described with minor modifications listed in the Supplemental Methods (Rhee and Pugh 2012; Serandour et al. 2013).
Chromatin fractionation
The frozen pellet of a 10-mL culture with an OD600 of 1.0 was lysed with 0.25- to 0.5-mm glass beads (Roth) and the BioSpec FastPrep-24 bead beater (MP-Biomedicals), with eight cycles at 6.5 m/sec for 30 sec and 3 min on ice. The chromatin fraction was separated from the soluble fraction by centrifugation at 21,000g for 20 min. DNA sample was treated with Proteinase K and RNase A, and the RNA sample was treated with Proteinase K and DNase I. Reverse transcription was performed with specific primers for tlh and tdh1. For more details, see the Supplemental Material.
3×Flag-Caf1 RNA IP with chromatin fractionation
Caf1-associated RNA in the chromatin and the nonchromatin fraction was isolated like the RNA IP described above. Just before sonication, the sample was centrifuged for 20 min at 21,000g at 4°C. The supernatant was taken as “soluble fraction”; the pellet was washed twice with lysis buffer and then resuspended in lysis buffer, which formed the “chromatin fraction.” IP was performed with anti-Flag M2-magnetic beads (Sigma-Aldrich).
Analysis of sequencing data
Single end sequencing of libraries was performed on an Illumina GA IIX sequencer at the LAFUGA core facility of the Gene Center, Munich. The Galaxy platform was used to demultiplex the obtained reads with Je-Demultiplex-Illu (Goecks et al. 2010).
Demultiplexed Illumina reads were mapped to the S. pombe genome, allowing a two-nucleotide mismatch to the genome using Novoalign (http://www.novocraft.com). The h90 S. pombe genome was assembled using the mat sequence from PomBase and imported in IGV. Small RNA reads mapping to multiple locations were randomly assigned. By using custom Perl scripts (see Supplemental Table S4; Scripts.tar.bz), the data sets were normalized to the number of reads per million (rpm) sequences for small RNA-seq or rpm mapping to coding sequences for total RNA-seq, poly(A) RNA-seq, Caf1-RIP-seq, and H3 RIP-seq. ChIP data were either normalized by rpm if variation in read amounts was low; if centromeric heterochromatin was lost for example, ChIP-seq data were normalized to regions that were not changed in different mutants. Caf1 ChIP and DRIP reads were summed in a window of 100 nt and divided by a corresponding control to display the fold-change using the Integrative Genomics Viewer (IGV) (http://www.broad.mit.edu/igv). Sequencing data were done in two replicates or the data were confirmed by another method like qRT-PCR, with the exception of small RNA sequencing, which was done only once. Sequenced strains are listed in the Supplemental Table S3.
Statistical analysis
Two-sided t-test for two independent data sets with high variance was used to calculate the P-value. The P-value is displayed with asterisks: <0.05 (*), <0.01 (**), and <0.001 (***).
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE94129.
Supplementary Material
Acknowledgments
We thank Sigrun Jaklin for excellent technical assistance. We thank Silvija Bilokapic for comments on the manuscript and undergraduate students Alrun Basfeld, Maike Becker, Adriana Savova, and Marco Rinn for assistance with molecular biology. We thank Fuyuki Ishikawa for strains. This work was supported by the BioSysNet (Bayerisches Staatsministerium für Wissenschaft, Forschung und Kunst), the European Research Council, ERC-smallRNAhet-309584, and the Deutsche Forschungsgemeinschaft, SFB646.
Author contributions: C.B. and M.H. designed the experiments. C.B., L.S., M.Z., and I.U. performed the experiments. C.B., L.S., and M.H. analyzed the data. C.B. and M.H. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.216986.116.
References
- Allshire RC. 1995. Elements of chromosome structure and function in fission yeast. Semin Cell Biol 6: 55–64. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Ekwall K. 2015. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb Perspect Biol 7: a018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basquin J, Roudko VV, Rode M, Basquin C, Séraphin B, Conti E. 2012. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell 48: 207–218. [DOI] [PubMed] [Google Scholar]
- Böhmdorfer G, Wierzbicki AT. 2015. Control of chromatin structure by long noncoding RNA. Trends Cell Biol 25: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond DM, Baulcombe DC. 2014. Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol 24: 100–107. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819. [DOI] [PubMed] [Google Scholar]
- Castel SE, Ren J, Bhattacharjee S, Chang A-Y, Sánchez M, Valbuena A, Antequera F, Martienssen RA. 2014. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 159: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, García-Muse T, Aguilera A. 2013. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 52: 583–590. [DOI] [PubMed] [Google Scholar]
- Chalamcharla VR, Folco HD, Dhakshnamoorthy J, Grewal SIS. 2015. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc Natl Acad Sci 112: 15548–15555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang Y-C, Denis CL. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA. 2016. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip Rev RNA 7: 438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO. 2012. The Ccr4–not complex. Gene 492: 42–53. [DOI] [PubMed] [Google Scholar]
- Cotobal C, Rodríguez-López M, Duncan C, Hasan A, Yamashita A, Yamamoto M, Bähler J, Mata J. 2015. Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenetics Chromatin 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K, Griffith JD. 1980. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res 8: 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Babbarwal V, Fu J, Brunke-Reese D, Libert DM, Willis J, Reese JC. 2015. Ccr4-not and TFIIS function cooperatively to rescue arrested RNA polymerase II. Mol Cell Biol 35: 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Braun CR, Gygi SP, Moazed D. 2014. Post-transcriptional regulation of meiotic genes by a nuclear RNA silencing complex. RNA 20: 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SEJ, Pan Q, Breen PC, Qi Y, Shi Z, Zhang C, Ruvkun G. 2013. Multiple small RNA pathways regulate the silencing of repeated and foreign genes in C. elegans. Genes Dev 27: 2678–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Garcia JF, Dumesic PA, Hartley PD, El-Samad H, Madhani HD. 2010. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev 24: 1758–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J, Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D. 2010. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140: 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-I, Ayoub N, Cohen A, Grewal SIS. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hansen KR, Ibarra PT, Thon G. 2006. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res 34: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakata S, Siomi MC. 2016. piRNA biogenesis in the germline: from transcription of piRNA genomic sources to piRNA maturation. Biochim Biophys Acta 1859: 82–92. [DOI] [PubMed] [Google Scholar]
- Hiriart E, Vavasseur A, Touat-Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, et al. 2012. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J 31: 2296–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T, Makino S. 2014. Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation. Front Genet 5: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SIS. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976. [DOI] [PubMed] [Google Scholar]
- Jonstrup AT, Andersen KR, Van LB, Brodersen DE. 2007. The 1.4-Å crystal structure of the S. pombe Pop2p deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acids Res 35: 3153–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. 2005. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. 2008. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 18: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. 2011. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev 25: 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell JG, Goodrich KJ, Bähler J, Cech TR. 2005. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J Biol Chem 280: 5249–5257. [DOI] [PubMed] [Google Scholar]
- Marasovic M, Zocco M, Halic M. 2013. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell 52: 173–183. [DOI] [PubMed] [Google Scholar]
- Martienssen R, Moazed D. 2015. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 7: a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, Reese JC. 2012. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47: 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgunova V, Akulenko N, Radion E, Olovnikov I, Abramov Y, Olenina LV, Shpiz S, Kopytova DV, Georgieva SG, Kalmykova A. 2015. Telomeric repeat silencing in germ cells is essential for early development in Drosophila. Nucleic Acids Res 43: 8762–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong E-JE, Li X, Gerber S, Denison C, Gygi S, Moazed D. 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. 2012. DNA–RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 17: 218–233. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2012. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol Chapter 21: Unit 21.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi L, Prasad P, Ekwall K, Cohen A, Svensson JP. 2015. The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO Rep 16: 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Y, Huarte M. 2013. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther 23: 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 65: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Brown GD, Cohen JD, Carroll JS. 2013. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol 14: R147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. 2014. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SIS. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Thillainadesan G, Chalamcharla VR, Meng Z, Balachandran V, Dhakshnamoorthy J, Zhou M, Grewal SIS. 2016. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol Cell 61: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F, Jia S. 2013. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev 27: 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JF, Ohle C, Schermann G, Bendrin K, Zhang W, Fischer T, Zhang K. 2016. A novel epigenetic silencing pathway involving the highly conserved 5′-3′ exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe. PLoS Genet 12: e1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. 2011. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell 41: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier L, Taglini F, Barrales RR, Webb S, Urano T, Braun S, Bayne EH. 2015. Global regulation of heterochromatin spreading by Leo1. Open Biol 5: pii:150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wahba L, Costantino L, Tan FJ, Zimmer A, Koshland D. 2016. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev 30: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Winkler GS. 2013. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta 1829: 561–570. [DOI] [PubMed] [Google Scholar]
- Wang J, Cohen AL, Letian A, Tadeo X, Moresco JJ, Liu J, Yates JR, Qiao F, Jia S. 2016. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev 30: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Shichino Y, Yamamoto M. 2016. The long non-coding RNA world in yeasts. Biochim Biophys Acta 1859: 147–154. [DOI] [PubMed] [Google Scholar]
- Yu R, Jih G, Iglesias N, Moazed D. 2014. Determinants of heterochromatic siRNA biogenesis and function. Mol Cell 53: 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marín L, Chang A-Y, Goto D, et al. 2011. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SIS. 2011. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science 331: 1624–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocco M, Marasovic M, Pisacane P, Bilokapic S, Halic M. 2016. The Chp1 chromodomain binds the H3K9me tail and the nucleosome core to assemble heterochromatin. Cell Discov 2: 16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SIS. 2012. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.