Abstract
Formation of the Hepatitis B (HBV) nucleocapsid (NC) is an essential step in the viral lifecycle but its assembly is not fully understood. We report the discovery of sequence-specific interactions between the viral pre-genome and HBV core protein (Cp) that play roles in defining the NC assembly pathway. Using RNA SELEX and bioinformatics we identified multiple regions in the pre-genomic RNA with high-affinity for Cp dimers. These RNAs form stem-loops with a conserved loop motif that trigger sequence-specific assembly of virus-like particles (VLPs) at much higher fidelity and yield than in the absence of RNA. The RNA oligos do not interact with preformed RNA-free VLPs, so their effects must occur during particle assembly. Asymmetric cryo-EM reconstruction of the T=4 VLPs assembled in the presence of one of the RNAs reveals a unique internal feature connected to the main Cp shell via lobes of density. Biophysical assays suggest that this is a complex involving several RNA oligos interacting with the C-terminal arginine-rich domains of Cp. These Cp-RNA contacts may play a role(s) in regulating the organization of the pre-genome during nucleocapsid assembly, facilitating subsequent reverse transcription and acting as a nucleation complex for NC assembly.
Introduction
The WHO reports that HBV has infected >2 billion people worldwide1. In adults most infections are acute. However, ˜240 million live with a chronic infection that can ultimately lead to liver failure, cirrhosis, or cancer, resulting in >700,000 deaths annually2. The availability of an effective vaccine3 has decreased the spread of HBV but is not curative for chronic infections. Standard treatment using nucleos(t)ide analogues directed against the viral polymerase rarely leads to a cure, and is thus a lifelong therapy4. Better understanding of HBV will help identify and characterize additional drug targets that could lead to new curative therapies.
HBV is a para-retrovirus, i.e. a DNA virus that initially packages an RNA form of its genome, the pre-genome5,6. In an infected cell, the basis of infection is viral, covalently closed, circular DNA (cccDNA) in the nucleus, a persistent, chromatinized episome whose protein complement also includes HBV core or capsid protein (Cp)7,8. It is 3200 bp long and encodes four overlapping reading frames for Polymerase (P), surface proteins (three different sizes are translated, collectively referred to as HBsAg for surface antigen), the cell regulatory factor protein X, and the core and pre-core proteins (HBcAg and HBeAg, respectively) (Fig 1a). The P, Cp, and HBeAg proteins are translated from the same RNA, the positive-sense, pre-genomic RNA (pgRNA), which also serves as the template for the reverse transcription reaction. The pgRNA is a terminally redundant transcript covering about 3500 nucleotides, but is otherwise a typical mRNA. Most of the pgRNA is not spliced concomitant with export from the nucleus9,10, suggesting a novel export mechanism, presumably involving the nuclear import and export signals on Cp11–13.
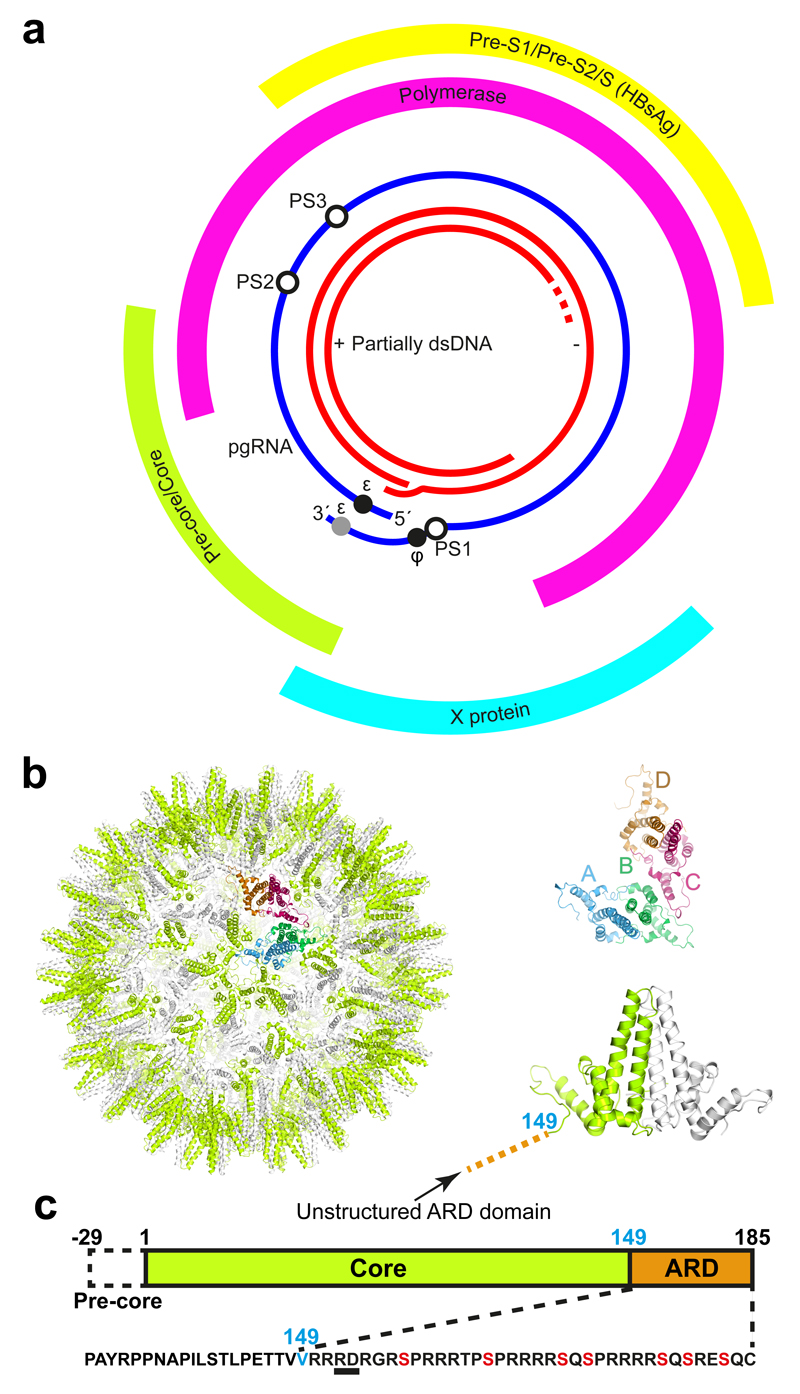
Figure 1. The Hepatitis B Virus.
(a) The genetic map of HBV showing the partially dsDNA genome and the four open reading frames of the virally encoded proteins: Pre-core/core (Cp), which forms the nucleocapsid (NC) shell; Pre S1/PreS2/S, the envelope embedded HBV antigen (HbsAg); X, which plays a role in numerous aspects of the HBV life-cycle within the cell; the polymerase (P), and the pgRNA with the positions of the 5′ ε, the redundant 3′ ε (grey circle), ϕ and the preferred sites (PSs) studied here highlighted by circles. (b) The HBV NC (left) comprises either 90 (T=3) or 120 Cp dimers (T=4 shown). Cp dimers form characteristic four-helix bundles, two from each monomer, that appear as spikes on the surface (right bottom). The two conformers of the HBV Cp dimer (A/B & C/D) that are needed to create the T=4 particle are also shown (right top). The HBV capsid and protein dimer were obtained from PDB (3J2V)58. (c) The Cp of the isolate used here is 185 amino acids long (RD dipeptide insertion underlined), with an alpha-helical rich region (149 amino acids long), and a C-terminal ARD. The 149th amino acid, V, is labelled blue for clarity. ARD is rich in both basic amino acids and serines. The latter, highlighted in red, are known sites for phosphorylation, which are thought to play roles in NC assembly.
In vivo assembly of an HBV nucleocapsid (NC) begins with a pgRNA-P protein complex that is required for pgRNA packaging. A correctly folded P and a functional stem-loop, termed epsilon (ε) located near the 5´ end of pgRNA are necessary for this process14–19. Cp phosphorylation is associated with RNA packaging20–22. Once encapsidated, P protein begins reverse transcription by priming DNA synthesis, adding the first 3-4 deoxynucleotides whilst bound to ε, before jumping to the 3´ end of the genome to complete synthesis of the minus strand. Three such template transfers are required for synthesis of the relaxed circular, dsDNA of mature HBV within the NC. Most of the RNA template is digested by the RNaseH domain of P protein during minus strand DNA synthesis. A sequence, phi (ϕ), at the 3´ end of the pre-genome complementary with ε23,24 is believed to facilitate strand transfer. Low resolution structural studies show that pre-genomic RNA forms a thin shell associated with the inner surface of the NC and that P protein is internal, suggesting that it travels on an RNA track to complete DNA reverse transcription25,26. The mature virion is enveloped by a host-derived membrane containing embedded HBsAg, which encloses an icosahedral NC with either T=4 (ca. 95%) or T=3 (5%) quasi-symmetry27. Similar ratios of T=4 to T=3 capsids are observed in many expression systems and following in vitro assembly28,29. NC is composed of dimers of the ˜183 residue Cp (Fig. 1b & c), organised as a shell-forming N-terminal domain of 149 residues connected via a linker region to a C-terminal arginine-rich domain (ARD).
Although HBV is ostensibly a DNA virus, we reasoned that the physics and functions of Cp-RNA interaction in HBV would resemble those found in RNA viruses. We recently uncovered a previously unsuspected principle of the assembly mechanisms of positive-sense, single-stranded RNA viruses that challenges the prevailing view that genomic RNAs are merely passive passengers in a process driven by viral coat proteins30. Instead, it appears that many viral genomes encompass cryptic, sequence-degenerate, dispersed RNA Packaging Signals (PSs). PSs have affinity for their cognate coat proteins, and can act collectively to ensure encapsidation of cognate genomic RNA, whilst building capsids rapidly and with great fidelity at low concentrations. Mathematical modelling of such PS-mediated assembly31 suggests that it confers many selective advantages and would therefore be expected to occur widely throughout nature. This appears to be the case for viruses infecting humans32,33, plants34 and bacteria35.
As HBV packages a pgRNA during assembly, we hypothesize that similar mechanistic constraints may contribute to formation of its NC. HBV RNA must be packaged in a manner that supports reverse transcription, and this could be facilitated by PS-like RNA motifs. In HBV we may more accurately redefine a PS as a preferred site for Cp binding. We therefore investigated whether the HBV pgRNA also encodes such PSs. Due to their nature, PS sites are difficult to identify by sequence analysis alone. We therefore developed a novel approach that combines experimental and bioinformatics methods. We used RNA SELEX against HBV Cp to generate a library of sequences with affinity for Cp. These aptamer sequences were then aligned across the cognate viral pgRNA, revealing genomic regions with sequence similarity to the aptamer pool capable of forming stem-loop structures, i.e. potential PSs. These sites are conserved across strain variants and each displays a RGAG sequence motif in the loop (R = purine). Individual genomic fragments encompassing these PSs show high affinity, sequence-specific interaction with Cp as demonstrated by their ability to induce formation of closed VLPs in vitro. Asymmetric cryo-EM reconstruction of these VLPs suggests that they contain a group of PS oligonucleotides interacting with Cp principally via the C-terminal ARDs. PS-like sites in the pre-genome may therefore play a role in favouring formation of an assembly competent form of Cp, effectively creating an assembly initiation complex for NC and specifying the quasi-symmetry of the capsid. Inhibiting formation of this complex could therefore be an anti-viral strategy.
Results
The HBV pgRNA contains preferred Cp binding sites
HBV VLPs assembled from (full-length) Cp subunits expressed in E. coli were purified as described36 (Supplementary Fig. 1a & Supplementary Table 1). They form a mixture of T=3 and predominantly T=4 shells. These were immobilised onto magnetic beads, disassembled by treatment with guanidinium chloride and then washed to remove host RNA, resulting in immobilised Cp dimers36 with their ARDs accessible. RNA SELEX was carried out using our standard protocols (Supplementary Fig. 1b) and the aptamer pool from the 10th round analysed by NextGen DNA sequencing (Methods).
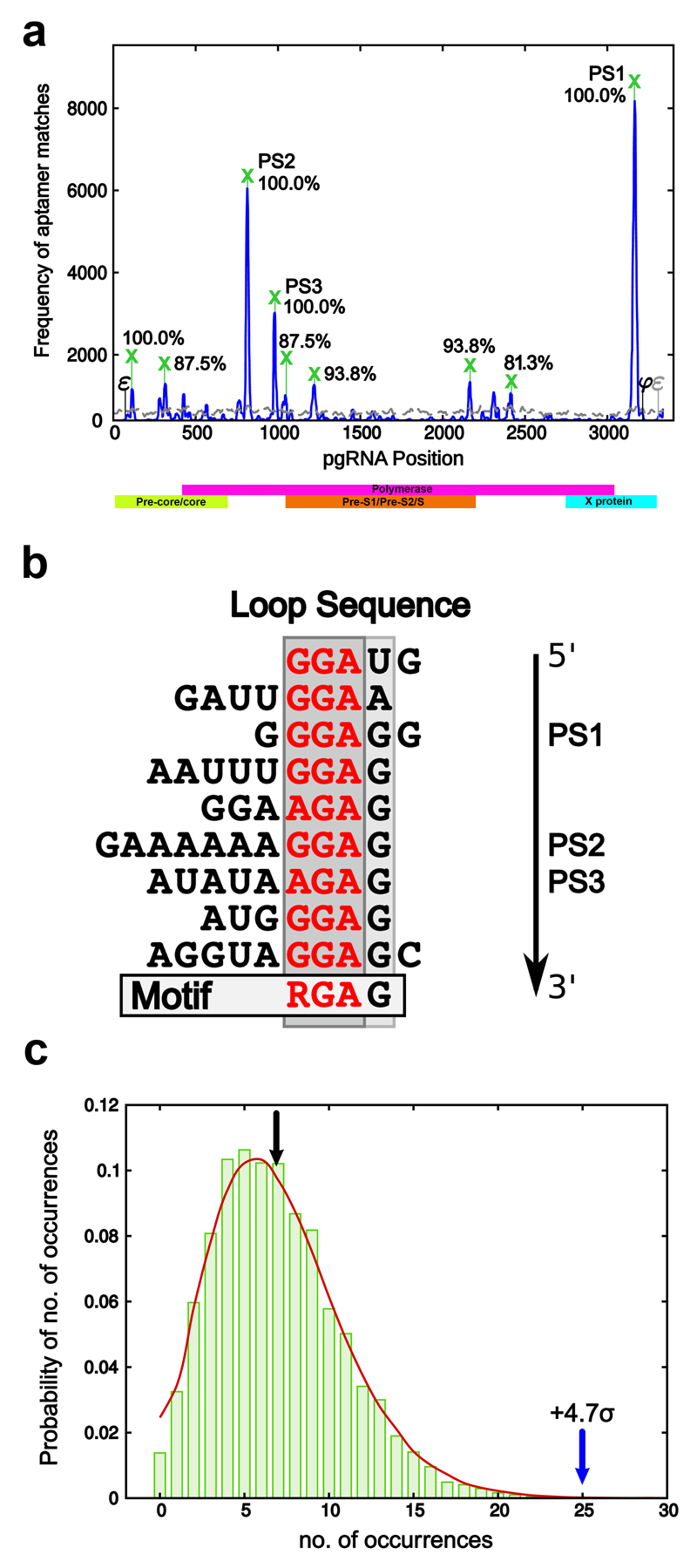
The RNA sequences that bind Cp in the selected library were aligned to the HBV pre-genome most closely related to the protein used for the SELEX experiments (the laboratory strain, GenBank Seq id NC_003977.125). Statistically significant matches (a Bernoulli score of 12 or more, Methods) to the pgRNA of this strain (the blue peaks in Fig. 2a) were benchmarked against an alignment of the unselected library (grey curve in Fig. 2a) to identify peaks that occur with significant frequency. This identifies multiple sites dispersed across the pgRNA having similar sequences/structures to Cp binding aptamers, consistent with our expectation for PS-like sites across the genome. We applied the same procedure to 14 randomly selected HBV strain variants from GenBank, the current NCBI HBV reference strain (GenBank Seq ID NC_003977.2) as well as the laboratory strain (GenBank Seq ID NC_003977.1) and identified all those peaks that are conserved in at least 80% of these strains (marked with green crosses in Fig. 2a). These genomic regions are thus likely to encompass PSs. The three peaks with the highest conservation (100%) and peak heights, the latter indicating how many aptamers matched these sites, are labelled PS1, PS2 and PS3 in Fig. 2a. For the nine sites with high conservation between strains, we extracted 30 nts 5´ and 3´ to the peak nucleotide in the genomic sequences of three representative strain variants, including the laboratory strain and the reference genome, and considered all their possible secondary structure folds with negative free energy via Mfold (Methods). A similarity analysis of primary and secondary structure revealed the predicted existence of stem-loops sharing a purine-rich loop recognition motif, RGAG (Fig. 2b).
Figure 2. Identification of conserved PS motifs in the pgRNA.
(a) Matches between aptamers from the HBV core selected library and the reference strain (NC_003977.1) with a Bernoulli score of at least 12 (i.e. all non-contiguous alignments with at least the same probability as a contiguous matching alignment of 12 nucleotides) are shown as a frequency plot (solid blue line). The equivalent frequency plot for the naïve library, i.e. the library before selection has taken place, is shown for comparison (grey dashed line). Peaks occurring in at least 80% of the tested strains are marked by a green cross, with conservation levels indicated as percentages. The peaks with the highest frequency and level of conservation are labelled PS1, PS2 & PS3. (b) Alignment of the loop sequences of representative stem-loops in regions of the genome overlapping with the nine conserved Bernoulli peaks reveals a conserved RGAG motif. (c) Probability distribution showing the proportion of sequences containing a given number of stem-loops with an RGAG containing loop across 10,000 randomised versions of genome. The green bars correspond to such randomised versions of the reference strain, whilst the red line gives corresponding probabilities across all five strains marked by an asterisk in Methods. The black arrow indicates the average number of occurrences over all randomised versions of the reference strain (= 6.85), whilst the blue arrow points to the number of occurrences in the reference strain (= 25), a 4.68 standard deviation from the average. The other tested strains exhibit similar levels of occurrence.
We computed the frequency of this motif in stem-loops across the 16 HBV strains analysed. Across all strains, the RGAG motif occurs in stem-loops on average ˜25.4 times (precisely 25 times in the laboratory strain). Compared to 10,000 randomised versions of the pgRNAs, the frequency of occurrence of RGAG in the actual genome is 4.68 standard deviations above the average (Fig. 2c), strongly implying a functional role(s).
pgRNA oligonucleotides trigger VLP formation in vitro
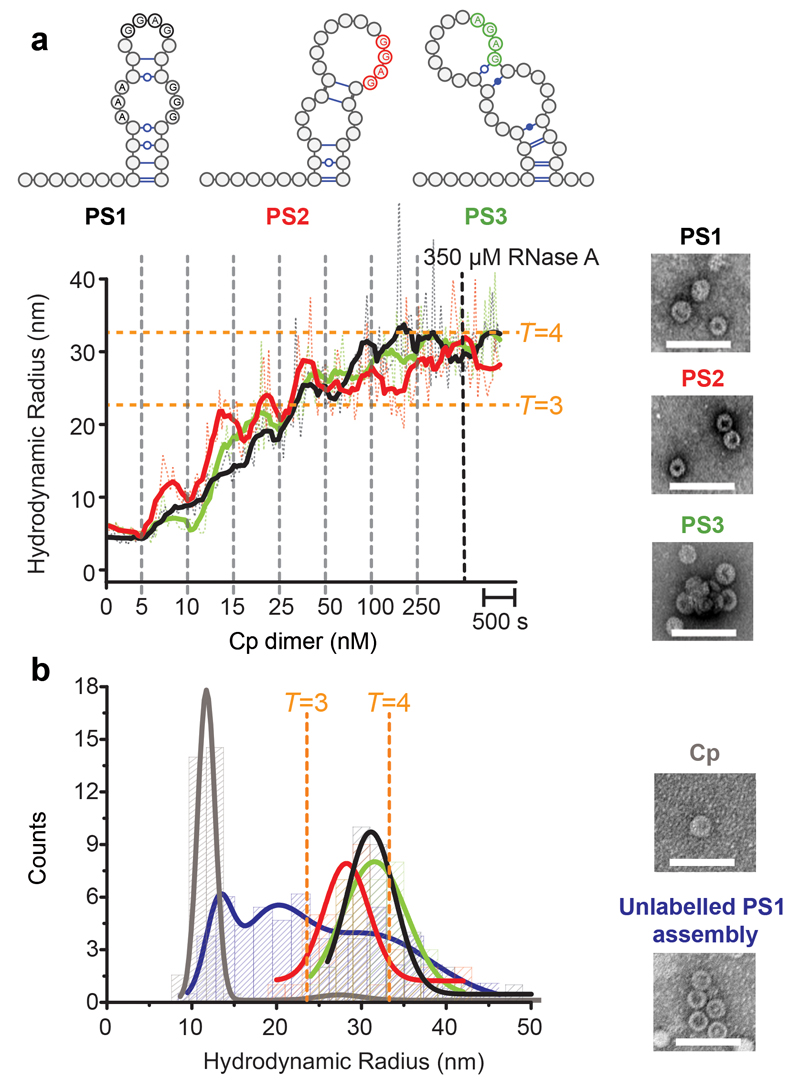
PS1, 2 & 3 oligonucleotides (Supplementary Fig. 2a), were tested for their ability to bind Cp dimers using single molecule fluorescence correlation spectroscopy (smFCS) (Fig. 3 & Supplementary Fig. 2b). This technique yields a real time estimate of the hydrodynamic radius (Rh) of dye-labelled species. Importantly, it allows reactions to be followed at low nanomolar concentrations, where we have shown that binding specificity more closely reflects the situation in vivo compared to most in vitro reactions. The latter are typically carried out at higher (e.g. 0.1-0.8 μM) concentrations36, where the specificity of PS-mediated assembly is reduced or lost. In order to avoid electrostatic effects due to differing oligo lengths, each PS was produced as part of a 47 nt long fragment, each dye-labelled at its 5´ end (Methods 34). The labelled oligos (˜15 nM) were then titrated with increasing amounts of Cp (5-250 nM Cp dimer) and the Rh values tracked over time (Fig. 3a). After each addition there was a pause of ˜10 min to allow reactions to equilibrate. The titrations lead to distortions in the data collection and the averaging, which is visible in the plots as noisy signals. After equilibration at 250 nM Cp, RNase was added to each reaction and the Rh values monitored for ˜10 min. If these declined steeply, it was assumed that the VLPs produced were incomplete. Negative stain EM images were obtained for the samples before RNase addition, and the sizes of the complexes present at this point were also assessed by calculation of Rh distribution plots (Supplementary Fig. 2c and Fig. 3b, respectively).
Figure 3. PSs trigger sequence-specific VLP assembly.
(a) Dye end-labelled RNA oligos encompassing PS1 (black), PS2 (red) or PS3 (green) were each assessed for their ability to bind Cp and form VLPs at nanomolar concentrations using smFCS. All reactions contained 15 nM of RNA dye-labelled as described in Methods. Vertical dotted lines indicate points where Cp was added with the final concentrations shown in nM. Samples were allowed to equilibrate between additions. The faint trace represents real time, raw signal, while the thick line represents smoothed data. EM images were recorded of the samples prior to RNase A addition (right). Scale bars represent 100 nm. (b) Hydrodynamic radial distributions of the reactions in (a) were taken following the last addition of Cp (here and throughout). The amount of Cp assembling beyond dimer in the absence and presence of RNA (unlabelled) was compared. At the end of these reactions, Cp was labelled with Alexa Fluor-488 (Methods) and the resulting Rh distributions quantitated for the Cp only (grey) and Cp plus unlabelled PS1 (blue) scenarios. Note, dye-labelling of the Cp dimer prevents it from assembling, implying that this is an end-point measurement. A sample of each was taken for analysis by TEM. smFCS and TEM were repeated in triplicate.
Each of the PS fragments stimulates assembly of both T=3 and T=4 complete VLPs with roughly equal efficiency under these conditions (Fig. 3a & b), with the latter being the dominant product, as expected29. Addition of Cp >250 nM does not increase the Rh values obtained, implying that by this stage all the RNAs have been incorporated into VLPs. In order to assess whether these effects are a direct consequence of Cp-PS interaction, we carried out a number of controls. Dye-labelled PS fragments do not bind to preformed VLPs and remain RNase sensitive in their presence (Supplementary Table 2), implying that the PSs only get internalised in assembling VLPs. To determine if the RNA triggers assembly, we compared assembly efficiency of Cp with and without PS RNA present by adding a protein modifying dye after incubation of Cp alone or completion of a titration of unlabelled PS1. The Rh distribution plots are shown in Fig. 3b. In the absence of RNA, <5% of Cp assembles under these conditions, in contrast to >80% of the Cp for assembly in the presence of RNA. It appears that Cp-PS interaction triggers an increase in the assembly efficiency. This effect varies with the age of the Cp, consistent with oxidation of an assembly-inhibiting disulphide at the dimer interface37. Comparative statements here are based on the results of both positive and negative control experiments with each batch of Cp.
We then probed the RNA sequence-specificity of these reactions (Supplementary Fig. 3a). Test oligos comprised the epsilon stem-loop, as well as loop and bulge variants of PS1. This included a variant in which the bulge region was fully base-paired. In similar assays to the PS1-3 reactions the Rh values for all three RNAs remain sensitive to nuclease action, implying that assembly of closed shells requires a specific RNA sequence/structure. EM images and distribution plots confirm this interpretation. The sequence sensitivity of the assembly reaction is further highlighted by additional PS1 variants (Supplementary Figs. 3b & c; Supplementary Table 3). Their effects on assembly confirm the importance of the bulge and/or sequences within it, and the loop RGAG (here a GGAG) motif. A DNA oligonucleotide encompassing the PS1 sequence (Supplementary Fig. 3d) elicits aggregation, showing that faithful assembly is a specific property of the PS in its RNA form, i.e. with an A helical duplex stem, as well as the Cp-recognition motif in the loop.
The C-terminal ARD of the HBV Cp is believed to mediate interactions with the pgRNA, and the 1-149 Cp fragment that lacks the ARD readily assembles in the absence of nucleic acid38. We therefore assessed the ability of Cp149 to respond to PSs in the smFCS assay. No RNA-dependent assembly, or PS binding by Cp149, occurs under these conditions (Supplementary Fig. 4a), although EM images show that the truncated Cp alone readily assembles, confirming that the ARD is essential for the interaction with RNA. The ARD is extensively phosphorylated in vivo, although the responsible cellular kinase remains unknown39. Lowering the positive charge on the C-terminus of Cp should reduce its ability to bind PS RNAs. We phosphorylated Cp in vitro40 (Supplementary Table 1) and tested its properties. EM images show that modified Cp readily assembles but does not bind to PS1 in smFCS assays (Supplementary Fig. 4b).
HBV NC assembly is triggered by formation of a sequence-specific RNA-Cp complex
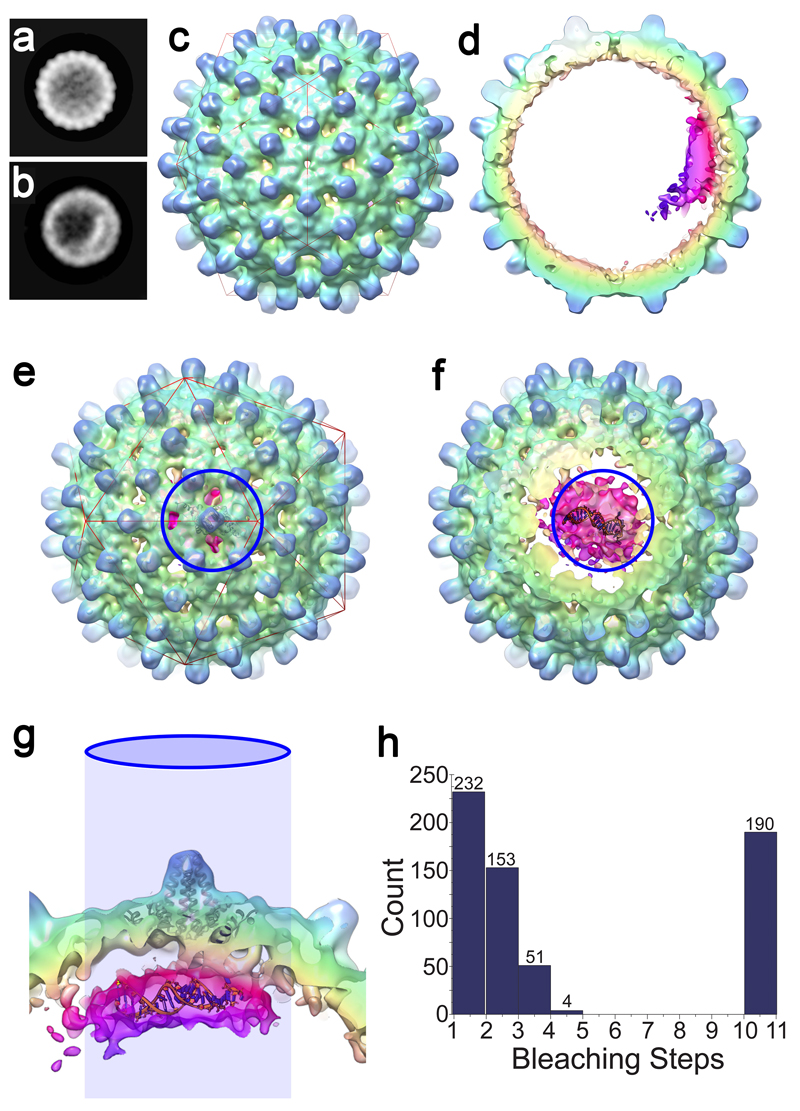
The VLPs assembled around PS1 were purified on a larger scale and their structures determined by cryo-EM, yielding icosahedrally-averaged reconstructions of the T=3 and T=4 particles (Fig. 4). A significant fraction (˜25%) of the T=4 particles also contained an asymmetric feature located just below the protein shell. An asymmetric reconstruction of these particles was also calculated (Fig. 5). The result suggests the asymmetric feature represents a complex between PS1 oligonucleotides and the ARD domains of the overlying Cp subunits.
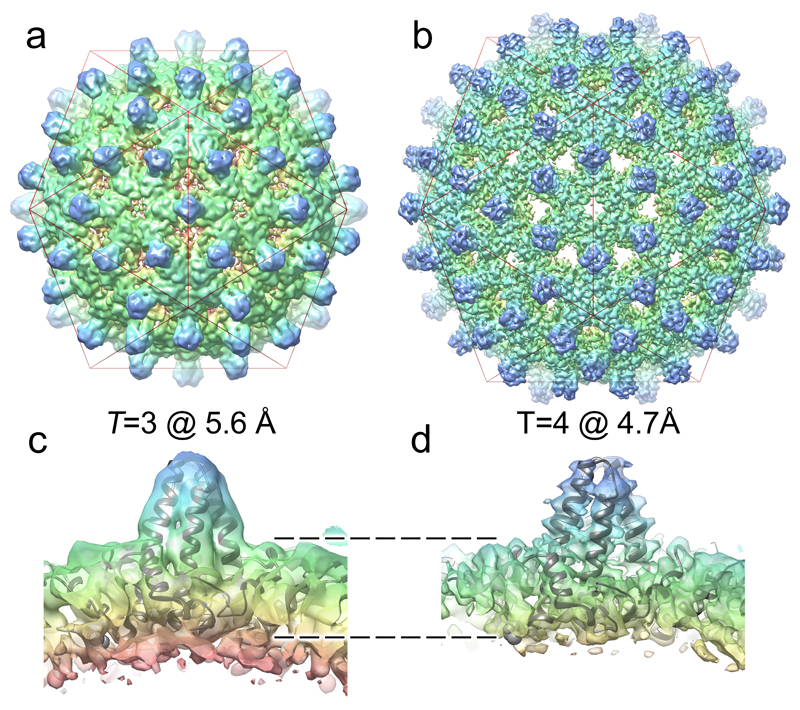
Figure 4. The structures of T=3 and T=4 HBV VLPs suggest a mechanism for the specification of their quasi-conformations.
The icosahedrally-averaged cryo-EM structures of (a) T=3 and (b) T=4 HBV VLPs at 5.6 Å and 4.7 Å resolution, respectively. A red icosahedron is included to assist interpretation of the two reconstructions, which are shown in the same orientation. (c & d) show ˜30 Å thick slabs through the structure of each particle, with a fitted Cp-dimer in each case. The T=3 shell is thicker, indicating that density corresponding to the ARDs is resolved in the T=3, but not the T=4, structure. Rendering both structures at equivalent resolution does not change this interpretation (Supplementary Fig. 5).
Figure 5. Asymmetric RNA feature in T=4 HBV VLPs.
(a & b) 2D views of 42,411 T=4 particles were calculated by maximum-likelihood-based classification in RELION. An asymmetric RNA feature is visible in a subset of these particles (b). (c) An asymmetric 3D reconstruction at 11.5 Å resolution of 10,851 particles containing the asymmetric feature. The asymmetric density for the protein shell is icosahedral, despite the lack of any symmetry averaging. (d) An approximately 40 Å thick slab through the asymmetric HBV VLP reconstruction shows the asymmetric feature bound to one region of the Cp shell, revealing density ascribed to RNA and ARDs within the protein shell (bright cerise, magenta and purple). The figures were rendered in a radial colour scheme (Blue=165Å; Cyan=152Å; Green=139Å; Yellow=126Å; Pink=113Å) using USCF Chimera. (e) The asymmetric RNA density is centred beneath a Cp dimer surrounding one of the 5-fold vertices of the T=4 particle (indicated by the blue circle). A single Cp dimer is fitted as a ribbon diagram into the appropriate position using the ‘Fit in map’ function in UCSF Chimera. (f) As the front of the map is slabbed away, the density within is revealed. Shown and manually fitted is a single copy of PS1 as a ribbon diagram (modelled in RNA Composer). (g) Side-view of the same portion of the map, with the view oriented by the projected blue circle. Discrete fingers of density are visible between the Cp layer and RNA density, which is large enough to accommodate 2-4 RNA oligonucleotides. (h) Histogram of photobleaching steps from 630 individual fluorescent spots on a grid containing PS1 HBV VLPs. Spots containing >10 steps resulted from traces exhibiting exponential decay, which were assumed to be aggregates in which multiple bleaching steps occur simultaneously. Photobleaching was performed in duplicate.
From the EM map at this resolution it is not possible to determine the number of PS oligonucleotides present in the complex. The A260/280 ratio of the purified VLP suggests that the RNA content, assuming T=4 morphology, is ˜5 oligos/particle 41. An additional estimate of this stoichiometry was obtained by studying photobleaching of PS1 VLPs (Fig. 4, Methods). VLPs show multiple bleaching steps, confirming that there are multiple oligos within each shell. Given the labelling efficiency of the oligos, the data are consistent with 2-4 oligos/VLP. We built a 3D model of PS1 and manually positioned it within the EM map (Fig. 4f, Methods). From the relative volume of the asymmetric density and the size of the PS1 oligo, it appears that at least two copies of the PS are present within the density. We cannot exclude the possibility that other RNA molecules are bound to the protein shell elsewhere, but are not visible due to mobility or an irregular location with respect to the ordered RNA density. The biochemical and structural data are consistent with the asymmetric structure being an assembly initiation complex, where an RNA preferred site(s) has initiated assembly culminating in the formation of the T=4 NC.
The cryo-EM data hint at a further insight into HBV biology. A minority of HBV particles, whether from assembly reactions or wild-type virus infections, assemble with T=3 quasi-symmetry and both types of particles are visible in our cryo-EM data. Using 2D and 3D classification the T=3 (˜11%) and T=4 (89%) particles are readily separable. Figure 4 shows 3D reconstructions of the two particles with imposed icosahedral symmetry at 5.6 Å and 4.7 Å resolution, respectively. In addition to the obvious differences in size and number of Cp dimers that the two VLP structures contain, the T=4 and T=3 maps are different in the features visible on their inner surfaces, where the ARDs are located and where RNA binding occurs. As might be expected for icosahedrally-averaged maps of a sub-stoichiometrically occupied VLP, both structures are essentially devoid of density attributable to RNA. The capsid shell of the T=4 structure is visibly thinner than the T=3 equivalent, however, and closer examination of the T=3 map suggests that additional density corresponding to ordered segments of the ARDs is visible (Fig. 4), which is absent in the T=4 structure (Fig. 4 c & d). This difference persists when the T=4 map is Fourier filtered to be at a similar resolution as the T=3 (Supplementary Fig. 5). This is consistent with previous studies that showed that the Cp C-terminal region, including the ARD, plays a role(s) in determining capsid geometry29,42.
Discussion
Previously we identified multiple RNA PSs within the genomes of positive-sense ssRNA viruses that play essential roles in their assembly34,35. Here we explored whether similar sequence-specific Cp-RNA interaction sites exist within the pre-genomic RNA of HBV. Many such sites emerge from this analysis, encompassing stem-loop structures presenting variations of a loop motif likely to be the Cp-recognition sequence. This motif is highly conserved across all strain variants, and is statistically strongly over-represented within the HBV genome. Three of these sites bind Cp in a sequence-specific manner as RNA stem-loops, promoting efficient, high-fidelity assembly into predominantly T=4 VLPs, with assembly properties similar to the PSs of ssRNA viruses. This sequence-specificity has not been observed previously in in vitro reassembly reactions 36, which suggest that both pre-genomic and non-genomic RNA get packaged co-operatively. However, under similar conditions Cp alone forms capsid shells, albeit with lower efficiency. This is in marked contrast to what we observe here at low nanomolar concentrations, perhaps mimicking in vivo assembly conditions. Under these conditions Cp appears stable as dimers, but in the presence of RNA assembles into higher order structures forming closed icosahedral shells in a sequence-specific fashion. Such reactions likely mimic events in the cell, providing new insight into the genome packaging specificity of HBV.
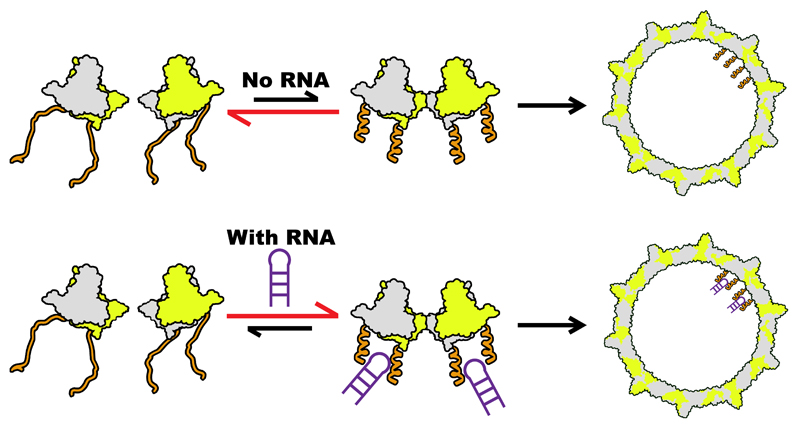
In bona fide ssRNA viruses, PSs regulate assembly by facilitating the formation of the protein-protein interactions of the (nucleo)capsid, simultaneously collapsing the conformational ensemble of the genomic RNA30,34. Individual PSs can also trigger VLP formation akin to the results seen here34. HBV pgRNA by itself, i.e. without bound polymerase, is insufficient to trigger packaging in vivo. Earlier observations indicate that activation and inactivation of assembly is sensitive to Cp conformation and is allosterically triggered43,44. This is consistent with the findings here. Cp149 assembles at low concentration without RNA, in contrast to Cp185, implying that the ARD is inhibitory for assembly under these conditions. This inhibition is removed for Cp185 either by binding PS RNA or by phosphorylation. Both these routes reduce the net charge on each ARD, implying that electrostatic repulsion might be the origin of the inhibition. It is therefore possible to postulate an assembly pathway (Fig. 6), that accounts for the known properties of HBV Cp. The Cp exists as a dimer with positively charged C-terminal ARDs. The latter create an electrostatic repulsion inhibiting formation of Cp complexes larger than dimer. This barrier is not absolute and some dimers of dimers can form, their concentration increasing with Cp concentration. If that higher order species is required to trigger NC assembly based on Cp-Cp contacts then reassembly of Cp, alone or in non-specific RNA-Cp complexes at higher concentrations, can be explained. At low concentration the Cp binds specific PSs within the pre-genome, triggering formation of the critical higher order species and hence NC formation. That species is likely to correspond to the structure seen in Fig. 5. In the pre-genome the PS sites forming the initiation complex would be different PSs, each folding into an SL presenting the recognition motif rather than the multiple identical copies of PS1 as seen here. The efficient assembly of the closed T=4 shell with PS1 suggests, however, that the assembly initiation step mediated by the nucleation complex would be similar.
Figure 6. Proposed model of HBV NC assembly.
ARD (orange) within a Cp dimer (green and grey) inhibit formation of a dimer of dimers, the first intermediate on the pathway to NC assembly. Reducing the net charge on the ARD by phosphorylation or PS RNA (purple, bottom) binding allows this structure to form more easily, triggering NC formation. At concentrations higher than those mimicking in vivo conditions as used here, the unmodified dimer of dimers forms and particles self-assemble without RNA or will bind RNA non-specifically to produce the same outcome.
HBV is not a ssRNA virus and has a much more complex lifecycle. Therefore the roles fulfilled by specific Cp-RNA interactions may also be distinct. Evidence suggests that the polymerase-ε complex plays a critical role in pgRNA selection and NC assembly. Conversely, the PS sequences identified in this work are highly conserved and demonstrably have specific affinity for Cp. For correct assembly the virus needs to ensure the following: 1) identify full-length pgRNA; 2) assemble a quasi-equivalent shell of Cp around that RNA; 3) complete reverse transcription of the pgRNA using the encapsidated P protein whilst degrading the template; and 4) complete copying of the negative ssDNA strand, creating a partially dsDNA genome. Evidence suggests the polymerase translocates extensively on the pgRNA during these processes 25. The 5´ ε (Fig. 1a) can base-pair with 3´φ, effectively circularising the pgRNA, an interaction that may play a role(s) during both packaging and template transfers. The φ site, at nucleotides 3172-3190, is adjacent to PS1. It is therefore possible that the polymerase:ε/φ complex favours the folding of PS1 to present its recognition motif contributing to assembly initiation. Such a mechanism would ensure that Cp assembly only occurs on a pre-genome that has recruited polymerase. It would also permit co-localisation of P with both ends of the pgRNA, imposing a defined position with respect to the encapsidated genome. The presence of the multiple PS sites would then result in formation of a defined, non-entangled path for the RNA within the NC, i.e. corresponding to the track along which the polymerase must travel. The HBV pre-genome has many fewer PS sites than are seen in ssRNA viruses, consistent with the need to have most of the RNA readily available for reverse transcription. There may also be other roles for these specific PS-Cp interactions in HBV. For instance, specific interaction of Cp with pgRNA in the nucleus may facilitate export of unspliced RNAs using the nuclear export signals on the Cp13.
Previous in vitro studies of empty capsid assembly have suggested that Cp conformational change is needed to trigger nucleation43,45. Candidate small molecule antiviral therapeutics are known that act as allosteric effectors driving assembly of HBV46,47. In addition, structural studies have revealed the breadth of HBV Cp conformational flexibility, suggesting that small molecules and/or genomic sequences could restrict an ensemble of structures to particular active, or inactive, forms44. The preferred RNA-Cp contacts identified here open new insights into regulation of assembly around a genome that must be reversed transcribed, and therefore offer additional therapeutic targets.
Methods
Cloning, expression and purification of proteins used
We obtained an E.coli Cp-expressing plasmid (a gift of Prof. Nicola Stonehouse), known to produce assembled HBV VLPs containing host RNAs48. The Cp encoded has the following amino acid sequence differences compared to the current GenBank reference strain (NC_003977.2): A61, E77-FAGAS (single letter amino acid code) -D78 insertion, S92N, F102I, I121L, R156-RD-R157 insertion. Since the wild-type C61 has been implicated in assembly37, this was restored to the gene before expression in a PET28b plasmid in BL21(DE3) E.coli cells. The inserted FAGAS epitope was also removed. Induction with 1 mM IPTG at 0.6 OD was followed by growth for 20 hrs at 21°C. Cells were lysed using a Soniprep 150 with 5x 30 sec bursts on ice. The lysate was then clarified by spinning at 11,000 g for 1 hr. VLPs were then pelleted by centrifugation at 120,000 g for 14 hr, resuspended in 20 mM Hepes (pH 7.5), 250 mM NaCl, and 5 mM DTT and applied to an XK50 column packed with 25 ml of Capto™core 700 resin (GE Life Sciences). Fractions containing VLPs were pooled and precipitated with 40%(w/v) ammonium sulphate. The Cp appeared pure on SDS-PAGE and its identity, and that of variants, was confirmed by mass spectrometry (Supplementary Table 1). Cp lacking the ARD, i.e. Cp149, was produced by mutagenesis (Q5 site-directed mutagenesis kit, NEB) and prepared similarly. Note, the Cp149 VLP expressed in E.coli lacks significant encapsidated cellular RNA. VLPs were visualised by negative stain transmission electron microscopy (TEM). Full length Cp VLPs were additionally purified by sucrose density gradient before dye-labelling using Alexa Fluor-488 SDP ester (Invitrogen) over 4 hrs at room temperature in 200 mM sodium carbonate buffer (pH 8.3), followed by desalting over a NAP5 column. There were two over-lapping VLP peaks on the gradient and it was impossible to separate them. TEM and smFCS confirm that they are the expected T=3 and T=4 shells, with the latter the predominant form (Supplementary Fig. 1a). The Cp region 140-148 has been shown to be a determinant of morphology, the shorter versions producing more T=3 shells29. It is possible that the dipeptide insertion adjacent to the linker region at position 157 may alter the properties of the Cp. However, when we removed the RD insertion, yielding Cp183, we found no differences with Cp185, either in RNA binding, ability to form VLPs with PS RNAs or preference for the dominant quasi-conformer shell formed. Since longer Cp was used for SELEX and the high resolution EM work, those are the data shown throughout.
All HBV variants used for assembly assays were dissociated from VLPs into protein dimers as previously described36, with the exception that dissociation was at pH 9.5, as opposed to 7.5. This was done in the presence of Complete Protease Inhibitor Tablets (Thermofisher Scientific). HBV core dimer concentration was determined by UV absorbance. Fractions with an A260:A280 ratio of approximately 0.6 or lower were used in assembly assays. SRPK∆ kinase was expressed and purified from a pRSETb plasmid, as previously described40.
SELEX protocol
Purified HBV capsids (˜360 μg) were immobilised onto 6 mg of M270 carboxylic acid Dynabeads (Thermofisher Scientific) following the manufacturer’s protocol. Beads were washed twice with selection buffer (25 mM Hepes, pH 7.5, 250 mM NaCl, 2 mM DTT, EDTA-free complete protease inhibitor) and unreacted N-hydroxysuccinamide blocked with a 15 m 50 mM Tris-HCl pH 7.4 wash. Beads were washed a further three times with selection buffer. Immobilised capsids were dissociated with a 30 minute incubation of 2 M guandinium chloride in 0.5 M LiCl2. Beads were then washed three times with B&W buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 2 M NaCl) and then washed three times with selection buffer. Beads were resuspended in selection buffer so that concentration of beads was 10 mg/mL. Negative selection beads were also prepared in the same manner but with no capsids. Ten rounds of SELEX were performed in vitro using a synthetic, combinatorial N40 2´OH RNA library (~1024 potential sequences) as described previously49. The amplified DNA of round 10 was then subjected to Next Generation Sequencing on an Illumina MiSeq platform. This yielded ˜1.6M sequence reads, in which one sequence occurs 65,802 times and there are 1149 aptamers with a multiplicity of 100 or higher. The overall frequencies of the four nucleotides in this aptamer pool is A34.30%; C9.09%; G40.97% & U15.64%, and compares with the same data for the unselected naïve library of A26.10%; C22.03%; G24.64% & U27.22%. The highest multiplicity for sequences in the latter pool is 4. These data confirm that selection from the naïve pool occurred, and that the base composition of the selected aptamers is consistent with the RGAG motif identified within the HBV genomes.
PS identification
PS identification was carried out using the laboratory HBV strain (*NC_003977.1). The aptamer library contained 1,664,890 unique sequences, each 40 nts in length that have been aligned against the genome as follows: Each aptamer sequence was slid along the genome in increments of 1 nt. For each such position of the reference frame, the subset of the aptamer sequence with the best alignment to the genome was identified according to the Bernoulli score B, which benchmarks the probability of a non-contiguous alignment to that of a contiguous alignment of B nucleotides. The Bernoulli scores for all reference frames of a given aptamer sequence in the library were rank-ordered starting from the largest score, and all matches with the genome up to a Bernoulli score of 12 counted. The procedure was then repeated for the other aptamer sequences and corresponding matches added, resulting in the peaks in Fig. 2a.
Identification of a consensus motif
HBV genome sequences with the following accession numbers were randomly extracted from 750 complete HBV genomes found in GenBank: KCS10648.1; *AF223955.1; AY781181.1; *AB116266.1; AB195943.1; KR014086.1; *KR014072.1; KR014055.1; KR013939.1; KR013921.1; KR013816.1; KR013800.1; EU796069.1; AB540582.1, and the NCBI HBV reference strain (GenBank Seq ID *NC_003977.2) and the laboratory strain (GenBank Seq ID NC_003977.1) were added to the ensemble. Sequences used for the statistical analysis in Fig. 2c are marked by an asterisk. Bernoulli peaks, which occurred within at most 10nts of each other in at least 80% of these 16 HBV strain variants, were marked by a green cross in Fig. 2a to indicate their conservation. To identify the putative PS recognition motif, we extracted sequences of 60 nts, centred around the peak nucleotide of each Bernoulli peak, from three representative strains (AF223955.1, NC_003977.1, & NC_003977.2) and determined all possible stem-loops of negative free energy via Mfold50. We carried out a similarity analysis of these stem-loops, comparing both sequence and structure elements, we identified for each peak area that representative that has the highest degree of similarity both with secondary structure elements in the other peak areas in the same genome and stem-loops corresponding to the same peak area in the other strains. This returned a stem-loop for each peak. An alignment of the corresponding loop sequences is shown in Fig. 2b.
RNA dye-labelling
PS1, PS2 and PS3 (47 nucleotides long) were purchased from Integrated DNA Technologies with a 5′ C6-amino group. To label RNA, 6 μL of RNA (200 μM) was mixed with 1 μL 1 M sodium borate buffer, pH 8 and 3 μL 10 mM Alexa-488-SDP (Thermofisher Scientific) and rolled at room temperature for 4 hours. 10 μL of 2x denaturing loading dye was then added to the RNA, boiled for 5 minutes and loaded onto a pre-warmed denaturing PAGE. RNA was gel extracted, isopropanol precipitated and finally re-suspended in DEPC-H2O and frozen at -80°C until needed.
Assembly Assays
Assembly reactions were performed by adding HBV Cp in dissociation buffer (50 mM Tris (pH 9.5), 1.5 M GuHCl, 500 mM LiCl and 5 mM DTT) to 15 nM Alexa-488 labelled RNA in a reassembly buffer containing 20 mM Hepes (pH 7.5), 250 mM NaCl, 5 mM DTT and 0.05%(v/v) Tween-20 at 25°C. Successive additions of dimer were performed until assembly was deemed complete by the measured Rh value plateauing, but never exceeded 10% of total reaction volume. Each addition of Cp is marked by a vertical dashed grey line in the titration plots and the expected hydrodynamic radii of T=3 and T=4 particles (as determined for dye-labelled particles expressed in E.coli) are marked by an orange horizontal dashed line within figures.
Manual mixing throughout the reactions caused an approximate 1 min delay at the start of FCS data collection. FCS measurements were made using a custom-built FCS setup with 30 sec data accumulation per autocorrelation function (CF). Individual CFs were decomposed into triplet state relaxation and diffusion (characterized by diffusion time, TD) components, and the latter was converted into an apparent hydrodynamic radius, Rh51. Samples for TEM were taken at the end of each measurement. Plots of Rh over time (thin dashed line) were smoothed (thick solid line) using the FFT filter in Origin Pro-8 with a cutoff percentage of 35%. Plots of Rh distribution were also fitted using Origin Pro-8 software, to a normal single or multiple peak Gaussian function. Samples taken for negative stain TEM analysis were placed on to a glow discharged carbon coated formvar 300 mesh Cu grid. Grids were stained with 2% uranyl acetate and dried.
Assembled particle labelling
Assembly was carried out as in smFCS experiments. In particular, Cp was titrated into reassembly buffer with and without 15 nM unlabelled PS1 to a final concentration of 250 nM. This was allowed to incubate at room temperature for 1 hour, and then buffer exchange was carried out via dialysis to remove guanidinium hydrochloride present. Labelling of protein was then carried out by adding Alexa Fluor-488 SDP ester (1:50 ratio of dye to Cp dimer) and incubating overnight at 4 °C. The resulting sample was then measured via smFCS in 30 s bins for 100 min and the Rh data plotted as above in a hydrodynamic radial distribution plot. A sample was then removed for analysis via TEM. Post labelling, Cp dimer became assembly incompetent, therefore Cp could not be tracked during real time assembly.
Photobleaching
HBV VLPs containing Alexa-488 labelled PS1 were assembled as described in smFCS assembly assays. Under those conditions all RNA is bound to protein as judged from fluorescence quenching and photon counting in the FCS experiments. VLPs were then added to two glow discharge-irradiated Carbon/Formvar 300-mesh grids (Agar Scientific), and one grid stained with 2% (w/v) uranyl acetate and viewed with a Jeol 1400 microscope at 40,000x magnification. The remaining, unstained grid was positioned Formvar side down onto a clean microscope coverslip and mounted onto an inverted TIRF microscope. The laser (Coherent Sapphire, 488 nm, 25 mW) power was adjusted to excite and photobleach the labelled RNA within the time frame of several minutes. Sequential images were taken with an emCCD camera (Andor iXon) with 0.2 sec exposures and em gain of 200. An unexposed field of view was used for each series.
Fluorescent spots were identified in the collected frames using previously described procedures and converted into time traces52. These were then inspected and classified according to the number of photobleaching steps. Frequencies of traces with a defined number of steps were collated in a histogram. Several bright spots per field of view exhibited continuous intensity decay, presumably representing larger aggregates. These were used to estimate the overall photobleaching rate (0.003 per frame) and formally included in the histogram as representing 10 steps. The histogram without the bin representing continuum events was modelled as a weighted sum of binomial distributions for up to quadruple occupancy and probability of labelling of 0.56 estimated from UV-Vis spectra.
Electron microscopic reconstructions
Large scale VLP preparation
smFCS experiments were scaled up into 96 well plates. Two 96 well plates (Non-Binding Surface, Corning) were used. PS1 RNA was labelled and gel purified as described earlier and HBV dimer was purified as described above. Each well contained 200 μL of 15 nM PS1 in re-assembly buffer. As in smFCS, ten 2 μL injections of 2.5 μM dimer in dissociation buffer were performed. A Perkin-Elmer Envision plate reader was used to carry out the injections and record the anisotropy of the PS1 RNA (FITC excitation and emission filters). VLPs were purified away from free RNA and capsid using a 1.33 g/mL caesium chloride gradient and spun at 113,652 x g for 90 hours using an SW40Ti rotor. A single band was observed and fractionated. The band was dialysed into reassembly buffer to remove caesium chloride. The 2 mL fraction of VLP was concentrated to 200 μL using an Amicon 100 kDa MWCO spin concentrator.
CryoEM specimen preparation
After recovery of the PS1-containing VLPs and removal of caesium chloride by dialysis, their structures were analysed using single-particle cryo-EM. VLPs were vitrified. 200 mesh EM grids with Quantifoil R 2/1 support film and an additional ˜ 5 nm continuous carbon film were washed using acetone and glow discharged for 40 s prior to use. CryoEM grids were prepared by placing 3μl of ˜ 3.2 mg/ml HepB VLP on the grid, before blotting and plunge freezing using a Leica EM GP freezing device. Chamber conditions were set at 8 °C and 95 % relative humidity, with liquid ethane temperature at -175 °C. Data was collected on a FEI Titan Krios (eBIC, Diamond Light Source, UK) transmission electron microscope at 300 keV using an electron dose of 27 e-/Å2/s, 2.5 s exposure, yielding a total electron dose of 67.5 e-/ Å2. Data was recorded on a 17 Hz FEI Falcon II direct electron detector. The dose was fractionated across 33 frames. Final object sampling was 1.34 Å per pixel. A total of 2397 micrographs were recorded using EPU (FEI) automated data collection software.
Single particle image processing
2397 micrographs were motion corrected and averages of each movie were generated using MotionCorr53, and contrast transfer function (CTF) parameters for each were determined using CTFFIND454. Micrographs with unacceptable astigmatism or charging, as determined by examining the output from CTFFIND4, were discarded leaving a total dataset of 1710 micrographs. All particle picking, classification and alignment was performed in RELION 1.355.
Approximately 57,000 particles were manually picked and classified using reference-free 2D classification in RELION 1.3. This classification confirmed the initial visual impression that although the VLPs were purified as a single band on a caesium gradient, two sizes of VLPs were present. A selection of resulting 2D class averages were used as templates for automated particle picking. The particle stack generated using auto-picking was subject to 2D classification to separate T=3 and T=4 particles, and to remove particles not corresponding to VLPs. The subsequent particle stacks (5589 for T=3, 42,411 for T=4) were subject to 3D classification, using a sphere with the approximate diameter of the VLP as a starting model. Subsets of the data were reconstructed including data out to the Nyquist frequency using the 3D autorefine option in RELION with I3 symmetry imposed to generate all structures presented in this work. Within the T=4 42,411 particle dataset it was clear that a further subset (10,851 particles) of the data contained a significant asymmetric feature inside the Cp shell where RNA binding would be expected to occur. An asymmetric (C1) reconstruction was performed on a relatively homogenous set of 10,851 such particles, giving the reconstruction at 11.5 Å resolution.
The 3D model of PS1 RNA was made using RNA Composer56. The cryoEM figures were rendered using USCF Chimera57.
Supplementary Material
Acknowledgments
We thank the UK MRC (MR/N021517/1) and the Universities of Leeds and York for financial support for parts of this work, which was also supported in part by grants from the Wellcome Trust (089311/Z/09/Z; 090932/Z/09/Z & 106692). PGS and RTw also thank The Wellcome Trust for financial support for virus work (Joint Investigator Award Nos. 110145 & 110146), & RTw acknowledges funding via a Royal Society Leverhulme Trust Senior Research Fellowship (LT130088) and EPSRC grant EP/K028286/1 for R.J.B. E.C.D. acknowledges funding via an Early Career Leverhulme Trust Fellowship (ECF-2013-019). AZ acknowledges funding from NIH grant R01-AI118933. We also thank the eBIC for collection time on the Titan Krios microscopes.
Footnotes
Data availability
The data that supports the findings of this study are available from the corresponding authors upon request. Correspondence and requests for materials should be addressed to P.G.S. The cryoEM reconstructions were deposited in the Electron Microscopy Databank (EMDB) with the following accession codes: EMD-3714 (asymmetric T=4 HBV VLP), EMD-3715 (T=4 HBV VLP with I3 symmetry imposed) and EMD-3716 (T=3 HBV VLP with I3 symmetry imposed).
Author contributions
P.G.S. and R.Tw conceived the project. N.P. performed smFCS and photobleaching experiments and he and R.T. analysed the data. S.J.W., R.F.T. and N.A.R. collected and analysed the cryoEM data. S.J.W. performed SELEX, and R.B., E.D., E.U.W, and R.Tw. analysed the resulting sequences to identify the PSs. N.P., S.J.W. and D.P.M. purified HBV VLPs. All authors contributed to the writing and editing of the manuscript.
Competing financial interest
AZ is a co-founder and consultant of Assembly BioSciences.
Research in the Zlotnick lab is supported by the NIH and Assembly.
No Assembly BioSciences employee contributed to Dr. Zlotnick’s contribution to this work.
References
- 1.WHO. Weekly epidemiological record. WHO. 2009;84:405–420. [Google Scholar]
- 2.Tillmann HL. Antiviral therapy and resistance with hepatitis B virus infection. World Journal of Gastroenterology. 2007;13:125–140. doi: 10.3748/wjg.v13.i1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray K, et al. Protective immunisation against hepatitis B with an internal antigen of the virus. J Med Virol. 1987;23:101–107. doi: 10.1002/jmv.1890230202. [DOI] [PubMed] [Google Scholar]
- 4.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. Vol. 2. Fields Virology; 2007. pp. 2977–3209. [Google Scholar]
- 6.Selzer L, Zlotnick A. Assembly and Release of Hepatitis B Virus. Vol. 5. Cold Spring Harbor Laboratory Press; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock CT, et al. Structural organization of the hepatitis B virus minichromosome. Journal of Molecular Biology. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 8.Guo Y-H, Li Y-N, Zhao J-R, Zhang J, Yan Z. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics. 2011;6:720–726. doi: 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- 9.Günther S, Sommer G, Iwanska A, Will H. Heterogeneity and Common Features of Defective Hepatitis B Virus Genomes Derived from Spliced Pregenomic RNA. Virology. 1997;238:363–371. doi: 10.1006/viro.1997.8863. [DOI] [PubMed] [Google Scholar]
- 10.Abraham TM, Lewellyn EB, Haines KM, Loeb DD. Characterization of the contribution of spliced RNAs of hepatitis B virus to DNA synthesis in transfected cultures of Huh7 and HepG2 cells. Virology. 2008;379:30–37. doi: 10.1016/j.virol.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh CT, Liaw YF, Ou JH. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J Virol. 1990;64:6141–6147. doi: 10.1128/jvi.64.12.6141-6147.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckhardt SG, Milich DR, McLachlan A. Hepatitis B virus core antigen has two nuclear localization sequences in the arginine-rich carboxyl terminus. J Virol. 1991;65:575–582. doi: 10.1128/jvi.65.2.575-582.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H-C, et al. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathogens. 2010;6:e1001162. doi: 10.1371/journal.ppat.1001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch RC, Lavine JE, Chang L, Varmus HE, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 18.Pollack JR, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knaus T, Nassal M. The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 1993;21:3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan YT, Li J, Liao W, Ou J. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology. 1999;259:342–348. doi: 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]
- 21.Gazina EV, Fielding JE, Lin B, Anderson DA. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J Virol. 2000;74:4721–4728. doi: 10.1128/jvi.74.10.4721-4728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köck J, Nassal M, Deres K, Blum HE, von Weizsäcker F. Hepatitis B virus nucleocapsids formed by carboxy-terminally mutated core proteins contain spliced viral genomes but lack full-size DNA. J Virol. 2004;78:13812–13818. doi: 10.1128/JVI.78.24.13812-13818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abraham TM, Loeb DD. Base pairing between the 5' half of epsilon and a cis-acting sequence, phi, makes a contribution to the synthesis of minus-strand DNA for human hepatitis B virus. J Virol. 2006;80:4380–4387. doi: 10.1128/JVI.80.9.4380-4387.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oropeza CE, McLachlan A. Complementarity between epsilon and phi sequences in pregenomic RNA influences hepatitis B virus replication efficiency. Virology. 2007;359:371–381. doi: 10.1016/j.virol.2006.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JC-Y, Nickens DG, Lentz TB, Loeb DD, Zlotnick A. Encapsidated hepatitis B virus reverse transcriptase is poised on an ordered RNA lattice. Proc Natl Acad Sci USA. 2014;111:11329–11334. doi: 10.1073/pnas.1321424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JC-Y, Dhason MS, Zlotnick A. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. PLoS Pathogens. 2012;8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stannard LM, Hodgkiss M. Morphological irregularities in Dane particle cores. J Gen Virol. 1979;45:509–514. doi: 10.1099/0022-1317-45-2-509. [DOI] [PubMed] [Google Scholar]
- 28.Crowther RA, et al. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 29.Zlotnick A, et al. Dimorphism of Hepatitis B Virus Capsids Is Strongly Influenced by the C-Terminus of the Capsid Protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
- 30.Borodavka A, Tuma R, Stockley PG. Evidence that viral RNAs have evolved for efficient, two-stage packaging. Proc Natl Acad Sci USA. 2012;109:15769–15774. doi: 10.1073/pnas.1204357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykeman EC, Stockley PG, Twarock R. Solving a Levinthal's paradox for virus assembly identifies a unique antiviral strategy. Proc Natl Acad Sci USA. 2014;111:5361–5366. doi: 10.1073/pnas.1319479111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart H, et al. Identification of novel RNA secondary structures within the hepatitis C virus genome reveals a cooperative involvement in genome packaging. Scientific Reports. 2016;6:22952–22952. doi: 10.1038/srep22952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shakeel S, et al. Genomic RNA folding mediates assembly of human parechovirus. Nat Commun. 2017;8:5. doi: 10.1038/s41467-016-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel N, et al. Revealing the density of encoded functions in a viral RNA. Proceedings of the National Academy of Sciences. 2015;112:2227–2232. doi: 10.1073/pnas.1420812112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stockley PG, et al. A Simple, RNA-Mediated Allosteric Switch Controls the Pathway to Formation of aT = 3 Viral Capsid. Journal of Molecular Biology. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porterfield JZ, et al. Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J Virol. 2010;84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selzer L, Katen SP, Zlotnick A. The hepatitis B virus core protein intradimer interface modulates capsid assembly and stability. Biochemistry. 2014;53:5496–5504. doi: 10.1021/bi500732b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludgate L, et al. Cyclin-dependent kinase 2 phosphorylates s/t-p sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids. J Virol. 2012;86:12237–12250. doi: 10.1128/JVI.01218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aubol BE, et al. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proceedings of the National Academy of Sciences. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porterfield JZ, Zlotnick A. A simple and general method for determining the protein and nucleic acid content of viruses by UV absorbance. Virology. 2010;407:281–288. doi: 10.1016/j.virol.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watts NR, et al. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 2002;21:876–884. doi: 10.1093/emboj/21.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Packianathan C, Katen SP, Dann CE, Zlotnick A. Conformational changes in the hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol. 2010;84:1607–1615. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatakrishnan B, et al. Hepatitis B Virus Capsids Have Diverse Structural Responses to Small-Molecule Ligands Bound to the Heteroaryldihydropyrimidine Pocket. J Virol. 2016;90:3994–4004. doi: 10.1128/JVI.03058-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilmer JK, Zlotnick A, Bothner B. Conformational Equilibria and Rates of Localized Motion within Hepatitis B Virus Capsids. Journal of Molecular Biology. 2008;375:581–594. doi: 10.1016/j.jmb.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourne C, et al. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J Virol. 2008;82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katen SP, Chirapu SR, Finn MG, Zlotnick A. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol. 2010;5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holmes K, et al. Assembly Pathway of Hepatitis B Core Virus-like Particles from Genetically Fused Dimers. J Biol Chem. 2015;290:16238–16245. doi: 10.1074/jbc.M114.622035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bunka DHJ, et al. Degenerate RNA packaging signals in the genome of Satellite Tobacco Necrosis Virus: implications for the assembly of a T=1 capsid. Journal of Molecular Biology. 2011;413:51–65. doi: 10.1016/j.jmb.2011.07.063. [DOI] [PubMed] [Google Scholar]
- 50.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podjarny A, Dejaegere AP, Kieffer B. Biophysical Approaches Determining Ligand Binding to Biomolecular Targets: Detection, Measurement and Modelling. Ch 5. Royal Society of Chemistry; 2011. p. 165. [Google Scholar]
- 52.Sharma A, et al. Domain movements of the enhancer-dependent sigma factor drive DNA delivery into the RNA polymerase active site: insights from single molecule studies. Nucleic Acids Res. 2014;42:5177–5190. doi: 10.1093/nar/gku146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SHW. Semi-automated selection of cryo-EM particles in RELION-1.3. J Struct Biol. 2015;189:114–122. doi: 10.1016/j.jsb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112–e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Yu X, Jin L, Jih J, Shih C, Zhou ZH. 3.5Å cryoEM structure of hepatitis B virus core assembled from full-length core protein. PLoS ONE. 2013;8:e69729. doi: 10.1371/journal.pone.0069729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.