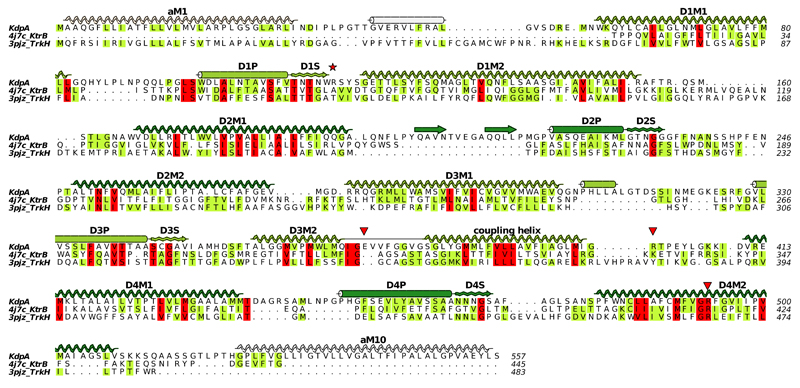
Extended Data Figure 6. Sequence alignment of KdpA with other SKT transporters TrkH and KtrB.
This structure-based alignment was done by the promals3d server using PDB depositions 4J7C for KtrB12 and 3PJZ for TrkH11. Sequence conservation is shown by coloring, red being most conserved and yellow least conserved. Secondary structure based on KdpA is shown with the coloring scheme in Fig. 1c. The four M1PM2 motifs have been distinguished as D1-D4, with S indicating the selectivity filter. The red star indicates the Q116R mutation and red triangles indicate residues implicated in coupling with KdpB, namely Glu370 and Arg393 that reside at the cytoplasmic side of the selectivity filter (Fig. 2c) as well as Arg400 that forms a salt bridge with the P-domain (Fig. 3b). This figure as well as those in Extended Data Figs 7, 8 and 9 were made with ALINE59.