Abstract
Plants maximise their fitness by adjusting their growth and development in response to signals such as light and temperature. The circadian clock provides a mechanism for plants to anticipate events such as sunrise and adjust their transcriptional programmes. However, the underlying mechanisms by which plants coordinate environmental signals with endogenous pathways are not fully understood. Using RNA-seq and ChIP-seq experiments, we show that the evening complex (EC) of the circadian clock plays a major role in directly coordinating the expression of hundreds of key regulators of photosynthesis, the circadian clock, phytohormone signalling, growth and response to the environment. We find that the ability of the EC to bind targets genome-wide depends on temperature. In addition, co-occurrence of phytochrome B (phyB) at multiple sites where the EC is bound provides a mechanism for integrating environmental information. Hence, our results show that the EC plays a central role in coordinating endogenous and environmental signals in Arabidopsis.
Plants are sensitive to their environment, and the distribution and phenology of plants has already altered in response to climate change1,2. Such growth and developmental changes require the integration of multiple environmental signals, such as light and temperature, into endogenous gene expression programmes. The circadian clock plays a key role in this process by enabling plants to anticipate future events such as sunrise and darkness as well as gating responses to environmental information according to time of day3–5. The expression levels of many circadian clock genes vary in response to changes in the environment6, but the underlying mechanisms by which these signals are integrated are not known. The circadian clock in Arabidopsis contains multiple interlocking loops with transcriptional and post-translational regulation. Three circadian clock genes, EARLY FLOWERING3, 4 (ELF3 and 4) and the MYB transcription factor LUX ARRYTHMO (LUX) together comprise the evening complex (EC)7,5, and are expressed at the end of the day. The EC coordinates elongation growth in Arabidopsis seedlings, as it directly represses the expression of the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR 4 (PIF4)7–9. PIF4 is necessary for warm temperature-mediated elongation growth10, and pif4 and pif4,5 mutants also display a reduced induction of flowering in response to warm temperature11–14. Natural variation in the activity of the EC is responsible for differences in thermal responsiveness, and warmer temperatures reduce the binding of the EC to target promoters, resulting in increased PIF4 expression15,16.
Of the core EC components, only LUX is recognisable as a transcription factor and has been shown to have direct DNA binding activity17, suggesting that the genome-wide targeting of the EC is likely dependent on the cognate LUX Binding Sites (LBS). Indeed, it has been shown that several known targets of LUX possess LBS in their promoters where binding of LUX has been demonstrated using chromatin immunopurification (ChIP)7,17,18
Although the role of the EC has been studied at specific circadian loci, the system-wide impact of the EC is not known, since its global binding pattern and regulatory effects are not yet characterised. In addition, the mechanisms by which the EC is able to provide environmental responsiveness to target loci and integrate environmental signals into the circadian clock are poorly understood. In this study we demonstrate that the EC regulates key nodes controlling photosynthesis, the circadian clock, growth, phytohormones and temperature signals. We show that the EC directly integrates temperature information and we describe a mechanism by which the co-binding of phytochrome B (phyB) with the EC to target loci enables environmental signals to be directly integrated into the circadian clock.
Results
G-box (CACGTG) motifs are highly enriched at EC binding sites
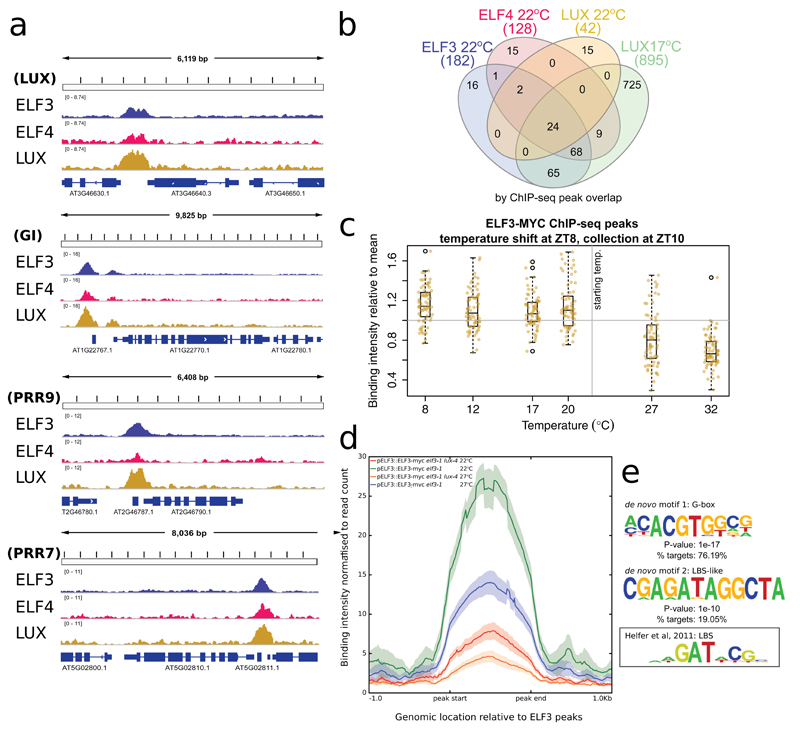
To understand how the EC may control plant responses to the environment, we sought to determine its binding genome-wide. Although the LUX Binding Site (LBS) is found at more than 10,000 sites in the Arabidopsis genome, typically only a subset of the theoretical transcription factor binding sites are occupied in vivo. It is also not known to what extent the target sites of the separate EC proteins overlap genome-wide. To resolve these questions we mapped the LUX, ELF3 and ELF4 target sites in vivo using chromatin immunopurification coupled to sequencing (ChIP-seq) using epitope tagged versions of these genes expressed under their own promoters (see Fig. S1 for westerns, Fig. S2 for complementation and Table S1 for sequencing read counts).
In the early evening (ZT10, 2 hours after darkness) at 22ºC, there is a large overlap in the binding sites of LUX, ELF4, and ELF3 (Fig. 1A,B and S3, Table S2). Additionally, the LUX ChIP at 22ºC has fewer significant binding sites than the other EC components, with 42 peaks being detected for LUX at 22ºC, compared to 182 for ELF3 (Fig. 1B). The reduced number of LUX peaks may be due to ELF3/ELF4 binding independently of LUX, or it may reflect a reduced detection resulting from a lower LUX ChIP efficiency.
Fig. 1.
To distinguish between these two possibilities, we sought to perform ChIP under conditions that would maximize the stability of LUX binding to improve the ChIP efficiency. Since it has been shown that the binding of EC components (i.e. ELF3 and ELF4) to the promoters of PIF4, LUX and PRR9 decreases with warmer temperature15, we performed an additional LUX ChIP-seq experiment at constant 17ºC (also at ZT10) to determine if additional targets could be identified. These experiments considerably increased the list of detectable LUX binding sites (Fig. 1B, Table S2). These additional LUX peaks at 17ºC overlapped even more substantially with the ELF3 and ELF4 peaks at 22ºC: e.g. 92% of ELF3 binding sites now overlap with other EC components. Since there are expected to be over 60,000 non-overlapping accessible regions for transcription factor binding in the Arabidopsis genome19, this is a highly significant overlap (see ‘VennDiagramStats’ in Table S3). These results are consistent with LUX binding to most if not all EC targets.
To determine whether the strength of binding of LUX changes globally in response to warmer temperatures, seedlings grown at 17ºC were shifted to 27ºC at ZT8 and collected at ZT9 and ZT10 (Fig. S3A). The strength of the LUX binding signal was weaker at the higher temperature at both time points (Fig. S3B), consistent with previous observations showing ELF3 binding to target loci decreases at 27ºC15.
We next analyzed ELF3 binding to its targets in plants grown at 22°C until ZT8 (dusk) and then shifted to a range of temperatures from 8 to 32ºC and measured at ZT10. There is a strong decline in ELF3 binding strength with increasing temperature across this range (Fig. 1C), consistent with previous observations that binding of ELF3 and ELF4 is temperature dependent20. LUX has been shown to provide DNA binding specificity for the EC at a number of loci, and we observe a large global reduction in ELF3 occupancy in the lux-4 background, confirming the importance of LUX in recruiting ELF3 to target loci (Fig. 1D). Interestingly, ELF3 binding is still highly responsive to temperature in lux-4, consistent with a role for ELF3 in responding to temperature (Fig. 1D).
Finally, we wished to determine whether LUX and ELF3 binding were consistent across multiple time points. Two additional LUX ChIP-seq experiments were conducted at 22ºC immediately prior to darkness (ZT8) and four hours after darkness (ZT12), to complement the previous ChIP-seq data at ZT10 (Fig. S3C). In addition, a time course ChIP-seq experiment was conducted for ELF3 with samples collected every four hours for a 24-hour cycle at 22ºC (Fig. S3D). These results suggest that LUX binding at ZT8 and ZT10, the two time points with greatest LUX expression, are very similar; however, there is a reduction in binding at ZT12 (Fig. S3C). Similarly, ELF3 binding near EC targets is high at dusk and in the early night (ZT8 to ZT16), with a reduction in binding during the day (Fig. S3D). This pattern of binding intensity is consistent with the expression patterns for LUX and ELF3, suggesting that their binding strength correlates with transcript and protein levels.
These results indicate that in vivo LUX mostly binds in complex with other EC components in the early evening. However, we also observed strong LUX binding to many of the same binding sites in an elf3-1 background, indicating that LUX binding in vivo can occur genome-wide independently of ELF3 (Fig. S4). This is in agreement with in vitro studies showing the ability of LUX to bind targets is independent of ELF3 and ELF4 (Silva et al under submission).
De novo motif analysis revealed two enriched elements under LUX binding sites at 22ºC at ZT10. Firstly, as expected, many peaks contain either perfect LBS sequences17 or a motif with strong similarity to it, motif 2 (Fig. 1E). Interestingly, G-boxes are even more enriched, being found in 76% of LUX binding sites at 22ºC, ZT10 (Fig. 1E, Table S4), including in the promoters of many different circadian clock genes, such as CCA1, GI, and PRR7 and 9. This suggests that one or more G-box binding transcription factor may play a significant role in EC regulation and function. De novo motif analysis was also conducted for the other datasets, and is available in Table S4 and Fig. S5—in all cases the G-box was the most enriched motif. We observe that the G-boxes lie near the centre of the EC peaks, and the data obtained by DNA Affinity Purification sequencing (DAP-seq)21 indicates multiple bZIP and bHLH TFs as candidates for co-binding with the EC (Fig. S6).
The EC is a node regulating photosynthesis, light, hormone, circadian and temperature signaling
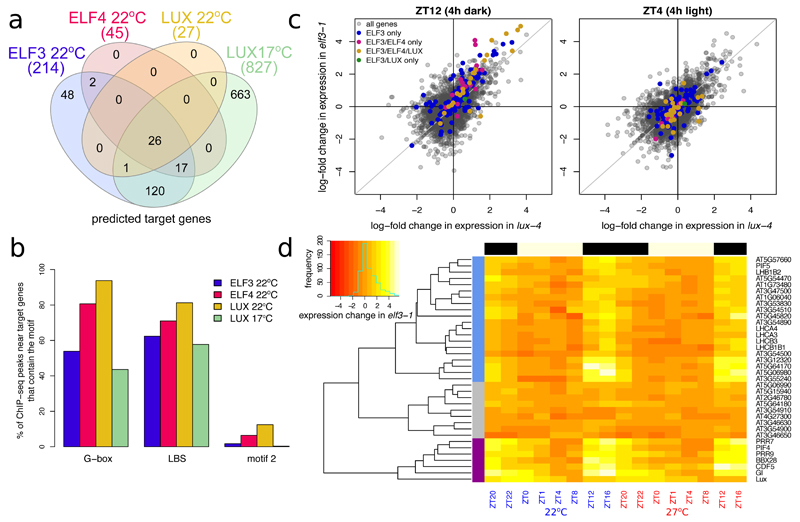
Since the EC has been proposed to act as a transcriptional regulator, we sought to identify the functional targets of the EC: genes that are both bound by EC components and also show a transcriptional response in EC mutant backgrounds. As the EC regulates transcription in a diurnal fashion we performed a gene expression time-course experiment, sampling every 4 h over a 24 h short day cycle (Table S5). Because the EC has been shown to integrate temperature information8,15,22, we performed this analysis at both 22ºC and 27ºC.
This enabled us to identify a set of genes present within 3000 bp of an EC binding peak that also show differential expression in at least one time point in either the elf3-1 or lux-4 backgrounds compared to Col-0 (Table 1, Table S6). This set of functional target genes for each EC component shows even more overlap with each other than do the original ChIP-seq peaks themselves, indicating the validity of this approach (Fig. 2A). Each gene referred to in the rest of the text as a potential ‘EC target’ has perturbed expression in at least one time point in elf3-1 or lux-4 and has at least two EC components binding within 3000 bp as determined by ChIP-seq.
Fig. 2.
Next, we searched for enrichment of the cis-elements already identified (Fig. 2B, Table S4) in ChIP-seq peaks in the vicinity of the functional targets. Interestingly, we observed an increase in the frequency of sequences corresponding to the true LBS, which are now more enriched than the related motif 2, suggesting that the true LBS is more important for a functional EC binding event compared to motif 2 (Fig. 2B). Additionally, we see that the occurrence of perfect G-boxes is increased in the subset of binding sites near EC functional targets. Together, these results suggest that functional EC binding is enriched for the co-occurrence of both LBS and G-box motifs, see Fig. S6 and Table S3. Furthermore, we wished to determine whether G-boxes were truly enriched under the EC peaks or whether they were simply enriched in the promoters of EC gene targets. We could find very few instances of G-boxes flanking our EC peaks (Table S3), which suggests that these G-boxes are in fact enriched under the EC peaks. Such a combinatorial requirement is likely to contribute to the specificity of EC action genome-wide.
The EC is established as a regulator of circadian responses, particularly growth, and we observe the described binding of EC components to the promoters of PRR9, LUX, PIF4 and PIF57,17,18 (Table 1, Fig. S7-8). Additionally, there is extensive interaction between the EC and other clock master regulators, including GI, CCA1, and PRR7, highlighting the high degree of cross-talk within the loops of the circadian clock23. Also, the EC appears to target a number of genes related to environment-dependent signaling and growth. In fact, most enriched GO terms amongst the EC targets are for genes involved in photosynthesis (Table S7, Fig. S8), including many photosystem light harvesting proteins and chlorophyll A/B binding proteins. In addition to altering the expression of nuclear encoded chloroplast genes, the EC also binds to the promoter of GUN5, which is necessary for retrograde signaling from the chloroplast to the nucleus24. Large-scale shutdown of photosynthesis genes upon the onset of darkness (as in Fig. S8) is presumably adaptive. Since the EC is not active in late night, these genes can then be reactivated prior to dawn in anticipation of light and photosynthesis. In addition to the described targets PIF4 and PIF5, the EC binds the promoters of other light signaling genes such as EARLY LIGHT INDUCIBLE PROTEIN, and EARLY PHYTOCHROME RESPONSIVE1.
In addition to the enrichment in photosynthesis and light response genes, the EC may regulate a number of genes implicated in heat response such as DNAJ11 and CXIP1 (see Fig. S8), which protects against oxidative damage caused by heat stress25, and the cold response (CBF1,2,3 and DREB2 class genes). Previously it has been shown that the promoter of LUX is bound by the master regulator of the cold response, CBF126, suggesting that the EC and CBFs form a self-regulating loop.
Finally, a major role of the EC is to gate growth, and this has been connected to the regulation of PIF4 and 5, master regulators of the elongation response. Additionally, we observe extensive binding of the EC to a number of targets involved in controlling phytohormone signaling—such as the bHLH growth regulators BANQUO1 and 2 that function downstream of the brassinosteroid responsive transcription factor BZR127,28 and the Myb-like transcription factor REVEILLE1, which controls free auxin levels29. Recently it has been shown that plants deficient in cytokinins are hypersensitive to circadian stress30. Interestingly, we see extensive connections between the EC and cytokinin signaling, since the EC represses the genes ARR6, ARR7, CYTOKINE OXIDASE5, CRF4 and CRF5.
Crucially, almost all of the proposed target genes show elevated levels of expression in elf3-1 and lux-4 in the early evening, but not the day, which is consistent with the EC’s role as an early evening repressor (Fig. 2C, S8-10, see ‘Scatterplots Stats’ in Table S3 for p-value calculations). To further understand the temporal dynamics of these EC targets, the ELF4 targets were clustered based on the log-fold change of expression in elf3-1 (Fig. 2D) (ELF4 targets were chosen as these have the largest degree of overlap with ELF3 and LUX, making them by this measure the best predictor of EC presence). Three clusters emerged—the largest cluster (shown in blue, which included PIF5 and many members of the photoharvesting system) had increased expression in the early evening. A second cluster (shown in purple)—which included all of the key circadian targets as well as PIF4—had increased expression in elf3-1 in the early morning, as well as at night. The last cluster had no clear pattern of expression in elf3-1, and may include genes that are ‘false-positive’ candidates. However, this third cluster (grey) only contains 9 genes, suggesting that we have successfully identified a high quality list of global EC gene targets.
About 40 of the direct targets of the EC are transcription factors, suggesting that the EC is a major node regulating large-scale transcriptional responses in the cell. For LUX at 17ºC, the GO term “transcription, DNA-regulated” is enriched (PANTHER, p-value 4.98E-09). Consistent with this, in elf3-1 and lux-4 we observed large-scale perturbation to the transcriptome extending beyond the evening period when the EC is most active. Many of these genes mis-regulated in elf3-1 and lux-4 are not bound by the EC, indicating that they are instead indirect targets (Fig. S11).
ELF3 and LUX are both required for EC repression of circadian targets
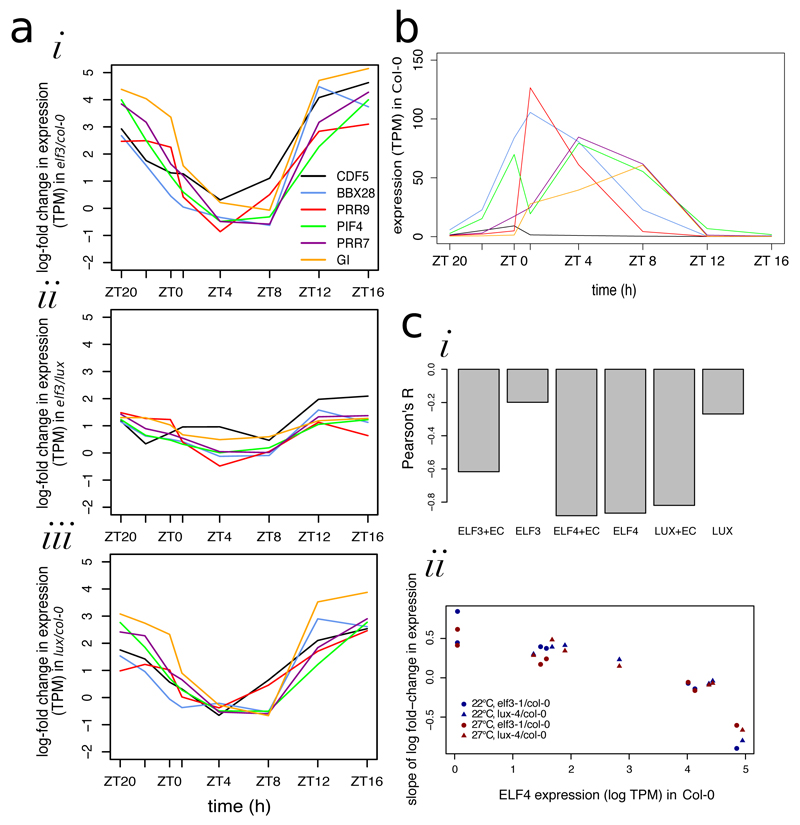
Because of the critical role the EC plays in controlling the circadian clock, we decided to further investigate the expression pattern of the cluster composed mainly of circadian genes from Fig. 2D. Firstly, we wished to determine whether these genes could only be regulated by the entire EC or whether LUX was capable of regulating these genes in the absence of ELF3. Fig. 3A demonstrates that these genes have increased expression at night in both the elf3-1 and lux-4 backgrounds compared to Col-0, but that these genes have almost identical patterns of expression in elf3-1 and lux-4. This suggests that both LUX and ELF3 are required to regulate the circadian targets of the EC. Strikingly, even though the circadian targets have very distinctive gene expression patterns in Col-0 (Fig. 3B), they all have highly similar log-fold changes of expression in elf3-1 and lux-4 (Fig. 3A), suggesting that the EC provides the same degree of repression to all of its circadian targets. This indicates that the EC may act by a single mechanism, and that this repression is achieved independently of the other factors influencing expression of the target genes.
Fig. 3.
The genes encoding EC components show diurnal patterns of expression (Fig. S12). We therefore investigated whether the expression of EC components is sufficient to account for the degree of EC target repression. As gene expression networks often assume that the concentration of a transcription factor influences the rate of change of gene expression of its downstream targets, we generated a set of linear models to predict the slopes of Fig. 3Ai,iii based on the log-fold change in concentration of LUX, ELF3, or ELF4 (Fig. S13). If we assume that all three EC components are required for EC function, the ‘effective concentrations’ of LUX in elf3-1 and of ELF3 in lux-4 should then both be zero—models that incorporate this assumption are labeled as +EC in Fig. 3Ci. In all cases, models that incorporate this assumption perform better than ones that do not, further suggesting that both ELF3 and LUX must be present for the EC to serve as a repressor. The ELF4 and LUX+EC models were remarkably accurate—the Pearson’s R-values were less than -0.8 (Fig. 3Ci). Therefore, the amount of repression of EC circadian targets can be accurately predicted using only information about either LUX or ELF4 expression levels—an example of the correlation of the ELF4+EC model is shown in Fig. 3Cii. This suggests a quantitative model in which the concentration of the EC determines the rate of repression of the circadian targets—a mechanism that could explain the expression of circadian targets at night. It is notable that the expression pattern of ELF3 is very broad, with a much-reduced amplitude compared to that of ELF4 and LUX, which show a very sharp peak of expression at ZT8 (Fig. S12). Because ELF4 and LUX have very similar patterns of expression, we cannot determine which is more important for circadian regulation using the modeling framework in Fig. 3. Finally, it is important to note that while this model is sufficient to explain our observations, the model may not be predictive.
The EC controls the night-time warm temperature transcriptome
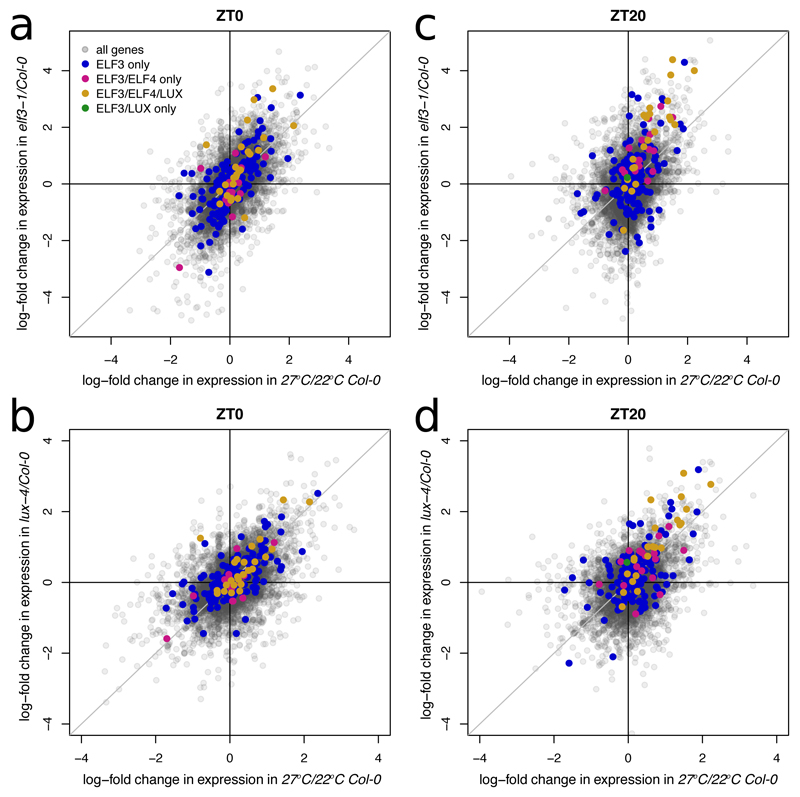
The EC has been shown to control warm temperature mediated induction of PIF4 and LUX15, and here we determined that the EC may also directly regulate key temperature response genes like CBFs. We therefore sought to see if the EC might play a global role in the warm temperature response. Comparing the transcriptomes of either elf3-1 or lux-4 with the warm temperature transcriptome reveals a significant positive correlation during the night (Fig. 4A,B, S14, see ‘Scatterplot Stats’ in Table S3 for p-value calculations). Interestingly, this correlation is stronger earlier in the night for those genes that are directly bound by the EC, while the correlation is strongest at the end of the night for all genes (Fig. S14). This suggests that the EC conveys temperature information to its direct targets at night, and that this signal cascades to a wider set of genes by dawn (Fig. 4C,D, S14).
Fig. 4.
The EC associates with phytochromes on target gene promoters
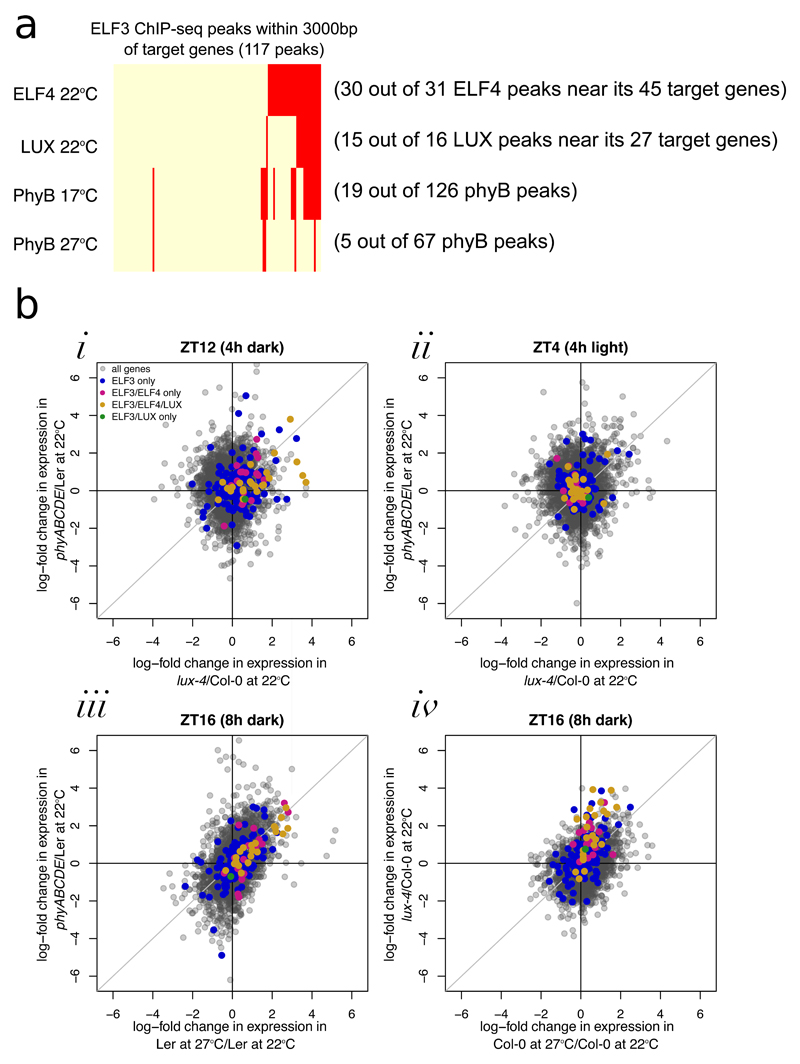
To understand how the EC might integrate temperature information, we investigated other proteins that have been demonstrated to interact with the EC31. Of these, phytochromes have recently been shown to function as thermosensors32,33, binding to target promoters in a temperature dependent manner. We therefore investigated whether the peaks bound by EC proteins are also bound by phytochromes. There is significant overlap between temperature responsive loci that are bound by two or more members of the EC and phyB (Fig. 5A, see ‘Fig5A’ sheet in Table S3 for p-value calculations, Table S8 for raw values, and Fig. S16 for specific examples). This suggests a mechanism whereby phytochromes may transmit light and temperature information directly to the evening expressed component of the clock.
Fig. 5.
Consistent with a direct role for phytochromes in controlling EC function, EC target genes have increased expression in phyABCDE at night, but not in the day (Fig. 5Bi,ii and S14). Furthermore, EC targets have similar expression in phyABCDE, elf3-1, and lux-4 at 22ºC compared to wild type, as they do when the temperature is raised to 27ºC. Phytochromes not only co-localise with the EC, but they can also change the expression pattern of EC targets in a similar way as a change in temperature (Fig 5Biii,iv and S14). A key question is whether the EC and phytochromes both relay temperature information, or whether a single component is sufficient. Since phyB is only detectable at a subset of the EC bound sites, and ELF4 targets do not completely lose their night-time temperature-dependent expression changes in either lux-4 or phyABCDE (Fig. S17), this suggests that both the EC and phytochromes contribute to the temperature transcriptome. A candidate for the transmission of this temperature signal in the EC is ELF3, since even in the absence of LUX, ELF3 binding responds to temperature (Fig. 1D).
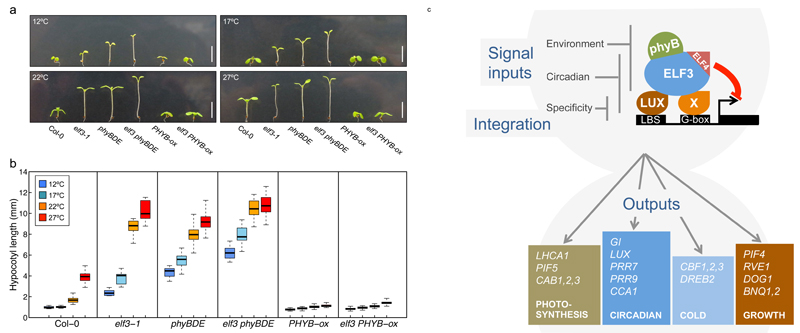
To understand the consequences and nature of the interactions between the EC and phytochromes with temperature, we analyzed hypocotyl elongation, a phenotypic parameter that is very responsive to temperature. elf3-1 phyBDE quadruple mutants display greater hypocotyl elongation than either elf3-1 or phyBDE seedlings, consistent with previous studies showing that phyB and elf3 exhibit additive effects34 (Fig. 6A,B). The difference between the elf3-1 and elf3-1 phyBDE decreased at higher temperatures, which is expected since higher temperatures reduce the activity of both ELF3 and phytochromes. Interestingly, overexpressing PHYB is sufficient to largely abolish both the effect of elf3-1 on hypocotyl elongation as well as thermal responsiveness. This is consistent with the role of phytochromes as thermosensors32,33, and indicates that a sufficient level of phyB Pfr (the active state of phyB) is sufficient block growth regardless of temperature, presumably at least in part through directly inhibiting the action of PIF4 and PIF5. Taken together, these results indicate that phytochromes and the EC bind to common promoter binding sites, directly relaying temperature information to genes involved in growth and development.
Fig. 6.
Discussion
The circadian clock allows plants to anticipate changes and coordinate multiple processes, including photosynthesis, growth and temperature responses with the environment. The EC, a core component of the circadian clock, has been shown to be important in coordinating responses such as growth, but the underlying mechanism of action has not been clear. We have found that the EC achieves specificity of target binding through a combinatorial mechanism, requiring both LBS and G-box motifs to be present at functional sites. Since none of the core EC components have been described as having G-box binding activity, this suggests an additional transcription factor plays a key role in EC specificity. Interestingly, the bHLH transcription factor PIF7, which recognises G-boxes, interacts with the EC in a phyB dependent manner31. Analysis of the elf3-1 and lux-4 transcriptomes enabled us to identify the set of high confidence direct EC functional target genes. This list reveals how extensively the EC is integrated into major transcriptional programmes in the cell, particularly photosynthesis, the circadian clock, growth, phytohormones and temperature signalling (Fig. 6B). Even in the lux-4 background, we observe reduced but significant binding of ELF3 to EC targets, supporting a role for the related TF, BROTHER OF LUX ARRHYTHMO (BOA) within the evening complex35. Finally, EC function may also be controlled by post-translational mechanisms.
Not only is the EC able to coordinate the circadian regulation of thousands of transcripts by repressing targets in the early evening, but it also transmits temperature information to these genes. Indeed, we see direct EC targets are globally up-regulated in elf3-1 and lux-4, resembling a warm grown plant. This signal propagates over the course of the night, to affect the global transcriptome. Strikingly, we observe that the recently described thermosensor phyB is recruited to many EC loci. This suggests a direct mechanism whereby phyB Pfr (which also acts as a transcriptional repressor) may directly provide temperature and light information to the EC. The recruitment of phyB to large transcriptional complexes has recently been observed at the FT locus36, suggesting this may represent a major mechanism for integrating environmental signals into gene expression. Consistent with previous observations34, it appears that the interactions between elf3-1 and phytochrome mutants are additive, and it is likely that additional temperature information is also transmitted by the EC. As well as co-binding to specific promoters, it is also apparent that the EC and phytochromes have independent functions, likely explaining their additive genetic interactions. It may seem contradictory that phytochromes and the EC interact physically, yet still display an additive phenotype. However, it is likely that the EC and phytochromes do not always function in complex—there are EC bound sites without phyB binding and vice versa. An intriguing line of future investigation would be to determine whether the EC and phytochromes influence each other’s binding pattern—do these two components genetically interact by affecting each other’s binding pattern or by affecting their activity? Environmental temperature is a variable signal, and it may be that measuring it independently over multiple time-scales and integrating this information enables more robust decision-making. The EC has been shown to be central in controlling key agricultural traits37, so understanding how this system transmits environmental information is particularly relevant during a period of climate change which threatens food security38.
Methods
Plant material and growing methods
The elf3-1 and lux-4 mutants have been described previously15. The phyBDE mutant in Columbia (Col) background was provided by P. Cerdán39. The PHYB-ox transgenic plant, in which the PHYB-GFP gene fusion is overexpressed in the phyB-9 background, was obtained from F. Nagy40. An initial ChIP-seq experiment was conducted on 35S::cELF3-HA— the CDS of ELF3 was cloned into pEG201 containing c-terminal HA tag and transformed into the elf3-1 line, as described in Box et al, 201615.The ELF3pro::ELF3-myc transgenic plant (ELF3-MYC) was used in the remaining ChIP-seq experiments, and was constructed by amplifying a 7.8 kb genomic fragment of ELF3 including its promoter with primers 6495 (5’-CACCGCTTCTTGTGTTAGTGACTTTCCTC) and 6497 (5’-AGGCTTAGAGGAGTCATAGCGTTTA). The PCR products were subcloned into pENTR vector (ThermoFisher) according to the manufacturer’s procedure. The resultant entry plasmid was recombined with LR clonase into the Gateway binary pJHA212K containing a C-terminal 5 copies of myc tag sequences. The binary construct was transformed into the elf3-1 mutant by floral dipping method. The ELF3-MYC transgenic plant was isolated by kanamycin selection and propagated to obtain single insertion lines rescuing the long hypocotyl phenotype of elf3-1. The complementation experiment for the ELF3-MYC line is shown in Fig. S2. The LUXpro::LUX-GFP lux-4 (LUX-GFP) and ELF4pro::ELF4-HA elf4-2 (ELF4-HA) transgenic lines used for ChIP-seq experiments have been described previously7. The LUX-GFP elf3-1 and ELF3-MYC lux-4 were generated by crossing the LUX-GFP to the elf3-1 and the ELF3-MYC to the lux-4, respectively. Their homozygous F3 generations were used for ChIP-seq experiments. Arabidopsis seeds were sterilized and sown on ½ X Murashige and Skoog-agar (MS-agar) plates at pH 5.7 without sucrose. Sterilized seeds were stratified for 3 days at 4ºC in the dark and allowed to germinate for 24 hours at 22ºC under cool-white fluorescent light at 170 μmol/m2s. The plates were then transferred to short-day conditions (8-h light and 16-h dark) at different temperatures for assays.
ChIP-seq
Please note that the phyB ChIP-seq data used in Figure 4 are previously published32. All ChIP-seq raw data is deposited in the NCBI Sequenced Read Archive (SRA) in PRJNA384110. The primary set of ChIP-seq experiments came from growing seedlings under short day (SD) conditions (8 hours light, 16 hours dark) on ½ MS agar plates without sucrose at constant 22ºC with samples collected at ZT10 (2 hours after darkness). These are the primary ChIP-seq experiments analyzed throughout the paper and there is only 1 replicate (Fig. 1-4, S3C [ZT10], S7-10,13,14,16,17).
In order to determine whether temperature affected the stability of LUX, a temperature shift experiment was conducted under SD conditions in MS agar plates, in which plants were grown at 17ºC for 11 days, and then a subset of these plants were shifted to 27ºC at the end of the light period (ZT8) and collected after 1h or 2h (ZT9 or ZT10), and control (constant 17ºC) plants were also collected from the same time points. Two replicates were conducted for this experiment. The experimental design of the temperature shift experiment is shown in Fig S1A and the results are in S1B. The seedlings grown at constant 17ºC and collected at ZT10 are the ones shown in Fig. 1B, 2A,B, S3, S10).
Also, to determine whether temperature affected the stability of ELF3, a temperature shift experiment was conducted in ELF3-MYC transgenic plants under SD conditions in ½ MS agar plates in which plants were grown at 22ºC at ZT8, and then a subset of these plants were shifted to 8ºC, 12ºC, 17ºC, 20ºC, 27ºC, or 32ºC at ZT10 (Fig. 1C).
Next, to evaluate the consistency of Evening Complex binding sites across the time points of the RNA-seq experiment, a ChIP-seq was conducted on ELF3-MYC in SD conditions at constant 22ºC with plants harvested at ZT0, 4, 8, 12, 16, and 20. In addition, the LUX-GFP ChIP-seq was done for samples collected at ZT8 and ZT12 that were grown at constant 22ºC. Only one replicate was performed. The results are shown in Fig. S3C,D.
Finally, to determine if LUX-GFP is capable of binding to DNA independent of ELF3, a LUX-GFP ChIP-seq was conducted in an elf3-1 background at ZT8—only one replicate was performed, and these results are shown in Fig. S4. Similarly, an ELF3-MYC ChIP-seq was performed in lux-4 at ZT10 with one replicate (Fig. 1D).
In all of these ChIP-seq experiments, 3 g seedlings for each treatment were fixed under vacuum for 20 min in 1xPBS (10 mM PO43−, 137 mM NaCl, and 2.7 mM KCl) containing 1% Formaldehyde (F8775 SIGMA). The reaction was quenched by adding glycine to a final concentration of 62 mM. Chromatin immunoprecipitation (ChIP) was performed as described41, with the exception that 100 µl of anti-c-Myc agarose affinity gel antibody was used (A7470 SIGMA-Aldrich ®) per sample for seedlings expressing the appropriate epitope tagged protein, Monoclonal Anti-HA-Agarose (A2095 SIGMA-Aldrich ®) for ELF4-HA seedlings, or GFP antibody from Abcam (ab290) with Dynabeads® Protein A and G were used for Immunoprecipitation of LUX-GFP seedlings. Sequencing libraries were prepared using TruSeq ChIP Sample Preparation Kit (Illumina IP-202-1024) or using NEBNext® Ultra™ II DNA Library Prep Kit and samples were sequenced on either the Illumina HiSeq or the Illumina NextSeq 500 platforms, as indicated in our SRA submission.
ChIP-seq bioinformatics processing
A standard ChIP-seq bioinformatics pipeline was used to process the fastq files: reads were mapped to the TAIR10 Arabidopsis genome using bowtie242, duplicate reads were removed, the reads were sorted and indexed using samtools43, and finally the read counts were normalized to the genome-wide coverage. This pipeline is available on Github at https://github.com/ezer/EC.
Peaks were identified using MACS244, using matching INPUT control samples. We used the following macs2 callpeak parameters for each ChIP-seq sample: “--keep-dup 1 --nomodel -p 0.05 --extsize <predictd>”, where <predictd> is the estimated fragment size (using the macs2 predictd command). The returned peaks were further filtered using the following criteria: fold-change > 5 (for 17ºC LUX data) or fold-change >3 (for everything else), and qvalue < 0.001. The 17ºC LUX data had many more peaks identified than the 22ºC even with the more stringent threshold for fold-change.
The Venn diagram for peak overlap was calculated using bedtools: ‘merge’ was used to create a single bed file, and each bed file was intersected to this45 (see Fig. S18). There are fewer peaks in the Venn diagram than appear in our tables—if we observe two peaks of one transcription factor overlapping with a single peak in the other transcription factor, we will consider this to be a single peak overlapping event.
De novo motifs were predicted using Homer246, using permuted sequence as background.
RNA-seq experiment
The Col-0, elf3-1, and lux-4 backgrounds were used for time-course RNA-seq experiments. Seedlings of the indicated genotypes were grown for 7 days at 22ºC and 27ºC and sampled at intervals over the diurnal cycle: ZT = 0, 1, 4, 8, 12, 16, 20 and 22 h. Total RNA was isolated from 30 mg of ground seedlings using the MagMAX-96 Total RNA Isolation kit (Ambion, AM1830), following the manufacturer’s instructions. RNA quality and integrity was assessed on the Agilent 2200 TapeStation. Library preparation was performed using 1 μg of high integrity total RNA (RIN>8) using the TruSeq RNA Library Preparation Kit v2 (Illumina, RS-122-2101 and RS-122-2001), following the manufacturer’s instruction. The libraries were sequenced on a HiSeq2000 using paired-end sequencing of 100 bp in length at the Beijing Genomics Institute (BGI) sequencing center.
The same pipeline was used to map these sequences as described in previously32, with the exception that the sequences were mapped to the TAIR10 genome. To analyze the sequence reads: First, adapters were trimmed with Trimmomatic-0.3247. Then, Tophat48 was used to map to the TAIR10 annotated genome, duplicates were removed and the read counts were normalized by genome-wide coverage. Raw counts were determined by HTseq-count49, and cufflinks was used to calculate Fragments Per Kilobase Million (FPKM), which was then converted into Transcripts Per Million (TPM). The RNA-seq data using the phyABCDE and Ler backgrounds, analyzed in Fig. 4, were published previously32.
All RNA-seq raw data is deposited in the NCBI SRA (PRJNA384110).
Identification of predicted target genes
All scripts used to identify predicted target genes are available in: https://github.com/ezer/EC. We predicted four distinct lists of prospective target genes (for LUX, ELF3, and ELF4 at 22ºC and for LUX at 27ºC) using the following criteria: i) TAIR10 annotated genes that are within 3000bp of ChIP-seq peaks using bedmap. ii) and that are within the top 5% most significant differentially expressed genes in at least one time point in either elf3-1 vs. Col-0 or lux-4 vs. Col-0, which was calculated using the edgeR50 package, using raw read counts and the exactTest with dispersion 0.1.
Next, we could use this list of predicted target genes to annotate some of the ChIP-seq peaks as “high confidence peaks”. These were peaks that were within 3000bp of genes that were on the prospective target gene lists. This strategy helped us filter out many of these peaks, which are likely false positives. GO analysis of our lists of predicted target genes was conducted using Panther51 and goatools52.
Analysis and modelling of log-fold change in expression
In all figures, log-fold change in expression refers to the natural logarithm of the transcripts per million (TPM) of the designated sample, minus the natural logarithm of the TPM in the associated wild type at 22ºC at the same time point. The associated wild type for elf3-1 and lux-4 is Col-0, while the associated wild type for phyABCDE is Ler. For instance, the log-fold change of expression in lux-4 can be expressed as: ln(TPMlux-4)-ln(TPMCol-0).
All of the plots related to log-fold change in expression were drawn in R. All of the hierarchical clustering uses default parameters from heatmap.2. Note that the heatmap in Fig. S11 uses z-scores of the TPM, rather than log-fold change in expression.
We also attempt to determine whether it is possible to predict the rate of repression of the EC from the expression of EC components—the problem statement is described in more depth in Fig. S13. The models were fit by least-squares regression (lm in R). Note that since the slope is calculated across two points and there are four time course experiments with eight time points each, there are a total of 28 data-points (7 slopes per time-course). Since this is a small number of points, we decided that each model should not have more than one fitted parameter, to decrease the likelihood of over-fitting our data. Also note that for each time course experiment there are 8 collection time points (and therefore 8 LUX, ELF4, or ELF3 concentrations per time course), but only 7 slopes: because of this, we chose to take the average TPM value between every consecutive pair of collection points.
Hypocotyl length measurements
To examine how phytochrome and EC signalling pathways interact for the thermal regulation of hypocotyl elongation, the phyBDE, and PHYB-ox were crossed with elf3-1 and the resultant homozygous F3 generations were used for hypocotyl length measurements at different temperatures, as described previously15. Seedlings, which were grown for 8 days under SD conditions with light intensity of 80 μmol/m2s, were photographed and analyzed using ImageJ software (http://rsbweb.nih.gov/ij/).
Western blots
Seedlings were surface sterilized and grown on plates (0.5 x Murashige and Skoog, 0.7% agar) for 10 days under short day conditions (8 h light/16 h dark) in Panasonic growth chambers. Proteins were extracted and separated by SDS-PAGE, and transferred to PVDF membrane using Trans-Blot Turbo Transfer System (Bio-Rad). ELF4 was detected using HA antibody coupled to horse radish peroxidase (HRP) (1:500, 12013819001 Roche), LUX using mouse anti-GFP (1:1000, 632380, Clontech Laboratories) and ELF3 using mouse anti-myc (1:1000, 05-724, Millipore), and secondary goat derived anti-mouse conjugated to HRP (1:3000, sc-2005, Santa Cruz Biotechnology). PVDF membranes were developed using Pierce ECL western blotting substrate (32106, Thermo Scientific), and scanned using an Amersham Imager 600 (GE Healthcare Life Sciences). Loading controls were obtained by staining membranes with Ponceau S solution (P7170, Sigma).
Detailed experimental set up is described below for each line used. (A), a) The LUX-GFP elf3-1 transgenic seedlings were grown continuously at 22 °C or 27 °C and harvested at ZT12. Aliquots for Western blot analysis were taken during Chromatin Immunoprecipitation (ChIP) preparation; b) The LUX-GFP transgenic seedlings were grown at 17 ºC and harvested at ZT8 or for second collection point seedlings were shifted on rafts to 27ºC or kept at 17ºC and harvested at ZT10. Nuclei enriched samples were prepared as for ChIP and loaded with SDS buffer. (B), The ELF3-MYC and ELF3-MYC lux-4 transgenic seedlings were grown at 22ºC, sampled and shifted at ZT8 to 27ºC or kept at 22ºC and sampled at ZT10. Aliquots for Western blot analysis were taken during chromatin IP preparation. (C), The ELF4-HA transgenic seedlings were grown at 22ºC and shifted at ZT8 to 27ºC or kept at 22°C and sampled at ZT10 and whole plant extract was grinded in SDS buffer and supernatant loaded. (D), The PHYBpro::PHYB-myc phyB-9 (PHYB-MYC) transgenic seedlings were grown at 17ºC and harvested at ZT8, for second collection points seedlings were shifted on rafts to 27ºC or kept at 17ºC and harvested at ZT10 and whole plant material after grinding was boiled in SDS buffer and supernatant loaded. (E), The ELF3-MYC transgenic seedlings were grown at 17ºC and harvested at ZT8, for second collection points seedlings were shifted on rafts to 27ºC or kept at 17ºC and harvested at ZT10 and whole plant extract was boiled in SDS buffer and the supernatant loaded. Col-0 was added as negative control in all western blots.
Statistical tests
In all cases where statistical tests are used in the text, a Fisher exact test is applied using R and either a Bonforroni or Holms-Bonferroni correction for multiple hypothesis testing is used as described in Table S3.
Supplementary Material
Fig. 7.
Acknowledgements
We thank members of the Wigge laboratory for feedback and discussions. This work was supported by the Biotechnology and Biology Research Council [RG80054 to P.A.W.]; P.A.W’s laboratory is supported by a Fellowship from the Gatsby Foundation [GAT3273/GLB]. Funding for open access charge: [Gatsby Foundation/GAT3273/GLB]. We thank Steve Kay for providing us with the gLUX-GFP lux-4 and gELF4-HA elf4-2 transgenic plants.
Footnotes
Data Availability
All data is available in SRA (PRJNA384110). All code used to generate the figures is available on github (https://github.com/ezer/EC).
Author Contributions
Daphne Ezer. Wrote a large proportion of the manuscript, lead researcher on all analysis. Involved in experimental design, prepared Figures 1, 2, 3 and 4.
Jae-Hoon Jung. Experimental design, generated Fig. 5, created lines used in the study.
Hui Lan. Extensive Bioinformatics analysis: mapped and analysed most of the data sets for the ChIP experiments. Performed motif searching etc.
Surojit Biswas. Performed the initial mapping of the first ELF3 ChIP and made insights into rhythmical gene expression using clustering that were instrumental in the development of the project.
Laura Gregoire. Collaborating group: performed the first analysis of EC binding and motif analysis.
Mathew S. Box. Generated the RNA-seq time course datasets.
Varodom Charoensawan. Generated the RNA-seq time course datasets.
Sandra Cortijo. Generated the RNA-seq time course datasets.
Dorothee Stöckle. Helped perform ChIP-seq experiments
Chloe Zubieta. PI. Collaborator, supervisor of Laura Gregoire, made key structural biology and experimental design contributions. Helped write the paper.
Katja E. Jaeger. PI. Performed all the ChIP-seq experiments the paper is based on. Writing the paper and experimental design.
Philip A. Wigge. PI. Experimental design and discussions, helped write the paper.
References
- 1.Willis CG, Ruhfel B, Primack RB, Miller-Rushing AJ, Davis CC. Phylogenetic patterns of species loss in Thoreau’s woods are driven by climate change. Proc Natl Acad Sci U S A. 2008;105:17029–17033. doi: 10.1073/pnas.0806446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitter AH, Fitter RS. Rapid changes in flowering time in British plants. Science (80-.) 2002;296:1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- 3.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 4.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 5.Huang H, Nusinow DA. Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 2016 doi: 10.1016/j.tig.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 2015;16:598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 7.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thines B, Harmon FG. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci U S A. 2010;107:3257–3262. doi: 10.1073/pnas.0911006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filo J, et al. Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5. Plant Signal Behav. 2015;10:e992707. doi: 10.4161/15592324.2014.992707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 11.Kumar SV, et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thines BC, Youn Y, Duarte MI, Harmon FG. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J Exp Bot. 2014;65:1141–51. doi: 10.1093/jxb/ert487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández V, Takahashi Y, Le Gourrierec J, Coupland G. Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J. 2016;86:426–440. doi: 10.1111/tpj.13183. [DOI] [PubMed] [Google Scholar]
- 14.Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat Plants. 2016;2:16055. doi: 10.1038/nplants.2016.55. [DOI] [PubMed] [Google Scholar]
- 15.Box MS, et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol. 2015;25:194–9. doi: 10.1016/j.cub.2014.10.076. [DOI] [PubMed] [Google Scholar]
- 16.Raschke A, et al. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol. 2015;15:197. doi: 10.1186/s12870-015-0566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helfer A, et al. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signaling & Behavior. 2012;7:170–173. doi: 10.4161/psb.18766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu C-P, Lin J-J, Li W-H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci Rep. 2016;6:25164. doi: 10.1038/srep25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Box MS, et al. ELF3 Controls Thermoresponsive Growth in Arabidopsis. Curr Biol. 2014;25:194–199. doi: 10.1016/j.cub.2014.10.076. [DOI] [PubMed] [Google Scholar]
- 21.O’Malley RC, et al. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuno T, et al. Ambient Temperature Signal Feeds into the Circadian Clock Transcriptional Circuitry Through the EC Night-Time Repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014;0:1–19. doi: 10.1093/pcp/pcu030. [DOI] [PubMed] [Google Scholar]
- 23.Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends Plant Sci. 2014;19:240–9. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci U S A. 2001;98:2053–8. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng N-H, Liu J-Z, Brock A, Nelson RS, Hirschi KD. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem. 2006;281:26280–8. doi: 10.1074/jbc.M601354200. [DOI] [PubMed] [Google Scholar]
- 26.Chow BY, et al. Transcriptional Regulation of LUX by CBF1 Mediates Cold Input to the Circadian Clock in Arabidopsis. Current Biology. 2014;24 doi: 10.1016/j.cub.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai M-Y, Fan M, Oh E, Wang Z-Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 2012;24:4917–29. doi: 10.1105/tpc.112.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell. 2012;24:4483–97. doi: 10.1105/tpc.112.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawat R, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci U S A. 2009;106:16883–8. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nitschke S, et al. Circadian Stress Regimes Affect the Circadian Clock and Cause Jasmonic Acid-Dependent Cell Death in Cytokinin-Deficient Arabidopsis Plants. Plant Cell. 2016;28:1616–39. doi: 10.1105/tpc.16.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang H, et al. Identification of Evening Complex Associated Proteins in Arabidopsis by Affinity Purification and Mass Spectrometry. Mol Cell Proteomics. 2016;15:201–217. doi: 10.1074/mcp.M115.054064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung J-H, et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- 33.Legris M, et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science (80-.) 2016;354 doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- 34.Reed JW, et al. Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 2000;122:1149–60. doi: 10.1104/pp.122.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai S, et al. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell. 2011;23:961–72. doi: 10.1105/tpc.111.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiserli E, et al. Integration of Light and Photoperiodic Signaling in Transcriptional Nuclear Foci. Dev Cell. 2015;35:311–21. doi: 10.1016/j.devcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendix C, Marshall CM, Harmon FG. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol Plant. 2015;8:1135–52. doi: 10.1016/j.molp.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science (80-.) 2009;323:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- 39.Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci U S A. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medzihradszky M, et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell. 2013;25:535–44. doi: 10.1105/tpc.112.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25:820–33. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang H, et al. GOATOOLS: Tools for Gene Ontology. doi: 10.5281/ZENODO.31628. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.