Abstract
Purpose
Next-generation sequencing (NGS) identifies potentially targetable alterations by US Food and Drug Administration (FDA)–approved drugs and/or by available experimental agents that may not have otherwise been contemplated. Many targeted drugs have been developed for diverse solid cancers; a smaller number of genomically targeted drugs have been approved for lymphoid malignancies.
Materials and Methods
We analyzed NGS results from 60 patients with various lymphoid malignancies and found 224 alterations (median per patient, three alterations).
Results
Forty-nine patients (82%) had potentially actionable alterations with the use of FDA-approved drugs and/or experimental therapies; only 11 patients (18%) had no theoretically actionable alterations. Only three patients (5%) had an alteration for which an approved drug in the disease is available (on label); 45 patients (75%) had an alteration for which an approved drug is available for another disease (off label). The median number of alterations per patient potentially actionable by an FDA-approved drug was one. Of note, 19 (32%) of 60 patients had intermediate to high tumor mutational burden, which may predict response to certain immunotherapy agents.
Conclusion
NGS identifies alterations that may be pharmacologically tractable in most patients with lymphoid malignancies, albeit with drugs that have usually been developed in the context of solid tumors. These observations merit expanded exploration in the clinical trials setting.
INTRODUCTION
The lymphoid malignancies have diverse biologic and clinical behavior and typically are treated with multiagent chemotherapy. Many therapeutic regimens for B-cell non-Hodgkin lymphomas and leukemias also incorporate the anti-CD20 monoclonal antibody rituximab, which has improved patient outcomes.1 Treatment of metastatic solid tumors, like lymphoid malignancies, has also relied heavily on the use of cytotoxic chemotherapy. However, over the past decade, the treatment paradigm for metastatic solid tumors has shifted away from chemotherapy toward matching oncogenic driver mutations with targeted therapy (precision medicine).2-4 For example, in patients with BRAF-mutated metastatic melanoma, the BRAF inhibitor vemurafenib markedly increases overall survival compared with chemotherapy.5 Numerous targeted therapies are now approved by the US Food and Drug Administration (FDA) for patients with metastatic solid tumors across a wide array of histologies (Data Supplement). Occasionally, targetable alterations are found that change the treatment paradigm for a disease, which is notable for solid tumors such as EGFR-mutated non–small-cell lung cancer. The first example of targeted therapeutic efficacy is the hematologic disorder chronic myelogenous leukemia, which has been transformed by the use of agents that affect Bcr-Abl kinase activity.
Despite the rapid success in development, approval, and use of small-molecule–targeted agents in patients with solid malignancies, a paucity of such therapies approved for use in lymphoid malignancies remains (Data Supplement). For many patients with lymphoid malignancies, oral small-molecule–targeted therapeutics are not an available option.
The rapid technological advances in next-generation sequencing (NGS) have allowed oncologists to sequence tumor exomes in a clinically meaningful period of time.6-8 Some studies have shown that patients with solid malignancies treated with matched therapy have improved outcomes,9 and meta-analyses in approximately 85,000 patients have supported this finding.10-12 The targeting of alterations such as BRAF can result in responses across a wide variety of cancers, including lymphoid malignancies (eg, hairy cell leukemia).13,14 Although not all malignancies that harbor BRAF mutations will respond equally well to BRAF inhibition, the strategy of cross-cancer basket trials has been established as highly worthwhile.
NGS accurately identifies substitutions, indels, copy number alterations, and gene fusions in hematologic malignancies.15 In this report, we use this technology to analyze the genomic alterations in a cohort of 60 patients with various lymphoid malignancies to estimate the frequency of theoretically actionable alterations. These results may help to inform further development of clinical trials in this field.
MATERIALS AND METHODS
Patients
We retrospectively reviewed the medical charts of 220 patients with hematologic malignancies who had undergone NGS. Only patients with lymphoid malignancies were selected for additional review. Patients were seen at the University of California San Diego Moores Cancer Center from October 2012 until March 2016. This study was performed and consents obtained in accordance with University of California San Diego institutional review board guidelines.
NGS
Tumor samples from tissue (Table 1) or peripheral blood were collected from 60 patients and submitted for NGS to Foundation Medicine, a clinical laboratory improvement amendments–certified laboratory for NGS. The FoundationOne Heme panel was used, which is a hybrid capture–based NGS test.16 The methods used in this assay have been described in detail in previous reports.7,15 The FoundationOne Heme assay simultaneously detects all genomic alterations, including base pair substitutions, indels, copy number alterations, and select gene rearrangements, in 405 cancer-related genes. For tumor mutational burden (TMB), the number of somatic mutations detected on NGS are quantified, and that value was extrapolated to the whole exome by using a validated algorithm described in detail in earlier publications.17,18 Alterations with known and likely effects on functional status are not counted.
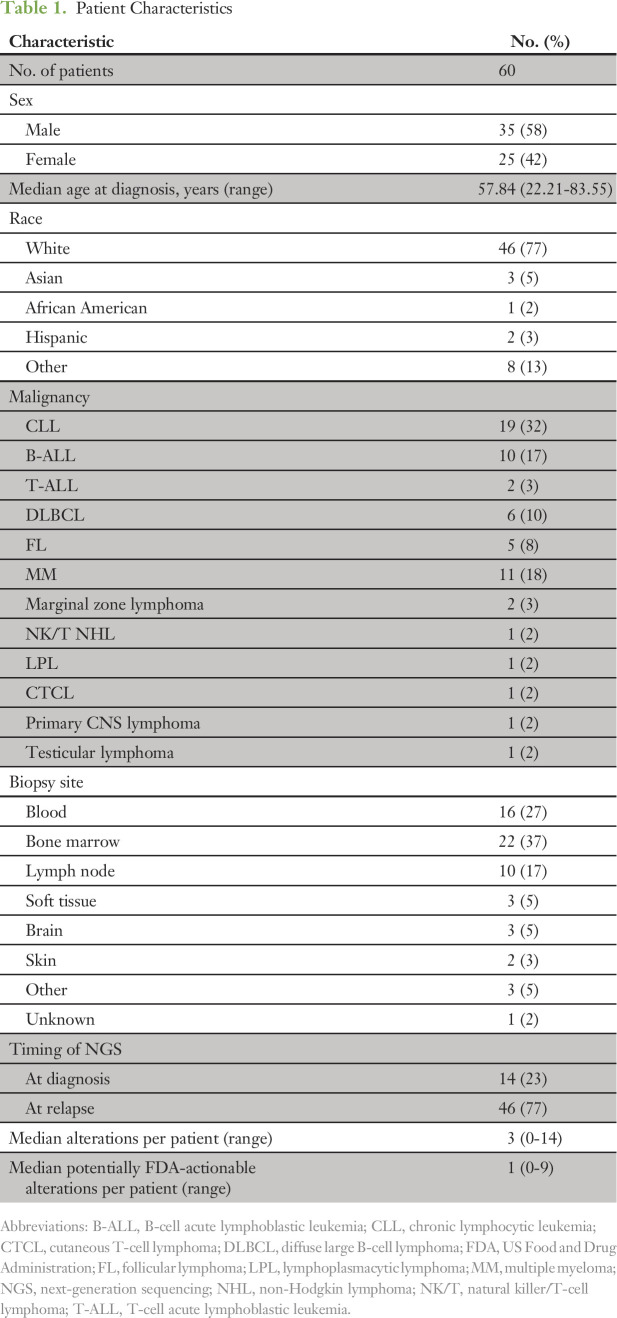
Table 1.
Patient Characteristics
Definition of Actionable Alteration
An alteration was defined as potentially actionable if its protein product is a component of a molecularly defined pathway for which there is at least one available FDA-approved drug or investigational drug that may affect the function of the protein product of the alteration or the immediate downstream effectors of the protein product or that differentially recognizes the protein in tumor versus normal cells. The protein products of genomic alterations were considered to be functional if the genomic alterations have been previously identified as relevant to cancer in the COSMIC database,19 which catalogs recurrent somatic alterations in cancer. Novel base substitution, indel, and rearrangement alterations that result in truncations and homozygous copy number deletions that occur in tumor suppressor genes were considered to have likely functional implications. Novel genomic alterations that occur at the same position as known alterations as well as alterations with conflicting evidence with regard to implication for function were subject to review by an internal panel of subject matter experts to determine functional status of the relevant protein product on the basis of all available evidence, including, but not limited to, the ExAC, dbSNP, and ClinVar databases.20-22
Data and Statistical Analyses
Patient characteristics were obtained through electronic medical record review. Descriptive statistics were used, including medians, ranges, and frequencies.
RESULTS
Patient Characteristics
Sixty patients (35 men [58%] and 25 women [42%]) with lymphoid malignancies were identified (Table 1). Forty-six patients (77%) were white. The most common malignancy in the cohort was chronic lymphocytic leukemia (CLL; 32%), followed by acute lymphoblastic leukemia (ALL; 20%), multiple myeloma (18%), diffuse large B-cell lymphoma (DLBCL; 10%), follicular lymphoma (8%), and other lymphoid neoplasms (5%). The most common site for obtaining tissue used for NGS was bone marrow (37%) followed by peripheral blood (27%) and lymph nodes (17%).
NGS Results
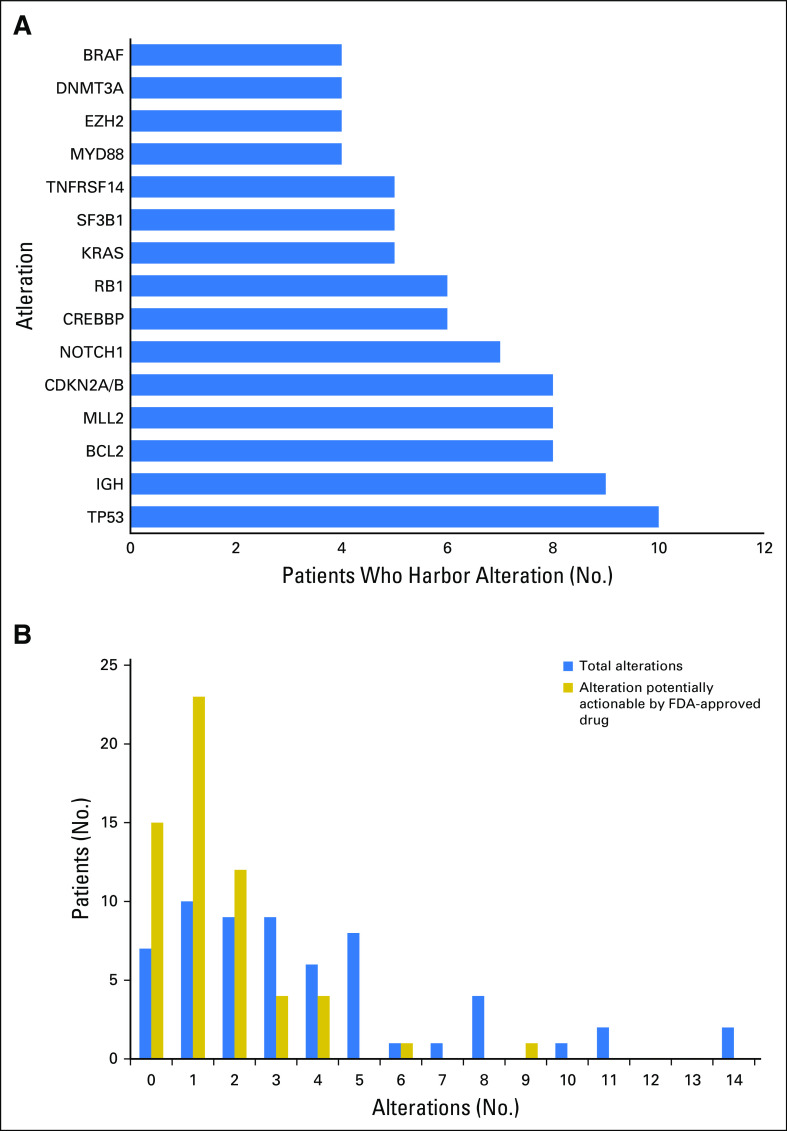
Two hundred twenty-four alterations were identified by NGS in the entire cohort of 60 patients. Types of alterations identified were substitutions, indels, copy number alterations, and gene fusions. Fig 1A shows the 15 most frequent alterations among the cohort, with TP53 mutations (10 patients), IGH translocations (nine patients), loss of CDKN2A/B (eight patients), and BCL2 mutations (eight patients) within the top five. All alterations identified in the cohort are listed in the Data Supplement.
Fig 1.
(A) Frequency and type of various molecular alterations found among 60 individuals with lymphoid malignancies. Only the 15 most common alterations are shown. The full list of alterations is shown in the Data Supplement. (IGH refers to IGH translocations that involve a partner gene.) (B) The number of patients with the designated number of total alterations and the number of patients with the designated number of potentially actionable alterations by an US Food and Drug Administration (FDA) –approved drug (on or off label).
All patients but two had a unique portfolio of alterations. One patient with CLL and one with multiple myeloma each had a solo NRAS mutation. However, the actual alteration in NRAS differed between the two patients (NRAS G13D v NRAS Q61R).
The median number of alterations detected per patient was three (range, zero to 14). As demonstrated in Figure 1B, seven patients (12%) had no reportable alterations, 10 (17%) had one alteration, and 43 (71%) had two or more alterations. The maximum number of alterations identified was 14, which was observed in two patients (3%), one with CLL, the other with DLBCL. Of note, all patients with DLBCL had five or more alterations.
Actionable Alterations
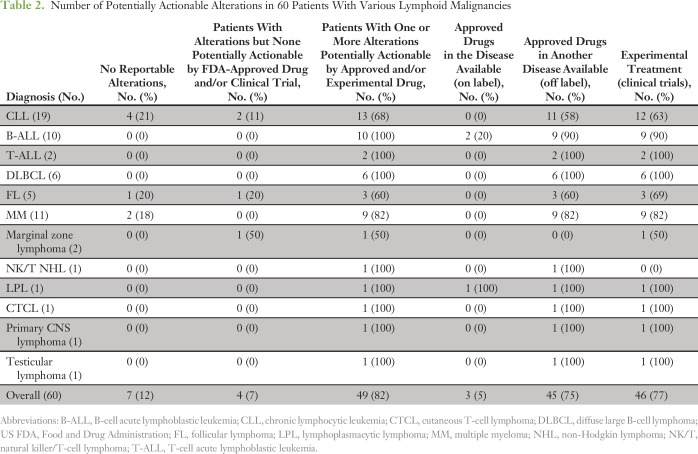
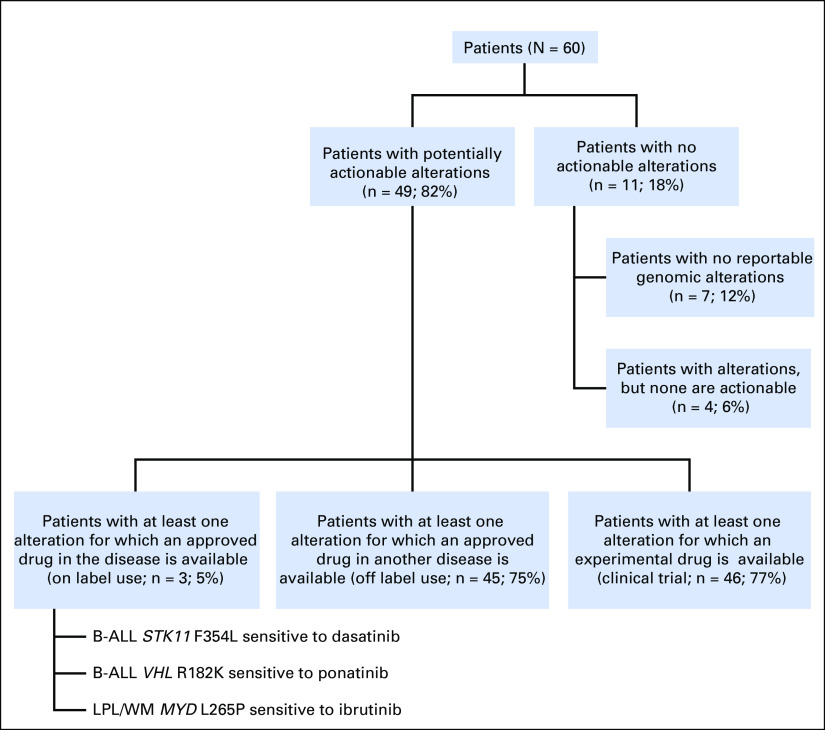
Potentially actionable alterations were identified in all disease subtypes (Table 2). All disease subtypes, except marginal zone lymphoma, had alterations targetable by FDA-approved drugs. Forty-nine patients (82%) had potentially actionable alterations with FDA-approved drugs and/or experimental therapies (clinical trials), whereas 11 patients (18%) had no theoretically actionable alterations.
Table 2.
Number of Potentially Actionable Alterations in 60 Patients With Various Lymphoid Malignancies
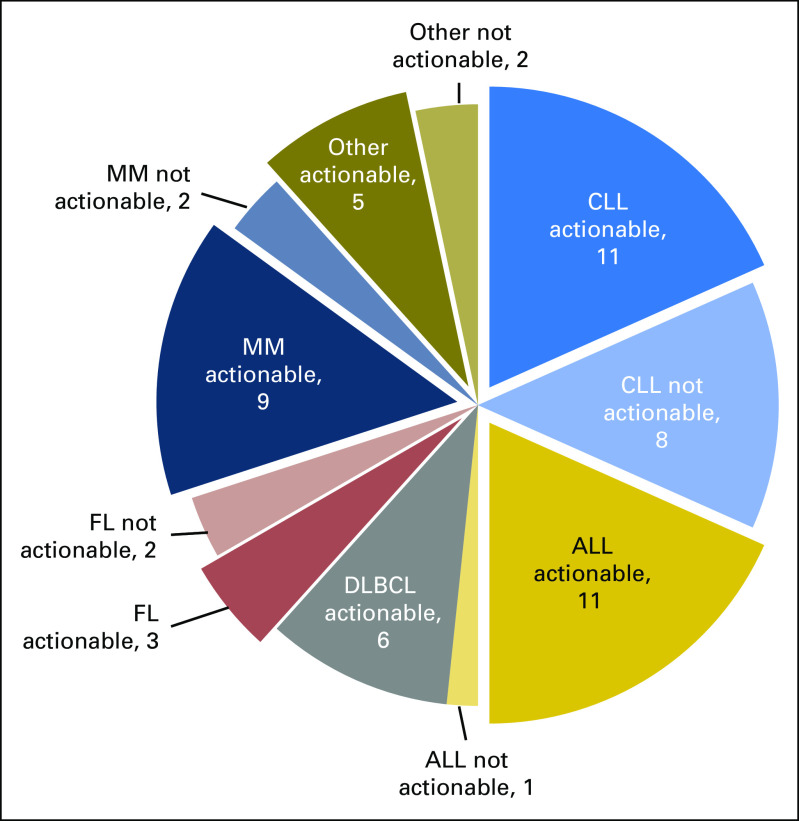
Depicted in Figure 2 is the number of patients with potentially FDA actionable alterations per disease group. For example, 11 (92%) of 12 patients with ALL and 11 (58%) of 19 patients with CLL had at least one theoretically targetable alteration. All patients with DLBCL had targetable alterations. The median number of potential FDA actionable alterations detected per patent was one. Twenty (33%), 15 (25%), and 10 (17%) patients had one, two, and three or more hypothetically targetable alterations, respectively.
Fig 2.
Number of patients with and without potentially US Food and Drug Administration–actionable alterations in each malignancy type. ALL, acute lymphoblastic leukemia; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; MM, multiple myeloma.
Figure 3 demonstrates that of the 49 patients (82%) with potentially targetable mutations, 45 had alterations that were targetable by FDA-approved drugs. Only three patients had an alteration for which an approved drug in the disease is available (on label), whereas 45 patients (75%) had an alteration for which an approved drug is available in another disease (off label). Forty-six patients (77%) had at least one alteration theoretically targetable by an experimental therapy (clinical trial). Eleven patients had no targetable alterations; these patients included seven with no detected genomic alterations.
Fig 3.
Breakdown of patients with potentially actionable alterations. B-ALL, B-cell acute lymphoblastic leukemia; LPL, lymphoplasmacytic lymphoma; WM, Waldenström macroglobulinemia.
Two patients with B-cell ALL (B-ALL) and one with Waldenström macroglobulinemia had alterations targetable with on-label–approved drugs (Fig 3). One patient with B-ALL was Philadelphia chromosome positive and already receiving dasatinib before testing, whereas the patient with Waldenström macroglobulinemia was switched to ibrutinib upon finding the MYD88 L265P mutation.
TMB
TMB ranges in a large cohort were defined as one or fewer to five (low), six to 19 (intermediate), and 20 or more (high) mutations per megabase of sequenced DNA.23 In the current cohort, TMB ranged from one or fewer to 140 (Data Supplement). High TMB was seen in five patients (8%), intermediate in 14 (23%), and low in 40 (67%). TMB was not available for one patient (2%). Intermediate to high TMB was noted across almost all histologies (except for marginal zone lymphoma, natural killer/T-cell lymphoma, and cutaneous T-cell lymphoma). One patient with CLL had a TMB of 140, whereas two patients with DLBCL had a TMB of 20. Two patients with B-ALL had an intermediate level of TMB (both 11). Three patients with multiple myeloma had an intermediate level of TMB.
DISCUSSION
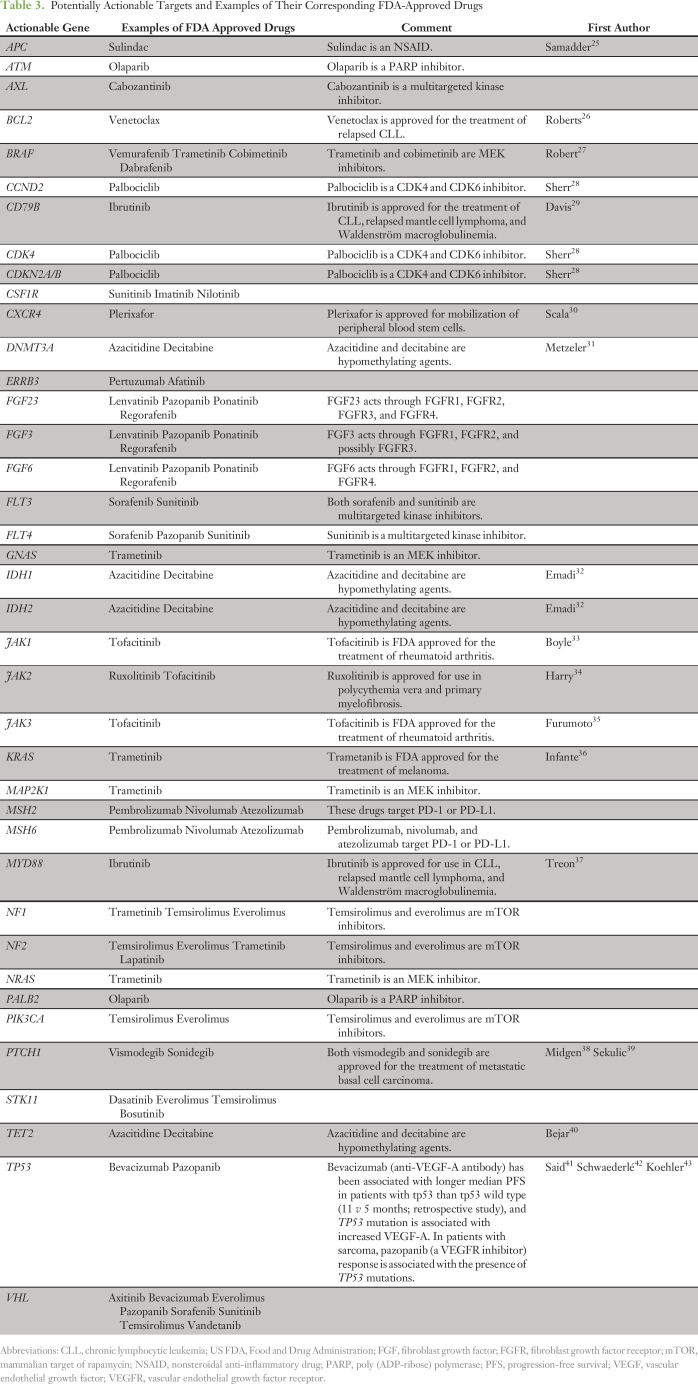
This study demonstrates that in the majority of patients (82%) with lymphoid malignancies whose disease was interrogated by NGS, alterations were found that might be pharmacologically tractable, a percentage similar to the 77% reported by He et al15 for diverse hematologic malignancies. Another study reported that 70% of patients with various solid tumors had an alteration that was theoretically actionable with an approved drug.24 This finding is similar for lymphoid malignancies, with 75% of the current study patients having an alteration potentially actionable by an approved drug. Table 3 lists therapeutics with their corresponding targets that were identified in the patient cohort.
Table 3.
Potentially Actionable Targets and Examples of Their Corresponding FDA-Approved Drugs
One of the major obstacles to performing comprehensive genomic profiling of clinical specimens is the necessity for an adequate tumor sample. Unlike solid malignancies where invasive biopsy is almost always required to obtain a tissue sample, comprehensive genomic profiling of lymphoid malignancies often can be performed with the use of peripheral blood and/or bone marrow. In this cohort, 54% of patients had specimens obtained from blood and/or bone marrow.
Tumors can acquire new mutations as they progress, which underscores the importance of obtaining new tissue when available for sequencing at the time of therapeutic decision making. For instance, the hallmark of chronic myelogenous leukemia is accumulation of genomic alterations with disease progression.44 Similarly, patients with lung cancer and EGFR mutations that are sensitive to first-generation EGFR inhibitors will acquire secondary genomic alterations in EGFR (eg, EGFR T790M) that are resistant to these agents but sensitive to third-generation inhibitors.45 In solid malignancies, NGS often is performed on tissue available from diagnosis to avoid repeat biopsy. In one study, 42% of patients did not have metastatic disease at the time of their biopsy.24 Lymphoid malignancies are more amenable to repeat biopsy, with more than one half of the current cohort having tissue available from the blood and/or bone marrow. In addition, 77% of patients had NGS performed on tissue from relapse as opposed to diagnosis. This accessibility facilitates repeat NGS to guide therapy.
Over the past decade, 26 orally administered targeted therapies have been FDA approved for the treatment of 13 different solid malignancies (Data Supplement). Over the same period, only five orally administered targeted therapies have been approved for the treatment of lymphoid malignancies (Philadelphia chromosome–positive ALL, CLL, follicular lymphoma, mantle cell lymphoma, and Waldenström macroglobulinemia). The current data demonstrate that 45 patients (75%) had an alteration that theoretically could have been targeted with an approved, albeit off-label, drug, whereas only three patients (5%) had an alteration for which an approved on-label drug was available. In a similar study in patients with solid malignancies, 20% had an alteration targeted by on-label agents, whereas 67% had an alteration targetable by off-label use.24 (Of note, the study used the same definitions for actionability as were used in this analysis.) Currently, a paucity of orally administered targeted therapy is available for on-label use in lymphoid malignancies. However, as our findings suggest, the majority of cases likely will have potentially targetable alterations.
The mutational landscape for numerous lymphomas has now been well characterized by several whole-exome sequencing studies.46,47 For example, MLL2, CREBBP, and TP53 alterations in DLBCL, as described by Pasqualucci et al,46 were some of the most common recurrent mutations in the current cohort of patients with DLBCL. In the 19 patients with CLL, the most prevalent aberrations were alterations in NOTCH1 found in five (26%). This alteration was also the most common (12%) in a cohort of patients with CLL sequenced by Puente et al.47 Furthermore, 82% of our patients had alterations that were pharmacologically tractable, similar to the 77% reported by He et al15 in diverse hematologic malignancies. These similar findings in other cohorts provide further support of the generalizability of our findings to other large cohorts of lymphoid malignancies.
MYD88, an adapter protein used by toll-like receptors, was shown to be mutated (MYD88 L265P) in > 90% of patients with Waldenström macroglobulinemia (lymphoplasmacytic lymphoma).48 This mutation not only is relatively specific for the disease but also predicts clinical presentation and survival.49 MYD88 signaling is important in lymphomagenesis and signals through BTK.50 In a phase II trial, ibrutinib produced an overall response rate of 91% in a group of previously treated patients with Waldenström macroglobulinemia, which led to its FDA approval.37 Furthermore, response rates to ibrutinib were significantly higher in patients with MYD88 mutations versus wild-type MYD88.51 In the current cohort, MYD88 L265P mutation was found in the one patient with Waldenström macroglobulinemia. However, MYD88 alterations were also identified in one patient with DLBCL (MYD88 L265P), one with follicular lymphoma (MYD88 S219C), and one with primary CNS lymphoma (MYD88 L265P). MYD88 mutations have been discerned in marginal zone B-cell lymphoma,52 CLL,53 DLBCL,54 and primary CNS lymphoma.55 Preliminary data from a phase I trial of single-agent ibrutnib in four patients with CNS lymphoma demonstrated responses in two of the three patients evaluated. The current data further confirm that mutations in MYD88 are readily identified by NGS and that trials in patients with lymphoid malignancies and MYD88 mutations with BTK inhibitors are warranted.
One of the most common recurrent alterations in the current cohort was loss of CDKN2A/B. CDKN2A/B encodes for the p16INK4A protein, which is a negative regulator of the cyclin D–dependent protein kinases CDK4 and CDK6 important in cell cycle progression from the G1 to S phase.28 Theoretically, this alteration can be targeted by palbociclib, a CDK4/6 inhibitor FDA approved for use in hormone receptor–positive/human epidermal growth factor 2–negative metastatic breast cancer in combination with letrozole56 and fulvestrant.57 Mantle cell lymphoma is a disease characterized by t(11;14)(q13;q32) translocation, which places the CCND1 gene under control of the IGH locus. High levels of CCND1 should upregulate CDK4/6. In a phase I trial of 17 patients with relapsed mantle cell lymphoma treated with single-agent palbociclib, five (18%) achieved progression-free survival of > 1 year, with one complete and two partial responses.58 The high percentage of CDKN2A/B alterations in the current study population gives further rationale for designing trials with CDK inhibitors in lymphoid malignancies.
Activating mutations in BRAF were found in three patients with CLL (two with BRAF G469A and one with BRAF V600E) and one patient with multiple myeloma (BRAF V600E). These mutations are potentially targetable by the BRAF inhibitors vemurafenib and dabrafenib and the MEK inhibitors trametinib and cobimetinib.5,27 BRAF alterations have been identified as a driver mutation and as a biomarker for sensitivity to BRAF inhibition in both hairy cell leukemia and Erdheim-Chester disease.59-61 BRAF alterations have been found in approximately 3% of patients with CLL.62 In the current study, two patients had mutations that led to an alanine substitution for a glycine at position 469. This mutation is activating in melanoma and confers sensitivity to BRAF inhibition.63 To our knowledge, this mutation has not been described in CLL before this report. Further studies that assess the role of BRAF inhibitors in patients with CLL who harbor BRAF mutations are warranted.
In addition to numerous theoretically targetable alterations, NGS identified mutations of prognostic significance in many of the tumor types studied. For example, alterations with known prognostic significance in CLL were identified, including TP53, SF3B1, BIRC3, NOTCH1, and ATM.64 Furthermore, NGS identified mutations in BTK and PLCγ2, which confer resistance to ibrutinib and would require a change in therapy.65 Recently, studies identified a group of BCR-ABL–negative B-ALL with a Philadelphia chromosome–like gene expression signature that carries an inferior prognosis.66 Mutations associated with this gene signature include IKZF1 deletions or mutations, kinase fusions, JAK2 mutations, and CRLF2 mutations. In the 10 patients with B-cell ALL in the current study, IKZF1 deletions and JAK2 were identified in three and two, respectively. NGS readily identified Philadelphia chromosome–like B-ALL and can assist in selecting patients for clinical trials of targeted agents aimed at improving the poor outcome of these patients.15
Higher neoantigen burden, which may be largely predicted by TMB, has been associated with higher objective response rates and progression-free survival in patients treated with PD-1 blockade.67,68 Melanoma, lung cancer, and renal cell carcinoma, all of which are highly responsive to PD-1 blockade, have been shown to have a high degree of mutational burden from whole-genome and -exome sequencing studies.69 In the current cohort, TMB ranged from one or fewer to 140 (Data Supplement). Intermediate to high TMB was noted across almost all histologies (except for marginal zone lymphoma, natural killer/T-cell lymphoma, and cutaneous T-cell lymphoma). For lymphoid malignancies, high TMB was observed in 8% of patients. In comparison, high TMB has been reported in approximately 10% of adenocarcinomas of the lung23 and approximately 42% of melanomas.70 Dramatic responses to PD-1 blockade have already been reported in Hodgkin lymphoma and have led to FDA approval of nivolumab for relapsed/refractory Hodgkin lymphoma.71 TMB has begun to be validated as a marker of response to immunotherapy in several different solid tumors.16,17,67 On the basis of the current findings, TMB likely merits investigation as a marker of response to immunotherapy in lymphoid malignancies as well.72
This study has several limitations. The cohort was small, and not all lymphoid malignancies were represented. The majority of the patients had CLL, multiple myeloma, and ALL, whereas other non-Hodgkin lymphomas were less well represented. Although the majority of patients had actionable mutations theoretically targeted by FDA-approved drugs, in practice, insurance approval for off-label drug use often is difficult to obtain.73 In addition, no standard definition exists for a targetable alteration, and the level of evidence needed to support this is a matter of debate. The number of actionable alterations discussed in this article may be overestimated, but even so, these patients should still be directed toward clinical trials that target these alterations so that the responsiveness or lack thereof can be determined. Recent guideline papers, such as Li et al,74 have begun to address this issue and formulate standardized criteria for the definition of a targetable alteration. Numerous additional clinical trials with a standardized definition of what constitutes a targetable alteration are needed to determine the extent to which patients respond when an alteration is theoretically druggable. Currently, a number of such trials are ongoing (ClinicalTrials.gov identifiers NCT02534675, NCT00851032, and NCT02465060).
In conclusion, we found that most patients with lymphoid malignancies have unique and complex molecular portfolios. In > 80% of patients were one or more genomic alterations that are potentially actionable with existing drugs. Therefore, patients with lymphoid malignancies who have exhausted standard therapy or who are unable to tolerate chemotherapy may be excellent candidates for matched targeted therapies ideally administered in the context of a clinical trial.
Footnotes
Supported in part by the Joan and Irwin Jacobs Fund and National Cancer Institute grant P30 CA016672 (to R.K.).
AUTHOR CONTRIBUTIONS
Conception and design: Aaron M. Goodman
Administrative support: Caitlin Costello, Razelle Kurzrock
Provision of study materials or patients: Michael Choi, Matthew Wieduwilt, Carolyn Mulroney, Caitlin Costello
Collection and assembly of data: Aaron M. Goodman, Michael Choi, Matthew Wieduwilt, Carolyn Mulroney, Caitlin Costello, Garrett Frampton
Data analysis and interpretation: Aaron M. Goodman, Garrett Frampton, Vincent Miller, Razelle Kurzrock
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Next-Generation Sequencing Reveals Potentially Actionable Alterations in the Majority of Patients With Lymphoid Malignancies
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Aaron M. Goodman
No relationship to disclose
Michael Choi
Consulting or Advisory Role: Abbvie/Genentech, Novartis
Speakers' Bureau: Gilead Sciences, Pharmacyclics, Abbvie/Genentech
Research Funding: Abbvie (Inst), Pharmacyclics (Inst)
Matthew Wieduwilt
Honoraria: Alexion Pharmaceuticals
Research Funding: Sigma-Tau (Inst), Shire (Inst)
Carolyn Mulroney
Stock and Other Ownership Interests: Bristol-Myers Squibb/Pfizer
Consulting or Advisory Role: Pharmacyclics
Travel, Accommodations, Expenses: Pharmacyclics
Caitlin Costello
No relationship to disclose
Garrett Frampton
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Vincent Miller
Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Receive periodic royalties related to T790M patent awarded to Memorial Sloan Kettering Cancer Center
Razelle Kurzrock
Leadership: CureMatch
Stock and Other Ownership Interests: Actuate Therapeutics, CureMatch
Honoraria: Cedars-Sina, Jubilant Biosys, NCCN, Janssen, The Dedham Group, Frankel Group, Segal Cancer Center, Sequenom, AACR, Global Biomarkers Consortium, UCSD Genomics Program Talk, Casdin Capital, NXT Health Strategies, Seminars in Oncology, Actuate Therapeutics, Yale Cancer Center, Northwestern University, XBiotech, PANCAN, Sylvester Comprehensive Cancer Center, Mayo Clinic Cancer Center, Kaiser Permanente, Health Advances
Consulting or Advisory Role: Jubilant Biosys, Janssen, CTRC-EAB San Antonio, Merck, AACR, SAIC NCI CCCT IDSE, Sequenom, Actuate Therapeutics, XBiotech, EMD Serono, Centocor Ortho Biotech, GlaxoSmithKline, Exelixis, Novartis, Merck, Roche/Genetech, Genentech Mypath, Foundation Medicine, Pfizer, Guardant Health, Merck Serono
Patents, Royalties, Other Intellectual Property: Four patents
Travel, Accommodations, Expenses: WIN, EMD Serono, Gateway Research Advisory Committee, ICRP, WIN 2015, Caris Centers of Excellence, AACR, Association of American Cancer Institutes, EMD Serono & Quintiles, Global Biomarkers Consortium, Guardant Health, Global Source Ventures/Novena Therapeutics, ASCPT, Meyers Consulting, FDA-OCRA, Genentech, Orbimed/Global Source Ventures, Sylvester Cancer Center, Journal of Precision Medicine, CureMatch, Lynx Group, Mayo Clinic Cancer Center, Kaiser Permanente, PANCAN, Cedars-Sinai, MedImmune/JK Associates Medical Communications Group
REFERENCES
- 1.Sehn LH, Donaldson J, Chhanabhai M, et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J Clin Oncol. 2005;23:5027–5033. doi: 10.1200/JCO.2005.09.137. [DOI] [PubMed] [Google Scholar]
- 2.Wheler JJ, Janku F, Naing A, et al. Cancer therapy directed by comprehensive genomic profiling: A single center study. Cancer Res. 2016;76:3690–3701. doi: 10.1158/0008-5472.CAN-15-3043. [DOI] [PubMed] [Google Scholar]
- 3.Helsten T, Kato S, Schwaederle M, et al. Cell-cycle gene alterations in 4,864 tumors analyzed by next-generation sequencing: Implications for targeted therapeutics. Mol Cancer Ther. 2016;15:1682–1690. doi: 10.1158/1535-7163.MCT-16-0071. [DOI] [PubMed] [Google Scholar]
- 4. Schwaederle M, Husain H, Fanta PT, et al: Use of liquid biopsies in clinical oncology: Pilot experience in 168 patients. Clin Can Res 22:5497-5505, 2016. [DOI] [PubMed]
- 5.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wheler J, Lee JJ, Kurzrock R. Unique molecular landscapes in cancer: Implications for individualized, curated drug combinations. Cancer Res. 2014;74:7181–7184. doi: 10.1158/0008-5472.CAN-14-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janku F, Wheler JJ, Naing A, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73:276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwaederle M, Parker BA, Schwab RB, et al. Precision oncology: The UC San Diego Moores Cancer Center PREDICT experience. Mol Cancer Ther. 2016;15:743–752. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 10.Schwaederle M, Zhao M, Lee JJ, et al. Association of biomarker-based treatment strategies with response rates and progression-free survival in refractory malignant neoplasms: A meta-analysis. JAMA Oncol. 2016;2:1452–1459. doi: 10.1001/jamaoncol.2016.2129. [DOI] [PubMed] [Google Scholar]
- 11.Jardim DL, Schwaederle M, Wei C, et al. Impact of a biomarker-based strategy on oncology drug development: A meta-analysis of clinical trials leading to FDA approval J Natl Cancer Inst 107djv253, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwaederle M, Zhao M, Lee JJ, et al. Impact of precision medicine in diverse cancers: A meta-analysis of phase II clinical trials. J Clin Oncol. 2015;33:3817–3825. doi: 10.1200/JCO.2015.61.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turski ML, Vidwans SJ, Janku F, et al. Genomically driven tumors and actionability across histologies: BRAF-mutant cancers as a paradigm. Mol Cancer Ther. 2016;15:533–547. doi: 10.1158/1535-7163.MCT-15-0643. [DOI] [PubMed] [Google Scholar]
- 14.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373:726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He J, Abdel-Wahab O, Nahas MK, et al: Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 127:3004-3014, 2016. [DOI] [PMC free article] [PubMed]
- 16. Foundation Medicine: What is FoundationOne? http://www.foundationone.com.
- 17. Rosenberg JE, Hoffman-Censits J, Powles T, et al: Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 387:1909-1920, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4:959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wellcome Trust Sanger Institute, Genome Research Limited: COSMIC: Catalogue of somatic mutations in cancer. http://cancer.sanger.ac.uk/cosmic.
- 20. Broad Institute. ExAC Browser (Beta) Exome Aggregation Consortium. http://exac.broadinstitute.org.
- 21. National Center for Biotechnology Information: dbSNP Short Genetic Variations. https://www.ncbi.nlm.nih.gov/projects/SNP.
- 22. National Center for Biotechnology Information: ClinVar. https://www.ncbi.nlm.nih.gov/clinvar.
- 23. Spigel DR, Schrock AB, Fabrizio D, et al: Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol 34, 2016 (suppl; abstr 9017) [Google Scholar]
- 24.Schwaederle M, Daniels GA, Piccioni DE, et al. On the road to precision cancer medicine: Analysis of genomic biomarker actionability in 439 patients. Mol Cancer Ther. 2015;14:1488–1494. doi: 10.1158/1535-7163.MCT-14-1061. [DOI] [PubMed] [Google Scholar]
- 25.Samadder NJ, Neklason DW, Boucher KM, et al. Effect of sulindac and erlotinib vs placebo on duodenal neoplasia in familial adenomatous polyposis: A randomized clinical trial. JAMA. 2016;315:1266–1275. doi: 10.1001/jama.2016.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts AW, Davids MS, Pagel JM, et al: Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med 374:311-322, 2016. [DOI] [PMC free article] [PubMed]
- 27.Robert C, Karaszewska B, Schachter J, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 28.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scala S. Molecular pathways: Targeting the CXCR4-CXCL12 axis—Untapped potential in the tumor microenvironment. Clin Cancer Res. 2015;21:4278–4285. doi: 10.1158/1078-0432.CCR-14-0914. [DOI] [PubMed] [Google Scholar]
- 31.Metzeler KH, Walker A, Geyer S, et al. DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia. 2012;26:1106–1107. doi: 10.1038/leu.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emadi A, Faramand R, Carter-Cooper B, et al. Presence of isocitrate dehydrogenase mutations may predict clinical response to hypomethylating agents in patients with acute myeloid leukemia. Am J Hematol. 2015;90:E77–E79. doi: 10.1002/ajh.23965. [DOI] [PubMed] [Google Scholar]
- 33.Boyle DL, Soma K, Hodge J, et al. The JAK inhibitor tofacitinib suppresses synovial JAK1-STAT signalling in rheumatoid arthritis. Ann Rheum Dis. 2015;74:1311–1316. doi: 10.1136/annrheumdis-2014-206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harry BL, Eckhardt SG, Jimeno A. JAK2 inhibition for the treatment of hematologic and solid malignancies. Expert Opin Investig Drugs. 2012;21:637–655. doi: 10.1517/13543784.2012.677432. [DOI] [PubMed] [Google Scholar]
- 35.Furumoto Y, Gadina M. The arrival of JAK inhibitors: Advancing the treatment of immune and hematologic disorders. BioDrugs. 2013;27:431–438. doi: 10.1007/s40259-013-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Infante JR, Fecher LA, Falchook GS, et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: A phase 1 dose-escalation trial. Lancet Oncol. 2012;13:773–781. doi: 10.1016/S1470-2045(12)70270-X. [DOI] [PubMed] [Google Scholar]
- 37.Treon SP, Tripsas CK, Meid K, et al. Ibrutinib in previously treated Waldenström’s macroglobulinemia. N Engl J Med. 2015;372:1430–1440. doi: 10.1056/NEJMoa1501548. [DOI] [PubMed] [Google Scholar]
- 38.Migden MR, Guminski A, Gutzmer R, et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): A multicentre, randomised, double-blind phase 2 trial. Lancet Oncol. 2015;16:716–728. doi: 10.1016/S1470-2045(15)70100-2. [DOI] [PubMed] [Google Scholar]
- 39.Sekulic A, Migden MR, Oro AE, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366:2171–2179. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bejar R, Lord A, Stevenson K, et al. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood. 2014;124:2705–2712. doi: 10.1182/blood-2014-06-582809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Said R, Hong DS, Warneke CL, et al. P53 mutations in advanced cancers: Clinical characteristics, outcomes, and correlation between progression-free survival and bevacizumab-containing therapy. Oncotarget. 2013;4:705–714. doi: 10.18632/oncotarget.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwaederlé M, Lazar V, Validire P, et al: VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: Implications for anti-angiogenesis therapy. Cancer Res 75:1187-1190, 2015. [DOI] [PubMed]
- 43. doi: 10.1093/annonc/mdv598. Koehler K, Liebner D, Chen JL: TP53 mutational status is predictive of pazopanib response in advanced sarcomas. Ann Oncol 27:539-543, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Bubnoff N, Schneller F, Peschel C, et al. BCR-ABL gene mutations in relation to clinical resistance of Philadelphia-chromosome-positive leukaemia to STI571: A prospective study. Lancet. 2002;359:487–491. doi: 10.1016/S0140-6736(02)07679-1. [DOI] [PubMed] [Google Scholar]
- 45.Jänne PA, Yang JC-H, Kim D-W, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 46.Pasqualucci L, Trifonov V, Fabbri G, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treon SP, Xu L, Yang G, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367:826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 49.Treon SP, Cao Y, Xu L, et al. Somatic mutations in MYD88 and CXCR4 are determinants of clinical presentation and overall survival in Waldenström macroglobulinemia. Blood. 2014;123:2791–2796. doi: 10.1182/blood-2014-01-550905. [DOI] [PubMed] [Google Scholar]
- 50.Rossi D. Role of MYD88 in lymphoplasmacytic lymphoma diagnosis and pathogenesis. Hematology (Am Soc Hematol Educ Program) 2014;2014:113–118. doi: 10.1182/asheducation-2014.1.113. [DOI] [PubMed] [Google Scholar]
- 51.Treon SP, Xu L, Hunter Z. MYD88 mutations and response to ibrutinib in Waldenström’s macroglobulinemia. N Engl J Med. 2015;373:584–586. doi: 10.1056/NEJMc1506192. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Lopez A, Curiel-Olmo S, Mollejo M, et al. MYD88 (L265P) somatic mutation in marginal zone B-cell lymphoma. Am J Surg Pathol. 2015;39:644–651. doi: 10.1097/PAS.0000000000000411. [DOI] [PubMed] [Google Scholar]
- 53.Baliakas P, Hadzidimitriou A, Agathangelidis A, et al. Prognostic relevance of MYD88 mutations in CLL: The jury is still out. Blood. 2015;126:1043–1044. doi: 10.1182/blood-2015-05-648634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi J-W, Kim Y, Lee J-H, et al. MYD88 expression and L265P mutation in diffuse large B-cell lymphoma. Hum Pathol. 2013;44:1375–1381. doi: 10.1016/j.humpath.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 55.Gonzalez-Aguilar A, Idbaih A, Boisselier B, et al. Recurrent mutations of MYD88 and TBL1XR1 in primary central nervous system lymphomas. Clin Cancer Res. 2012;18:5203–5211. doi: 10.1158/1078-0432.CCR-12-0845. [DOI] [PubMed] [Google Scholar]
- 56.Finn RS, Crown JP, Lang I, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 57.Turner NC, Ro J, André F, et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373:209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 58.Leonard JP, LaCasce AS, Smith MR, et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood. 2012;119:4597–4607. doi: 10.1182/blood-2011-10-388298. [DOI] [PubMed] [Google Scholar]
- 59.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiacci E, Park JH, De Carolis L, et al. Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. N Engl J Med. 2015;373:1733–1747. doi: 10.1056/NEJMoa1506583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munoz J, Schlette E, Kurzrock R. Rapid response to vemurafenib in a heavily pretreated patient with hairy cell leukemia and a BRAF mutation. J Clin Oncol. 2013;31:e351–e352. doi: 10.1200/JCO.2012.45.7739. [DOI] [PubMed] [Google Scholar]
- 62.Jebaraj BMC, Kienle D, Bühler A, et al. BRAF mutations in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1177–1182. doi: 10.3109/10428194.2012.742525. [DOI] [PubMed] [Google Scholar]
- 63.Flaherty KT, McArthur G. BRAF, a target in melanoma: Implications for solid tumor drug development. Cancer. 2010;116:4902–4913. doi: 10.1002/cncr.25261. [DOI] [PubMed] [Google Scholar]
- 64.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127:2122–2130. doi: 10.1182/blood-2015-07-659144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Woyach JA, Furman RR, Liu T-M, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roberts KG, Gu Z, Payne-Turner D, et al: High frequency and poor outcome of Philadelphia chromosome–like acute lymphoblastic leukemia in adults. J Clin Oncol 35:394-401, 2017. [DOI] [PMC free article] [PubMed]
- 67.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. George TJ, Frampton GM, Sun J, et al: Tumor mutational burden as a potential biomarker for PD1/PD-L1 therapy in colorectal cancer. J Clin Oncol 34, 2016 (suppl; abstr 3587)
- 69. doi: 10.1038/nature12477. Alexandrov LB, Nik-Zainal S, Wedge DC, et al: Signatures of mutational processes in human cancer. Nature 500:415-421, 2013 [Erratum: Nature 502:258, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johnson DB, Frampton GM, Rioth MJ, et al: Hybrid capture-based next-generation sequencing (HC NGS) in melanoma to identify markers of response to anti-PD-1/PD-L1. J Clin Oncol 34, 2016 (suppl; abstr 105) [Google Scholar]
- 71.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. doi: 10.1038/nrclinonc.2016.168. Goodman A, Patel SP, Kurzrock R: PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol 14:203-220, 2017. [DOI] [PubMed] [Google Scholar]
- 73.Schwaederle M, Parker BA, Schwab RB, et al. Molecular tumor board: The University of California-San Diego Moores Cancer Center experience. Oncologist. 2014;19:631–636. doi: 10.1634/theoncologist.2013-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]