Abstract
During the maternal-to-zygotic transition (MZT), transcriptionally silent embryos rely on post-transcriptional regulation of maternal mRNAs until zygotic genome activation (ZGA). RNA-binding proteins (RBPs) are important regulators of post-transcriptional RNA processing events, yet their identities and functions during developmental transitions in vertebrates remain largely unexplored. Using mRNA interactome capture, we identified 227 RBPs in zebrafish embryos before and during ZGA, hereby named the zebrafish MZT mRNA-bound proteome. This protein constellation consists of many conserved RBPs, some of which are potential stage-specific mRNA interactors that likely reflect the dynamics of RNA–protein interactions during MZT. The enrichment of numerous splicing factors like hnRNP proteins before ZGA was surprising, because maternal mRNAs were found to be fully spliced. To address potentially unique roles of these RBPs in embryogenesis, we focused on Hnrnpa1. iCLIP and subsequent mRNA reporter assays revealed a function for Hnrnpa1 in the regulation of poly(A) tail length and translation of maternal mRNAs through sequence-specific association with 3′ UTRs before ZGA. Comparison of iCLIP data from two developmental stages revealed that Hnrnpa1 dissociates from maternal mRNAs at ZGA and instead regulates the nuclear processing of pri-mir-430 transcripts, which we validated experimentally. The shift from cytoplasmic to nuclear RNA targets was accompanied by a dramatic translocation of Hnrnpa1 and other pre-mRNA splicing factors to the nucleus in a transcription-dependent manner. Thus, our study identifies global changes in RNA–protein interactions during vertebrate MZT and shows that Hnrnpa1 RNA-binding activities are spatially and temporally coordinated to regulate RNA metabolism during early development.
Early embryogenesis in all metazoan species is marked by transcriptional silence of the zygotic genome (Tadros and Lipshitz 2009; Langley et al. 2014). Therefore, the reductive cell divisions that follow fertilization are solely supported by the activity of maternally deposited RNAs and proteins. Subsequent stages of development are governed by the expression of the zygotic genome (Lee et al. 2014), which begins at zygotic genome activation (ZGA). This switch in developmental control from maternal to zygotic instructions is known as maternal-to-zygotic transition (MZT). Recent studies have provided insight into embryonic transcriptomes during MZT, identifying the earliest zygotic transcripts and revealing dynamic, highly orchestrated changes in maternal mRNA stability and translation that are important for establishment of ZGA (Thomsen et al. 2010; Aanes et al. 2011; Pauli et al. 2012; Harvey et al. 2013; Lee et al. 2013; Paranjpe et al. 2013; Heyn et al. 2014; Subtelny et al. 2014; Eichhorn et al. 2016; Lim et al. 2016).
How maternal mRNA is regulated at the levels of translation and stability is the subject of intense investigation. Cytoplasmic polyadenylation represents a conserved mechanism for translational enhancement of dormant mRNAs with short poly(A) tails (Bettegowda and Smith 2007). Strong coupling between poly(A) tail length and translation efficiency (TE) has been observed in the cleavage embryos of many species (Subtelny et al. 2014; Eichhorn et al. 2016; Lim et al. 2016), although the molecular mechanisms that enable this coupling remain unclear. Recent studies have begun to determine a variety of mechanisms through which maternal mRNA clearance is accomplished (Giraldez et al. 2006; Tadros et al. 2007; Lund et al. 2009; Tadros and Lipshitz 2009; Barckmann et al. 2015; Bazzini et al. 2016; Mishima and Tomari 2016). An important role has been ascribed to the evolutionarily conserved miRNA, miR-430/427, that mediates translational repression and clearance of a subset of maternal mRNAs in zebrafish and Xenopus embryos during MZT (Giraldez et al. 2006; Lund et al. 2009). Finally, a strong reliance on the splicing machinery at ZGA has been demonstrated (König et al. 2007; Strzelecka et al. 2010; Lee et al. 2013), yet little is known about how splicing and other pre-mRNA processing events are regulated in the early embryo.
Given the dependence of early embryos on RNA-based regulation of gene expression, major roles for RNA-binding proteins (RBPs) in developmental transitions are anticipated. RBPs regulate every step of RNA life through their association with RNA sequences and/or secondary structures (Glisovic et al. 2008; Jankowsky and Harris 2015). Several RBPs that regulate maternal mRNA degradation, a hallmark of MZT in all metazoans (Tadros and Lipshitz 2009), have been identified. For example, Smaug protein is the master regulator of maternal mRNA degradation in Drosophila melanogaster embryos (Tadros et al. 2007). Nonviable Smaug mutants show cell cycle defects and failure to activate the zygotic genome (Benoit et al. 2009). The mouse tristetraprolin protein ZFP36L2 down-regulates AU-rich element (ARE)-containing mRNAs and is involved in the degradation of maternal mRNAs (Lai et al. 2000). Lack of ZFP36L2 protein causes embryonic arrest at the two-cell stage (Ramos et al. 2004). These two cases exemplify the importance of RBPs in developmental transitions. However, the identity and contribution of other RBPs in shaping early embryonic transcriptome and developmental processes remain elusive.
Recently, mRNA interactome capture has been used to identify mRNA-binding proteins (mRBPs) in mammalian cells, yeast, C. elegans, and Drosophila (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013; Mitchell et al. 2013; Beckmann et al. 2015; Matia-Gonzalez et al. 2015; Sysoev et al. 2016; Wessels et al. 2016). This method uses UV crosslinking to induce the formation of covalent bonds between mRNA and RBPs, enabling stringent purification of RBPs on oligo(dT) beads. This purification scheme is ideal for vertebrate embryos that contain abundant yolk protein contaminants like vitellogenin, which comprises ∼90% of total protein weight (Jorgensen et al. 2009). Here, we uncover the repertoire of vertebrate mRBPs during MZT, by adapting mRNA interactome capture to living zebrafish embryos at specific time points before and during ZGA. To determine the contribution of an identified mRNA interactome component—the known splicing factor Hnrnpa1—to the regulation of maternal and zygotic RNAs, individual nucleotide resolution crosslinking and immunoprecipitation (iCLIP) (König et al. 2010) was also established in zebrafish embryos. Follow-up experiments reveal unique roles and dynamic activities of Hnrnpa1 in regulation of RNA metabolism during embryogenesis.
Results
Identification of the zebrafish embryonic mRNA-bound proteome before and during ZGA
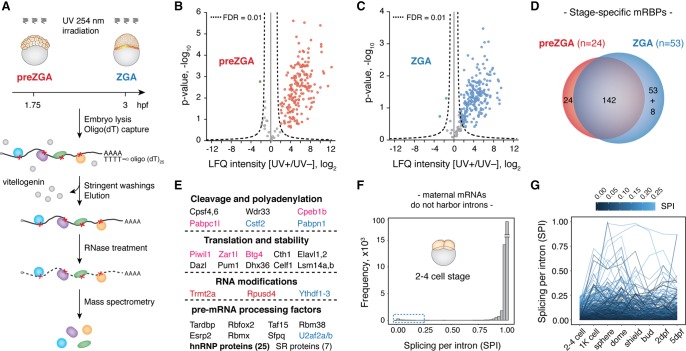
To purify mRBPs during zebrafish MZT, we adapted the mRNA interactome capture method to two stages of zebrafish development, before ZGA (32–64 cell stage, hereafter called preZGA) and at the onset of ZGA (512–1k cell stage, hereafter called ZGA). RNA–protein crosslinking was carried out by irradiating living embryos with 254 nm UV light, after which the embryos were lysed and mRNPs purified on oligo(dT) beads. Proteins were subsequently released by RNase treatment and analyzed by LC-MS/MS (Fig. 1A). To control for nonspecific binders, we simultaneously performed mRNP purifications from UV nonirradiated (UV−) embryos. In total, mass spectrometry quantified 262 and 312 proteins from preZGA and ZGA samples, respectively, with clear separation of UV+ and UV− samples in two distinct clusters (Supplemental Fig. S1A,B; Supplemental Table S1). Furthermore, 200 and 247 proteins were quantified from at least two replicates in UV+ purifications, whereas 176 and 212 proteins were enriched in UV+ samples with a false discovery rate (FDR) of 1% in preZGA and ZGA, respectively (Fig. 1B,C; Supplemental Table S2A,B). After removing vitellogenin and zona pellucida contaminants, we report 166 preZGA and 203 ZGA high-confidence mRNA-interacting proteins. We refer to the full constellation of 227 proteins from both embryonic stages combined (Supplemental Table S3) as the zebrafish MZT mRNA-bound proteome (mRBPome).
Figure 1.
The zebrafish MZT mRNA-bound proteome is conserved, dynamic, and rich in pre-mRNA processing factors. (A) Schematic of the method for in vivo identification of mRBPs from early zebrafish embryos. Experiments were performed at two developmental time points: preZGA (32–64 cell stage) and ZGA (512–1k cell stage) in three biological replicates: (hpf) hours post-fertilization. (B,C) Volcano plots display the enrichment of detected proteins between UV+ and UV− samples (x-axis) and their significance (y-axis). Hyperbolic, dashed lines represent the applied statistical threshold of FDR = 0.01. Data points beyond this threshold refer to proteins enriched in UV+ (blue and red) and UV− (green) conditions. Nonenriched proteins are shown in gray. (D) Venn diagram displaying differences between preZGA (red) and ZGA (blue) mRBPomes. Note that eight proteins within the ZGA-specific group were also detected at the preZGA stage, yet below the enrichment threshold. (E) MZT mRBPome components grouped with respect to their major known functions. Names of the selected proteins are color-coded based on their embryo- and stage-specific detection: (black) both stages; (red) preZGA-specific; (blue) ZGA-specific; (pink) embryo-specific. (F) Histogram displaying splicing score per intron (SPI) of all expressed maternal mRNAs at the two- to four-cell stage shows that the majority (97.5%) of maternal transcripts are efficiently spliced. (G) Line plot tracking introns with low SPI values (0–0.25) from the two- to four-cell stage to 5 d post-fertilization (dpf) suggests the majority of these introns are never spliced out.
Gene Ontology (GO) and protein domain analyses of the zebrafish MZT mRBPome revealed significant enrichment of RNA binding and nucleic acid binding, as well as translational regulation annotations (Supplemental Fig. S1C–E). Canonical RNA-binding domains were highly represented protein modules with the lowest P-values for RRM and KH protein domains (Supplemental Fig. S1F). Although only 91 of 227 proteins were annotated as RNA, mRNA, and poly(A) RNA binding in zebrafish, comparison of the remaining proteins with human orthologs increased this number to a total of 198, or 87% of all identified proteins (Supplemental Table S3). Pairwise comparison with mammalian mRNA interactomes revealed that the majority of proteins were shared (Supplemental Table S4), indicating that mRBPomes are conserved among vertebrates. Interestingly, 20 proteins in the zebrafish MZT mRBPome were not previously detected by mRNA interactome capture in mammalian cells (Supplemental Table S3), indicating potential embryo-specific mRNA association. Indeed, this group includes proteins with restricted expression and/or elevated functions during early embryogenesis among which were embryonic poly(A)-binding protein (Pabpc1l) and cytoplasmic polyadenylation element binding protein (Cpeb1b), regulators of embryonic cytoplasmic mRNA polyadenylation (Charlesworth et al. 2013); the paralog of the maternal-effect gene Zar1 (Wu et al. 2003); and Btg4 and Piwi proteins, negative regulators of maternal mRNA stability in mouse oocytes and fly embryos, respectively (Barckmann et al. 2015; Yu et al. 2016). Finally, comparison of the preZGA and ZGA mRBPomes revealed 24 and 53 proteins that were specifically detected at preZGA and ZGA, respectively (Fig. 1D; Supplemental Table S3). To test whether these differences in the mRNA-bound fraction arose from changes in protein expression during MZT, we analyzed changes in the translational efficiency (TE) of mRNAs encoding respective proteins as an indirect measure of their differential expression over development (Supplemental Fig. S1G,H). We found that the expression of identified proteins was not substantially different between 2 and 4 hpf, suggesting that changes in the levels of mRBPs cannot fully account for the observed differences in the mRNA-bound fraction between the examined developmental time points. Taken together, we conclude that the zebrafish MZT mRBPome mostly consists of known and highly conserved mRNA interactors, some of which appear to be specific for early embryogenesis and developmental stage.
The zebrafish MZT mRNA-bound proteome is rich in conserved pre-mRNA processing factors
Inspection of individual RBPs detected in the MZT mRBPome enables their grouping into different classes (Fig. 1E). For example, core components of the cytoplasmic mRNA polyadenylation complex were detected, consistent with the regulation of maternal mRNA translational efficiency by cytoplasmic polyadenylation (Subtelny et al. 2014). Interestingly, several constituents of the nuclear polyadenylation machinery, e.g., nuclear poly(A)-binding protein (Pabpn1) and nuclear cleavage factor (Cstf2), were ZGA-specific, consistent with their requirement for 3′-end processing at the beginning of zygotic transcription. Many positive and negative regulators of mRNA stability and/or translation were shared between the two developmental stages, some of which appear as embryo-specific mRNA-interactors. A number of stage-specific writers and readers of RNA modifications were also identified, suggesting that RNA modifications may play important roles in the regulation of the embryonic transcriptome.
Surprisingly, pre-mRNA processing factors were notably enriched at both stages of development, although we did not expect pre-mRNA to be present in transcriptionally inactive embryos. SR proteins and heterogeneous ribonucleoprotein particle (hnRNP) proteins—both known for prominent roles in pre-mRNA splicing—represented the most numerous protein groups in the zebrafish MZT mRBPome after ribosomal proteins (Fig. 1E). In somatic cells, hnRNP proteins bind to introns of nascent transcripts to globally regulate pre-mRNA processing events, namely pre-mRNA packaging, splicing, and mRNA export (Huelga et al. 2012; Müller-McNicoll and Neugebauer 2013). A potential explanation for the association of hnRNP proteins with mRNAs before ZGA could be the persistence or even regulatory presence of introns in maternal transcripts. The delayed splicing of specific introns serves as a potent mechanism for the translational regulation of maternal mRNA in fern, Marsilea versita (Boothby et al. 2013). To determine whether retained introns are present in the maternal mRNA pool, we analyzed poly(A)+ RNA-seq data from two- to four-cell-stage embryos (Pauli et al. 2012). Transcriptome-wide calculation of a splicing score based on exon–intron and exon–exon junction reads (Herzel and Neugebauer 2015) was carried out for each annotated intron and revealed that 97.5% of introns are fully removed from zebrafish maternal mRNAs (Fig. 1F). A small subset of introns that displayed partial retention (Fig. 1F, blue dashed box) were further analyzed for changes in splicing levels from fertilization to 5 d of development (Fig. 1G). Among these, the majority maintained a low level of splicing, suggesting that these retained introns are either incorrectly annotated or always inefficiently spliced. Therefore, we conclude that zebrafish maternal mRNAs are fully spliced.
Hnrnpa1 regulates poly(A) tail length and translation before ZGA through prominent 3′ UTR binding
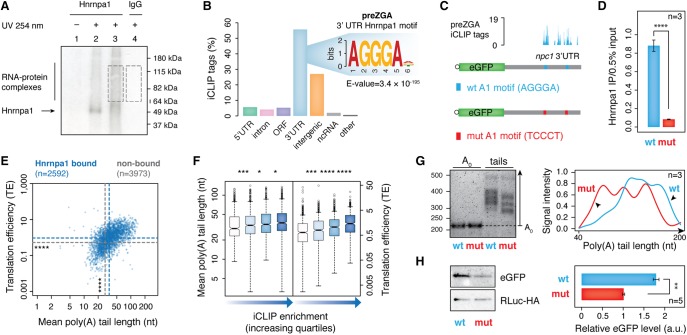
The abundance of hnRNP proteins in the preZGA mRBPome in the absence of unspliced transcripts suggested that these factors may take on splicing-independent roles in transcriptionally silent embryos. To investigate the type of regulation hnRNP proteins exert on maternal mRNAs, we utilized iCLIP to identify endogenous RNA targets and binding sites for one representative hnRNP protein family member, Hnrnpa1. Zebrafish genome contains two hnrnpa1 genes (hnrnpa1a and hnrnpa1b) that arose from the teleost-specific genome duplication (Howe et al. 2013). These genes encode two highly similar Hnrnpa1a and Hnrnpa1b proteins, both of which were detected in our zebrafish MZT mRBPome (Supplemental Table S3). We refer to both of these protein variants as Hnrnpa1 protein throughout the study. First, we confirmed that the monoclonal antibody used in this study specifically reacts with endogenous zebrafish Hnrnpa1 protein (Supplemental Fig. S2A). After adaptation of the iCLIP protocol to zebrafish embryos (Supplemental Fig. S2B), Hnrnpa1-RNA complexes were immunopurified at preZGA, and isolated RNA was converted into a size-selected and amplified cDNA library for RNA-seq (Fig. 2A; Supplemental Fig. S2C). Three independent biological replicates were highly reproducible and therefore pooled for further identification of significant iCLIP tags (FDR < 0.01). Nonimmune IgG pulldowns yielded significantly fewer iCLIP tags that poorly correlated with the Hnrnpa1 data, as expected for background (Supplemental Fig. S2D; Supplemental Table S5). Analysis of Hnrnpa1 association with genic and intergenic regions showed that most Hnrnpa1 iCLIP tags mapped to protein-coding genes. In contrast to the preferential association with introns in mammalian cells (Huelga et al. 2012), 55% of all iCLIP tags were assigned to 3′ UTR regions of maternal mRNAs (Fig. 2B). Furthermore, motif analysis revealed a significant enrichment of the AGGGA hexanucleotide sequence that resembles previously reported Hnrnpa1 consensus binding motif UAGGGA/U (Burd and Dreyfuss 1994), suggesting that the observed Hnrnpa1 association with 3′ UTRs was sequence-specific (Fig. 2B).
Figure 2.
Before ZGA, Hnrnpa1 regulates poly(A) tail length and translation of maternal mRNAs by binding 3′ UTRs. (A) Representative autoradiograph showing Hnrnpa1–RNA complexes immunopurified from zebrafish embryos at 2 hpf, resolved by SDS-PAGE and transferred to nitrocellulose. Lane 2: high RNase I treatment (1:50 dilution); lanes 1,3,4: low RNase I treatment (1:1000). RNA within the RNP complexes indicated by dashed boxes was purified and used for cDNA library preparation. (B) Bar plot displaying the proportion of Hnrnpa1 iCLIP tags in different genic and intergenic regions indicated below each bar. Inset shows significantly enriched hexanucleotide Hnrnpa1 binding motif in 3′ UTRs. (C) Schematic diagram of two constructs consisting of eGFP coding sequence and 1.1-kb-long 3′ UTR of an Hnrnpa1-bound mRNA (npc1) with either wild-type ([WT] blue) or mutated ([mut] red) Hnrnpa1 motif. Hnrnpa1 binding sites from iCLIP data are shown with blue bars above the npc1 3′ UTR annotation. (D) RT-qPCR on the α-Hnrnpa1 immunopurified RNA from UV-irradiated embryos, previously injected with mRNA reporters. The assay shows differential binding of Hnrnpa1 to the WT and mut reporter mRNAs in vivo. y-axis, the amount of immunopurified mRNA relative to 0.5% input; error bars represent SEM; n = 3. (****) P < 0.0001 (two-sided t-test). (E) Scatterplot showing mutual dependence of poly(A) tail length (x-axis) and TE (y-axis) for Hnrnpa1-bound maternal mRNAs. Blue and gray dashed lines refer to the median of poly(A) tail length and TE for Hnrnpa1-bound and nonbound transcripts, respectively: (****) P < 2.2 × 10−16 (Wilcoxon rank test). (F) Box plots showing the distribution of poly(A) tail length and TE of Hnrnpa1 targets with respect to the extent of Hnrnpa1 binding to maternal transcripts (iCLIP enrichment). Box plots refer to iCLIP enrichment quartiles in an increasing order for each mRNA feature, as shown by the blue-colored gradient arrows: (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001 (Wilcoxon rank test). (G) 1.5% agarose gel (left) showing different poly(A) tail lengths of the WT and mut mRNA reporters at 64-cell stage measured by ePAT. The right panel represents the average distribution of poly(A) tail signal intensities from three independent biological replicates. (H) Western blots show RLuc-HA and eGFP protein levels from 64-cell stage embryos injected with WT and mut mRNA reporters. The adjacent bar plot presents quantification of eGFP levels normalized to the RLuc-HA expression (injection control). Error bars show SEM. Five embryos were loaded per lane (n = 5). (**) P < 0.01 (two-sided t-test).
To test whether the iCLIP-derived motif plays a role in Hnrnpa1 binding in vivo, we created two mRNA reporters consisting of a common eGFP coding sequence followed by the 1.1-kb-long 3′ UTR of an Hnrnpa1 target (npc1) with either its wild-type ([WT] AGGGA) or a mutated ([mut] TCCCT) Hnrnpa1 motif (Fig. 2C). These mRNA reporters were injected into one-cell stage embryos, and the differential binding of Hnrnpa1 was assayed 2 h later. RT-qPCR analysis on the RNA from UV-crosslinked and immunopurified Hnrnpa1-RNA complexes revealed that Hnrnpa1 associated 10.6-fold less with the mRNA reporter carrying the mutated motif (Fig. 2D). This shows that the Hnrnpa1 AGGGA motif is necessary for proper Hnrnpa1 binding to RNA targets in vivo.
3′ UTRs are common targets of RBPs and miRNAs that regulate mRNA stability and translation (Matoulkova et al. 2012). In transcriptionally silent embryos, maternal mRNA translation is tightly controlled by regulation of poly(A) tail length, whereas mRNA stabilities remain largely unaffected (Subtelny et al. 2014; Eichhorn et al. 2016). Analysis of available ribosome and poly(A) tail profiling data (Lee et al. 2013; Subtelny et al. 2014) revealed that Hnrnpa1 targets harbor significantly longer poly(A) tails (median length: 34.4 and 28.1, P = 7 × 10−60) and display higher TE than nonbound mRNA counterparts (median TE: 0.94 and 0.53, P = 5.02 × 10−59) (Fig. 2E). Therefore, we sought to determine whether the extent of Hnrnpa1 binding predicts poly(A) tail length and translational output of maternal transcripts. First, we quantified Hnrnpa1 binding to individual transcripts by normalizing iCLIP tag density to mRNA expression levels (iCLIP enrichment) (Supplemental Fig. S2E). The calculated iCLIP enrichment was subsequently compared with poly(A) tail length and TE of all Hnrnpa1 mRNA targets. These analyses uncovered a moderate correlation between the extent of Hnrnpa1 binding to maternal mRNAs and both mRNA features (Fig. 2F).
Collectively, the preceding data suggested that Hnrnpa1 may be directly involved in the regulation of poly(A) tail length and translation of maternal mRNAs in preZGA embryos. To test this possibility, we injected established mRNA reporters into one-cell-stage embryos along with HA-tagged Renilla luciferase RNA (Rluc-HA RNA, injection control) and measured polyadenylation and translation 2 h after injection. To measure poly(A) tail length, we implemented the extension poly(A) test (ePAT) (Supplemental Fig. S3A; Janicke et al. 2012), which showed that poly(A) tails of transcripts with the WT 3′ UTR were significantly longer than the poly(A) tails of the same transcript in which the Hnrnpa1 binding site had been mutated (Fig. 2G; Supplemental Fig. S3B). Moreover, semiquantitative Western blot analysis revealed a significant 1.7-fold decrease in eGFP protein expression from the mutated mRNA reporter (Fig. 2H; Supplemental Fig. S3C). Because the levels of each mRNA remained unchanged over the course of both experiments (Supplement Fig. S3D–F), these effects of Hnrnpa1 binding cannot be ascribed to differential stabilities of the injected mRNAs. Taken together, the biochemical data and bioinformatic analysis indicate that Hnrnpa1 controls poly(A) tail length and translation in vivo by binding to AGGGA consensus sequences in the 3′ UTRs of maternal transcripts before ZGA.
Hnrnpa1 displays dynamic RNA-binding activities and cellular relocalization during MZT
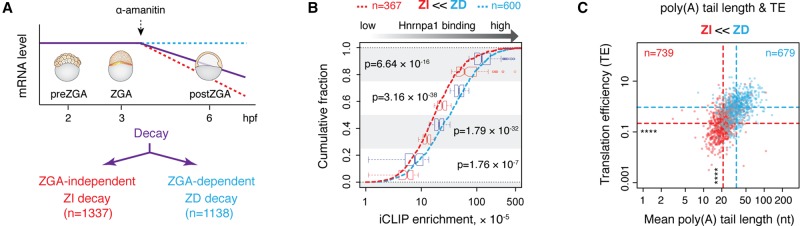
Another scorable feature of MZT is maternal mRNA stability. Maternally provided mRNAs undergo destabilization either via transcription-independent (ZGA-independent [ZI]) or transcription-dependent (ZGA-dependent [ZD]) pathway. Using processed RNA-seq data (Mishima and Tomari 2016), we identified all decaying maternal transcripts based on the significant drop in their expression (greater than twofold) between 2 and 6 hpf. These decaying mRNAs were subsequently divided into two cohorts depending on their expression response to α-amanitin treatment; genes whose mRNA levels significantly decreased upon transcription block were considered ZI decay genes, whereas those whose mRNA expression became stable upon α-amanitin injection were classified as ZD decay genes (Fig. 3A). Interestingly, Hnrnpa1 was found to bind to a substantially higher number of ZD decay mRNAs and to a significantly higher extent than to ZI decay transcripts (ZD decay, 52.7% and ZI decay, 27.4%) (Fig. 3B). Consistent with preceding findings (Fig. 2E,F), we also demonstrated that ZD decay mRNAs harbor significantly longer poly(A) tails (median length: 34.8 and 21 nt, P = 5.62 × 10−119) and show higher TE (median TE: 0.93 and 0.21, P = 1.29 × 10−88) at preZGA than ZI decay counterparts (Fig. 3C). Thus, we conclude that a high level of Hnrnpa1 binding represents a feature of long-tailed and highly translated maternal transcripts that will be targeted for degradation hours later in a ZGA-dependent manner.
Figure 3.
Hnrnpa1 preferentially associates with ZGA-dependent decay genes. (A) Schematic representation of two distinct decaying gene groups. All decaying maternal mRNAs are shown by a purple line. Among those, ZGA-independent (ZI) decay genes (red dashed line) do not require zygotic transcription for their destabilization and continuously decay upon α-amanitin treatment. ZGA-dependent (ZD) decay genes (blue dashed line) require zygotic transcriptional activity for their clearance, thereby remaining stable in α-amanitin treated embryos. The total number of identified ZI and ZD decay genes is indicated. (B) Cumulative plot showing the fraction of ZI (red dashed line) and ZD (dashed blue line) decay genes (y-axis) and their respective Hnrnpa1 iCLIP enrichment (x-axis). Box plots in the background show the distribution of iCLIP enrichment per each quartile for ZI and ZD decay genes. Indicated P-values were derived from pairwise comparison of ZI (red) and ZD (blue) decay genes within each quartile (Wilcoxon rank test). (C) Scatterplot showing differences in poly(A) tail length and TE of ZI and ZD decay genes. Red and blue dashed lines refer to the median of poly(A) tail length and TE for ZI and ZD decay genes, respectively. The number of genes in each group is indicated: (****) P < 2.2 × 10−16 (Wilcoxon rank test).
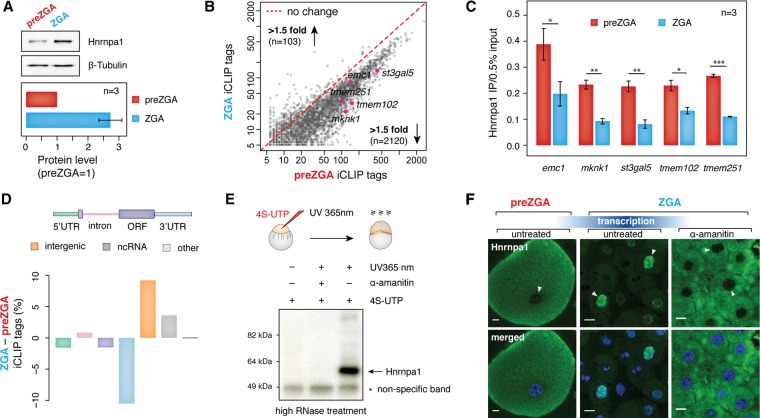
If Hnrnpa1 is important for maternal mRNA stability during MZT, it may change its association with maternal RNA at ZGA. First, we tested whether Hnrnpa1 levels change during MZT by performing semiquantitative Western blot analysis and measured a ∼2.5-fold increase in Hnrnpa1 between preZGA and ZGA (Fig. 4A; Supplemental Fig. S4A). To examine if the elevated Hnrnpa1 expression corresponds to its increased association with maternal transcripts at ZGA, an additional iCLIP experiment was conducted at ZGA in four biological replicates (Supplemental Fig. S4B–D). Although some differentially bound transcripts were detected, the majority of Hnrnpa1 targets were shared between preZGA and ZGA (Supplemental Fig. S4E). To precisely determine the level of Hnrnpa1 binding to the overlapping set of transcripts, we quantified Hnrnpa1 iCLIP tags mapping to individual maternal mRNAs at two different time points. Since maternal mRNAs do not change their expression levels between preZGA and ZGA, as evidenced by our RT-qPCR analysis (Supplemental Fig. S4F) and several previous studies (Voeltz and Steitz 1998; Subtelny et al. 2014; Eichhorn et al. 2016; Mishima and Tomari 2016), we utilized the absolute number of iCLIP tags as a measure of the Hnrnpa1 binding to individual mRNAs. Surprisingly, a comparison of iCLIP tag numbers revealed a >1.5-fold decrease in Hnrnpa1 association with 70% of mRNAs at the ZGA stage (Fig. 4B). The observed loss of Hnrnpa1 interaction with maternal mRNAs at ZGA was validated by UV crosslinking, immunoprecipitation, and RT-qPCR on five endogenous genes (Fig. 4C). It is also noteworthy that the proportion of iCLIP tags generally decreased within protein-coding transcripts at ZGA, in particular within 3′ UTR regions (Fig. 4D). Thus, Hnrnpa1 seems to bind less, not more, to maternal mRNAs during MZT.
Figure 4.
Hnrnpa1 acquires different RNA-binding activities and localization at ZGA. (A) Western blots showing Hnrnpa1 expression level at 2 hpf (preZGA) and 3 hpf (ZGA). Beta-tubulin served as a loading control. A single embryo was loaded per lane. Bar plot displayed below shows quantified levels of Hnrnpa1 expression at the preZGA and ZGA stages. Hnrnpa1 expression at preZGA was set to 1. Error bar shows SEM; n = 3. (B) Scatterplot showing a decrease in the number of significant Hnrnpa1 iCLIP tags per mRNA at the ZGA stage. The number of iCLIP tags for the overlapping set of Hnrnpa1 mRNA targets was plotted (x-axis, ZGA stage; y-axis, preZGA stage). The number of genes with a <1.5-fold increase and decrease in Hnrnpa1 iCLIP tags at ZGA is indicated. Red dashed line refers to no change in the number of iCLIP tags between two developmental stages. mRNAs assayed in Figure 4C are highlighted in pink. (C) RT-qPCR on the RNA recovered from in vivo UV crosslinked and immunopurified Hnrnpa1-RNA complexes shows different levels of Hnrnpa1 binding to five endogenous maternal mRNAs at preZGA and ZGA: (y-axis) amount of immunopurified transcripts relative to 0.5% input; error bars represent SEM; n = 3. (D) Bar plot displaying global changes in the proportion of Hnrnpa1 iCLIP tags between preZGA and ZGA developmental stages. The color of each bar corresponds to the colored annotation above the bar plot. Negative and positive values on y axis indicate decrease and increase of the iCLIP tag percent in the respective regions at ZGA. (E) PAR-CLIP test showing Hnrnpa1 association with newly synthesized zygotic transcripts at ZGA. Presented autoradiograph shows the nitrocellulose membrane with immunopurified and radiolabeled zygotic RNA species in complex with Hnrnpa1 at 3 hpf. Note that the complexes were detected only in 4S-UTP-injected and UV365 nm-irradiated embryos. (F) Fluorescence microscopy images show a change of Hnrnpa1 subcellular localization during the zebrafish MZT. Immunostaining was performed on fixed embryos at the developmental stages indicated above the images. Nuclei were stained with Hoechst (blue) and Hnrnpa1 localization revealed by Alexa 488 dye (green). Transcription block was induced by microinjections of 0.4 ng α-amanitin into one-cell stage embryos. Nuclei are indicated with white arrowheads: (scale bars) 10 µm. Note the change in cell size due to the reductive cell divisions.
As expected, the loss of Hnrnpa1 binding to mRNA coincided with a small, yet significant gain of iCLIP tags in introns (P < 0.01, two-sided t-test), consistent with de novo transcription of intron-containing genes and binding of Hnrnpa1 to newly produced zygotic transcripts. Moreover, we observed a substantial increase of iCLIP tags within noncoding and intergenic regions (Fig. 4D; Supplemental Fig. S5A). Interestingly, the increase of intergenic iCLIP tags could be largely ascribed to Chromosome 4, which harbors a significantly higher number of noncoding genes and repetitive elements than any other chromosome in zebrafish (Supplemental Fig. S5B,C; Howe et al. 2013). Moreover, among noncoding iCLIP tags that increased at the ZGA stage, we found preferential binding of Hnrnpa1 to nuclear U1 and U6 snRNAs, whose genomic loci mainly localize in clusters on Chromosome 4 (Supplemental Fig. S5D,E). To test de novo association of Hnrnpa1 with zygotic transcripts in vivo, we performed photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) in the presence and absence of α-amanitin. After metabolic labeling of newly produced transcripts by 4S-UTP injections and embryo UV irradiation with long wavelength UV light (UV 365 nm), Hnrnpa1-RNA complexes were immunopurified, radiolabeled, and resolved by SDS-PAGE. Autoradiography detected robust Hnrnpa1 binding to newly synthesized zygotic RNA at ZGA (Fig. 4E).
Taken together, the global decrease in iCLIP tags in protein-coding genes and simultaneous increase in introns and intergenic and noncoding regions suggested that Hnrnpa1 functions may shift from the cytoplasmic to the nuclear compartment following ZGA. This hypothesis was tested through Hnrnpa1 immunostaining of preZGA and ZGA embryos, in which Hnrnpa1 was detected exclusively in the cytoplasm before ZGA and intensely in some of the nuclei during ZGA (Fig. 4F). Because cell divisions are asynchronous at the ZGA time point, a likely explanation for the dimly stained cells lacking nuclear staining is that they are in or close to mitosis. In agreement with the notion that Hnrnpa1 relocalization to the nucleus depends on ZGA, we blocked transcription with α-amanitin and observed Hnrnpa1 cytoplasmic retention (Fig. 4F). Immunostaining with monoclonal antibody 16H3, which recognizes a shared epitope on four SR proteins and numerous components of the spliceosome (Neugebauer et al. 1995), revealed a similar shift from cytoplasmic to nuclear localization (Supplemental Fig. S6), indicating the generality of this phenomenon for pre-mRNA splicing factors.
Hnrnpa1 binds pri-mir-430 and regulates its processing in the nucleus
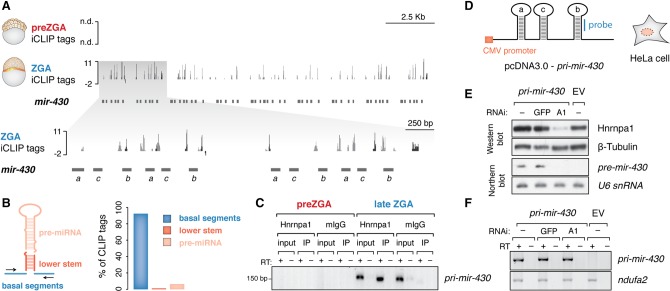
Analysis of the ZGA iCLIP tags that were mapped to intergenic and noncoding RNA showed their presence across the miR-430 genomic region (Fig. 5A). Despite a comprehensive understanding of the mode of miR-430 action during MZT and its importance for development (Giraldez et al. 2005; Bazzini et al. 2012; Takacs and Giraldez 2015), little is known about the regulation of miR-430 processing. The zebrafish mir-430 locus is a 17-kb-long genomic region on Chromosome 4 that harbors more than 50 repeated miR-430 sequences. The majority (85%–92%) of Hnrnpa1 iCLIP tags mapping to the mir-430 locus were located in the basal segments that surround individual miR-430 hairpins and belong exclusively to primary miR-430 sequences, whereas only 10%–13% mapped to sequences within the pre-miRNA intermediates (Fig. 5B; Supplemental Fig. S5F). This pattern differs from Hnrnpa1 binding to conserved terminal loops in several human miRNAs (Guil and Cáceres 2007; Michlewski et al. 2008; Michlewski and Cáceres 2010). We confirmed Hnrnpa1 association with pri-mir-430 by UV crosslinking and immunoprecipitation followed by RT-PCR (Fig. 5C). These data suggest a role for Hnrnpa1 in the nuclear cleavage of pri- to pre-mir-430 through binding to regions at the base of pri-miRNA stems.
Figure 5.
Hnrnpa1 binds nuclear pri-mir-430 transcripts and regulates their processing at ZGA. (A) ZGA-specific Hnrnpa1 binding across the mir-430 locus. Significant iCLIP tags (FDR < 0.01) derived from unique and multimapped reads are shown in dark and light gray, respectively. The inset displays an enhanced view of iCLIP tags in the 5′ end of the mir-430 locus (gray shaded box). Annotations of individual mir-430 genes are indicated with horizontal dark gray bars (mir-430a, mir-430c, mir-430b): (n.d.) not detected; negative values in the tracks represent iCLIP tags mapped to the opposite (−) strand. (B) Bar plot shows relative abundance of Hnrnpa1 iCLIP tags derived from multimapped reads across different types of pri-mir-430 regions. Each bar color corresponds to the colored region in the schematic depiction of the pri-miRNA structure on the left. (C) RT-PCR on RNA extracted from in vivo crosslinked and immunopurified Hnrnpa1–RNA complexes shows specific association of the protein with pri-mir-430 transcripts only at the time of its expression. Note a lack of RNA detection at the preZGA stage. (D) Schematic of the cytomegalovirus (CMV) promoter-driven construct harboring three consecutive miR-430 hairpins (a, c, b) from the zebrafish miR-430 cluster utilized for transient HeLa cell transfection. Location of the DNA probe used for Northern blot analysis is shown in blue. (E) Western blot showing levels of Hnrnpa1 in cells transfected with shRNA-expressing constructs against Hnrnpa1 (A1), GFP (GFP), and empty RNAi vector (−). Cells were subsequently transfected with pri-mir-430 construct and empty vector (EV). Beta-tubulin served as a loading control. Northern blot showing a decrease in pre-mir-430 levels upon Hnrnpa1 depletion; U6 snRNA served as a loading control. Note that the human ortholog of mir-430 is not endogenously expressed (−/EV). (F) RT-PCR on total RNA from HeLa cells shows unchanged pri-mir-430 levels upon Hnrnpa1 depletion.
To determine whether Hnrnpa1 is required for miR-430 biogenesis, an in vivo experimental assay was established. We found that depletion of Hnrnpa1 from early embryos using translation-blocking morpholinos was not possible due to the large maternal contribution of protein (data not shown). We therefore devised a heterologous assay system that relies on the evolutionary conservation of Hnrnpa1 and core proteins involved in the miRNA biogenesis pathway (Akindahunsi et al. 2005; Batista et al. 2014; Ha and Kim 2014). Because human HeLa cells lack expression of the human mir-430 ortholog, MIR302, which is also restricted to early development (Rosa et al. 2009; Cai et al. 2013), introduction of zebrafish pri-mir-430 transcripts in HeLa cells could provide a test for the contribution of Hnrnpa1 to the nuclear processing of zebrafish pri-mir-430. Therefore, a construct containing three consecutive miR-430 hairpins derived from the zebrafish miR-430 polycistron was transfected into HeLa cells (Fig. 5D). Under control conditions, pri-mir-430 was efficiently processed to pre-mir-430. Upon depletion of Hnrnpa1, pre-mir-430 production was strongly decreased, whereas pri-mir-430 levels remained unchanged (Fig. 5E,F). Together with the ZGA iCLIP data from zebrafish, these findings indicate that the nuclear pool of Hnrnpa1 regulates pri-mir-430 processing (Fig. 6).
Figure 6.
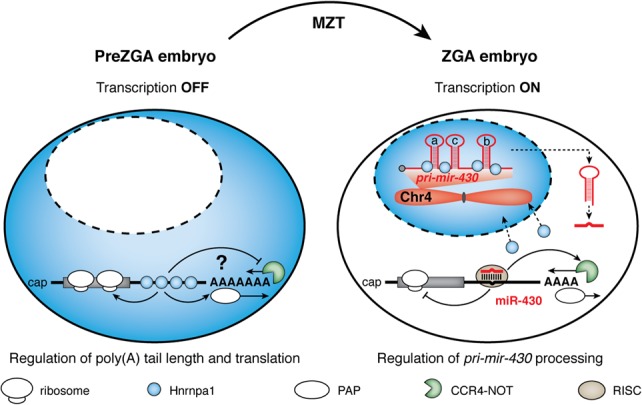
Working model for dynamic activities of Hnrnpa1 during zebrafish MZT. Before ZGA, cytoplasmically localized Hnrnpa1 selectively binds to 3′ UTRs of maternal mRNAs that undergo ZGA-dependent (ZD) decay and positively regulates poly(A) tail length and translation via an unknown mechanism (either by inhibiting active deadenylation or by promoting polyadenylation). Following ZGA, Hnrnpa1 dissociates from maternal transcripts and translocates to the nucleus, where it binds noncoding RNAs derived from zebrafish Chromosome 4, including pri-mir-430. Nuclear Hnrnpa1 promotes pri-mir-430 processing, thereby indirectly contributes to consequential CCR4-NOT-mediated deadenylation and clearance of ZD decay mRNAs.
Discussion
The sole dependence of early development on post-transcriptional control of gene expression before ZGA defines early embryos as a rich biological context for RNA regulation. Although RNA-binding proteins are the acknowledged drivers of post-transcriptional regulatory networks, their identities and functions during developmental transitions in vertebrates are largely unknown. By using mRNA interactome capture, we identify a constellation of 227 high-confidence mRBPs from two developmental stages encompassing zebrafish MZT. mRNA interactome capture was originally developed to comprehensively annotate mRNA binding proteins expressed by mammalian tissue culture cells, including HeLa and HEK293 (Baltz et al. 2012; Castello et al. 2012). Subsequently, the approach was extended to yeast, mouse ES cells, human hepatocytic HuH-7 cells, whole C. elegans animals (Kwon et al. 2013; Mitchell et al. 2013; Beckmann et al. 2015; Matia-Gonzalez et al. 2015), and more recently to early Drosophila embryos (Sysoev et al. 2016; Wessels et al. 2016). Despite a large number of shared components, these mRBPomes contain potentially unique mRNA interactors, raising the possibility that differences between cell types may in part be explained by distinct regulation of RNA metabolism by cell-type–specific mRBPs. Consistent with this, a comparison with mammalian mRBPomes revealed that a fraction of our zebrafish MZT mRBPome (9%) has not been previously detected in other mRNA interactomes. Indeed, it has been shown that the depletion of maternal-effect genes, Zar1 and Zar1l, causes embryonic arrest in early mouse embryos (Wu et al. 2003; Hu et al. 2010). Moreover, lack of maternal Piwi protein in fly embryos causes developmental defects, such as abnormal chromosome morphology and cell cycle arrest (Mani et al. 2014). One possibility is that these mRBPs (e.g., ZAR1 and Piwi) escaped detection in other mRNA interactome capture experiments. Alternatively, our observations suggest that these unique mRBPs are restricted to and important for RNA metabolism in early embryogenesis.
Our subsequent findings suggest that the activities of shared proteins between different mRBPomes may be cell-type–specific or developmental-stage–specific. First, numerous factors that stimulate mRNA degradation in mammalian systems were associated with zebrafish maternal transcripts hours before the major wave of maternal mRNA degradation, suggesting that maternal mRNA stability may be regulated by changes in the activities of these proteins during embryogenesis. Second, the enrichment of nuclear pre-mRNA processing factors, such as hnRNP proteins, before ZGA suggests that these proteins may take on unconventional functions, because preZGA zebrafish embryos are transcriptionally silent and maternal mRNAs lack introns. The detection of pre-mRNA splicing factors in the preZGA fly mRBPome underscores this possibility (Sysoev et al. 2016; Wessels et al. 2016), which we investigated in depth through studies of the MZT mRNA interactome component, Hnrnpa1.
Here, we provide several lines of evidence that Hnrnpa1 regulates poly(A) tail length and translation of maternal mRNAs before ZGA. First, our preZGA iCLIP data detect an unusual sequence-specific association of Hnrnpa1 with 3′ UTRs of long-tailed and highly translated maternal mRNAs. Second, the extent of Hnrnpa1 binding was found to correlate with poly(A) tail length and TE, two tightly connected mRNA features in preZGA embryos. Third, we show that the mutation of the AGGGA hexanucleotide Hnrnpa1 binding motif in the 3′ UTR of an Hnrnpa1 target decreases Hnrnpa1 binding, shortens the poly(A) tail, and decreases translational output of a eGFP reporter. It has been proposed that selective deadenylation of maternal mRNAs is likely a major determinant of the differences in poly(A) tail length and TE between maternal transcripts in fly embryos, whereas CPE-mediated cytoplasmic polyadenylation creates these differences in vertebrates (Eichhorn et al. 2016). Thus, Hnrnpa1 likely protects transcripts from active deadenylation or promotes their polyadenylation in zebrafish. However, the precise mechanism underlying Hnrnpa1-mediated regulation of poly(A) tail length still remains to be uncovered (Fig. 6).
Application of mRNA interactome capture at two developmental stages enabled us to determine differences in preZGA and ZGA mRBPomes. PreZGA-specific (24) and ZGA-specific (53) mRBPs likely reflect the dynamic nature of mRNA control through regulated changes in both RBP abundance and their RNA-binding affinities. This is consistent with recently described global changes in the mRBPome during Drosophila MZT (Sysoev et al. 2016). The detection of certain zebrafish proteins at ZGA (e.g., Pabpn1, Cstf2) may be explained by their selective association with newly produced zygotic transcripts. ZGA-specific detection of the YTH group of proteins implicates RNA modification in the regulation of embryonic transcriptomes and thereby control of developmental transitions. YTH proteins regulate mRNA stability and translation through their direct recognition of N6-methyladenosine (m6A) RNA modifications (Dominissini et al. 2012; Wang et al. 2014, 2015). Importantly, m6A RNA modification has been implicated in the control of cell fate transition in mammalian embryonic stem cells (Batista et al. 2014).
We provide evidence that RBPs shared among preZGA and ZGA mRBPomes undergo dynamic localization and RNA-binding activities to differentially regulate maternal and zygotic transcripts. The observed transcription-dependent nuclear accumulation of Hnrnpa1 reflects a shift from cytoplasmic to nuclear RNA targets during MZT, consistent with global decrease in Hnrnpa1 association with maternal mRNAs at ZGA and simultaneous increase in binding to introns and intergenic and noncoding regions on Chromosome 4. Within Chromosome 4, nuclear U1, U6 snRNAs and pri-mir-430 represent the most prominent new binding substrates of Hnrnpa1 (Fig. 6). Robust transcription of snRNA genes and the mir-430 locus at ZGA has previously been reported in Xenopus and zebrafish embryos, respectively (Newport and Kirschner 1982; Mattaj and Hamm 1989; Heyn et al. 2014), indicating interactions of Hnrnpa1 with de novo synthesized zygotic RNAs. The relatively small, yet significant increase in intron binding at ZGA may be explained either by the limited number of intron-containing genes expressed (Heyn et al. 2014) and/or short half-life of introns due to rapid splicing and degradation. Since the transcription-dependent nuclear accumulation of Hnrnpa1 coincides with its reduced association with maternal mRNAs and de novo binding to zygotic transcripts, we propose that transcriptional activity of the embryo causes Hnrnpa1 dissociation from maternal mRNAs in the cytoplasm to support nuclear RNA processing events (Fig. 6).
We report a developmentally essential nuclear function of Hnrnpa1 at ZGA—the regulation of miR-430 biogenesis. MiR-430 comprises an evolutionarily conserved miRNA family. Failure to produce miR-430 disrupts neural tube development and brain morphogenesis (Giraldez et al. 2005; Takacs and Giraldez 2015). Transcriptional induction of the mir-430 locus is achieved through translational control of pluripotency factors prior to ZGA (Lee et al. 2013). However, regulation of miR-430 processing has not been addressed. In contrast to mammalian cells, where Hnrnpa1 regulates microprocessor efficiency through binding to pri-miRNA terminal loops (Michlewski et al. 2008), we show that Hnrnpa1 binds basal, single-stranded segments of pri-mir-430. Previous findings that pri-miRNA sequence has important roles in miRNA gene definition and processing efficiency (Auyeung et al. 2013) are consistent with the notion that Hnrnpa1 binding to pri-mir-430 transcript regions regulates miR-430 expression in our heterologous in vivo pri-miRNA processing assay.
The described cytoplasmic to nuclear shift of Hnrnpa1 activities likely applies to other maternally provided splicing factors. We show that other proteins in this category, such as SR proteins and spliceosomal components, also translocate to the nucleus during MZT. We speculate that the redistribution of pre-mRNA processing factors during MZT may have a global impact on changes in maternal mRNA stabilities following ZGA. For example, Hnrnpa1-associated ZGA-dependent decay genes are abandoned by Hnrnpa1 following zygotic transcriptional onset, and their stability is indirectly compromised by nuclear accumulation of Hnrnpa1 and its concomitant involvement in miR-430 processing, a major direct instability determinant of this group of transcripts (Fig. 6; Mishima and Tomari 2016). Therefore, our results illustrate the dynamic changes in the mRBPome during MZT and show that pre-mRNA processing factors adopt unique roles in global regulation of embryonic transcriptomes through their changing interactions with maternal and zygotic RNAs.
Methods
Zebrafish embryo maintenance and treatments
Embryos were obtained from WT AB zebrafish line (BIOTEC) and maintained in accordance with AAALAC research guidelines, under the protocol approved by MPI-CBG and Yale University, IACUC. Embryos were incubated in E3 medium at 28°C and staged as previously described (Kimmel et al. 1995). Microinjections were performed on one-cell-stage embryos. α-Amanitin (0.4 ng) was injected for immunostaining experiments. Scrambled control and Hnrnpa1 and b morpholinos (MOs) were injected in four different concentrations (0.625, 1.25, 2.5, and 5 ng) for α-Hnrnpa1 antibody validation. For Hnrnpa1 binding, ePAT and translation assays, 100 ng eGFP-3′ UTR, and 100 ng HA-tagged Renilla luciferase mRNA reporters were used.
In vivo isolation of mRBPs from zebrafish embryos
The mRNA interactome capture protocol was performed as previously described (Baltz et al. 2012; Castello et al. 2012) with modifications. Briefly, 200 zebrafish embryos at 32–64 cell stage (1.75–2 hpf, preZGA) and 512–1k cell stage (2.75–3 hpf, ZGA) were irradiated twice with 0.8 J/cm2 UV (254 nm) and snap frozen in liquid N2. Nonirradiated embryos served as negative controls. The embryos were subsequently lysed in Lysis/binding buffer (100 mM Tris HCl, pH 7.5, 500 mM LiCl, 10 mM EDTA pH 8.0, 1% [w/v] lithium-dodecylsulfate (LiDS), 5 mM DTT, Complete Mini EDTA-free protease inhibitors [Roche]) and subjected to three rounds of 1-h mRNA capture at room temperature using 120 μL magnetic oligo(dT)25 beads (Invitrogen). After two rounds of 20-min washes with Lysis/binding buffer, beads were washed three times with NP40 wash buffer (50 mM Tris HCl, pH 7.5, 140 mM LiCl, 2 mM EDTA pH 8.0, 0.5% NP40, 0.5 mM DTT). mRNPs were heat eluted from the beads in low salt buffer (10 mM Tris HCl, pH 7.5), and the eluates from two samples from the same developmental stage were combined. Proteins were released from mRNPs by 1-h RNase I (Ambion) treatment at 37°C in low salt buffer supplemented with 1 mM MgCl2 and subjected to mass spectrometry analyses. For further information about the mass spectrometry measurements and subsequent data analysis, see Supplemental Methods.
iCLIP
The iCLIP experiment was performed as previously described (König et al. 2010). For a single biological replicate, 400 zebrafish embryos at 32–64 cell stage (1.75–2 hpf, preZGA) and 512–1k cell stage (2.75–3 hpf, ZGA) were irradiated twice with 0.8 J/cm2 (Stratalinker 2400, Stratagene), lysed, and subjected to mild RNA fragmentation. Crosslinked RNA–proteins complexes were immunopurified using monoclonal α-Hnrnpa1 antibody (a kind gift from Douglas Black). RNA was extracted and converted into cDNA using experiment-specific RT primers containing 9-nt barcodes. Following size selection and PCR amplification, cDNA libraries were subjected to high-throughput sequencing on the Illumina HiSeq2000 platform to generate 75-nt single-end reads. For more details about the iCLIP procedure, see Supplemental Methods.
Data analysis
Analysis of the RNA-seq data from iCLIP libraries was conducted using iCOUNT (König et al. 2010) and motif analysis was performed using DREME (Bailey 2011). For more information concerning iCLIP data analysis, see Supplemental Methods. Reannotation of the zebrafish transcriptome was accomplished by 3′ UTR extensions as previously described (Ulitsky et al. 2012). Translation efficiency (TE) was calculated using the processed data from Lee et al. (2013) as ribosome protected fragments (RPF)/mRNA expression levels in input. Mean poly(A) tail length of mRNAs expressed at 2 hpf was retrieved from Subtelny et al. (2014).
Indirect immunofluorescence
Immunostaining was performed on staged embryos as previously described (Strzelecka et al. 2010). In brief, fixed and permeabilized embryos were incubated 1 h with primary antibodies (α-Hnrnpa1 and 16H3 in 1:5 and 1:4 dilutions, respectively) in 3% BSA-MgPBS, supplemented with 0.2% glycerol phosphate. Following three washing steps with MgPBS, secondary antibodies (mouse IgG-Alexa 488 and mouse IgG-TRITC for Hnrnpa1 and 16H3, respectively; Jackson Immunoresearch Laboratories) in 3% BSA-MgPBS and 0.2% glycerol phosphate, were added and incubated 1 h at room temperature. Embryos were mounted in 80% glycerol supplemented with 25 mg/mL anti-fading agent, DABCO (Sigma-Aldrich) and 2 μg/mL Hoechst 3342 (Sigma-Aldrich). Immunostained embryo specimens were imaged on the Zeiss LSM710 Duo NLO microscope using 63× Oil NA: 1.4 objective (Zeiss) and Zen2010 built-in software. Single-plane images were taken and further analyzed using ImageJ software. Figures were prepared using Adobe Illustrator CS5.1 (Adobe).
RT-qPCR
Two micrograms total RNA from different developmental stages were used for reverse transcription using a hexanucleotide mix (Roche). Expression levels of each mRNA was normalized to unchanging nd3-nd4l mitochondrial precursor RNA (Heyn et al. 2014). Experiments were performed in three independent biological replicates. DNA oligos are listed in Supplemental Table 6.
Cell culture and treatments
HeLa Kyoto cells were cultured in DMEM GlutaMax medium (Thermo), supplemented with penicillin/streptomycin (PAA) and fetal bovine serum (FBS; Thermo) at 37°C and in a 5% CO2 atmosphere. For plasmid transfections in six-well plates, 2 μg shRNA-expressing or empty pSUPER plasmids and 5 μL Lipofectamine 2000 (Thermo) were used per one well, containing cells grown to 70% confluency. Forty-eight hours later, 2 μg pcDNA3.0-pri-mir-430 construct or pcDNA3.0 empty plasmid were transfected using 5 μL Lipofectamine 2000 reagent. After another 48h, cells were trypsinized, harvested, and washed three times with 1× PBS. Forty percent of the cells was sonicated in 100 μL RIPA buffer and used for Western blot analyses. Total RNA was extracted from the remaining cells using TRIzol reagent (Thermo) and used for Northern blot and RT-PCR analyses. For more information regarding plasmids and Western and Northern blot analysis, see Supplemental Methods.
Data access
Hnrnpa1 iCLIP raw and processed data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://ncbi.nlm.nih.gov/geo/) under accession number GSE76212. Mass spectrometry proteomics data from this study have been submitted to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al. 2016) under the data set identifier PXD006098.
Supplementary Material
Acknowledgments
We thank J.A. Steitz, N.L. Vastenhouw, A.J. Giraldez and their laboratories for helpful discussions. Douglas L. Black (UCSF, San Francisco) and Javier F. Cáceres (MRC, Edinburgh) made generous gifts of the monoclonal α-Hnrnpa1 antibody and shRNA-expressing plasmids, respectively. We thank T. Curk for his help on iCOUNT-based iCLIP data analysis. We thank Neugebauer laboratory members, especially M. Machyna and P. Heyn, for sharing their knowledge about iCLIP and zebrafish work, and S. Oyadomari for her helpful feedback on the manuscript. This work was supported by funding to K.M.N. from the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, where this work was initiated, by the Deutsche Forschungsgemeinschaft (NE909/2-2 to K.M.N.), and by the Foundation for the National Institutes of Health (NIH) R01GM112766 from the NIGMS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Work in the Butter group was partially supported by the Rhineland Palatinate Forschungsschwerpunkt GeneRED.
Author contributions: V.D. and K.M.N. designed the experiments and wrote the manuscript. V.D. performed all the experiments, with technical assistance from K.S. with tissue culture. M.D. and F.B. performed mass spectrometry analyses, and V.D. further analyzed MS data. M.G., J.K., and J.Z. reannotated zebrafish 3′ UTRs, remapped iCLIP data for custom annotation, and performed motif analysis under the supervision of M.B.G. V.D. performed all downstream iCLIP data analysis. L.H. performed bioinformatic analysis on splicing of embryonic transcripts.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.215954.116.
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. 2011. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res 21: 1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akindahunsi AA, Bandiera A, Manzini G. 2005. Vertebrate 2xRBD hnRNP proteins: a comparative analysis of genome, mRNA and protein sequences. Comput Biol Chem 29: 13–23. [DOI] [PubMed] [Google Scholar]
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. 2013. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell 152: 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. 2011. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. [DOI] [PubMed] [Google Scholar]
- Barckmann B, Pierson S, Dufourt J, Papin C, Armenise C, Port F, Grentzinger T, Chambeyron S, Baronian G, Desvignes JP, et al. 2015. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep 12: 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. 2014. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. 2012. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35: 2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W, et al. 2015. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun 6: 10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. 2009. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 136: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda A, Smith GW. 2007. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci 12: 3713–3726. [DOI] [PubMed] [Google Scholar]
- Boothby TC, Zipper RS, van der Weele CM, Wolniak SM. 2013. Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev Cell 24: 517–529. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J 13: 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N, Wang YD, Zheng PS. 2013. The microRNA-302-367 cluster suppresses the proliferation of cervical carcinoma cells through the novel target AKT1. RNA 19: 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, Meijer HA, de Moor CH. 2013. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA 4: 437–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. 2016. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 5: e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308: 833–838. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. 2006. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79. [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Cáceres JF. 2007. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14: 591–596. [DOI] [PubMed] [Google Scholar]
- Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524. [DOI] [PubMed] [Google Scholar]
- Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D, Smith JC. 2013. Identification of the zebrafish maternal and paternal transcriptomes. Development 140: 2703–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L, Neugebauer KM. 2015. Quantification of co-transcriptional splicing from RNA-Seq data. Methods 85: 36–43. [DOI] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. 2014. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep 6: 285–292. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang F, Zhu X, Yuan Y, Ding M, Gao S. 2010. Mouse ZAR1-like (XM_359149) colocalizes with mRNA processing components and its dominant-negative mutant caused two-cell-stage embryonic arrest. Dev Dyn 239: 407–424. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. 2012. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA 18: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Harris ME. 2015. Specificity and nonspecificity in RNA–protein interactions. Nat Rev Mol Cell Biol 16: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Steen JA, Steen H, Kirschner MW. 2009. The mechanism and pattern of yolk consumption provide insight into embryonic nutrition in Xenopus. Development 136: 1539–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. [DOI] [PubMed] [Google Scholar]
- König H, Matter N, Bader R, Thiele W, Müller F. 2007. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 131: 718–729. [DOI] [PubMed] [Google Scholar]
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20: 1122–1130. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J Biol Chem 275: 17827–17837. [DOI] [PubMed] [Google Scholar]
- Langley AR, Smith JC, Stemple DL, Harvey SA. 2014. New insights into the maternal to zygotic transition. Development 141: 3834–3841. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Lee M, Son A, Chang H, Kim VN. 2016. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev 30: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. 2009. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15: 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SR, Megosh H, Lin H. 2014. PIWI proteins are essential for early Drosophila embryogenesis. Dev Biol 385: 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matia-Gonzalez AM, Laing EE, Gerber AP. 2015. Conserved mRNA-binding proteomes in eukaryotic organisms. Nat Struct Mol Biol 22: 1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E, Michalova E, Vojtesek B, Hrstka R. 2012. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol 9: 563–576. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Hamm J. 1989. Regulated splicing in early development and stage-specific U snRNPs. Development 105: 183–189. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Cáceres JF. 2010. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol 17: 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Cáceres JF. 2008. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell 32: 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Tomari Y. 2016. Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61: 874–885. [DOI] [PubMed] [Google Scholar]
- Mitchell SF, Jain S, She M, Parker R. 2013. Global analysis of yeast mRNPs. Nat Struct Mol Biol 20: 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-McNicoll M, Neugebauer KM. 2013. How cells get the message: dynamic assembly and function of mRNA–protein complexes. Nat Rev Genet 14: 275–287. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Stolk JA, Roth MB. 1995. A conserved epitope on a subset of SR proteins defines a larger family of Pre-mRNA splicing factors. J Cell Biol 129: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982. A major developmental transition in early xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30: 675–686. [DOI] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, Veenstra GJ. 2013. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics 14: 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. 2012. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro-Neto F, Blackshear PJ. 2004. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131: 4883–4893. [DOI] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH. 2009. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16: 517–527. [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. 2010. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17: 403–409. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoev VO, Fischer B, Frese CK, Gupta I, Krijgsveld J, Hentze MW, Castello A, Ephrussi A. 2016. Global changes of the RNA-bound proteome during the maternal-to-zygotic transition in Drosophila. Nat Commun 7: 12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 12: 143–155. [DOI] [PubMed] [Google Scholar]
- Takacs CM, Giraldez AJ. 2015. miR-430 regulates oriented cell division during neural tube development in zebrafish. Dev Biol 409: 442–450. [DOI] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. 2010. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol 11: R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. 2012. Extensive alternative polyadenylation during zebrafish development. Genome Res 22: 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Steitz JA. 1998. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol 18: 7537–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels HH, Imami K, Baltz AG, Kolinski M, Beldovskaya A, Selbach M, Small S, Ohler U, Landthaler M. 2016. The mRNA-bound proteome of the early fly embryo. Genome Res 26: 1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. 2003. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet 33: 187–191. [DOI] [PubMed] [Google Scholar]
- Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu B, Sun QY, et al. 2016. BTG4 is a meiotic cell cycle–coupled maternal-zygotic-transition licensing factor in oocytes. Nat Struct Mol Biol 23: 387–394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.