SUMMARY
Indoleamine 2,3-dioxygenase 1 (IDO1) plays a key role in the immune system by regulating tryptophan levels and T cell differentiation. Several tumor types overexpress IDO1 to avoid immune surveillance making IDO1 of interest as a target for therapeutic intervention. As a result, several IDO1 inhibitors are currently being tested in clinical trials for cancer treatment as well as several other diseases. Many of the IDO1 inhibitors in clinical trials naturally bear structural similarities to the IDO1 substrate tryptophan, as such, they fulfill many of the structural and functional criteria as potential AHR ligands. Using mouse and human cell-based luciferase gene reporter assays, qPCR confirmation experiments, and CYP1A1 enzyme activity assays, we report that some of the promising clinical IDO1 inhibitors also act as agonists for the aryl hydrocarbon receptor (AHR), best known for its roles in xenobiotic metabolism and as another key regulator of the immune response. The dual role as IDO antagonist and AHR agonist for many of these IDO target drugs should be considered for full interrogation of their biological mechanisms and clinical outcomes.
Keywords: IDO1, Aryl Hydrocarbon Receptor, IDO Inhibitors, AHR Agonists, Cancer, Clinical Trials
INTRODUCTION
The essential amino acid L-Tryptophan (Trp) is metabolized in a tissue-specific manner by the rate-limiting enzymes tryptophan 2,3-dioxygenase 2 (TDO2) and indoleamine 2,3-dioxygenase (IDO1 and IDO2) to produce L-Kynurenine (Kyn) (Austin and Rendina, 2015). IDO1 has garnered the most attention due to its key roles in inflammation, as Trp metabolites, particularly Kyn, act as regulators of immune cell differentiation and proliferation. As an example, IDO1 expression directs T cell polarization and decreases T cell proliferation (Taylor and Feng, 1991; Hwu et al., 2000; Jaronen and Quintana, 2014).
The Aryl Hydrocarbon Receptor (AHR) is a basic helix-loop-helix transcription factor that binds and responds to xenobiotics (Denison et al., 2002). The AHR is notable for its capacity to bind hundreds of identified ligands, the majority of which are exogenous environmental toxicants. Following ligand binding, the AHR translocates to the nucleus where it regulates the transcription of thousands of target genes in a ligand-specific manner (Karyala et al., 2004), the best known of which are a gene battery encoding proteins involved in xenobiotic metabolism.
T-cell polarization also involves the AHR (Mezrich et al., 2010; Quintana and Sherr, 2013). Exposure to some AHR agonists, including the potent environmental toxicant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), induces polarization of naïve T cells to an anti-inflammatory Treg status contributing to an immunosuppressive condition (Faith and Luster, 1979; Mohinta et al., 2015). The AHR responds to other ligands by promoting polarization of naïve T-cells to the pro-inflammatory Th17 cell phenotype (Veldhoen et al., 2008) demonstrating that the roles of the AHR in the immune response are complex and ligand-specific.
IDO1 gene expression is regulated by two pathways: an interferon gamma (IFNγ)-independent and an IFNγ-dependent pathway (Brandacher et al., 2007; Sarkar et al., 2007; Chen, 2011; Bessede et al., 2014). The AHR carries out its role in the IFNγ-independent pathway, in which the AHR binds the IDO1 promoter to directly regulate IDO1 transcription (Vogel et al., 2008; Mezrich et al., 2010), and second, by induction of the AHR-regulated miR-132/212 cluster, which also plays a role in T-cell polarization (Nguyen et al., 2010; Nguyen et al., 2013). Additionally, the major IDO1 metabolite Kyn is itself an AHR ligand (Mezrich et al., 2010) producing a likely positive regulatory feedback loop between the AHR and IDO1 (Julliard et al., 2014).
IDO1 is frequently overexpressed in cancer cells causing a depletion of Trp in the microenvironment and leading to the suppression of immune-surveillance (Friberg et al., 2002; Brandacher et al., 2006; Liu et al., 2009; Ferdinande et al., 2012). Thus, IDO1 is a popular target for chemotherapeutic intervention, with a number of IDO1 inhibitors currently being tested in cancer clinical trials (Vacchelli et al., 2014; Austin and Rendina, 2015; Rohrig et al., 2015). For example, INCB024360 (INCB) is involved in at least eight clinical trials, either as a single agent or in combination with other drugs for treatment of melanomas, reproductive tract cancers, and other solid tumors; while NLG919 (NLG) is in a phase I clinical trial for treatment of solid tumors. It should be noted that the inhibition of IDO1 is also the goal in clinical trials for Huntington’s disease (Mazarei and Leavitt, 2015), neurological disorders (Fujigaki et al.), and autoimmunity and other diseases (Yeung et al., 2015).
Most IDO1 inhibitors are structurally similar to Trp by possessing a planar, polycyclic molecular configuration that is favorable for binding to the promiscuous AHR (Murray et al., 2014). In this report, we show that some IDO1 inhibitors currently in testing in clinical trials can also act as AHR agonists. The prospect that a compound that inhibits IDO1 activity and reduces immune tolerance may also trigger a wide range of AHR-mediated effects, including altered immune cell differentiation and polarization and an upregulation of IDO1 expression, poses a significant clinical challenge. The question arises whether the observed clinical effects of an IDO-inhibiting drug are due to the inhibition of IDO activity and/or to the activation of AHR signaling.
MATERIALS AND METHODS
Materials
INCB024360 (INCB), NLG919 (NLG) (MCE, Monmouth Junction, NJ), Norharmane (Norh) (Santa Cruz Biotechnology, Dallas, TX), Kynurenine (Kyn), Kynurenic acid (KA), Quinolinic acid (QA), 2,3,7,8-Tetrachlorodibenzodioxin (TCDD), 1-Methyl-L-Tryptophan (1MLT), and 1-Methyl-D-Tryptophan (1MDT) (Sigma Aldrich, St. Louis MO) were dissolved in dimethylsulfoxide (DMSO) and used at the indicated concentrations. For the HepG2 (40/6) treatments, 1MLT and 1MDT were dissolved in 50% DMSO. The P450-Glo™ Assay kit specific for CYP1A1 (catalog# V8752) was purchased from Promega (Madison, WI).
Cell Culture
Murine Hepa-1c1c7 (Hankinson et al., 1991) and H1L7.5c3 hepatocyte cell lines, in which the latter is derived from Hepa-1c1c7 cells and has a stably transfected luciferase reporter gene regulated by a promoter with multiple AHR response elements (He et al., 2011) (courtesy of Dr. Michael Denison, University of California, Davis, CA), were cultured in alpha Minimum Essential Medium (Corning, Manassas, VA), supplemented with 10% FBS (Hyclone Laboratory, Logan, UT), 2mM L-glutamine, 0.2% penicillin/streptomycin, and 2.2g/l sodium bicarbonate (Sigma Aldrich). The HepG2 (40/6) human hepatoma stable cell line (Long et al., 1998) containing the stably integrated pGudluc 6.1 luciferase reporter construct under the control of the CYP1A1 enhancer were cultured in α-modified essential media (Sigma-Aldrich) supplemented with 8% fetal bovine serum (Hyclone Laboratories), 100 IU/ml penicillin/100μg/ml streptomycin (Sigma-Aldrich). The Hepa-1c1c7, H1L7.5c3, and HepG2 (40/6) cells were maintained at 37°C and 5% CO2. H1L7.5c3 cells were seeded in white-walled, white-bottomed 96-well plates (Corning, Manassas, VA) at 4000 cells/well and incubated for 24hr in culture medium. After the 24-hr incubation, the medium was removed, and the cells were washed once with Dulbecco’s Phosphate Buffered Saline (DPBS) (Corning). The Hepa-1c1c7 and H1L7.5c3 cells were treated for an additional 24hr with the reagents at the indicated concentrations. HepG2 (40/6) cells were seeded in 12-well plates and cultured to ~80% confluence before treatment for an additional 4 hr with the reagents at the indicated concentrations. DMSO did not exceed 0.1% concentration in the culture medium.
Luciferase Assays
Luciferase assays were carried out using the H1L7.5c3 and HepG2 (40/6) cells. At the conclusion of the indicated exposures, H1L7.5c3 cells were removed from incubation and allowed to equilibrate to room temperature for 15 min. After equilibration, the medium was removed and the cells were washed twice with at room temperature with DPBS. The cells were lysed with 20μl/well 1X Passive Lysis Buffer (Promega, Madison, WI) and shaken for 20 min at room temperature. Luciferase activity was recorded using an LMax Luminometer Microplate Reader (Molecular Devices, Sunnyvale, CA) programmed to inject 50μl of Luciferase Assay Reagent (Promega, Madison, WI) per well with a 10 sec integration of emitted luminescence. For the HepG2 (40/6) luciferase assays (Murray et al., 2010), cells were removed from incubation and lysed in 400μl of 25mM Tris-phosphate, pH 7.8, 2mM dithiothreitol, 2mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% (v/v) glycerol, and 1% (v/v) TritonX-100]. Lysate (20μl) was combined with 80μl of Luciferase Reporter Substrate (Promega, Madison, WI), and luciferase activity was measured with a TD-20e Luminometer (Turner Designs, Sunnyvale, CA).
Quantitative Real-Time Polymerase Chain Reaction Assays
To verify the microarray results, qPCR analysis was carried out using primers designed for mouse AHR target Cyp1a1 mRNA (Mm00487218_m1) and mouse reference Gapdh mRNA (Mm99999915_g1) purchased from ThermoFisher Scientific, Inc. (Waltham, MA). Approximately 5μg of total RNA from each H1L7.5c3 cell culture (three biological replicates per treatment) served as template for the cDNA synthesis. The cDNA was synthesized using TaqMan® assay kits with the Superscript III First-Strand Synthesis System (ThermoFisher Scientific, Inc.). The qPCR reactions were performed using the Fast Advanced Master Mix (ThermoFisher Scientific, Inc.) on a BioRad CFX96 System using version 3.1 software (BioRad, Hercules, CA) set at 40 cycles. Assays to determine levels of DNA contamination were carried out by omitting reverse transcriptase and mRNA template from the reactions.
For the HepG2 (40/6) cells, primers (Integrated DNA Technologies, Coralville, IA) for qPCR analysis (Murray et al., 2010) were selected to detect human CYP1A1 mRNA and ribosomal protein L13a mRNA as a reference (see Table 1 in Murray et al., 2010). PCR was performed on a MyiQ (Bio-Rad Laboratories, Hercules, CA) system using PerfeCTa SYBR Green reagent (Quanta Biosciences, Gaithersburg, MD). In all cases, melting point analysis revealed amplification of a single product. Data acquisition and analysis were carried out using MyIQ software (Bio-Rad Laboratories).
CYP1A1 Enzyme Activity Assays
Hepa-1c1c7 cells were plated in 24-well plates and incubated overnight as described above for the H1L7.5c3 hepatocytes to a growth confluency of 75–90%. All treatments were carried out in quadruplicate. The cells were transferred to a new 24-well plate and treated with DMSO vehicle, 10nM TCDD, and 10μM of the different IDO1 inhibitors for 12 hr. The cells in the vehicle and TCDD-treated wells were treated again with DMSO and the test cells treated again with 10μM of the different IDO1 inhibitors for an additional 12 hr. After the 24-hr treatments, the cells were washed and then incubated at 37°C for 3 hr in culture media with CYP1A1 substrate from the Luciferin-CEE P450-Glo assay kit. A 25μl-sample of supernatant was transferred from each well into wells of a white-walled 96-well plate (Corning, USA). After a 20-min incubation at room temperature, the plate was read on a Promega GloMax Multidetection System (integration = 0.5s).
Data Analysis
Differences were considered statistically significant with a p-value ≤0.05. Statistical significance was calculated using two-sided paired Student’s t-test, and dependent upon the data at hand, equal or unequal variance analysis was applied. Error bars represent standard error of the mean (SEM).
RESULTS
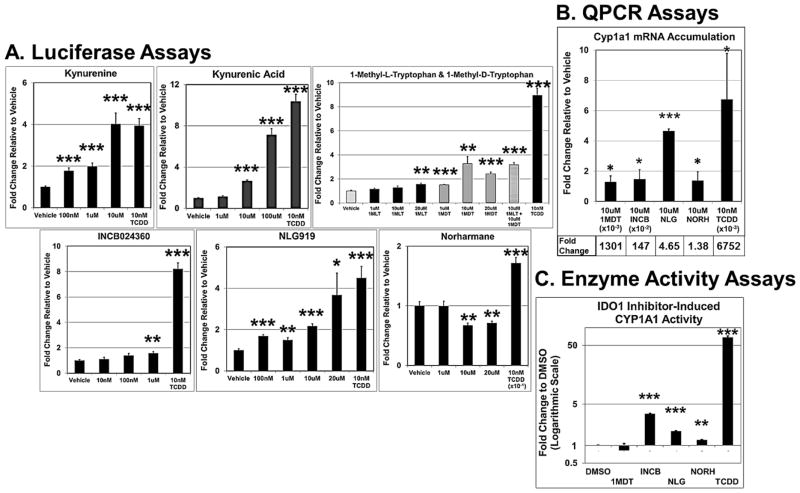
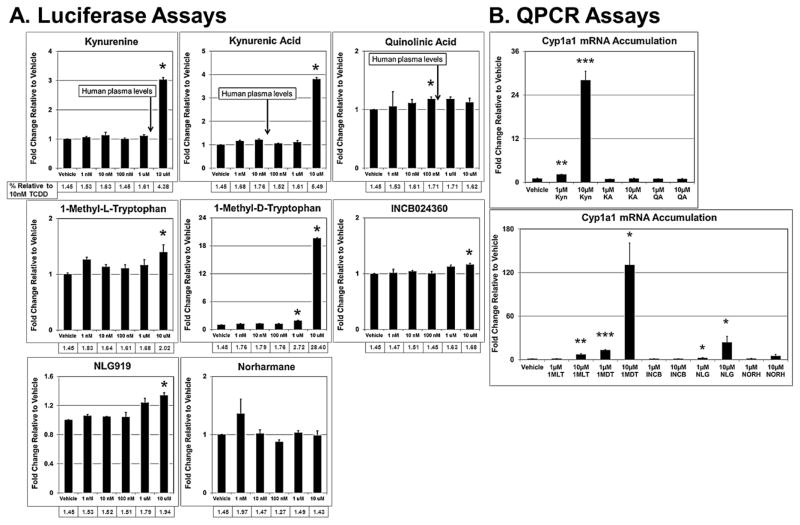
Mouse Hepa-1c1c7 cells and human HepG2 cells are commonly used in liver studies and were derived from a mouse and human hepatomas, respectively (Bernhard et al., 1973; Hankinson, 1979; Hankinson et al., 1991; Ihrke et al., 1993). The mouse H1L7.5c3 cell line variant was engineered by stable transfection of the pGudLuc7.5 plasmid into Hepa-1c1c7 cells (He et al., 2011) as was the HepG2 (40/6) cell line using pGudluc 6.1 and parental HepG2 cells (Long et al., 1998), in which both cell lines express an AHR-dependent, luciferase reporter gene system. We exposed replicate H1L7.5c3 (Fig. 1) and HepG2 (40/6) (Fig. 2) cell cultures to several IDO1 inhibitors (Table 1) and Trp metabolites to assess relative AHR transcriptional activity. The Trp metabolites Kyn, KA, and QA were included in our studies because components of the Kynurenine pathway are often implicated in gliomas (Adams et al., 2014) and presumably compete with the IDO1 inhibitors for binding to the AHR. Kyn and the tested IDO1 inhibitors operate at comparable plasma concentration levels (Table 2), and therefore, the concentrations used in our studies are of likely biological relevance. In agreement with previous studies (DiNatale et al., 2010; Mezrich et al., 2010; Bessede et al., 2014; Moyer et al., 2016), Kyn and KA induced AHR-directed luciferase activity in the mouse (Fig. 1A) and human (Fig. 2A) cells. Kyn elicited a greater effect in mouse cells to that of the human cells, causing a significant change at 100nM, 1μM, and 10μM, whereas in the human cells, a significant effect was not observed at the lower concentrations. Kyn also appeared to be a stronger AHR agonist than KA in the H1L7.5c3 cells but of near equal potency in the HepG2 (40/6) cells. At the tested concentrations, QA had no inductive effect on the AHR in the HepG2 (40/6) cells.
Fig. 1.
IDO inhibitors induce AHR signaling in mouse hepatocytes. Cells were incubated with the given test compound for 24hr at the indicated concentrations and assayed for luciferase activity in H1L7.5c3 cells (A); and in Hepa-1c1c7 cells, assayed for Cyp1a1 mRNA accumulation by QPCR (B) and CYP1A1 enzymatic activity (C). All values are the mean of four to six biological replicates. Error bars represent standard error of the mean. p-values relative to Vehicle (DMSO)-treated cells: *, ≤0.05; **, ≤0.01; ***, ≤0.001.
Fig. 2.
IDO inhibitors induce AHR signaling in human hepatocytes. HepG2 (40/6) cells were incubated with the given test compound for 24hr at the indicated concentrations and assayed for luciferase activity (A) and CYP1A1 mRNA accumulation by QPCR (B). All values are the mean of three to six biological replicates. Error bars represent standard error of the mean. p-values relative to Vehicle (DMSO)-treated cells: *, ≤0.05; **, ≤0.01; ***, ≤0.001.
Table 1.
IDO Inhibitors In Cancer Clinical Trials Induce AHR Transcriptional Activity
(Eguchi et al., 1984; Hou et al., 2007; Mautino et al., 2013; Adams et al., 2014)
| Compound | Structure | Mouse Hepa-1c1c7 Cells (Fold Change to DMSO Control) |
Human Hep-G2 Cells (Fold Change to DMSO Control) |
IDO1 Inhibition (ki) |
In Clinical Trials |
References | |||
|---|---|---|---|---|---|---|---|---|---|
| AHR-directed Luciferase Activity (10μM of compound) |
qPCR-based Endogenous Cyp1a1 mRNA Levels (10μM of compound) |
Endogenous CYP1A1 Activity (10μM of compound) |
AHR-directed Luciferase Activity (10μM of compound) |
qPCR-based Endogenous Cyp1a1 mRNA Levels (10μM of compound) |
|||||
| Tryptophan (TRP) |
|
Not Done | Not Done | Not Done | Not Done | Not Done | Not Applicable | No | Not Applicable |
| Kynurenine (Kyn) |
|
4.02*** | Not Done | Not Done | 3.03* | 28.05*** | Not Applicable | No | Bessede et al., 2014; Mezrich et al., 2010; Moyer et al., 2016 |
| Kynurenic Acid (KA) |
|
2.66*** | Not Done | Not Done | 3.80* | 1.03 | Not Applicable | No | DiNatale et al., 2010; Moyer et al., 2016 |
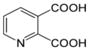
| Quinolinic Acid (QA) |
|
Not Done | Not Done | Not Done | 1.12 | 0.95 | Not Applicable | No | Adams et al., 2014 |
| 1-Methyl-L-Tryptophan (1MLT) |

|
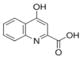
1.26** | Not Done | Not Done | 1.40* | 6.89** | 19–53μM | Yes | Hou et al., 2007; Rohrig et al., 2015 |
| 1-Methyl-D-Tryptophan (1MDT) |
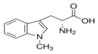
|
3.27** | 1301* | 0.82 | 19.60* | 130* | >100μM | Yes | Austin and Rendina, 2015; Hou et al., 2007; Vacchelli et al., 2014 |
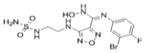
| INCB024360 (INCB) |
|
1.57* (1μM)1 | 147* | 3.43*** | 1.16* | 1.07 | Not Applicable | Yes | Austin and Rendina, 2015; Rohrig et al., 2015; Vacchelli et al., 2014 |
| NLG919 (NLG) |
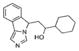
|
2.17*** | 4.65*** | 1.74*** | 1.34* | 23.59* | 7nM | Yes | Mautino et al., 2013; Rohrig et al., 2015; Vacchelli et al., 2014 |
| Norharmane (NORH) |
|
0.67 | 1.38* | 1.23** | 0.99 | 5.04 | 120μM | Yes | Austin and Rendina, 2015; Eguchi et al., 1984 |
| 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) |
|
~2–10*** (10nM)2 | 6752* (10nM)2 | 68.82*** (10nM)2 | ~82–100*** (10nM)2 | 100** (10nM) | Not Applicable | No | Not Applicable |
p- value ≤0.05;
p -value ≤0.01;
p -value ≤0.001
1 μM of compound was tested
10nM used as positive control
Table 2.
Reported Plasma Concentrations of the Tested Tryptophan Metabolites and IDO1 Inhibitors
(Aarsland et al., 2015), (Schwieler et al., 2016), (Wirthgen et al., 2016), (Soliman et al., 2016), (Shi et al., 2016), (Nayak et al., 2015)
| Tryptophan Metabolite | Steady State Plasma Levels (μM) | References |
|---|---|---|
| Kynurenine | 1.57–3.1 | Aarsland et al., 2015; Schwieler et al., 2016 |
| Kynurenic Acid | 0.046 | |
| Quinolinic Acid | 0.306 | |
| IDO1 Inhibitor | [Cmax] Plasma Levels (μM) | References |
| 1-Methyl-L-Tryptophan | ~30 (porcine) | Wirthgen et al., 2016 |
| 1-Methyl-D-Tryptophan | ~16 | Soliman et al., 2016 |
| INCB024360 | ~2 | Shi et al., 2016 |
| NLG919 | ~40 | Nayak et al., 2015 |
| Norharmane | ~ | no known reference |
Cmax, Concentration Maximum
In regard to the IDO1 inhibitors, the two racemic forms of 1-Methyltryptophan differentially inhibit the three Trp-metabolizing enzymes. 1MLT preferentially inhibits IDO1 (Opitz et al., 2011; Austin and Rendina, 2015) and 1MDT appears to inhibit TDO2, IDO1, and IDO2 to varying degrees (Metz et al., 2007; Löb et al., 2009; Austin and Rendina, 2015). 1MLT is a test compound in several clinical trials (Rohrig et al., 2015) (Table 1), and like the other tested IDO1 inhibitors, plasma concentrations in the course of treatment in clinical trials reaches the micromolar range (Table 2). Thus, focusing on the biologically relevant concentration of 10μM used in the luciferase assays, in H1L7.5c3 cells (Fig. 1A), 1MLT exhibited a non-significant 1.14-fold increase in AHR-driven luciferase activity to that of the media+DMSO vehicle control; whereas 10μM 1DMT, under the trade name Indoximod in development by NewLink Genetics Corporation and in several clinical trials (Vacchelli et al., 2014; Austin and Rendina, 2015) (Table 1), showed a significant 3.27-fold increase relative to control levels. In human HepG2 (40/6) cells (Fig. 2A), 1MLT showed a slight but significant increase in AHR induction (2.02-fold) while 1MDT caused the largest inductive effect of all the IDO1 inhibitors (28.4-fold).
The orally administered INCB developed by Incyte Corporation and in clinical trials for melanoma and other cancers (Vacchelli et al., 2014; Austin and Rendina, 2015; Rohrig et al., 2015) showed a 1.57-fold increase in AHR-directed luciferase activity in H1L7.5c3 cells at 1μM (Fig. 1A) and 1.16 fold in HepG2 (40/6) cells at 10μM (Fig. 1B). Similarly, 10μM NLG, a drug in development by NewLink Genetics and in clinical trials for solid tumors (Vacchelli et al., 2014; Rohrig et al., 2015), showed a 2.17-fold increase in luciferase activity in H1L7.5c3 cells (Fig. 1A) and 1.34 fold in HepG2 (40/6) cells (Fig. 1B). The less specific and indirect IDO1 inhibitor NORH (10μM) showed no significant increase in AHR-driven luciferase activity in either the mouse or human cells. CAY10581, another commercially available IDO1 inhibitor, was assayed and was found to be highly cytotoxic in H1L7.5c3 cells (data not shown). TCDD (10nM) was included as a positive control.
A series of qPCR assays were carried out in order to confirm the AHR-directed luciferase results (Figs. 1B and 2B). The qPCR results in H1L7.5C3 cells showed that 10μM 1MDT (1300 fold), 10μM NLG (4.65 fold), and 10μM INCB (147 fold) caused an accumulation of Cyp1a1 mRNA to significant levels relative to that of vehicle control cells (DMSO treated) (Fig. 1B). Although 10μM NORH (1.38 fold) also caused a significant increase in Cyp1a1 mRNA accumulation, the increase was relatively minimal and arguably biologically insignificant. TCDD (10nM) was again used as the positive control and produced a 6,751-fold increase. The qPCR data from the HepG2 (40/6) cells (Fig. 2B) were normalized to vehicle control and validated the luciferase results for 10μM Kyn (28.05 fold) and 10μM QA (0.95 fold) but not for 10μM KA (1.03 fold). The qPCR results reflected the luciferase data of the IDO1 inhibitors for 10μM 1MLT (6.89 fold), 10μM 1MDT (130 fold), and 10μM NLG (23.59 fold), but the data were not statistically significant for 10μM INCB (1.07 fold). Consistent with the luciferase results 10μM NORH (5.04 fold) was not statistically significant. Table 1 summarizes the luciferase and QPCR results.
A third set of experiments was carried out with Hepa-1c1c7 cells to demonstrate that the induction of AHR signaling by the IDO1 inhibitors led to physiological consequences. That is, as further confirmation that the IDO1 inhibitors acted as AHR agonists to ultimately lead to AHR-directed cellular protein activity, CYP1A1 enzyme activity was assayed. In the assay (P450-Glo), a CYP1A1-specifc substrate, an analog of luciferin, is converted to luciferin by CYP1A1 in the Hepa-1c1c7 cell lysate. The luciferin, in turn, reacts with luciferase to produce light, which is directly proportional to CYP1A1 activity. We found that the cells treated with INCB (3.43-fold), NLG (1.74 fold), and NORH (1.23 fold) caused a significant fold increase of CYP1B1 enzyme activity over the DMSO-treated control cells (Table 1 and Fig. 1C). To our surprise, 1MDT, which was the top inducer of AHR activity in the luciferase and qPCR assays, did not cause a significant change in CYP1A1 activity, and suggests that 1MDT treatment may also cause a rapid turnover of Cyp1a1 mRNA in Hepa-1c1c7 cells.
DISCUSSION
Our studies show that some IDO1 inhibitors, including at least two being tested as immunomodulating compounds in ongoing clinical trials, can act as AHR agonists. Because the AHR plays a key role in immune cell differentiation, the dual roles of the IDO1 inhibitors may be a relevant factor in understanding clinical trial outcomes and assessed side effects. That these compounds act as AHR agonists have not, to our knowledge, been previously reported or considered. Many but not all AHR agonists cause an immunosuppressive effect, frequently resulting in increased Treg cell production (Quintana and Sherr, 2013) and a counterproductive reaction for chemotherapeutics focused on driving immune-mediated tumor clearance. Our findings may also help explain some confusing and contradictory observations. For example, it was reported that IDO1-positive human cancer cells incubated with 1MDT increased rather than decreased Kyn production (Opitz et al., 2011). The 1MDT did not alter IDO1 enzymatic activity but instead caused an increase in IDO1 mRNA and protein levels. Because IDO1 gene expression is regulated in an AHR-dependent manner (Vogel et al., 2008; Mezrich et al., 2010), it may be that 1MDT, which we showed here to be an AHR agonist, stimulated AHR signaling to induce transcription of the IDO1 gene.
The results reported here demonstrate that potential AHR activation is worth considering as a factor in assessing IDO1 inhibitors as part of a suitable therapeutic approach. A number of techniques are available to assess AHR agonist activity, including conventional techniques for determining mRNA or protein expression levels of major AHR-regulated genes, such as CYP1A1 and CYP1B1 (Chang et al., 2003), as well as the high-throughput luciferase reporter assay used in this study.
It is our recommendation that an IDO1 inhibitor deemed a feasible therapeutic, particularly those with structural similarity to AHR ligands, i.e., planar and/or polycyclic (Denison and Nagy, 2003), be considered a potential AHR agonist and evaluated accordingly. We would recommend that the testing of a putative IDO1 inhibitor as a cancer therapeutic include AHR induction as part of the regimen in assessing suitability (Rohrig et al., 2015). The possibility that an IDO1 inhibitor may also activate the AHR to cause contradictory outcomes on immune cell differentiation and response, IDO1 status, and drug metabolism may not only call into question the drug’s efficacy but may also be an underlying mechanism for off-target effects.
In conclusion, based on the data compiled from the luciferase expression assays, induced CYP1A1 mRNA levels, and CYP1A1 activity levels from two different cell lines (Hepa-1c1c7 and HepG2 cells); we found that 1MLT, 1MDT, NLG, INCB, and even NORH induced AHR signaling in one or more assays (Table 1). The results for the IDO1 inhibitors were comparable or outperformed the tested endogenous AHR agonists Kyn, KA, and QA. In total, the results suggest that IDO1 inhibitors may act as AHR agonists and should be a consideration when using them in clinical trials.
Highlights.
Indoleamine-2,3-dioxygenase 1 (IDO1) inhibitors are in cancer clinical trials.
Some IDO1 inhibitors also potently activate AHR signaling.
The dual role of the IDO1 inhibitors may explain some past paradoxical findings.
AHR induction studies must be included in assessing clinical suitability.
Acknowledgments
We thank Dr. Michael Denison at UC Davis for the H1L7.5c3 cells. We thank the reviewers and editor for their thoughtful comments. We also thank Carrie Freitag and Drs. Mark Israel, Jason Moore, and Richard Rothstein for their support. This work was supported by funding from NIH/NCRR award 5P20RR024475-02, NIH/NIGMS award 8P20GM103534-02, Department of Medicine Research Award, and NIEHS award ES004869-27 (GHP).
ABBREVIATIONS
- 1MDT
1-Methy-D-Tryptophan
- 1MLT
1-Methyl-L-Tryptophan
- AHR
Aryl Hydrocarbon Receptor
- DMSO
Dimethylsulfoxide
- DPBS
Dulbecco’s Phosphate Buffered Saline
- IDO
Indoleamine 2,3-dioxygenase
- INCB
INCB024360
- Kyn
Kynurenine
- KA
Kynurenic Acid
- NLG
NLG919
- NORH
Norharmane
- QA
Quinolinic Acid
- TCDD
2,3,7,8 Tetrachlorodibenzo-p-Dioxin
- Trp
Tryptophan
Footnotes
AUTHOR CONTRIBUTIONS
BJM, IYT, HFH, GHP, and CRT conceived and designed the study. BJM, IYR, IAM, SL, and HFH performed the experiments and acquired the data. BJM, IYT, IAM, GHP, and CRT analyzed the data, and BJM, IYR, and CRT wrote and edited the manuscript. The authors approved the manuscript and declare they have no actual or potential competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarsland TI, Landaas ET, Hegvik TA, Ulvik A, Halmoy A, Ueland PM, Haavik J. Serum concentrations of kynurenines in adult patients with attention-deficit hyperactivity disorder (ADHD): a case-control study. Behav Brain Funct. 2015;11:36. doi: 10.1186/s12993-015-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S, Teo C, McDonald KL, Zinger A, Bustamante S, Lim CK, Sundaram G, Braidy N, Brew BJ, Guillemin GJ. Involvement of the Kynurenine Pathway in Human Glioma Pathophysiology. PloS one. 2014;9:e112945. doi: 10.1371/journal.pone.0112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CJ, Rendina LM. Targeting key dioxygenases in tryptophan-kynurenine metabolism for immunomodulation and cancer chemotherapy. Drug Discov Today. 2015;20:609–617. doi: 10.1016/j.drudis.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Bernhard HP, Darlington GJ, Ruddle FH. Expression of liver phenotypes in cultured mouse hepatoma cells: synthesis and secretion of serum albumin. Developmental biology. 1973;35:83–96. doi: 10.1016/0012-1606(73)90008-0. [DOI] [PubMed] [Google Scholar]
- Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, Iannitti R, Tissi L, Volpi C, Belladonna ML, Orabona C, Bianchi R, Lanz TV, Platten M, Della Fazia MA, Piobbico D, Zelante T, Funakoshi H, Nakamura T, Gilot D, Denison MS, Guillemin GJ, DuHadaway JB, Prendergast GC, Metz R, Geffard M, Boon L, Pirro M, Iorio A, Veyret B, Romani L, Grohmann U, Fallarino F, Puccetti P. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandacher G, Hoeller E, Fuchs D, Weiss HG. Chronic immune activation underlies morbid obesity: is IDO a key player? Curr Drug Metab. 2007;8:289–295. doi: 10.2174/138920007780362590. [DOI] [PubMed] [Google Scholar]
- Brandacher G, Perathoner A, Ladurner R, Schneeberger S, Obrist P, Winkler C, Werner ER, Werner-Felmayer G, Weiss HG, Gobel G, Margreiter R, Konigsrainer A, Fuchs D, Amberger A. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- Chang TKH, Chen J, Pillay V, Ho JY, Bandiera SM. Real-Time Polymerase Chain Reaction Analysis of CYP1B1 Gene Expression in Human Liver. Toxicological Sciences. 2003;71:11–19. doi: 10.1093/toxsci/71.1.11. [DOI] [PubMed] [Google Scholar]
- Chen W. IDO: more than an enzyme. Nat Immunol. 2011;12:809–811. doi: 10.1038/ni.2088. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Denison MS, Rogers JM, Rushing SR, Jones CL, Tetangco SC, Heath-Pagliuso S. Analysis of the aryl hydrocarbon receptor (AhR) signal transduction pathway. Curr Protoc Toxicol. 2002;Chapter 4(Unit4 8) doi: 10.1002/0471140856.tx0408s11. [DOI] [PubMed] [Google Scholar]
- DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS, Laurenzana EM, Omiecinski CJ, Perdew GH. Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol Sci. 2010;115:89–97. doi: 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi N, Watanabe Y, Kawanishi K, Hashimoto Y, Hayaishi O. Inhibition of indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase by beta-carboline and indole derivatives. Arch Biochem Biophys. 1984;232:602–609. doi: 10.1016/0003-9861(84)90579-4. [DOI] [PubMed] [Google Scholar]
- Faith RE, Luster MI. Investigations on the effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on parameters of various immune functions. Ann N Y Acad Sci. 1979;320:564–571. doi: 10.1111/j.1749-6632.1979.tb56634.x. [DOI] [PubMed] [Google Scholar]
- Ferdinande L, Decaestecker C, Verset L, Mathieu A, Moles Lopez X, Negulescu AM, Van Maerken T, Salmon I, Cuvelier CA, Demetter P. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br J Cancer. 2012;106:141–147. doi: 10.1038/bjc.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, Mellor AL, Munn DH, Antonia SJ. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int J Cancer. 2002;101:151–155. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- Fujigaki H, Yamamoto Y, Saito K. L-Tryptophan-kynurenine pathway enzymes are therapeutic target for neuropsychiatric diseases: Focus on cell type differences. Neuropharmacology. doi: 10.1016/j.neuropharm.2016.01.011. [DOI] [PubMed] [Google Scholar]
- Hankinson O. Single-step selection of clones of a mouse hepatoma line deficient in aryl hydrocarbon hydroxylase. Proc Natl Acad Sci U S A. 1979;76:373–376. doi: 10.1073/pnas.76.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O, Brooks BA, Weir-Brown KI, Hoffman EC, Johnson BS, Nanthur J, Reyes H, Watson AJ. Genetic and molecular analysis of the Ah receptor and of Cyp1a1 gene expression. Biochimie. 1991;73:61–66. doi: 10.1016/0300-9084(91)90075-c. [DOI] [PubMed] [Google Scholar]
- He G, Tsutsumi T, Zhao B, Baston DS, Zhao J, Heath-Pagliuso S, Denison MS. Third-Generation Ah Receptor–Responsive Luciferase Reporter Plasmids: Amplification of Dioxin-Responsive Elements Dramatically Increases CALUX Bioassay Sensitivity and Responsiveness. Toxicological Sciences. 2011;123:511–522. doi: 10.1093/toxsci/kfr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou DY, Muller AJ, Sharma MD, DuHadaway J, Banerjee T, Johnson M, Mellor AL, Prendergast GC, Munn DH. Inhibition of Indoleamine 2,3-Dioxygenase in Dendritic Cells by Stereoisomers of 1-Methyl-Tryptophan Correlates with Antitumor Responses. Cancer Research. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- Hwu P, Du MX, Lapointe R, Do M, Taylor MW, Young HA. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J Immunol. 2000;164:3596–3599. doi: 10.4049/jimmunol.164.7.3596. [DOI] [PubMed] [Google Scholar]
- Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. The Journal of Cell Biology. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaronen M, Quintana FJ. Immunological Relevance of the Coevolution of IDO1 and AHR. Front Immunol. 2014;5:521. doi: 10.3389/fimmu.2014.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julliard W, Fechner JH, Mezrich JD. The aryl hydrocarbon receptor meets immunology: friend or foe? A little of both. Front Immunol. 2014;5:458. doi: 10.3389/fimmu.2014.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyala S, Guo J, Sartor M, Medvedovic M, Kann S, Puga A, Ryan P, Tomlinson CR. Different global gene expression profiles in benzo[a]pyrene- and dioxin-treated vascular smooth muscle cells of AHR-knockout and wild-type mice. Cardiovasc Toxicol. 2004;4:47–73. doi: 10.1385/ct:4:1:47. [DOI] [PubMed] [Google Scholar]
- Liu P, Xie BL, Cai SH, He YW, Zhang G, Yi YM, Du J. Expression of indoleamine 2, 3-dioxygenase in nasopharyngeal carcinoma impairs the cytolytic function of peripheral blood lymphocytes. BMC Cancer. 2009;9:416. doi: 10.1186/1471-2407-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löb S, Königsrainer A, Zieker D, Brücher BDM, Rammensee HG, Opelz G, Terness P. IDO1 and IDO2 are expressed in human tumors: levo- but not dextro-1-methyl tryptophan inhibits tryptophan catabolism. Cancer Immunology, Immunotherapy. 2009;58:153–157. doi: 10.1007/s00262-008-0513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long WP, Pray-Grant M, Tsai JC, Perdew GH. Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol Pharmacol. 1998;53:691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- Mautino MR, Jaipuri FA, Waldo J, Kumar S, Adams J, Van Allen C, Marcinowicz-Flick A, Munn D, Vahanian N, Link CJ. Abstract 491: NLG919, a novel indoleamine-2,3-dioxygenase (IDO)-pathway inhibitor drug candidate for cancer therapy. Cancer Research. 2013;73:491–491. [Google Scholar]
- Mazarei G, Leavitt BR. Indoleamine 2,3 Dioxygenase as a Potential Therapeutic Target in Huntington’s Disease. Journal of Huntington’s disease. 2015;4:109–118. doi: 10.3233/JHD-159003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R, DuHadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC. Novel Tryptophan Catabolic Enzyme IDO2 Is the Preferred Biochemical Target of the Antitumor Indoleamine 2,3-Dioxygenase Inhibitory Compound d-1-Methyl-Tryptophan. Cancer Research. 2007;67:7082–7087. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohinta S, Kannan AK, Gowda K, Amin SG, Perdew GH, August A. Differential Regulation of Th17 and T Regulatory Cell Differentiation by Aryl Hydrocarbon Receptor Dependent Xenobiotic Response Element Dependent and Independent Pathways. Toxicological Sciences. 2015;145:233–243. doi: 10.1093/toxsci/kfv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer BJ, Rojas IY, Kerley-Hamilton JS, Hazlett HF, Nemani KV, Trask HW, West RJ, Lupien LE, Collins AJ, Ringelberg CS, Gimi B, Kinlaw WB, 3rd, Tomlinson CR. Inhibition of the aryl hydrocarbon receptor prevents Western diet-induced obesity. Model for AHR activation by kynurenine via oxidized-LDL, TLR2/4, TGFbeta, and IDO1. Toxicol Appl Pharmacol. 2016;300:13–24. doi: 10.1016/j.taap.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Flaveny CA, DiNatale BC, Chairo CR, Schroeder JC, Kusnadi A, Perdew GH. Antagonism of Aryl Hydrocarbon Receptor Signaling by 6,2′,4′-Trimethoxyflavone. Journal of Pharmacology and Experimental Therapeutics. 2010;332:135–144. doi: 10.1124/jpet.109.158261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, Patterson AD, Perdew GH. AH RECEPTOR LIGANDS IN CANCER: FRIEND AND FOE. Nature reviews. Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Hao Z, Sadek R, Dobbins R, Marshall L, Vahanian MN, Ramsey WJ, Kennedy E, Mautino M, Link C, Lin R, Royer-Joo S, Morrissey K, Mahrus S, McCall B, Pirzkall A, Munn DH, Janik JE, Khleif SN. (Published Poster) Phase 1a Study of the Safety, Pharmacokinetics, and Pharmacodynamics of GDC-0919 in Patients with Recurrent/Advanced Solid Tumors. Poster: European Society for Medical Oncology (ESMO) 2015 http://qfuse.com/client_downloads/ESMO2015_GDC-0919_Nayak_22Sep15.pdf.
- Nguyen NT, Hanieh H, Nakahama T, Kishimoto T. The roles of aryl hydrocarbon receptor in immune responses. Int Immunol. 2013;25:335–343. doi: 10.1093/intimm/dxt011. [DOI] [PubMed] [Google Scholar]
- Nguyen NT, Kimura A, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Opitz U, Sahm F, Ochs K, Lutz C, Wick W, Platten M. The indoleamine-2,3-dioxygenase (IDO) inhibitor 1-methyl-D-tryptophan upregulates IDO1 in human cancer cells. PLoS One. 2011;6:e19823. doi: 10.1371/journal.pone.0019823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrig UF, Majjigapu SR, Vogel P, Zoete V, Michielin O. Challenges in the Discovery of Indoleamine 2, 3-Dioxygenase 1 (IDO1) Inhibitors. J Med Chem. 2015 doi: 10.1021/acs.jmedchem.5b00326. [DOI] [PubMed] [Google Scholar]
- Sarkar SA, Wong R, Hackl SI, Moua O, Gill RG, Wiseman A, Davidson HW, Hutton JC. Induction of Indoleamine 2,3-Dioxygenase by Interferon-γ in Human Islets. Diabetes. 2007;56:72–79. doi: 10.2337/db06-0617. [DOI] [PubMed] [Google Scholar]
- Schwieler L, Samuelsson M, Frye MA, Bhat M, Schuppe-Koistinen I, Jungholm O, Johansson AG, Landen M, Sellgren CM, Erhardt S. Electroconvulsive therapy suppresses the neurotoxic branch of the kynurenine pathway in treatment-resistant depressed patients. J Neuroinflammation. 2016;13:51. doi: 10.1186/s12974-016-0517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JG, Chen X, Punwani NG, Williams WV, Yeleswaram S. Potential Under-Prediction of Warfarin Drug Interaction from Conventional Interaction Studies & Risk Mitigation: A Case Study with Epacadostat, an IDO1 Inhibitor. J Clin Pharmacol. 2016 doi: 10.1002/jcph.737. [DOI] [PubMed] [Google Scholar]
- Soliman HH, Minton SE, Han HS, Ismail-Khan R, Neuger A, Khambati F, Noyes D, Lush R, Chiappori AA, Roberts JD, Link C, Vahanian NN, Mautino M, Streicher H, Sullivan DM, Antonia SJ. A Phase I study of indoximod in patients with advanced malignancies. Oncotarget. 2016 doi: 10.18632/oncotarget.8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- Vacchelli E, Aranda F, Eggermont A, Sautes-Fridman C, Tartour E, Kennedy EP, Platten M, Zitvogel L, Kroemer G, Galluzzi L. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology. 2014;3:e957994. doi: 10.4161/21624011.2014.957994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Vogel CF, Goth SR, Dong B, Pessah IN, Matsumura F. Aryl hydrocarbon receptor signaling mediates expression of indoleamine 2,3-dioxygenase. Biochem Biophys Res Commun. 2008;375:331–335. doi: 10.1016/j.bbrc.2008.07.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthgen E, Kanitz E, Tuchscherer M, Tuchscherer A, Domanska G, Weitschies W, Seidlitz A, Scheuch E, Otten W. Pharmacokinetics of 1-methyl-L-tryptophan after single and repeated subcutaneous application in a porcine model. Exp Anim. 2016;65:147–155. doi: 10.1538/expanim.15-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung Amanda WS, Terentis Andrew C, King Nicholas JC, Thomas Shane R. Role of indoleamine 2,3-dioxygenase in health and disease. Clinical Science. 2015;129:601–672. doi: 10.1042/CS20140392. [DOI] [PubMed] [Google Scholar]