Fell et al. deleted KIF1Bβ in the mouse sympathetic nervous system and observed impaired sympathetic nervous function and misexpression of genes required for sympathoadrenal lineage differentiation. They discovered that KIF1Bβ is required for NGF-dependent neuronal differentiation through anterograde transport of the NGF receptor TRKA.
Keywords: KIF1Bβ, NGF, TRKA, chromosome 1p36 loss, neuroblast differentiation, neuroblastoma
Abstract
We recently identified pathogenic KIF1Bβ mutations in sympathetic nervous system malignancies that are defective in developmental apoptosis. Here we deleted KIF1Bβ in the mouse sympathetic nervous system and observed impaired sympathetic nervous function and misexpression of genes required for sympathoadrenal lineage differentiation. We discovered that KIF1Bβ is required for nerve growth factor (NGF)-dependent neuronal differentiation through anterograde transport of the NGF receptor TRKA. Moreover, pathogenic KIF1Bβ mutations identified in neuroblastoma impair TRKA transport. Expression of neuronal differentiation markers is ablated in both KIF1Bβ-deficient mouse neuroblasts and human neuroblastomas that lack KIF1Bβ. Transcriptomic analyses show that unfavorable neuroblastomas resemble mouse sympathetic neuroblasts lacking KIF1Bβ independent of MYCN amplification and the loss of genes neighboring KIF1B on chromosome 1p36. Thus, defective precursor cell differentiation, a common trait of aggressive childhood malignancies, is a pathogenic effect of KIF1Bβ loss in neuroblastomas. Furthermore, neuropathy-associated KIF1Bβ mutations impede cargo transport, providing a direct link between neuroblastomas and neurodegeneration.
Neuroblastoma is a childhood tumor arising from the peripheral nervous system. High-stage neuroblastoma has poor prognosis and is associated with loss of chromosomal regions at 1p36 and/or MYCN amplifications. Numerous studies have proposed CAMTA1, CASZ1, CHD5, P73, MIR34A, and KIF1Bβ, respectively, as 1p36-encoded tumor suppressors; however, no bona fide tumor suppressor function has yet been ascribed to any of these genes individually (Jiang et al. 2011).
Previous studies have demonstrated that KIF1Bβ is critical for apoptosis in sympathetic neuron progenitors in response to nerve growth factor (NGF) deprivation and that KIF1Bβ is silenced in 1p36 hemizygous-deleted neuroblastomas (Munirajan et al. 2008; Schlisio et al. 2008; Chen et al. 2014; Li et al. 2016). Furthermore, pathogenic KIF1B mutations previously identified in neuroblastoma and pheochromocytoma all fail to mediate apoptosis (Schlisio et al. 2008; Li et al. 2016), and genetic studies of familiar pheochromocytoma suggest that KIF1Bβ acts as a tumor suppressor by mediating developmental apoptosis (Yeh et al. 2008). Competition for growth factors, such as NGF, is a major event in the formation of the peripheral nervous system. However, the importance of KIF1Bβ for sympathoadrenal development has not been investigated in vivo.
KIF1B is a member of the kinesin 3 family (Lawrence et al. 2004). Proteins within this family act as molecular motors that transport cargo along microtubules toward their plus end to the cell periphery. KIF1B encodes two alternatively spliced isoforms: KIF1Bα and KIF1Bβ (Nagai et al. 2000). The recently identified role of KIF1Bβ in NGF-mediated neuronal apoptosis during sympathetic neuron development has been characterized as a motor-independent function (Chen et al. 2014). However, earlier work has suggested a role for KIF1Bβ in axon development, and a loss-of-function mutation in the motor domain of KIF1Bβ causes defects in axon development and myelination that mirror those seen in multiple sclerosis (Lyons et al. 2009). In addition, a mutation in the KIF1Bβ motor domain is associated with the hereditary neuropathy Charcot-Marie-Tooth (CMT) disease and was shown to impair axonal transport and cause peripheral neuropathy in mice (Zhao et al. 2001). These data led us to ask whether KIF1Bβ is required for neuronal differentiation and maintenance as well as developmental culling in the peripheral nervous system. Furthermore, would these dual roles depend on one or several distinct molecular actions performed by KIF1Bβ, and how might this be related to suppression of neuroblastoma?
Results
Loss of KIF1Bβ impairs sympathetic nervous function
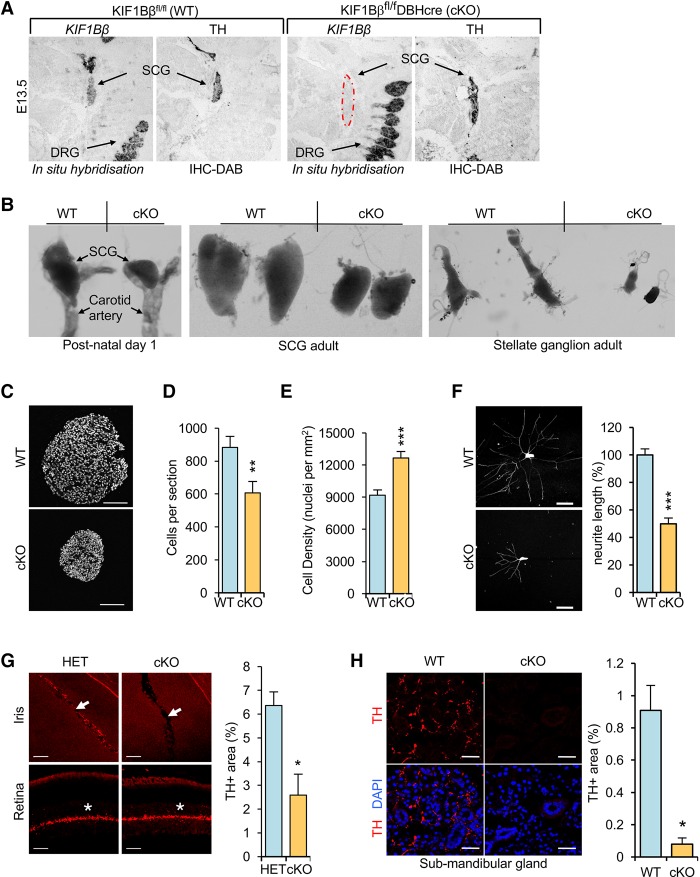
We generated mice in which KIF1Bβ is specifically inactivated in sympathetic neurons by Cre-recombinase expressed under the control of the DBH promoter. The resulting mice (KIF1Bβfl/flDBHCre; referred to here as KIF1BβcKO [conditional knockout]) cease to express the β isoform of KIF1B in sympathetic ganglia by embryonic day 13.5 (E13.5), before the onset of NGF competition and target innervation (Fig. 1A). Given the role of KIF1Bβ in NGF deprivation-induced apoptosis, we expected enlarged sympathetic ganglia in KIF1BβcKO mice due to impaired developmental culling. KIF1Bβ acts downstream from the prolyl hydroxylase EglN3 to induce apoptosis during NGF competition (Schlisio et al. 2008). However, the superior cervical ganglia (SCGs) from postnatal day 1 (P1) mice lacking KIF1Bβ were drastically smaller than those of their wild-type littermates (Fig. 1B) and not enlarged as in EGLN3−/− mice (Bishop et al. 2008). The SCGs of P1 KIF1BβcKO mice were normally located in the fork of the carotid artery with proximal nerve projections along the external and internal carotid artery (data not shown). The smaller size of KIF1BβcKO ganglia persists in adult mice (Fig. 1B) and is due to reduced cell number and greater cell density (Fig. 1C–E). Reduced sympathetic ganglion size was also evident in the stellate ganglia of KIF1BβcKO mice (Fig. 1B). Neurite outgrowth was reduced in primary cultures of KIF1BβcKO SCG neurons (Fig. 1F), suggesting that the smaller size of these ganglia might be due to defects in neurotrophic signaling causing impaired axonal outgrowth and target innervation. We hypothesized that this might decrease neuronal survival and/or differentiation and lead to smaller sympathetic ganglia.
Figure 1.
Loss of KIF1Bβ impairs sympathetic nervous function. (A) In situ hybridization using a probe for KIF1Bβ mRNA on sagittal sections of wild-type (KIF1Bβfl/fl WT) and KIF1Bβ conditional knockout (KIF1Bβfl/flDBH-CRE cKO) E13.5 embryos and immunohistochemistry for tyrosine hydroxylase (TH) on the adjacent section. Arrows point to either SCGs or dorsal root ganglia (DRGs). Note that DBH is not expressed in the DRGs, and thus deletion of KIF1Bβ is specific to the DBH-expressing and TH-expressing SCGs only. (B) Photographs of dissected mouse SCGs or stellate ganglia of the indicated genotype and age. (C) Nuclear-stained (DAPI) cryosections of P1 SCGs of the indicated genotypes. (D,E) Mean number of DAPI-stained cells per section (n = 9 sections and 3 animals per genotype) (D) and mean cell density (nuclei per square millimeter) (E) from P1 SCGs. (F) Immunofluorescent staining of TUJ1 in P1 SCG neurons cultured for 12 h with 10 ng/mL NGF and quantification of mean neurite length relative to wild type after 12 h in culture. n = 8 animals and 40 neurons per genotype. (G,H) TH immunostaining of the irises (G) and submandibular glands (H) of adult mice to visualize sympathetic nerve innervation. Arrows indicate the iris. An asterisk indicates TH expression in the retina (control). The TH-positive area was quantified and compared between groups. n ≥ 9 sections and 3 animals per genotype. Pairwise comparisons were made using two-sided unpaired Student's t-tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Error bars depict SEM. Bars: C,F, 100 µm; G, 50 µm; H. 30 µm.
Neurons from the SCGs innervate an array of targets in the head and neck, including the iris and submandibular salivary gland. KIF1BβcKO irides displayed a drastic reduction in sympathetic innervation compared with hemizygous littermates as measured by tyrosine hydroxylase (TH) staining, while TH in the retina served as control staining and appeared normal (Fig. 1G). Likewise, sympathetic innervation of the submandibular gland was significantly reduced in KIF1BβcKO mice (Fig. 1H). The reduction in sympathetic innervation in the iris and submandibular gland suggested a more general defect in sympathetic innervation and sympathetic nervous system function in KIF1BβcKO mice. One major function of the sympathetic system is maintenance of blood pressure through effects on vascular smooth muscle tone (Guyenet 2006). We found that KIF1BβcKO mice have very low mean arterial pressure compared with their wild-type counterparts, resulting in very low, below detection, and irregular measurements of hemodynamic parameters (Supplemental Fig. S1A) despite the presence of elevated levels of both noradrenaline and adrenaline in the blood plasma of these animals (Supplemental Fig. S1B,C). We interpret this increase in adrenaline and noradrenaline as a compensatory adrenal response to the markedly lower sympathetic vascular tone. Collectively, these results imply an important developmental role for KIF1Bβ in sympathetic nervous system formation and function.
Loss of KIF1Bβ affects sympathetic neuron identity
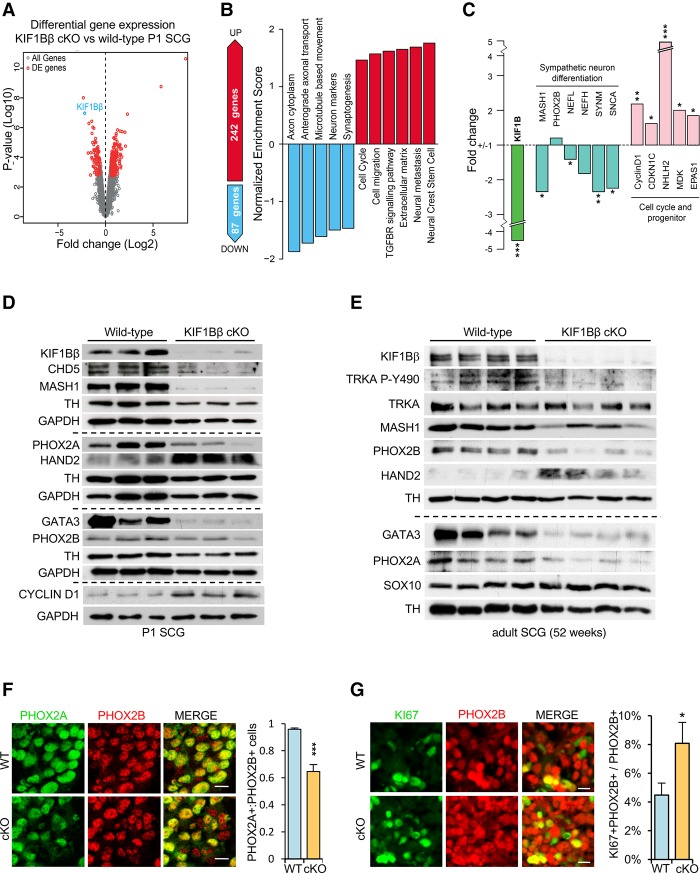
To investigate how loss of KIF1Bβ impairs sympathetic nervous system function, we performed gene expression profiling of P1 sympathetic ganglia (SCGs) by RNA sequencing (RNA-seq). Two-hundred-forty-two genes were significantly up-regulated and 87 genes were significantly down-regulated by a fold change of at least 1.5 in KIF1BβcKO ganglia (Fig. 2A,B; Supplemental Table S1). Gene set enrichment analysis (GSEA) revealed increased expression of gene sets for neural crest stem cells, neural metastasis, the extracellular matrix, the transforming growth factor β receptor signaling pathway, cell migration, and the cell cycle, while gene sets associated with neuronal characteristics such as axon cytoplasm, anterograde axonal transport, microtubule-based movement, neuron markers, and synaptogenesis were down-regulated in KIF1BβcKO SCGs (Fig. 2B).
Figure 2.
Loss of KIF1Bβ alters sympathetic neuron identity. (A) Volcano blot displaying differential gene expression by RNA-seq in KIF1BβcKO versus wild-type P1 SCGs. Genes in red are significantly differentially expressed by a fold change of at least 1.5 and adjusted P-value of <0.05. (B) GSEA from RNA-seq gene expression profiling of KIF1BβcKO versus wild-type P1 SCGs. (C) Expression of selected genes implicated in neuronal sympathetic function and differentiation shown as fold change in KIF1BβcKO. (D,E) Immunoblot analysis of P1 SCGs (D) and adult SCGs (E) of the indicated genotypes. (F,G) Immunofluorescence staining of endogenous PHOX2A (green) and PHOX2B (red) and quantification in the P1 SCGs from KIF1Bβ wild type (WT) and KIF1BβcKO (F) and staining and quantification of KI67-expressing (green) in PHOX2B-expressing (red) cells in KIF1BβcKO compared with wild-type SCGs at P1 (G). n = 9 sections and 3 animals per genotype. Pairwise comparisons were made using two-sided unpaired Student's t-tests. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Error bars depict SEM. The P-values indicated in C are adjusted P-values from VOOM limma differential expression analysis of RNA-seq data described in the Materials and Methods. Bars: F,G, 10 µm.
Expression of MASH1 (ASCL1), a transcription factor central to the differentiation of neuroblasts, was reduced 2.3-fold in KIF1BβcKO ganglia, while PHOX2B, the other main sympathoneurogenic transcription factor, was not affected (Fig. 2C). Reduction in the expression of neurofilament proteins (NEFL and NEFH) and synaptic markers (SYNM and SNCA) along with up-regulation of cell cycle regulators (CyclinD1 and CDKN1C) and markers associated with neural progenitor status (NHLH2 and EPAS1) collectively suggest that loss of KIF1Bβ might cause defects in the differentiation and function of sympathetic neurons (Fig. 2A–C). Immunoblot analysis of various markers of sympathetic neuron identity in P1 SCGs verified that transcription factors critical for sympathetic neuron differentiation are misexpressed in the conditional knockout. MASH1, PHOX2A, CHD5, and GATA3 proteins are reduced, while HAND2 protein is increased in KIF1BβcKO SCGs (Fig. 2D), and these differences persist in adult mice (Fig. 2E). CHD5, a putative 1p36 tumor suppressor involved in terminal neuronal differentiation (Egan et al. 2013), was reduced in KIF1Bβ-deficient ganglia. However, the adrenergic marker TH was unaffected by loss of KIF1Bβ, and PHOX2B protein was unchanged by loss of KIF1Bβ at P1 yet decreased in adult SCGs (Fig. 2D,E). MASH1, PHOX2A, GATA3, and HAND2 are all required for normal development and differentiation of sympathetic neurons in vivo, as indicated by mouse knockout models (Guillemot et al. 1993; Morin et al. 1997; Morikawa et al. 2007). The altered levels of sympathoadrenal differentiation factors observed in postnatal KIF1BβcKO sympathetic ganglia led us to speculate that KIF1Bβ may be required for differentiation of sympathetic neurons.
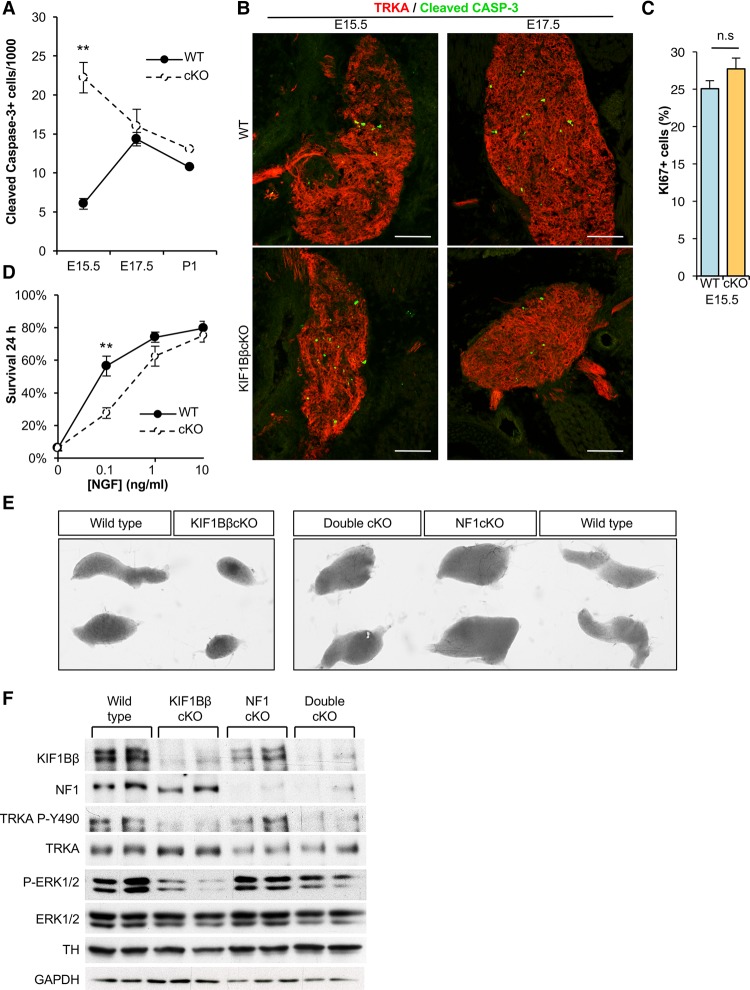
Alternatively, KIF1BβcKO ganglia might contain fewer neurons relative to other cell types, such as glial subtypes, when compared with wild-type ganglia. At P1, these two cell groups can be distinguished by mutually exclusive expression of either PHOX2B (neurons) or SOX10 (glia). The ratio of PHOX2B+ cells to SOX10+ cells was similar in wild-type and KIF1BβcKO ganglia (Supplemental Fig. S2A), indicating that the ratio of neurons to glia was not affected by loss of KIF1Bβ. Moreover, significantly fewer PHOX2B-positive neurons express PHOX2A in KIF1BβcKO ganglia (Fig. 2F), implying that the altered levels of sympathetic differentiation factors observed could indicate impaired neuronal differentiation. In addition, we observed a twofold increase in KI67-positive neurons in P1 KIF1BβcKO SCGs (Fig. 2G). Consistent with increased expression of cell cycle regulators such as Cyclin D1 (CCND1) and CDKN1A (P57kip2) (Fig. 2C,D), these data suggest that loss of KIF1Bβ might impair cell cycle exit in developing sympathetic neurons.
KIF1Bβ mediates NGF-induced neuronal differentiation
The correct location of the SCGs in KIF1BβcKO mice suggests that defects in neuroblast differentiation, rather than migration, account for the impaired nervous system function, axonal outgrowth, and target innervation. Secretion of NGF from sympathetic target tissues and NGF binding to TRKA on the neuronal cell membrane are required for sympathetic axon extension and target innervation. NGF binding to TRKA stimulates TRKA autophosphorylation, leading to the activation of neurotrophic signaling pathways. Mice that lack either NGF or TRKA have smaller sympathetic ganglia and ablated sympathetic target innervation (Crowley et al. 1994; Fagan et al. 1996; Glebova and Ginty 2004). These phenotypic similarities to KIF1BβcKO mice lead us to question whether KIF1Bβ is required for the response to NGF.
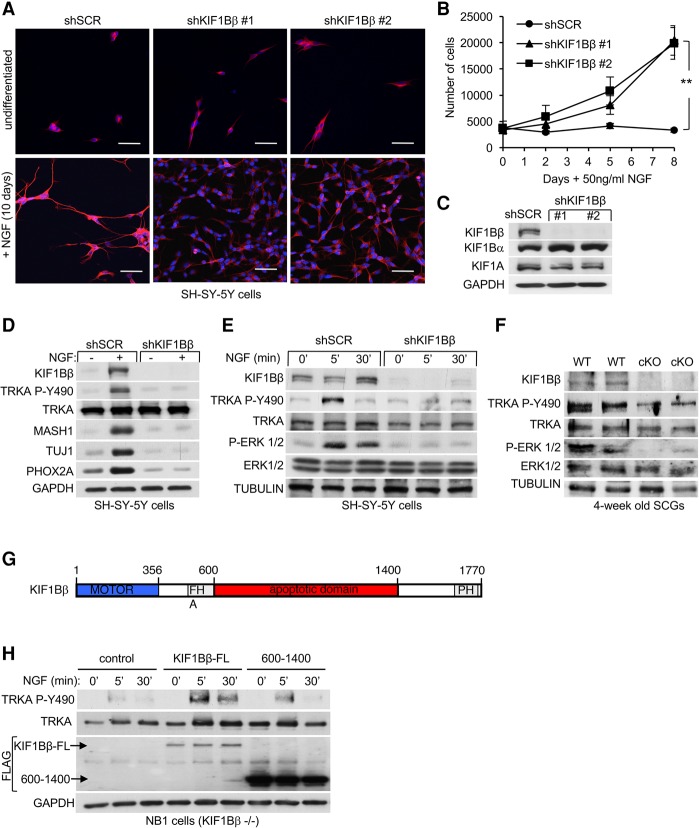
NGF stimulation of 1p36-positive (SH-SY-5Y) neuroblastoma cells (Fig. 3A) or PC12 cells (Supplemental Fig. S3A) induced differentiation within 10 d, as evinced by growth arrest, neurite outgrowth, and expression of neuronal markers, including TUJ1 (shSCR control in Fig. 3A,B). KIF1Bβ expression and neuronal differentiation markers such as MASH1, TUJ1, and PHOX2A increased after 10 d of NGF stimulation (shSCR in Fig. 3D). In contrast, cells harboring shRNAs targeting KIF1Bβ (shKIF1Bβ) failed to differentiate and continued to proliferate (Fig. 3A–C). Furthermore, MASH1, TUJ1, and PHOX2A were not induced after NGF stimulation in KIF1Bβ knockdown cells (Fig. 3D). The abundance of KIF1Bα and KIF1A was unchanged in these cells (Fig. 3C). TRKA autophosphorylation at Tyr490 in response to NGF binding is essential for downstream differentiation signaling through the MAP kinase pathway (Biarc et al. 2013). We observed that loss of KIF1Bβ reduced phosphorylation of TRKA on Tyr490 in SH-SY-5Y cells expressing KIF1Bβ shRNAs both after 10 d of exposure to NGF (Fig. 3D) and in response to acute NGF stimulation (Fig. 3E). KIF1Bβ knockdown impaired MAP kinase (ERK1/2) phosphorylation in response to NGF stimulation (Fig. 3E), implying that KIF1Bβ is required for NGF-mediated TRKA signaling. These observations were confirmed in vivo, where phosphorylation of TRKA (Tyr490) and ERK1/2 was reduced in KIF1BβcKO SCGs compared with wild-type littermates (Fig. 3F). Together, these findings demonstrate that KIF1Bβ is necessary for NGF–TRKA neurotrophic signaling both in vitro and in vivo.
Figure 3.
KIF1Bβ mediates NGF-induced neuronal differentiation. (A) Immunofluorescence images of undifferentiated or 10-d NGF-differentiated SH-SY-5Y cells (1p36+/+) with stable KIF1Bβ knockdown (shKIF1Bβ) or nontargeting control (shSCR) as indicated. TUJ1 (red) staining served as a terminal differentiation marker, and DAPI (blue) stained nuclei. Bars, 100 µm. (B) Growth curves of NGF-differentiated SH-SY-5Y cells during NGF differentiation. (C) KIF1Bβ knockdown efficiency is shown by anti-KIF1Bβ immunoblot. (D) Corresponding immunoblot of undifferentiated or 10-d NGF-differentiated cells as in A for neuronal differentiation and lineage markers as indicated. (E) Immunoblot of short-term NGF-stimulated SH-SY-5Y cells (5 and 30 min) as indicated. (F) immunoblot of dissected SCGs from 4-wk-old mice of the indicated genotypes. (G) Schematic of KIF1Bβ domain architecture. (H) Immunoblot of short-term NGF-stimulated NB1 cells (KIF1Bβ−/−) that were transduced with control lentivirus or virus encoding either Flag-KIF1Bβ full-length (FL) or Flag-KIF1Bβ600–1400 (proapoptotic domain) as indicated.
Recently, we demonstrated that KIF1Bβ is required for activation of the phosphatase calcineurin (CN), causing DRP1-mediated mitochondrial fission (Li et al. 2016). CN has been implicated previously in neuronal activity and differentiation (Graef et al. 2003; Cho et al. 2014), and mitochondrial dynamics have been proposed to mediate oncogenic ERK function (Kashatus et al. 2015). However, we observed that regulation of mitochondrial dynamics by knockdown of the critical CN enzymatic subunit PPP3CA or the CN-activating protein CALM2 by shRNA did not impair NGF-induced differentiation of PC12 cells, in contrast to KIF1Bβ knockdown (Supplemental Fig. S3A,B). This suggests that the requirement for KIF1Bβ in NGF-stimulated differentiation might depend on a novel KIF1Bβ function independent of its ability to regulate mitochondrial fission and CNA activation. KIF1B is not expressed in NB1 neuroblastoma cells due to a small homozygous chromosomal 1p36 deletion that includes KIF1B. NB1 (KIF1Bβ−/−) cells fail to differentiate in response to NGF (data not shown), and TRKA is not phosphorylated on Tyr490 in response to NGF treatment (Fig. 3H, control lanes). Stable reintroduction of wild-type KIF1Bβ (KIF1Bβ-FL) in NB1 cells enabled a robust phosphorylation of TRKA Tyr490 in response to NGF stimulation (Fig. 3H). We recently defined the KIF1B 600- to 1400-amino-acid region as necessary and sufficient to mediate apoptosis (Fig. 3G; Chen et al. 2014; Li et al. 2016). Reintroduction of KIF1Bβ600–1400 in NB1 cells failed to rescue TRKA Tyr490 phosphorylation to the level caused by wild-type KIF1Bβ (Fig. 3H). Importantly, KIF1Bβ600–1400 lacks the motor domain. These data suggest that the functions of KIF1Bβ in regulating apoptosis and NGF response involve distinct molecular mechanisms and that KIF1Bβ motor activity is required for mediating NGF–TRKA signaling.
KIF1Bβ mediates anterograde axonal transport of TrkA in sympathetic neurons
Earlier studies have shown that KIF1Bβ transports anterograde presynaptic vesicles and certain mRNAs toward the cell periphery (Niwa et al. 2008; Lyons et al. 2009). We speculated that KIF1Bβ might transport TRKA to the cell surface, where it is presented to ligands such as NGF.
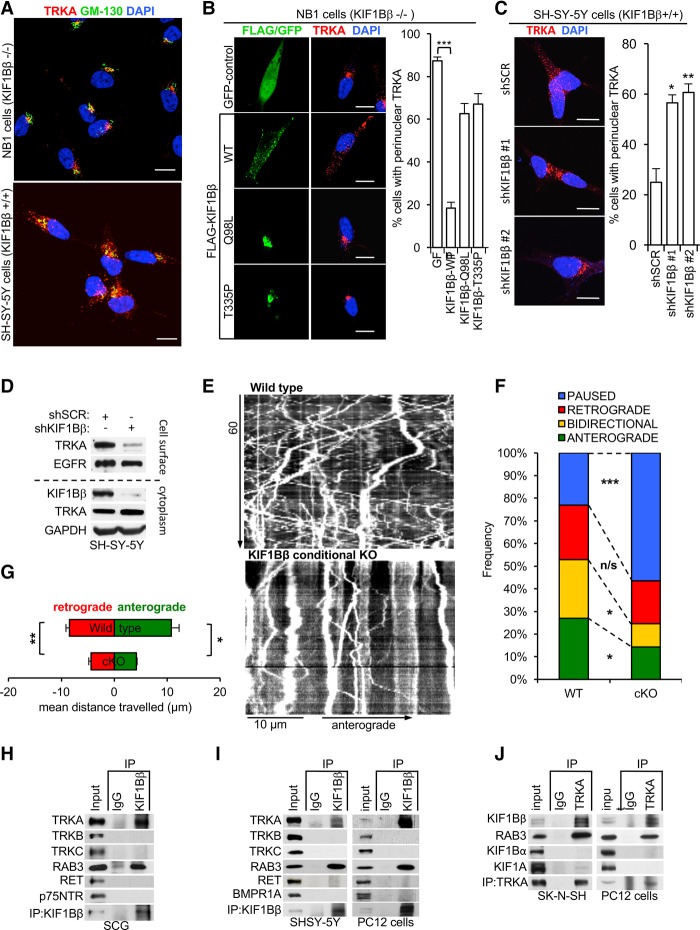
In KIF1Bβ-null cells (NB1 cells, 1p36−/−), we observed TRKA via immunofluorescence in small punctae that are highly concentrated in the perinuclear region in the vicinity of the Golgi apparatus (Fig. 4A). In contrast, TRKA is visible as more evenly dispersed punctae in KIF1Bβ-expressing SH-SY-5Y (1p36+/+) cells (Fig. 4A). Next, we reintroduced either KIF1Bβ or KIF1Bβ point mutants (Q98L and T335P) that harbor functionally deleterious mutations in the motor domain (Zhao et al. 2001; Lyons et al. 2009) into NB1 cells. TRKA was significantly relocalized by ectopic expression of wild-type KIF1Bβ and dispersed away from the perinuclear region. However, the KIF1Bβ motor mutants had minimal effect on TRKA localization (Fig. 4B). Both motor-defective KIF1Bβ mutants (Q98L and T335P) were highly concentrated, along with TRKA, in the perinuclear region, in contrast to wild-type KIF1Bβ that was more dispersed in the cytoplasm (Fig. 4B). Moreover, knockdown of KIF1Bβ by lentiviral-encoded shRNAs in SH-SY-5Y cells (KIF1Bβ+/+) caused a significant increase in the percentage of cells with TRKA concentrated in the perinuclear region (Fig. 4C), similar to KIF1Bβ-null cells (NB1). KIF1Bβ’s effect on TRKA localization appeared to be specific because KIF1Bβ did not affect the intracellular distribution of other transmembrane receptors tested, such as TRKC or EGFR (Supplemental Fig. S4A). Furthermore, we investigated whether adding back either wild-type or motor mutant KIF1Bβ by ectopic expression in KIF1Bβ knockdown cells could restore TRKA localization. We observed that adding back wild-type KIF1Bβ redistributed endogenous TRKA throughout the cells (Supplemental Fig. S4B) in a punctiform localization pattern similar to that seen in control SH-SY-5Y cells (shSCR) (Fig. 4C). Tellingly, KIF1Bβ motor domain mutants (Q98L and T335P) were unable to restore endogenous TRKA localization (Supplemental Fig. S4B). Thus, we conclude that KIF1Bβ motor function is required for TRKA distribution in these cells.
Figure 4.
KIF1Bβ mediates anterograde axonal transport of TrkA in sympathetic neurons. (A) Immunofluorescence imaging of endogenous TRKA (red) in SH-SY-5Y (KIF1Bβ+/+) or NB1 (KIF1Bβ−/−) cells. The Golgi apparatus is indicated by staining of GM-130 (green), and nuclei are indicated by DAPI stain (blue). (B) Endogenous TRKA (red) immunofluorescence in NB1 cells transfected with GFP control, Flag-KIF1Bβ wild-type (WT), or motor mutants Q98L and T335P (green). (C) TRKA (red) immunofluorescence images of SH-SY-5Y (KIF1Bβ+/+) cells transduced with lentivirus encoding shRNA targeting KIF1Bβ as indicated. (D) Immunoblot of cell surface membrane biotinylation and subsequent streptavidin pull-down to measure total surface proteins in SH-SY-5Y cells transduced with lentivirus encoding shKIF1Bβ or nontargeting control. (E) Representative kymographs of TRKA-mCherry vesicle movements in axons of KIF1Bβ+/+ and KIF1BβcKO SCG neurons. See also Supplemental Movies S1 and S2. (F) Frequency of TRKA-mCherry vesicles moving in the specified directions. (G) Mean cumulative distance travelled by motile vesicles in the indicated directions. (F,G) n = 394 vesicles, 12 cells, and 3 animals for wild-type; n = 450 vesicles, 18 cells, and 3 animals for conditional knockout. Error bars depict SEM. (H–J) Immunoprecipitation of vesicle-enriched fractions from cultured SCG neurons (SCG) (H) or SH-SY-5Y and PC12 cells (I) using anti-KIF1Bβ or anti-TRKA (J) with normal IgG as control. Immunoblots show coimmunoprecipitation of endogenous TRKA and endogenous KIF1Bβ. RAB3 served as a positive control. Bars, 10 µm.
We performed cell surface biotinylation assays to assess the effect of KIF1Bβ on TRKA presentation on the extracellular surface. Knockdown of KIF1Bβ (shKIF1Bβ) led to a robust reduction in the amount of TRKA on the cell surface of SH-SY-5Y cells (Fig. 4D). In contrast, cell surface abundance of another transmembrane receptor tyrosine kinase, EGFR, was not affected by KIF1Bβ knockdown (Fig. 4D), indicating that KIF1Bβ loss specifically alters TRKA localization to the cell surface.
To test whether KIF1Bβ effects movement of TRKA in neuronal processes, we transfected cultured SCG neurons with TrkA-mCherry and measured the movement of fluorescent vesicles by time-lapse microscopy and tracked vesicular movement over time with kymographs (Fig. 4E; Supplemental Movies S1, S2). Tracking of ∼400 vesicles in both KIF1Bβ-negative (conditional knockout) and wild-type neurons showed that anterograde and bidirectionally moving TRKA vesicles in axons were significantly less frequent in knockout neurons and that motile TRKA vesicles travelled shorter distances (Fig. 4F,G). Similar results were observed in differentiated PC12 cells when KIF1Bβ expression was abrogated by shRNA (Supplemental Fig. S4C–F), indicating that KIF1Bβ mediates anterograde transport of TRKA in neuronal processes.
Next, we immunoprecipitated endogenous KIF1Bβ from vesicle-enriched fractions of mouse sympathetic neurons (SCGs), PC12 cells, and neuroblastoma cells (SH-SY-5Y and SK-N-SH). TRKA and Rab3 coimmunoprecipitated specifically with KIF1Bβ (Fig. 4H,I; Supplemental Fig. S4G). RAB3 served as a control because it is known to be highly enriched in KIF1Bβ-bound presynaptic vesicles (Niwa et al. 2008). However, other members of the TRK receptor family (TRKB and TRKC) or other cell surface receptor proteins critical in sympathetic neuron development (P75NTR, RET, and BMPR1A) did not interact with KIF1Bβ (Fig. 4H,I; Supplemental Fig. S4G). Furthermore, reciprocal immunoprecipitation of endogenous TRKA coimmunoprecipitated endogenous KIF1Bβ. Importantly, no or negligible interaction between TRKA and the KIF1Bα isoform or KIF1A was observed (Fig. 4J; Supplemental Fig. S4H) despite recent reports that KIF1A can transport TRKA in sensory neurons of the dorsal root ganglia (Tanaka et al. 2016). This suggests that KIF1Bβ is the main kinesin responsible for TRKA transport in cells of sympathoadrenal origin. Collectively, these data indicate that RAB3-associated vesicular transport of TRKA by KIF1Bβ is critical for NGF–TRKA signaling. However, it is possible that other unknown KIF1Bβ cargos besides TRKA might also contribute to the observed phenotype in the KIF1BβcKO sympathetic ganglia.
Loss of Neurofibromin (NF1) rescues neuronal loss in KIF1BβcKO mice and restores RAS signaling
NGF-stimulated neurotrophic signaling through TRKA is required for survival and differentiation of sympathetic neurons and also for apoptosis during NGF competition from E17.5 to P3 (Bibel and Barde 2000; Nikoletopoulou et al. 2010). Thus, we asked whether the smaller size of the KIF1BβcKO ganglia is caused by increased apoptosis or decreased cell proliferation. TRKA is expressed in the SCGs from E13.5 (Fagan et al. 1996), a time when KIF1Bβ expression is already silenced in the KIF1BβcKO SCGs (Fig. 1A). Compared with control SCGs, we observed a significant increase in cleaved Caspase-3 in the KIF1Bβ-deficient SCGs at E15.5, before NGF competition (Fig. 5A,B). A similar increase in apoptosis has been reported in the TRKA−/− SCGs at E15.5 (Fagan et al. 1996). KIF1BβcKO SCGs were already visibly smaller at E17.5 (Fig. 5B), and, at E15.5, the proportion of cells in the cell cycle was similar in KIF1BβcKO and wild-type SCGs (Fig. 5C), indicating that the smaller size of the SCGs in the KIF1BβcKO is a consequence of increased apoptosis at E15.5.
Figure 5.
Loss of NF1 rescues neuronal loss in KIF1BβcKO mice and restores RAS signaling. (A,B) Quantification (A) and images (B) of immunofluorescence staining of cleaved Caspase-3 (green) in the SCGs of wild-type and KIF1Bβ−/− littermates at the indicated embryonic ages. n = at least 9 sections and 3 animals per genotype at each age. Endogenous TRKA (red) served as a counterstain. (C) Quantification of the percentage of KI67-positive cells in the SCGs of wild-type and KIF1Bβ−/− littermates at E15.5. n = 9 sections and 3 animals per genotype. (D) Quantification of sympathetic neuron survival after 24 h cultured with the specified concentrations of NGF. (E) Photographs of dissected mouse SCGs of the indicated genotypes and ages. (F) Immunoblot of SCGs from 6-mo-old wild-type, KIF1Bβ−/−, NF1−/−, and KIF1Bβ−/−;NF1−/− mice. In A, C, and D, mean values are depicted, and error bars correspond to SEM. Pairwise comparisons were made using two-tailed unpaired Student's t-tests. (**) P < 0.01. Bar, 100 µm.
However, unlike sympathetic neurons that lack either TRKA or NGF, which progressively shrink and completely degenerate after birth due to neuronal degeneration (Crowley et al. 1994; Fagan et al. 1996), KIF1Bβ-deficient SCG neurons survive (Fig. 1B), and apoptosis declines at E17.5 and P1 to a level comparable with wild type (Fig. 5A).
To understand whether the observed apoptosis in the KIF1BβcKO SCGs at E15.5 is due to lack of the trophic signal provided by NGF–TRKA, we cultured primary sympathetic neurons with different concentrations of NGF. Survival of KIF1Bβ-deficient sympathetic neurons cultured with low concentrations of NGF is much lower than in wild-type neurons (Fig. 5D), although they survive as wild type at higher concentrations of NGF. This suggests that the trophic effect of NGF is decreased in KIF1BβcKO neurons due to impaired anterograde transport of TRKA, leading to neuronal death and smaller sympathetic ganglia.
Activation of RAS proteins by NGF binding to TRKA leads to downstream activation of MAP kinases required for sympathetic neuronal survival and differentiation. We observed that MAP kinase activation in response to NGF depends on KIF1Bβ (Fig. 3E,F) To understand whether elevated RAS signaling could restore KIF1BβcKO SCGs to wild-type size, we crossed KIF1BβcKO mice with NF1fl/fl mice (Zhu et al. 2002). NF1 is a negative regulator of the RAS–MAP kinase signal, and loss of NF1 results in constitutive RAS activation (Cichowski and Jacks 2001). Loss of tumor suppressor NF1 has been associated with tumors originating from the neural crest, including neuroblastoma (The et al. 1993; Cichowski and Jacks 2001). We obtained the KIF1Bβfl/fl;Nf1fl/fl;DbhCre mouse (referred to here as KIF1Bβ;NF1cKO) after two generations at regular Mendelian ratios. Nf1fl/fl;DbhCre (NF1cKO) SCGs are larger than wild-type littermates (Fig. 5E). Importantly, KIF1Bβ;NF1cKO SCGs are much larger than KIF1BβcKO ganglia and are of similar or larger size compared with wild-type SCGs (Fig. 5E), indicating that loss of NF1 rescues the smaller size of KIF1Bβ-deficient SCGs. Despite reduced phosphorylation of TRKA on Y490 in both KIF1BβcKO and KIF1Bβ;NF1cKO SCGs, loss of NF1 in the KIF1Bβ;NF1cKO SCGs returned phospho-ERK1/2 to almost wild-type levels, demonstrating activated RAS signaling and that introduction of an alternative MAPK stimulus could compensate for the loss of NGF–TRKA trophic signaling and maintain neuronal survival in neurons that lack KIF1Bβ (Fig. 5F).
Cancer and neurodegenerative disease KIF1Bβ mutations are impaired in TRKA transport but differ in apoptotic capacity
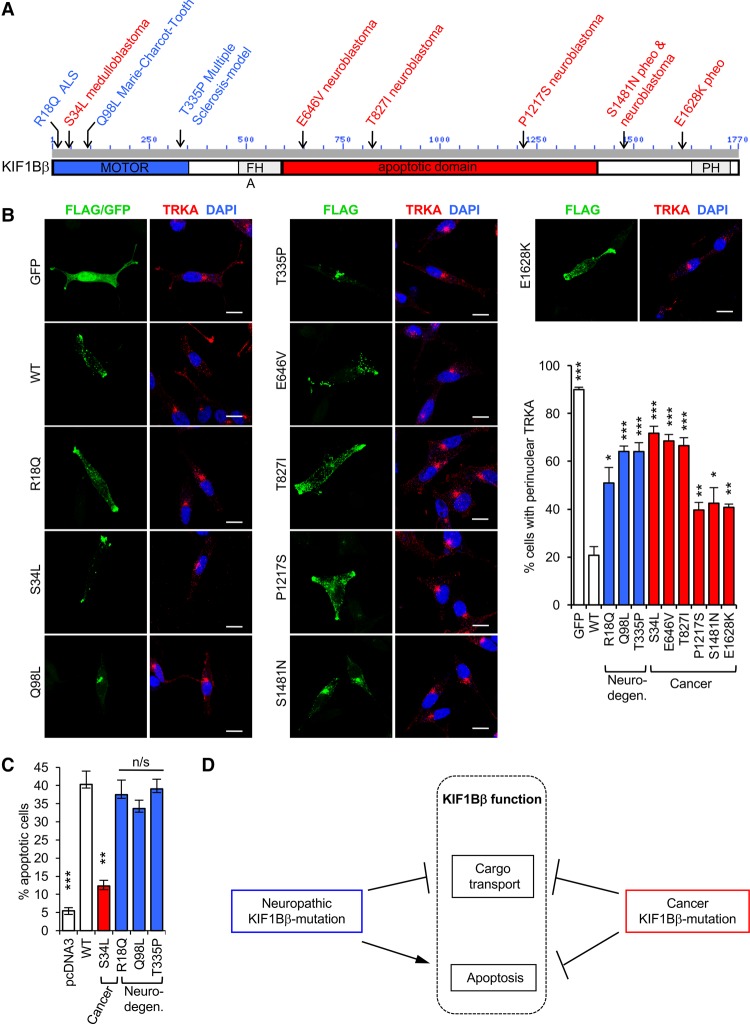
We recently identified pathogenic KIF1Bβ missense mutations in neuroblastoma and pheochromocytoma (Fig. 6A, in red; Schlisio et al. 2008). Mutations in KIF1Bβ are also associated with neurodegenerative disease. In particular, a mutation in KIF1Bβ (Q98L) defective in axonal transport has been reported previously to cause the common neuropathy CMT2A (Zhao et al. 2001). In addition, a coding variant in the KIF1Bβ motor domain (R18Q) segregating with amyotrophic lateral sclerosis (ALS) has been described (Herdewyn et al. 2012), and a mutation in the KIF1Bβ motor domain (T335P, commonly referred to as st43 in several studies) causes defects in axon development and myelination that resemble the pathology of multiple sclerosis (Fig. 6A, in blue). If loss of KIF1Bβ hinders neuronal differentiation through impaired transport of TRKA, then KIF1Bβ mutations associated with neuronal diseases such as cancer or neurodegeneration may exert their pathogenic effect through loss of this function. We tested disease-associated KIF1Bβ variants identified in neuroblastoma, pheochromocytoma, and neurodegenerative disease for their ability to affect TRKA localization in NB1 cells and observed that they are all significantly disabled in TRKA localization compared with wild type (Fig. 6B). Point mutations toward the N terminus, including the motor domain, reduced KIF1Bβ’s ability to affect TRKA localization to a greater extent than the three most C-terminal mutants (P1217S, S1481N, and E1628K) (Fig. 6B). Interestingly, all three of these mutants bind endogenous TRKA similar to wild-type KIF1Bβ in immunoprecipitation assays, while the other KIF1Bβ mutants failed to interact with TRKA (Supplemental Fig. S5A). These data together (Supplemental Fig. S5B) suggest that other factors, such as protein tertiary structure and interaction with adapter proteins (i.e., DENN/MADD, RAB3, and KIF1BP) (Hirokawa et al. 2010), potentially contribute to the pathogenic effects of disease-associated KIF1Bβ mutations on TRKA transport.
Figure 6.
Cancer and neurodegenerative disease KIF1Bβ mutations are impaired in TRKA transport but differ in apoptotic capacity. (A) Schematic of KIF1Bβ pathogenic mutations and locations identified in cancer and neurodegenerative disorders. (B) TRKA (red) immunofluorescence images of NB1 cells transfected with GFP control, Flag-KIF1Bβ wild type (WT), or pathogenic mutations (green) and quantification of TRKA localization. (C) Percentage of DAPI-stained GFP-positive primary rat sympathetic neurons exhibiting apoptotic changes after electroporation to produce GFP along with the indicated KIF1Bβ disease mutations. The values are mean ± SD from at least three individual experiments. (D) Schematic representing loss of KIF1Bβ function in both diseases, neurodegeneration (loss of KIF1Bβ motor function) and cancer (loss of both motor and apoptosis function), thus linking both types of disease. In B and C, pairwise comparisons were made using two-sided unpaired Student's t-tests and corrected for multiple testing using the Benjamini-Hochberg method. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. Error bars depict SEM, except in C. Bars, 10 µm.
Considering that all identified cancer-associated KIF1Bβ mutations have a reduced ability to induce apoptosis in sympathetic neurons (Schlisio et al. 2008; Li et al. 2016), we hypothesize that these mutations contribute to cancer malignancy by disrupting KIF1Bβ function in both apoptosis and differentiation. In contrast, it is possible that KIF1Bβ mutations associated with neurodegenerative disease primarily disable KIF1Bβ’s transport function and maintenance of neurotrophic support. Thus, we tested whether the neurodegenerative group of KIF1Bβ mutants is capable of inducing apoptosis similar to wild-type KIF1Bβ (Fig. 6C). As described previously, electroporation of wild-type KIF1Bβ into primary rat sympathetic neurons caused increased apoptosis (40%) compared with empty control or cancer-associated KIF1BβS34L (12%). However, KIF1Bβ mutants associated with neurodegenerative disease (R18Q, Q98L, and T335P) induced apoptosis similar to wild-type KIF1Bβ (Fig. 6C), indicating that they are not impaired in apoptotic function and are only defective in cargo transport. Given that the neuronal subtypes affected in these diseases do not necessarily express TRKA, it is possible that the malicious effects of these mutations are caused by failure to correctly transport other as yet unidentified KIF1Bβ cargos. Thus, KIF1Bβ provides a direct link between neurodegenerative disorders and cancer (Fig. 6D).
Loss of KIF1Bβ contributes to less differentiated and more aggressive neuroblastoma
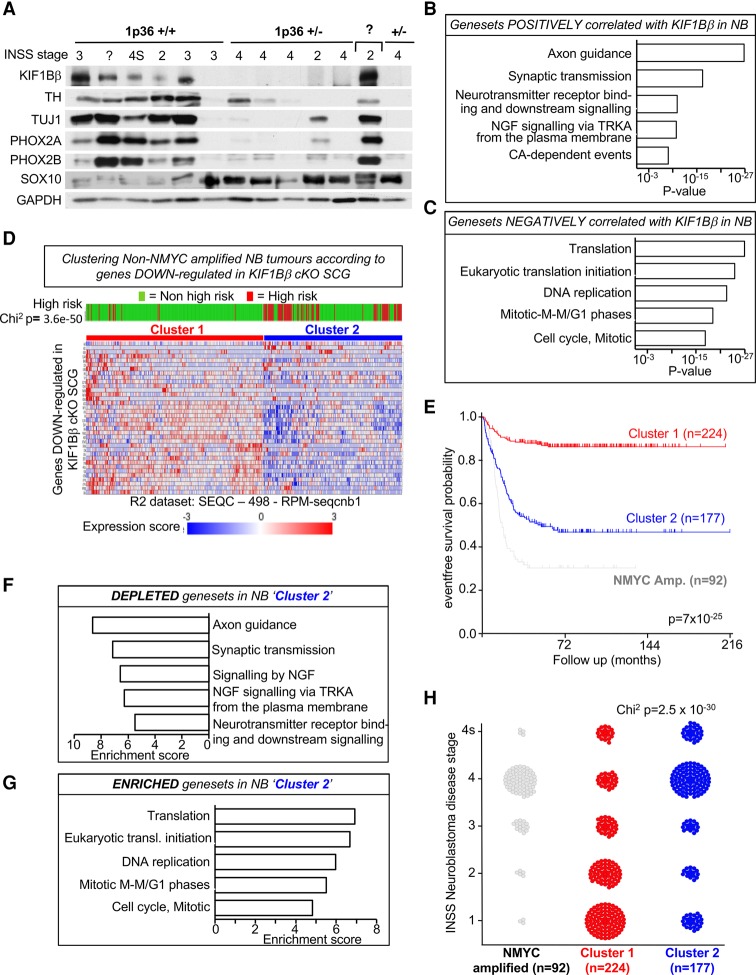
We observed previously that primary neuroblastoma tumors with hemizygous deletion of the 1p36 locus do not express KIF1Bβ protein (Li et al. 2016). We analyzed these tumors and observed very little or no protein expression of sympathetic neuron markers such as TH, TUJ1, and PHOX2A/B in KIF1Bβ-negative (1p36+/−) tumors in contrast to the 1p36-positive KIF1Bβ-expressing tumors. In contrast, KIF1Bβ-negative tumors expressed high levels of the neural crest stem cell protein SOX10 (Fig. 7A). We showed previously that low expression of KIF1Bβ correlates with poor prognosis in neuroblastoma (Li et al. 2016). In addition, individual analysis of a large gene expression database of neuroblastomas (R2: SEQC-498-RPM-1) confirmed that TUJ1 and TH expression positively correlates with KIF1Bβ expression and better prognosis in neuroblastoma, (Supplemental Fig. S6A,B). Importantly, expression of genes down-regulated in KIF1BβcKO SCGs, such as the synapse-related proteins SYNM and SNCA, also positively correlates with KIF1Bβ expression and better prognosis in neuroblastoma, (Supplemental Fig. S6C,D). Expression of genes that were up-regulated in KIF1BβcKO SCGs (such as ALK ligand MDK and NHLH2, both suspected oncogenes in neuroblastoma pathogenesis) (Brown et al. 1992; Aoyama et al. 2005; Isogai et al. 2011; Carpenter and Mosse 2012) negatively correlates with KIF1Bβ expression and is associated with worse prognosis in neuroblastoma (Supplemental Fig. S6E,F). Together, these data suggest that loss of KIF1Bβ is associated with a less differentiated and potentially more aggressive tumor phenotype. Furthermore, gene sets that positively correlate with KIF1Bβ expression in neuroblastoma (SEQC-498-RPM-1 data set)—“nervous system development,” “transmission of nerve impulse,” “synaptic transmission,” “cytoskeleton organization and biogenesis,” “microtubule based process,” and “neuron differentiation” (Fig. 7B)—mirror those cellular processes affected in the SCGs of KIF1BβcKO mice. Conversely, gene sets that are negatively correlated with KIF1Bβ expression in neuroblastoma describe increased translation and mitosis (Fig. 7C), suggesting increased cellular activity and proliferation in the absence of KIF1Bβ.
Figure 7.
Loss of KIF1Bβ contributes to less differentiated and more aggressive neuroblastoma. (A) Immunoblot analysis for key differentiation markers in primary neuroblastoma tumors. 1p36 status has been characterized and is shown as indicated. (B,C) Gene sets from the “pathway commons” collection that positively (B) and negatively (C) correlate with KIF1Bβ in the SEQC-498-RPM neuroblastoma RNA-seq expression data set (R2 platform, http://r2.amc.nl). (D) Clustering of non-MYCN-amplified neuroblastoma tumors (public SEQC-498 data set) according to genes that are down-regulated in KIF1BβcKO SCGs (cKO). (E) Kaplan-Meier overall survival curve for individuals of both clusters identified in D using the tumor neuroblastoma public SEQC-498 data set (R2 platform, http://r2.amc.nl). MYCN-amplified tumors from the same data set were included for comparison. (F,G) Parametric analysis of gene set enrichment showing gene sets depleted (F) and enriched (G) in tumors from cluster 2 that was defined in D. (H) International Neuroblastoma Staging System (INSS) disease stage distribution of tumors from clusters identified in D in comparison with MYCN-amplified tumors from the tumor neuroblastoma public SEQC-498 data set.
Loss or low expression of KIF1Bβ has been observed in neuroblastomas containing hemizygous deletion of chromosome 1p36 (Chen et al. 2014; Li et al. 2016). Deletion of 1p36 removes several genes simultaneously, making it hard to judge the contribution of each individual deleted gene to tumor pathogenesis. It is therefore possible that phenotypes associated with low expression of KIF1Bβ in neuroblastoma (Fig. 7B,C) are simply a consequence of the loss of other candidate tumor suppressor genes located on 1p36—a so-called “bystander effect.” Thus, we performed computational analyses to assess the specific effect of KIF1Bβ loss on neuroblastoma independent of other 1p36 genes that would be lost with 1p36 deletion.
Therefore, we used genes that were significantly down-regulated in the SCGs of KIF1BβcKO mice (Supplemental Table S1) and compared this mouse gene list with the gene expression profiles from a large data set of human neuroblastoma tumors from the R2 database SEQC-498_RPM-1 (Fig. 7D; Su et al. 2014). We used non-MYCN-amplified neuroblastomas in this analysis because MYCN amplification in neuroblastoma has a dominating effect on gene expression and frequently co-occurs with 1p36 loss. In an unsupervised clustering analysis (k-means clustering), we found that clustering tumors according to genes down-regulated in KIF1BcKO SCGs produced two distinct clusters of tumors: cluster 1 and cluster 2 (Fig. 7D).
Tumors in cluster 2 have low expression of genes that were down-regulated in KIF1BβcKO SCG (Fig. 7D). Thus, we infer that these cluster 2 tumors mirror, to some extent, what would happen if specifically KIF1Bβ was lost in neuroblastoma. These cluster 2 tumors are significantly more likely to be found in patients with high-risk neuroblastoma (Fig. 7D), poor survival outcome (Fig. 7E), and higher disease stage (Fig. 7H). The classification of these cluster 2 tumors by International Neuroblastoma Staging System (INSS) disease stage closely resembles that of MYCN-amplified tumors with a prevalence in advanced metastatic disease, stage 4 (Fig. 7H), and high-risk classification (Fig. 7D). Furthermore, the prognosis for cluster 2 tumors is far worse than tumors in cluster 1 and nears the low survival probability of MYCN-amplified tumors (Fig. 7E).
Gene sets depleted in cluster 2 involve “axon guidance” and “NGF signaling via TRKA” (Fig. 7F), while enriched gene sets include “DNA replication” and “cell cycle” (Fig. 7G). These gene sets are highly similar to those affected in KIF1BβcKO SCGs (Fig. 2B) and those that correlate with KIF1Bβ expression in neuroblastoma (Fig. 7B,C), suggesting that this clustering analysis did reflect specific loss of KIF1Bβ in neuroblastoma.
This analysis demonstrates the capacity of the KIF1BβcKO down-regulated genes to predict and cluster neuroblastomas into high-risk versus low-risk tumors, suggesting a specific effect of KIF1Bβ loss on neuroblastoma regardless of any potential “bystander” effects caused by concurrent loss of other 1p36 genes. In conclusion, this suggests that loss of KIF1Bβ has a specific pathogenic effect on neuroblastoma development independent of additional genes located on 1p36.
Discussion
Abnormal NGF signaling has been linked to sympathetic nervous system tumors, and expression of the NGF receptor TRKA is a strong favorable prognostic factor, especially in non-MYCN-amplified neuroblastoma (Kogner et al. 1993; Nakagawara et al. 1993; Brodeur et al. 2009). Recently, we reported that low KIF1Bβ expression correlates with worse outcome in non-MYCN-amplified neuroblastoma (Li et al. 2016). Here we generated mice deficient in KIF1Bβ in the sympathoadrenal cell lineage and found that clustering non-MYCN-amplified tumors according to expression of genes down-regulated in KIF1Bβ-deficient sympathetic ganglia was prognostically relevant in regard to risk classification, disease stage, and survival (Fig. 7D,E,H). Our analysis of neuroblastoma tumors here suggests that specific loss of KIF1Bβ contributes to less differentiated and more aggressive disease independent of MYCN amplification and the loss of genes neighboring KIF1B on 1p36. This was also reflected by immunoblot analysis comparing 1p36 hemizygous-deleted with 1p36 intact neuroblastomas. KIF1Bβ protein expression together with critical sympathoadrenal differentiation markers was ablated in 1p36-deleted tumors (Fig. 7A). Indeed, we observed that KIF1Bβ is required for neuron maturation and function in the mouse sympathetic nervous system. Together, these results suggest that KIF1Bβ may affect precursor cell differentiation in neuroblastoma. Pertinently, defective precursor cell differentiation could be a common trait or “hallmark” of pediatric malignancies.
We found that KIF1Bβ is required for NGF-mediated sympathetic neuron differentiation and proper sympathetic neuron target innervation by mediating axonal transport of the NGF receptor TRKA. Pathogenic KIF1Bβ mutations identified in neuroblastoma as well as loss of KIF1Bβ all impair anterograde transport of TRKA. Thus, pathogenic KIF1Bβ mutations, similar to loss of KIF1Bβ, might contribute to defective NGF-mediated differentiation and survival of developing sympathetic neurons. The smaller ganglia observed in KIF1BβcKO mice resemble those of TRKA−/− and NGF−/− mice, with less differentiated neurons and impaired axonal extension and target innervation. Moreover, we observed decreased protein expression of MASH1, PHOX2A, and GATA3, all of which are critical for sympathetic neuron differentiation in the KIF1BβcKO SCGs. PHOX2B, the other main sympathoneurogenic transcription factor, was not effected at P1. It has been established that MASH1 is essential for sympathoblast differentiation and that loss of MASH1 causes the absence of differentiated neurons in developing sympathetic ganglia (Guillemot et al. 1993; Hirsch et al. 1998). Previous studies have shown that NGF–TRKA signaling promotes MASH1 expression by alleviating the active repression of MASH1 by HES1 (Johnson et al. 1990; Strom et al. 1997). Furthermore, more recent work has suggested that activated ERK can phosphorylate MASH1, inducing MASH1 transcriptional activity (Li et al. 2014). Both of these mechanisms potentially regulate MASH1 in response to NGF–TRKA signaling in the developing sympathetic ganglia. Thus, loss of KIF1Bβ may lead to decreased MASH1 expression and transcriptional activity as a result of impaired NGF–TRKA signaling.
Decreased MASH1 could also contribute to the observed differences in PHOX2A and GATA3 expression (Hirsch et al. 1998; Tsarovina et al. 2004); however, further studies would be required to conclusively investigate these interactions. We speculate that lack of NGF neurotrophic signaling due to failure of TRKA transport might reduce MASH1 activity in the KIF1BβcKO SCG. Altered MASH1 activity could conceivably affect expression of other key transcription factors in the developing sympathetic ganglia, contributing to impaired neuronal maturation and survival and some of the functional defects in sympathetic nervous function observed in this study.
Importantly, KIF1BβcKO SCGs do not degenerate after birth as observed in the TRKA−/− or NGF−/− mice (Crowley et al. 1994; Fagan et al. 1996). The phenotype of KIF1Bβ-deficient ganglia resembles that reported for heterozygous loss of NGF, suggesting that KIF1Bβ deletion may impair NGF–TRKA signaling rather than completely ablate it. However, we and others reported previously that acute loss of KIF1Bβ protects cultured primary rat sympathetic neurons from apoptosis after NGF withdrawal (Munirajan et al. 2008; Schlisio et al. 2008; Chen et al. 2014; Li et al. 2016). Thus, loss of KIF1Bβ might protect neurons from developmental culling when they compete for target-derived NGF. Such protection from apoptosis could balance the negative effects of reduced trophic signaling and explain why TRKA−/− or NGF−/− neurons undergo complete degeneration (Fagan et al. 1996), in contrast to the KIF1Bβ−/− neurons that persist during adult life.
Our discovery that KIF1Bβ is important for NGF–TRKA neurotrophic signaling is highly relevant for certain neurodegenerative diseases. Previous studies (Pogoda et al. 2006; Lyons et al. 2009; Tanaka et al. 2016) have demonstrated that loss of KIF1Bβ can be neuropathic; however, it has been unclear mechanistically how loss of KIF1Bβ function might contribute to neurodegeneration. By mediating transport of TRKA to axon terminals, KIF1Bβ helps to maintain neurotrophic support critical for the survival of TRKA-expressing neurons. A growing body of evidence suggests that deficient axonal transport is a common feature of adult-onset neurodegenerative diseases (Millecamps and Julien 2013), such as ALS and multiple sclerosis, and congenital neuropathies, including CMT disease and hereditary sensory and autonomic neuropathies (HSAN) (Scherer 2006). Furthermore, congenital mutations in NGF (HSAN-V) and TRKA (HSAN-IV) impair neurotrophic signaling and contribute to loss of function and degeneration of sympathetic and other peripheral neurons in these diseases (Rotthier et al. 2012). Dysregulated TRKA signaling also contributes to degeneration of neurons in Alzheimer's disease, and clinical trials using NGF to abate neurodegeneration on the cortices of Alzheimer's patients have had some success (Tuszynski et al. 2015). The mechanistic data presented here suggest that loss of KIF1Bβ motor function has a neuropathogenic effect due to its role in axonal transport of TRKA.
Interestingly, we found that KIF1Bβ mutations associated with neuropathy have an unimpaired capacity to induce apoptosis, while cancer-associated KIF1Bβ mutations have both reduced apoptotic and TRKA transport capabilities. Thus, cancer-associated mutations in KIF1Bβ target both hallmarks of cancer, the failure to die and differentiate, whereas neuropathy-associated KIF1Bβ mutations impair only cargo transport required for neurotrophic support. Although cancer and neurodegeneration have apparently opposing disease ontologies (the former due to enhanced resistance to cell death and the latter due to premature cell death), we propose here that KIF1Bβ provides a direct link between these two apparently unrelated types of disease by mediating the axonal anterograde transport of TRKA.
Materials and methods
Plasmids, antibodies, proteins, and reagents
Expression plasmids for Flag-KIF1Bβ and mutants were described previously (Li et al. 2016). Plasmids encoding Flag- KIF1Bβ Q98L and Flag-KIF1Bβ T335P were generated using QuikChange II XL site-directed mutagenesis kit (Stratagene). Primers are listed in Supplemental Table S2. Lentiviral pLKO.1 plasmids encoding shRNAs for human KIF1Bβ were purchased from Sigma, and lentiviral pGFP-c-shLenti plasmids encoding shRNAs for rat KIF1Bβ were purchased from Origene target sequences (Supplemental Table S2). Plasmid encoding TRKA-mCherry (pEZ-M56-TRKA) was purchased from Genecopoeia, Inc.
Antibodies are listed in Supplemental Table S3. Fluorophore-conjugated secondary antibodies were from Molecular Probes, and HRP-conjugated antibodies were purchased from Thermo Scientific. Recombinant human β-NGF was purchased from Peprotech.
KIF1BβcKO mice
Creation of the KIF1Bβfl/fl;DbhCre mouse strain is described in Li et al. (2016), and NF1fl/fl mice were described recently in Zhu et al. (2002) and were obtained from the Jackson laboratory [mouse strain B6.129(Cg)-Nf1tm1par/J].
Cell culture
PC12 cells and neuroblastoma cell lines were maintained as described previously (Schlisio et al. 2008). NB1 cells were obtained from the Japanese Collection of Research Bioresources. SH-SY-5Y, SK-N-SH, and PC12 cell lines were obtained from American Type Culture Collection. The cell lines used were not further authenticated but were tested for mycoplasma contamination before use. Primary sympathetic neurons from P1 mice were isolated from the SCGs and cultured as described previously (Palmada 2002). PC12 cells cultured with 50 ng/mL NGF were counted using a hemocytometer, and the average of two counts was used to estimate cell number at each time point for each biological replicate.
Neuronal survival assay
Primary sympathetic neurons were cultured on poly-L-ornithine and laminin-coated coverslips engraved with a numbered grid in the indicated concentrations of 2.5S NGF (Harlan, catalog no. BT5017). The number of adherent phase-positive neurons in a defined area of the grid was counted 3 and 24 h after plating. The number of neurons surviving after 24 h was expressed as a percentage of the initial number of neurons (at 3 h). Approximately 100 neurons were counted for each of six biological replicates per genotype.
Light microscopy
Bright-field images of whole SCGs and tissue sections stained by in situ hybridization or immunohistochemically were captured on a Nikon C-DSS230 dissecting microscope with an SMZ2800 objective using Nikon “digital sight” hardware and NIS-elements software.
Immunofluorescence and confocal microscopy
Cultured cells were prepared for immunocyctochemistry as described previously (Li et al. 2016) except for primary cultures of sympathetic neurons that were blocked and permeabilized in a solution of 25% donkey serum and 0.2% Tween-20 in PBS. SCGs were dissected and immediately fixed in 4% paraformaldehyde for 90 min on ice, washed three times in PBS, incubated overnight in 30% sucrose, and, finally, embedded and frozen in OCT cryomount solution (Histolab). Twelve-micrometer serial sections were cut on a Microm HM500OM freezing cryostat, mounted directly onto Superfrost Plus (Menzel-Gläser) glass slides, and stained for immunohistochemistry essentially as described previously (Laguna et al. 2008). Littermates were used in all comparative experiments. Slides were mounted using Prolong Diamond anti-fade reagent (Invitrogen). Images were acquired and analyzed using a Zeiss LSM 5 Exciter or Zeiss LSM 510 Meta laser scanning confocal microscope together with Zeiss LSM 5 Exciter or Zeiss LSM 510 software, respectively. ImageJ (Schneider et al. 2012) and Adobe Photoshop 5 were used to process the images. Cells displaying positive staining for various markers were counted using the “count tool” in Adobe Photoshop. Cell density was calculated by dividing the number of DAPI-positive nuclei in sections of dissected SCGs by the cross-sectional area of the SCGs for the section. For quantification of protein intracellular localization, at least 100 cells were visually assessed and counted per treatment per experiment from at least three separate experiments.
RNA extraction from SCGs
Total RNA was isolated from snap-frozen SCGs using a modified Trizol extraction protocol as follows. SCG pairs from each animal were thawed and then suspended in 300 µL of Trizol (Invitrogen) containing 20 µg/mL proteinase K (Invitrogen) and incubated for 10 min at 55°C with shaking. Samples were then cooled on ice, and 2 µL of Pelletpaint (Novagen) was added followed by 60 µL of chloroform. Samples were then incubated for 5 min on ice and centrifuged at 20,000g for 10 min to separate phases. The upper (aquaeus) phase was then collected and combined 1:1 with isopropanol, mixed vigorously, incubated for 5 min on ice, and centrifuged at 20,000g for 30 min to precipitate RNA. After aspiration of supernatant, the RNA pellets were then washed with 300 µL of 75% ethanol and centrifuged for a further 10 min. After aspiration of the supernatant, pellets were resuspended in 100 µL of water, and 10 µL of 3 M sodium acetate was added followed by 250 µL of ice-cold 100% EtOH. Following incubation for 1 h at −20°C and centrifugation for 30 min, suparnatant was aspirated, and 300 µL of 75% ethanol was added to wash the RNA pellets, which were then centrifuged for a further 10 min. Finally, supernatant was completely aspirated from the pellets, which were then briefly left to dry at room temperature in a fume hood and then resuspended in 20 µL of water. RNA was then quantified using a Nanodrop and quality-controlled using RNA PICO chips on an Agilent Bioanalyzer. Only samples with an RNA integrity index of >7.5 were used for RNA-seq.
RNA-seq
RNA extracted from SCGs of four KIF1Bβfl/fl;DbhCre animals and four KIF1Bβfl/fl control animals was sequenced. Total RNA (180 µg) from quality-controlled samples was processed for mRNA-seq using TRUseq version 2 (Illumina protocol; polyA+, not strand-specific) and sequenced on an Illumina HiSeq 2000 with a 50-base-pair single-end read length. Reads were mapped using RNA-star (Dobin et al. 2013), and relative expression levels were estimated with RPKMforgenes (Ramskold et al. 2009). Differential gene expression was assessed using the VOOM limma package (Ritchie et al. 2015) for R (https://CRAN.R-project.org/package=nplr) with the following parameters: counts per million (CPM) >1, reads per kilobase per million mapped reads (RPKM) >1, padj >0.05, and fold change >1.5. These data are available on the NCBI Gene Expression Omnibus (accession no. GSE90952)
Gene ontology analysis was performed using GOrilla (Eden et al. 2009), with an input of all differentially expressed genes compared with a background list of expressed genes (CPM >1; RPKM >1).
We performed GSEAs using GSEA software and the Molecular Signature Database (MSigDB) (Subramanian et al. 2005) on the gene expression data described above. We used the standard statistical cutoffs for GSEA, which had a nominal P-value of <0.05 and false discovery rate of <0.25. Gene sets enriched in our data set and presented in Figure 2 of this study are shown in Table 1.
Table 1.
Gene sets either enriched or depleted in KIF1Bβfl/fl;DbhCre P1 SCGs
Analysis of neuroblastoma gene expression
Analysis of neuroblastoma gene expression databases was performed using the R2 Genomics Analysis and Visualization Platform (Koster et al. 2015). Using the neuroblastoma SEQC-498-RPM-seqcnb1 data set (Su et al. 2014) that consists of RNA-seq data from 498 neuroblastomas, we tested for genes that correlate with KIF1Bβ (NM_015074) with a Pearson's R cutoff of P < 0.01, excluding tumors with MYCN amplificiation. We then performed a GSEA on both of the genes that positively or negatively correlate with KIF1Bβ using “geneset_pathway_commons” gene set collections with a P-value cutoff of <0.05.
The list of differentially expressed genes in KIF1Bβfl/fl;DbhCre SCGs generated by RNA-seq analysis was used to group non-MYCN-amplified tumor samples from the SEQC-498-RPM-seqcnb1 data set according to their expression of genes that were either up-regulated or down-regulated in the KIF1Bβfl/fl;DbhCre SCGs. First, we filtered genes that were either up-regulated or down-regulated in the KIF1BβcKO SCGs based on whether they varied significantly with INSS disease stage in this tumor data set. We then used k-means iterative clustering analysis to group the tumors according to their expression of these genes (two groups, 10 × 10 iterations). We then saved the clusters as parameter “tracks” in the R2 interface and plotted these tumor clusters along with the MYCN-amplified tumors, which were not included in the clustering analysis, against INSS disease stage and event-free survival. The parameteric analysis of gene set enrichment (PAGE) tool was then used to identify gene sets that were enriched in the tumor clusters using the “geneset_broad_2015_go” collection.
Immunoprecipitation
One 100-mm plate of cells was rinsed twice in ice-cold PBS, scraped into a small volume of ice-cold PBS, collected by centrifugation at 1000g for 5 min, resuspended in lysis buffer (10 mM Tris at pH 7.4, 10% sucrose, 1 mM EDTA) containing protease inhibitors, and homogenized by six passages through a 22-gauge needle. The homogenate was centrifuged at 2000g for 5 min to pellet nuclei and unlysed cells, and the resulting supernatant was incubated overnight at 4°C with IgG, KIF1Bβ, or TrkA antibodies, respectively (Supplemental Table S2). Samples were incubated for 4 h at 4°C with 70 µL of protein G agarose slurry and washed twice with wash buffer (0.5% NP-40, 150 mM NaCl, 10 mM Tris-HCl). Samples were centrifuged and washed five times with wash buffer (0.5% NP-40, 150 mM NaCl, 10 mM Tris-HCl). Bound protein complexes were eluted with 20 mL of Laemmli buffer, boiled for 5 min, and centrifuged at 8000g for 30 sec. Eluted fractions were analyzed by immunoblot.
Quantification of sympathetic innervation
Twelve-micrometer serial sagittal sections of frozen paraformaldehyde-fixed eyes from 12-wk-old mice or submandibular glands from 17-wk-old mice were probed with an Pel-Freez rabbit anti-TH (1:1000) followed by Alexa fluor donkey anti-rabbit 555 (1:1000) secondary antibody. Images were taken with a 20× objective, and image capture parameters were kept constant for all images analyzed. Image analysis was performed in ImageJ (Schneider et al. 2012). Briefly, background staining levels in the iris were equalized, and images were transformed into positive binary images using the “threshold → select [otsu] auto” command. Iris area for each section was traced freehand and saved. “Area, area fraction, limit to threshold and display label” were then measured for each image, and the relative TH-positive area (“area fraction”) was compared between groups. Three to five sections were analyzed per animal and averaged. There were three animals in each group. Group means were compared using a two-tailed equal variance t-test.
TrkA biotinylation and internalization assay
Receptor biotinylation and endocytosis assays were performed by the cleavable biotin method ( Vickery and von Zastrow 1999) using a commercial kit (cell surface protein isolation kit, Pierce, catalog no. 89881). SHSY-5Y cells stably transduced with either scramble or KIF1Bβ targeting shRNA were pretreated with 100 nM Bafilomycin A1 (LC Laboratories) and 20 µM leupeptin (hemisulfate salt, Sigma Aldrich) for 2 h before assay to prevent lysosomal proteolysis of internalized receptors. Cell surfaces were biotinylated with 500 µg/mL cleavable Sulfo-NHS-SS-Biotin for 30 min at 4°C. After washing with PBS, cells were switched to the culture medium and stimulated with 50 ng/mL NGF for 40 min at 37°C. After ligand stimulation, cells were cooled to 4°C, and unendocytosed surface biotin was cleaved by incubating in the glutathione cleavage buffer (100 mM glutathione, 75 mM NaCl, 10 mM EDTA, 1% BSA at pH 7,5–8). Afterward, cells were lysed (50 mM Tris-HCl at pH 8, 120 mM NaCl, 0.5% NP-40, protease inhibitors) and disrupted by sonication. Lysates were cleared by centrifugation at 10, 000g for 10 min at 4°C. Biotin-labeled proteins were isolated with NeutrAvidin agarose beads from the cell lysates. The bound proteins were released by incubation with SDS-PAGE sample buffer containing 50 mM DTT. Biotinylated proteins were resolved by SDS-PAGE, transferred to a nitrocellulose filter, and immunoblotted with an anti-TrkA antibody (1:1000; Cell Signaling, catalog no. 2505), anti-EGFR antibody (1:1000; Cell Signaling, catalog no. 2232), or anti-TNFR1 antibody (1:1000; Cell Signaling, catalog no. 3736).
Neurite outgrowth
SCG neurons from P2 mice were incubated with 1 or 10 ng/mL NGF for 12 h, fixed, and incubated with TuJ1 primary antibody (Covance) followed by Alexa 488-conjugated secondary antibody (Molecular Probes). Images of complete branches were quantified by the NeuriteTracer plug-in (M. Pool) for ImageJ software (five neurons per animal). The average total length per neuron was calculated, and the mean value for each experiment was normalized to wild type (n = 4–6 animals).
Live imaging of vesicle movement
P1 SCG neurons were transfected using the Neon electroporation system (Invitrogen) with TrkA-mCherry and plated on poly-L-ornithine and laminin-coated glass coverslips. For imaging, the cells were placed in a CO2- and temperature-controlled incubator attached to the microscope. Time-lapse movies were recorded on a DeltatVision deconvolution microscope with a 60× oil objective. Images were captured every 0.5 sec for 1 min per neurite, and a total of ∼400 vesicles was analyzed for each genotype in 12–18 neurons. Independent cell cultures were prepared from each littermate animal, with at least three animals assayed per genotype. Vesicle movements were analyzed with ImageJ using the KymoToolBox plug-in. Time-lapse images were also taken of PC12 cells transfected (Lipofectamine 2000) with TrkA-mCherry. Briefly, PC12 cells were transfected with TRKA-mCherry-expressing and shRNA-expressing plasmid, split the following day into NGF-containing differentiation medium, and then imaged after 3 d of culture with 50 ng/mL NGF.
Statistics
Statistical comparisons between groups were made using ANOVA followed by pairwise comparisons using two-sided unpaired Student's t-tests with the assumption of nonequal variance unless otherwise stated. The Benjamini-Hochberg method was used to correct for multiple testing (P < 0.05 [*], P < 0.01 [**], and P < 0.001 [***]). In bar charts, the top of a bar depicts the mean, and error bars depict the SEM. Where ANOVA and t-tests did not reveal statistically significant differences, we checked that the power of the test was at least 0.8 or that the magnitude of the difference between treatment groups was likely to be biologically meaningful. Statistical analyses were performed using Microsoft Excel or R Core Team software. We did not exclude any samples post hoc. The investigator was blinded to group allocation during sample collection (Supplemental Fig. S1A–C), during measurement of hemodynamics (Supplemental Fig. S1A), in experiments involving counting of cells or micrographs (Figs. 1D–H, 2F,G, 3B,C, 4B–D, 5A–D, 6B,C; Supplemental Figs. S2A, S4B), and/or during image capture (Figs. 1F–H, 4E–G; Supplemental Fig. S4C–E) when automated quantification was used.
Ethical considerations
All available neuroblastoma tumors were from the Swedish NB Registry, and ethical permission was granted by the relevant authority (KI Forskningskommitee Nord). Ethical permits for animal studies were approved by the following appropriate local and national authorities: Jordbruksverket (Sweden) and the University of South Florida's Division of Research Integrity and Compliance Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
The Susanne Schlisio laboratory is supported by the Swedish Children Cancer Foundation, the Swedish Cancer Foundation, the Paradifference Foundation, the Swedish Research Council (VR), the Gösta Fraenckel Foundation, and the Ludwig Institute for Cancer Research (LICR).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.297077.117.
References
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. 2005. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res 65: 4587–4597. [DOI] [PubMed] [Google Scholar]
- Biarc J, Chalkley RJ, Burlingame AL, Bradshaw RA. 2013. Dissecting the roles of tyrosines 490 and 785 of TrkA protein in the induction of downstream protein phosphorylation using chimeric receptors. J Biol Chem 288: 16606–16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M, Barde YA. 2000. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev 14: 2919–2937. [DOI] [PubMed] [Google Scholar]
- Bishop T, Gallagher D, Pascual A, Lygate CA, de Bono JP, Nicholls LG, Ortega-Saenz P, Oster H, Wijeyekoon B, Sutherland AI, et al. 2008. Abnormal sympathoadrenal development and systemic hypotension in PHD3−/− mice. Mol Cell Biol 28: 3386–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM, Minturn JE, Ho R, Simpson AM, Iyer R, Varela CR, Light JE, Kolla V, Evans AE. 2009. Trk receptor expression and inhibition in neuroblastomas. Clin Cancer Res 15: 3244–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Espinosa R III, Le Beau MM, Siciliano MJ, Baer R. 1992. HEN1 and HEN2: a subgroup of basic helix-loop-helix genes that are coexpressed in a human neuroblastoma. Proc Natl Acad Sci 89: 8492–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EL, Mosse YP. 2012. Targeting ALK in neuroblastoma—preclinical and clinical advancements. Nat Rev Clin Oncol 9: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Wallis K, Fell SM, Sobrado VR, Hemmer MC, Ramskold D, Hellman U, Sandberg R, Kenchappa RS, Martinson T, et al. 2014. RNA helicase A is a downstream mediator of KIF1Bβ tumor-suppressor function in neuroblastoma. Cancer Discov 4: 434–451. [DOI] [PubMed] [Google Scholar]
- Cho A, Tang Y, Davila J, Deng S, Chen L, Miller E, Wernig M, Graef IA. 2014. Calcineurin signaling regulates neural induction through antagonizing the BMP pathway. Neuron 82: 109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. 2001. NF1 tumor suppressor gene function: narrowing the GAP. Cell 104: 593–604. [DOI] [PubMed] [Google Scholar]
- Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, McMahon SB, Shelton DL, Levinson AD, et al. 1994. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell 76: 1001–1011. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O'Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, et al. 2013. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell 26: 223–236. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Zhang H, Landis S, Smeyne RJ, Silos-Santiago I, Barbacid M. 1996. TrkA, but not TrkC, receptors are essential for survival of sympathetic neurons in vivo. J Neurosci 16: 6208–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebova NO, Ginty DD. 2004. Heterogeneous requirement of NGF for sympathetic target innervation in vivo. J Neurosci 24: 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 113: 657–670. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. 1993. Mammalian achaete–scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75: 463–476. [DOI] [PubMed] [Google Scholar]
- Guyenet PG. 2006. The sympathetic control of blood pressure. Nat Rev Neurosci 7: 335–346. [DOI] [PubMed] [Google Scholar]
- Herdewyn S, Zhao H, Moisse M, Race V, Matthijs G, Reumers J, Kusters B, Schelhaas HJ, van den Berg LH, Goris A, et al. 2012. Whole-genome sequencing reveals a coding non-pathogenic variant tagging a non-coding pathogenic hexanucleotide repeat expansion in C9orf72 as cause of amyotrophic lateral sclerosis. Hum Mol Genet 21: 2412–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68: 610–638. [DOI] [PubMed] [Google Scholar]
- Hirsch MR, Tiveron MC, Guillemot F, Brunet JF, Goridis C. 1998. Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development 125: 599–608. [DOI] [PubMed] [Google Scholar]
- Isogai E, Ohira M, Ozaki T, Oba S, Nakamura Y, Nakagawara A. 2011. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS One 6: e19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Stanke J, Lahti JM. 2011. The connections between neural crest development and neuroblastoma. Curr Topics Dev Biol 94: 77–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Anderson DJ. 1990. Two rat homologues of Drosophila achaete–scute specifically expressed in neuronal precursors. Nature 346: 858–861. [DOI] [PubMed] [Google Scholar]
- Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. 2015. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell 57: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogner P, Barbany G, Dominici C, Castello MA, Raschella G, Persson H. 1993. Coexpression of messenger RNA for TRK protooncogene and low affinity nerve growth factor receptor in neuroblastoma with favorable prognosis. Cancer Res 53: 2044–2050. [PubMed] [Google Scholar]
- Koster J, Molenaar JJ, Versteeg R. 2015. Abstract A2-45: R2: accessible Web-based genomics analysis and visualization platform for biomedical researchers. Cancer Res 10.1158/1538-7445.TRANSCAGEN-A2-45. [DOI] [Google Scholar]
- Laguna A, Aranda S, Barallobre MJ, Barhoum R, Fernández E, Fotaki V, Delabar JM, de la Luna S, de la Villa P, Arbonés ML. 2008. The protein kinase DYRK1A regulates Caspase-9-mediated apoptosis during retina development. Dev Cell 15: 841–853. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LSB, Goodson HV, Hirokawa N, Howard J, et al. 2004. A standardized kinesin nomenclature. J Cell Biol 167: 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mattar P, Dixit R, Lawn SO, Wilkinson G, Kinch C, Eisenstat D, Kurrasch DM, Chan JA, Schuurmans C. 2014. RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J Neurosci 34: 2169–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fell SM, Surova O, Smedler E, Wallis K, Chen ZX, Hellman U, Johnsen JI, Martinsson T, Kenchappa RS, et al. 2016. The 1p36 tumor suppressor KIF 1Bβ is required for calcineurin activation, controlling mitochondrial fission and apoptosis. Dev Cell 36: 164–178. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Naylor SG, Scholze A, Talbot WS. 2009. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet 41: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien JP. 2013. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci 14: 161–176. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, D'Autreaux F, Gershon MD, Cserjesi P. 2007. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev Biol 307: 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Cremer H, Hirsch MR, Kapur RP, Goridis C, Brunet JF. 1997. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron 18: 411–423. [DOI] [PubMed] [Google Scholar]
- Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M, Koda T, Hirota T, Ozaki T, Nakagawara A. 2008. KIF1Bβ functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem 283: 24426–24434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Ichimiya S, Ozaki T, Seki N, Mihara M, Furuta S, Ohira M, Tomioka N, Nomura N, Sakiyama S, et al. 2000. Identification of the full-length KIAA0591 gene encoding a novel kinesin-related protein which is mapped to the neuroblastoma suppressor gene locus at 1p36.2. Int J Oncol 16: 907–916. [DOI] [PubMed] [Google Scholar]
- Nakagawara A, Arima-Nakagawara M, Scavarda NJ, Azar CG, Cantor AB, Brodeur GM. 1993. Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med 328: 847–854. [DOI] [PubMed] [Google Scholar]
- Nikoletopoulou V, Lickert H, Frade JM, Rencurel C, Giallonardo P, Zhang L, Bibel M, Barde YA. 2010. Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature 467: 59–63. [DOI] [PubMed] [Google Scholar]
- Niwa S, Tanaka Y, Hirokawa N. 2008. KIF1Bβ- and KIF1A-mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/MADD. Nat Cell Biol 10: 1269–1279. [DOI] [PubMed] [Google Scholar]
- Palmada M. 2002. c-jun is essential for sympathetic neuronal death induced by NGF withdrawal but not by p75 activation. J Cell Biol 158: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda H-M, Sternheim N, Lyons DA, Diamond B, Hawkins TA, Woods IG, Bhatt DH, Franzini-Armstrong C, Dominguez C, Arana N, et al. 2006. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev Biol 298: 118–131. [DOI] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLos Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu YF, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthier A, Baets J, Timmerman V, Janssens K. 2012. Mechanisms of disease in hereditary sensory and autonomic neuropathies. Nat Rev Neurol 8: 73–85. [DOI] [PubMed] [Google Scholar]
- Scherer SS. 2006. Finding the causes of inherited neuropathies. Arch Neurol-Chicago 63: 812–816. [DOI] [PubMed] [Google Scholar]
- Schlisio S, Kenchappa RS, Vredeveld LCW, George RE, Stewart R, Greulich H, Shahriari K, Nguyen NV, Pigny P, Dahia PL, et al. 2008. The kinesin KIF1β acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev 22: 884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A, Castella P, Rockwood J, Wagner J, Caudy M. 1997. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix–loop–helix repressor of neuronal differentiation. Genes Dev 11: 3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Fang H, Hong H, Shi L, Zhang W, Zhang W, Zhang Y, Dong Z, Lancashire LJ, Bessarabova M, et al. 2014. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol 15: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Niwa S, Dong M, Farkhondeh A, Wang L, Zhou R, Hirokawa N. 2016. The molecular motor KIF1A transports the TrkA neurotrophin receptor and is essential for sensory neuron survival and function. Neuron 90: 1215–1229. [DOI] [PubMed] [Google Scholar]
- The I, Murthy AE, Hannigan GE, Jacoby LB, Menon AG, Gusella JF, Bernards A. 1993. Neurofibromatosis type 1 gene mutations in neuroblastoma. Nat Genet 3: 62–66. [DOI] [PubMed] [Google Scholar]
- Tsarovina K, Pattyn A, Stubbusch J, Müller F, van der Wees J, Schneider C, Brunet J-F, Rohrer H. 2004. Essential role of Gata transcription factors in sympathetic neuron development. Development 131: 4775–4786. [DOI] [PubMed] [Google Scholar]
- Tuszynski MH, Yang JH, Barba D, U HS, Bakay RA, Pay MM, Masliah E, Conner JM, Kobalka P, Roy S, et al. 2015. Nerve growth factor gene therapy: activation of neuronal responses in Alzheimer disease. JAMA Neurol 72: 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery RG, von Zastrow M. 1999. Distinct dynamin-dependent and -independent mechanisms target structurally homologous dopamine receptors to different endocytic membranes. J Cell Biol 144: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh IT, Lenci RE, Qin Y, Buddavarapu K, Ligon AH, Leteurtre E, Cao CD, Cardot-Bauters C, Pigny P, Dahia PLM. 2008. A germline mutation of the KIF1Bβ gene on 1p36 in a family with neural and nonneural tumors. Hum Genet 124: 279–285. [DOI] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, et al. 2001. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bβ. Cell 105: 587–597. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. 2002. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science 296: 920–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.