This Outlook discusses the study published in this issue by Ke et al., which clarifies the role of N6-methyladenosine (m6A) in mRNA biogenesis and function and shows that m6A methylation levels negatively correlate with transcript half-life but are not required for most pre-mRNA splicing events.
Keywords: m6A-CLIP, pre-mRNA, cell fractionation, mRNA turnover
Abstract
Post-transcriptional modification of RNA nucleosides has been implicated as a pivotal regulator of mRNA biology. In this issue of Genes & Development, Ke and colleagues (pp. 990–1006) provide insights into the temporal and spatial distribution of N6-methyladenosine (m6A) in RNA transcripts by analyzing different subcellular fractions. Using a recently developed biochemical approach for detecting m6A, the researchers show that m6A methylations are enriched in exons and are added to transcripts prior to splicing. Although m6A addition is widely thought to be readily reversible, they demonstrate in HeLa cells that once RNA is released from chromatin, the modifications are surprisingly static. This study integrates data from previous publications to clarify conflicting conclusions regarding the role of m6A in mRNA biogenesis and function. Ke and colleagues found that m6A methylation levels negatively correlate with transcript half-life but are not required for most pre-mRNA splicing events.
Although N6-methyladenosine (m6A) modification of RNA was discovered in the 1970s, the prevalence of this modification in mammalian mRNAs has become evident only recently, with the advent of transcriptome-wide m6A detection. Advances in the field have shown that m6A methylations are introduced into mRNA by METTL3 in complex with METTL14 and WTAP (Liu et al. 2014). Experiments manipulating the levels of these proteins and other proteins that interact with m6A have suggested that this modification negatively affects mRNA stability and alters splicing patterns (Dominissini et al. 2012; Wang et al. 2014). These findings motivated researchers to investigate further the importance of this modification to mRNA biology. However, the mechanisms modulating m6A modification and its precise role in mRNA biogenesis and regulation have remained debatable. In this issue of Genes & Development, Ke et al. (2017) clarify many of these questions by analyzing the presence of m6A in RNAs from different HeLa subcellular fractions and investigating how the distribution and extent of m6A modification relate to splicing and turnover of transcripts.
The precise mapping of m6A in cellular transcripts was facilitated by the development of m6A cross-linking immunoprecipitation (m6A-CLIP) (Ke et al. 2015). Single-nucleotide resolution of the positions of m6A modification is achieved by UV cross-linking immunoprecipitated RNA bound to anti-m6A antibody prior to high-throughput sequencing. The reduced background of m6A-CLIP enables researchers to identify the location of m6A modifications at unprecedented resolution. By analyzing the distribution of m6A in total RNA using this technique, Ke et al. (2015) confirmed that most m6A peaks are located in the last exon of a transcript such that nearly half of all m6As in mRNAs occur in 3′ untranslated regions (UTRs) (Meyer et al. 2012). Ke et al. (2017) furthered their investigation of RNA methylation by comparing the distribution of m6A in chromatin-associated, nucleoplasmic, and cytoplasmic RNA fractions obtained from HeLa cells. They found that even for incompletely spliced chromatin-associated pre-mRNA, the majority of m6A modifications is in exonic sequences. Additionally, the number and location of m6As do not change between nucleoplasmic and cytoplasmic RNAs (Fig. 1). Prior studies suggested that this modification is dynamic based on the presence of m6A demethylases in the nucleus (Jia et al. 2011); however, the findings of Ke et al. (2017) show that changes in the methylation state are minimal and occur at the pre-mRNA level prior to release from chromatin.
Figure 1.
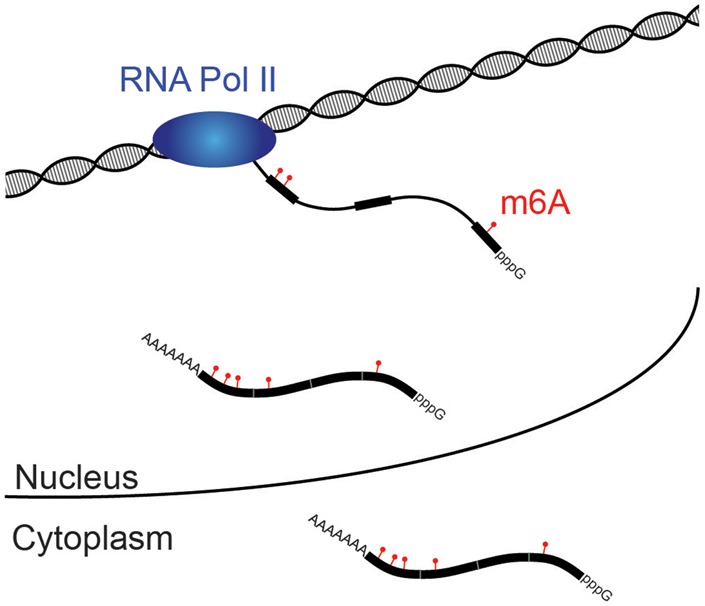
m6A modifications are added to exonic sequences before splicing and shorten mRNA half-life. RNA polymerase II (blue) synthesizes the pre-mRNA transcript, and, prior to the completion of splicing, a methylation complex containing METTL3 selectively adds m6A modifications to exonic sequences (thicker black bars). After release of spliced mRNA first into the nucleoplasm and ultimately into the cytoplasm, no further additions or subtractions of m6A are detected. The presence of m6A in an mRNA accelerates turnover.
After demonstrating that m6A methylation occurs before splicing is complete, the researchers assessed the number of m6A modifications in relation to splice sites. Although chromatin-associated RNA contains slightly more m6A modifications near 5′ and 3′ splice sites than the same regions in nucleoplasmic or cytoplasmic transcripts, a reduction in the density of methylation marks was observed within 50 nucleotides of splice sites in all fractions. Furthermore, no enrichment of this modification was found at either end of exons that can be alternatively spliced. Because this observation contradicts findings by other groups, Ke et al. (2017) reanalyzed previously published data using their own bioinformatic analysis pipeline. This reaffirmed their conclusion that m6A is not enriched at the ends of alternatively spliced exons. Finally, the researchers knocked out METTL3 in mouse embryonic stem cells and observed no significant changes in splicing patterns compared with those of wild-type cells. Once again, they were able to corroborate this observation by reanalyzing previously published data.
Several laboratories have concluded that the presence of m6A results in faster mRNA turnover (Sommer et al. 1978; Wang et al. 2014; Geula et al. 2015). Ke et al. (2017) confirmed this observation by studying the half-life of m6A-containing mRNAs after treating cells with actinomycin D. In general, mRNA transcripts containing multiple m6As have shorter half-lives than those containing a single m6A methylation. mRNAs that have m6As in both the coding sequence and the 3′ UTR have the shortest half-lives. Knocking out METTL3 resulted in longer half-lives of m6A-containing mRNAs, confirming that the differences in RNA life spans can be attributed to the presence of this RNA modification.
Taken together, the findings presented by Ke et al. (2017) provide a better understanding of the distribution of m6A at different stages of mRNA processing and demonstrate that the presence of m6A modification influences the half-life of an mRNA. However, these results raise several interesting new questions and highlight the importance of standardizing bioinformatic analyses to ensure that researchers can achieve reproducible results across the RNA modification field. The observation that the m6A distribution is constant in the nucleoplasmic and cytoplasmic fractions highlights the need to revisit the idea that m6A modification is dynamically regulated. The data presented do not preclude the possibility that m6A modification may be dynamic in certain stress conditions or developmental processes. The findings also call for the elucidation of the spatiotemporal relationship between the activities of RNA polymerase II, m6A methylases and demethylases, and the spliceosome with respect to the nascent RNA. The conclusion that m6A is added selectively to exons before the mRNA is spliced raises the questions of how exons are defined and whether m6A plays a role in establishing exon identity. This comprehensive analysis by Ke et al. (2017) will hopefully begin to unify the field so that these newly arising questions can be addressed.
Acknowledgments
N.A.R.-M. is supported by a Yale University Fellowship. J.B.W. is a Hope Funds for Cancer Research Fellow. J.A.S. is a Howard Hughes Medical Institute Investigator.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.302695.117.
References
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. 2015. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347: 1002–1006. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. 2015. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29: 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev (this issue) 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KCAD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Lavi U, Darnell JE. 1978. The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J Mol Biol 124: 487–499. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]


