Abstract
Scope
Tumor metastasis greatly contributes to the mortality of prostate cancer. The glucosinolate-derived PEITC has been widely documented to reduce the risk of prostate cancer by modulating multiple biologically relevant processes. Emerging evidence suggests that PEITC may exert its anti-cancer effects through epigenetic mechanisms including microRNAs. Altered levels of miRNA have been linked to tumor malignancy due to their capacity to regulate functional gene expression in carcinogenesis. Here, we assessed the effects of PEITC on miRNA expression which is related to PCa cell invasiveness.
Methods and results
Utilizing oligonucleotide microarray first identified the most affected miRNAs in LNCaP cells after PEITC treatment. Several top altered miRNAs were further validated using quantitative PCR. Interestingly, overexpression of miR-194 suppressed PC3 cell invasion in matrigel-coated Transwell chambers. Bone morphogenetic protein 1 (BMP1) was shown to be a direct target of miR-194. Downregulation of BMP1 by miR-194 or PEITC led to decreased expression of key oncogenic matrix metalloproteinases, MMP2 and MMP9. This in turn, resulting in the suppression of tumor invasion.
Conclusion
Our results indicate that miR-194 downregulates the expression of oncogenic MMP2 and MMP9 by targeting BMP1, which suggests a potential new mechanistic target by which PEITC suppresses prostate cancer cell invasiveness.
Keywords: phenethyl isothiocyanate, prostate cancer, microRNA, miR-194, cell invasion
1. Introduction
Prostate cancer (PCa) is one of the most commonly diagnosed malignancies in men in the U.S. Although it is potentially curable when it occurs inside the prostate, PCa bone metastasis causes severe pain and greatly contributes to disease mortality. Tumor metastasis is a complex multistep process that includes the escape of neoplastic cells, integrating intravasation, circulation and target-organ colonization [1]. From cellular and molecular aspects, the metastatic potential of a tumor cell depends on its interactions with homeostatic factors that are associated with tumor cell growth, survival, angiogenesis, invasion and metastasis [2]. Genetic and epigenetic factors together control gene transcription programming, which is crucial for maintaining tissue homeostasis. In addition to DNA methylation and histone modifications, microRNA (miRNA)-mediated regulation represents another level of epigenetic control of gene expression.
miRNAs are widely found as a class of short, non-coding RNAs of 20–22 nucleotides that post-transcriptionally repress their target gene expression through complementary pairing with the 3′-untranslated regions (UTRs) of cognate mRNAs [3, 4]. Altered miRNA levels have been linked to the development and progression of human PCa in clinical observations [5, 6]. Functionally, certain miRNAs have been categorized as either tumor suppressors or oncogenes [7]. In PCa, the miR-15a–miR-16-1 cluster [8], the Let-7 family [9], miR-34 [10], and miR-200 [11] have been identified as tumor suppressors, whereas miR-21 [12] and miR-125b [13] promote PCa tumorigenesis. miRNAs play an essential role in metastasis, possibly due to their regulatory functions in cell adhesion, neovascularization and tissue invasion networks [14].
Numerous epidemiological and pharmacological studies have suggested that dietary consumption of cruciferous vegetables confers substantial beneficial effects on reducing the risks of cancer in humans, including PCa [15, 16]. Therefore, a group of bioactive sulfur-containing compounds named glucosinolates, which are abundant in cruciferous vegetables, have become a great interest [17]. Their breakdown products, isothiocyanates (ITCs) such as sulforaphane (SFN) and phenethyl isothiocyanate (PEITC), have been extensively evaluated as chemopreventive agents [18]. Studies have shown that PEITC exerts promising anti-cancer effects in PCa cell lines and xenografts in vitro and several preclinical PCa animal models in vivo [19, 20]. More recently, emerging evidence has also shown that PEITC has the potential to modulate epigenetic events, such as DNA methylation [21], histone modifications [22], and miRNAs [23], all of which may be involved in carcinogenesis.
In this study, we examined the effects of PEITC treatment on miRNA expression in PCa cells. Among the most altered miRNAs, we found that miR-194 down-regulates oncogenic matrix metalloproteinase (MMP)2 and MMP9 expression by targeting bone morphogenetic protein 1 (BMP1), consequently accounting for the effects of PEITC on suppressing PCa cell metastasis.
2. Materials and methods
2.1 Cell culture and treatment
PCa LNCaP and PC3 cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere. PEITC was purchased from Sigma-Aldrich (St. Louis, MO, USA). Cells were grown to approximately 80% confluence and then treated with the medium containing PEITC at indicated concentrations for 24 h. DMSO at 0.1% was used as the vehicle control.
2.2 RNA isolation, miRNA profiling and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from the treated cells using miRCURY™ RNA Isolation Kits (Exiqon, Woburn, MA). miRNA profiling was performed by using the mean values from a pre-printed miRCURY LNA microRNA Array (Exiqon, Woburn, MA). miRNA samples were labeled using a miRCURY LNA microRNA Power Labeling Kit, Hy3/Hy5 (Exiqon, Woburn, MA). A reference miRNA was spiked in each microarray to facilitate comparisons among samples; miRNA samples were labeled with the Hy3, and the reference was labeled with Hy5. After enzyme inactivation, the samples were subjected to microarray hybridization in an HS4000-PRO platform (Tecan, Mannendorf, Switzerland) at 56 °C for 16 h and then scanned at 535 and 635 nm. Data analysis was performed using GeneSpring software version 7.2 (Agilent Technologies).
To measure individual miRNA levels, reverse transcription was performed with 500 ng total RNA and certain miRNA-specific primers using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Grand Island, NY). To determine gene mRNA expression levels, first-strand cDNA was synthesized from 500 ng total RNA using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Grand Island, NY) according to the manufacturer’s manual. The cDNA was used as the template for qPCR in the ABI7900HT system (Applied Biosystems). The sequences of the primers used for PCR amplifications are listed in Supplementary Table 1.
2.3 Transfection of has-miR-194-5p mimic and inhibitor
We utilized a miRCURY LNA microRNA Mimic/Inhibitor (Exiqon, Woburn, MA) of hsa-miR-194-5p to test the cellular functions of miR-194. For transfection experiments, cells were grown to 70% confluence and then treated with a 30 nM has-miR-194-5p mimic/inhibitor using Lipofectamine 3000 reagent (Life Technologies, Grand Island, NY) for 24 h. Negative control miRNA was used in parallel as a control.
2.4 Matrigel cell invasion assay
The invasion capacity of PC3 cells was measured using Matrigel-coated Transwell cell culture chambers (8 μm pore size). The cells treated with miR-194 mimic/inhibitor or PEITC, were maintained in serum-free medium for 24 h and were then seeded in the upper chamber of the Transwell insert (5*104 cells/well). Culture medium containing 10% FBS was placed in the lower chamber. Cells were incubated for 12 h at 37°C in a humidified 5% CO2 atmosphere. The cells penetrated through the Matrigel to the lower surface of the membrane were stained with a Differential Quik Stain Kit (Polysciences, Warrington, PA). Invasive cell numbers were counted under a computerized microscope system with the Nikon ACT-1 program (Version 2.20).
2.5 Western blotting
The treated cells from above were harvested using radioimmunoprecipitation assay (RIPA) buffer supplemented with a protein inhibitor cocktail (Sigma, St. Louis, MO). The clear supernatants from whole cell lysates were loaded (20 μg/lane) and separated by 4–15% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA). Then, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA) followed by blocking with 5% bovine serum albumin (BSA) in Tris-buffered saline-0.1% Tween 20 (TBST) buffer. Primary antibodies against MMP2 and MMP9 (Abcam, Cambridge, MA) and β-Actin (Santa Cruz Biotechnology, Santa Cruz, CA) were used. The blots were visualized by using a SuperSignal enhanced chemiluminiscence (ECL) detection system and recorded using a Gel Documentation 2000 system (Bio-Rad, Hercules, CA). Densitometry of the bands was analyzed using ImageJ (Version 1.48d, NIH).
2.6 Luciferase reporter activity assay
The 3′-UTR of BMP1 mRNA was reverse transcribed, amplified by PCR, then cloned into the pGL4.15 [luc2P/Hygro] (Promega, Madison, WI) luciferase reporter vector. The recombinant plasmid sequences were validated by direct sequencing (Genewiz, South Plainfield, NJ). For the luciferase activity assay, PC3 or HEK293 cells were seeded into 12-well plates until they reached 80% confluence and were then co-transfected with 30 nM miR-194-5p mimic/inhibitor and 300 ng of the reporter plasmid using Lipofectamine 3000 (Life Technologies, Grand Island, NY) according to the manufacturer’s manual; 100 ng of the pSV-β-Galactosidase vector was cotransfected as internal control. After 24 h of incubation, the cells were lysed in 1X Reporter Lysis Buffer (Promega, Madison, WI, USA). Luciferase activity was measured with the cell lysate using a Sirius luminometer (Berthold Technologies, Pforzheim, Germany). The luciferase activities were normalized with β-Galactosidase activity and were reported as fold changes compared with the negative control miRNA.
2.7 RNA interference
RNAi Duplex Oligos (siRNA) targeting human BMP1 were designed and synthesized by Integrated DNA Technologies (Coralville, IA). Transfection of siRNA was conducted using Lipofectamine 3000 (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. PC3 cells were seeded in 6-well plates until they reached 80% confluence and were then transfected with 30 nM BMP1 siRNA. After 24 h of incubation, the cells were collected for RNA and protein extraction. A universal negative control siRNA was similarly prepared in parallel and used as the control. The siRNA sequences are listed in Supplementary Table 2.
2.8 Statistical analysis
The data are presented as means ± SD. The statistical analyses were performed using a one-way analysis of variance (ANOVA). Statistically significant differences among the means were set at *p<0.05, **p<0.01, and #p<0.001.
3. Results
3.1 PEITC alters miRNA expression in PCa cells
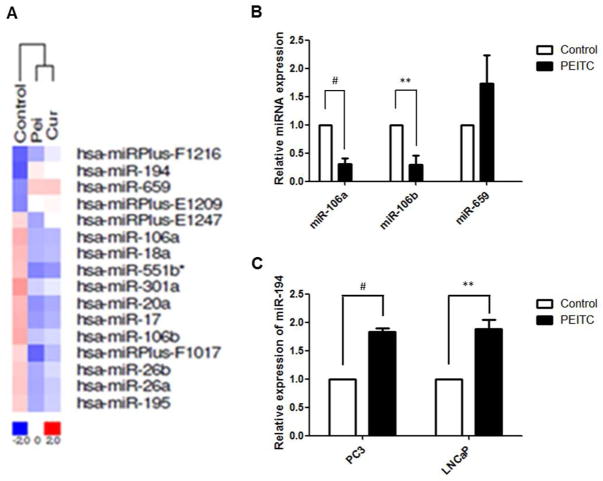
An oligonucleotide microarray was used to analyze the miRNA expression profiles in PCa LNCaP cells following PEITC treatment. A hierarchical cluster analysis was employed to evaluate differential miRNA expression among the samples. The clustering was performed on log2 (Hy3/Hy5) ratios that passed the filtering criteria for variations between each sample versus control. As shown in Fig 1A, samples treated with PEITC and curcumin (another widely investigated chemopreventive agent) clustered together on the right side. The names of top altered miRNAs are also listed in Fig 1A. Because we used a single sample without replications on the microarray, qPCR validation was performed on several top altered miRNAs. In agreement with the microarray results, PEITC treatment decreased miR-106a/b and miR-695 levels in LNCaP cells (Fig 1B), whereas miR-194 levels were up-regulated in both the LNCaP and the PC3 cells (Fig 1C).
Figure 1. PEITC alters the miRNA profile in PCa cells.
A) Hierarchical cluster analysis relative to the expression profiles of miRNAs in untreated group and cells treated with either PEITC or curcumin. miRNA expression intensity is represented according to a color scale ranging between blue (low) and red (high), only the most altered miRNAs are shown with their names; B) qPCR validation of miR106a, miR106b, miR-659 levels in LNCaP cells treated by PEITC; C) PEITC induces miR-194 levels in both LNCaP and PC3 cells.
3.2 miR-194 suppresses PC3 cell invasiveness in vitro
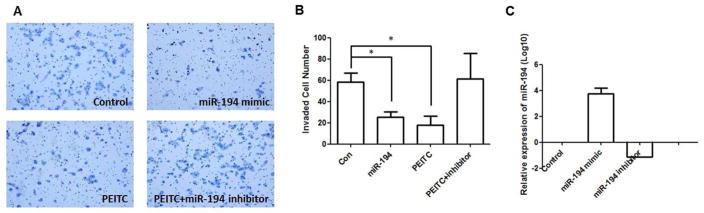
Among the miRNAs that were most frequently altered by PEITC treatment, miR-194 has been reported to be a potential tumor suppressor that is inversely associated with tumor invasion in gastric cancer [24]. In this study, we tested whether miR-194 contributes to the effects of PEITC in suppressing prostate cancer cell invasion, by using Matrigel-coated Transwell cell culture chambers. After 12 h of incubation, the cells that migrated from the upper chamber to the lower chamber on the basal side of the membrane were captured (Fig 2A) and counted (Fig 2B). Treatment with PEITC (2.5 μM) or the miR-194 mimic reduced the number of cells that penetrated the Matrigel by 55% and 52%, respectively. In contrast, the miR-194 inhibitor antagonized the inhibitory effect of PEITC on cell invasiveness. The transfection efficiency of miR-194 mimics and miR-194 inhibitors were confirmed in parallel before the cells were seeded in the Transwell chamber (Fig 2C).
Figure 2. miR-194 suppresses PC3 cells invasiveness.
Matrigel Transwell cell invasion assay was performed to test the effect of miR-194 on the invasive ability of PC3 cells. PC3 cells were treated with miR-194 mimic, 2.5μM PEITC, and a combination of miR-194 inhibitor and 2.5μM PEITC prior to seeding in the chamber. The cells that invaded the basal side of the membrane were captured under microscopy and the cell numbers were counted. A) representative imagines of the Transwell assays for cell migration; B) invasion ability of PC cells quantified by counting the number of cells that invaded the underside of the membrane; C) validation of transfection efficiency of miR-194 mimic and miR-194 inhibitor by qPCR. Data are presented as the mean ± SD from at least three independent experiments.
3.3 PEITC down-regulates MMP2 and MMP9 via miR-194
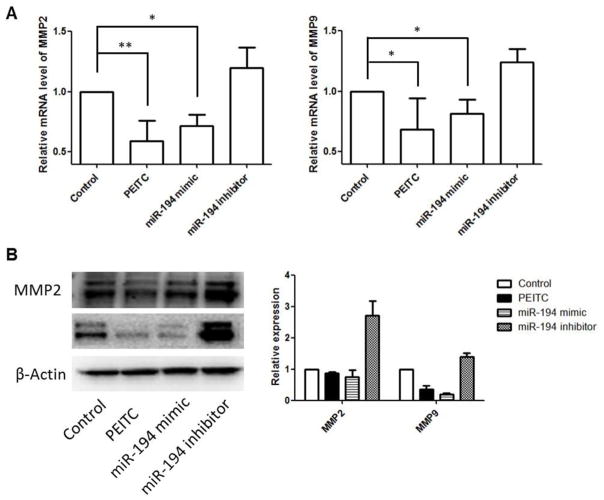
MMPs are enzymes capable of degrading extracellular matrix proteins. Among all MMPs, oncogenic MMP2 (gelatinase-A) and MMP9 (gelatinase-B) are deeply involved in tumor migration, invasion and metastasis for various human cancers [25]. We tested whether miR-194 would decrease the levels of MMP2 and MMP9 in prostate cancer cells underlying its invasion suppressive function. Upon PEITC treatment or transfection of miR-194 mimic, we found that both mRNA and protein levels of MMP2 and MMP9 decreased, whereas gene expression increased in the cells upon miR-194 inhibition (Fig 3A & B).
Figure 3. miR-194 decreases the expression of MMP2 and MMP9 in PCa cells.
The mRNA and protein levels of MMP2 and MMP9 were determined using qPCR and Western blotting following the indicated treatments, respectively. A) the relative mRNA levels of MMP2 and MMP9; B) representative immunoblotting bands of MMP2 and MMP9 protein levels. The relative abundance of each target protein was calculated by normalizing the band intensity to β-actin using ImageJ. The bar chart in the right panel presents the mean ± SEM of three independent experiments.
3.4 BMP1 is a direct target of miR-194
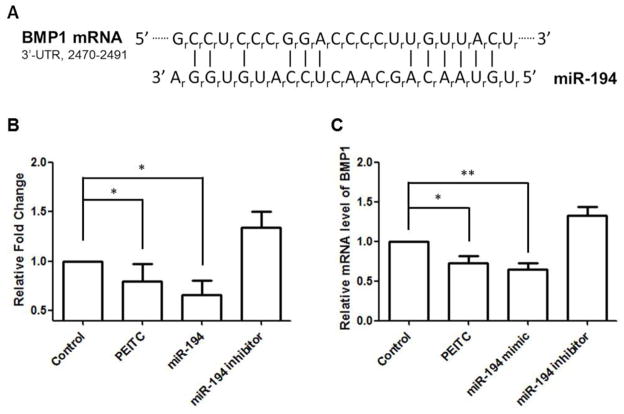
Each miRNA may naturally target a series of mRNAs [26]. To further examine the mechanism by which miR-194 regulates MMP2 and MMP9 expression, TargetScan [27] and PicTar [28] were used to discover the mRNA targets of miR-194. Surprisingly, neither MMP2 nor MMP9 was predicted to be a miR-194 target according to these two algorithms. Among the candidate mRNAs, we found that BMP1 may play a role in promoting cell migration and invasion through gene ontology analysis (Fig 4A). We then cloned the 3′-UTR of BMP1 into a luciferase reporter vector. Co-transfection of the miR-194 mimic and the luciferase construct reduced luciferase activity, indicating that miR-194 directly targets on the BMP1 3′-UTR. Moreover, corresponding with the elevated miR-194 levels mentioned above, PEITC treatment decreased the luciferase activity of the same vector (Fig 4B). In addition to the reporter assay, we performed qPCR to evaluate BMP1 mRNA levels after miR-194 mimic/inhibitor or PEITC treatment. We found that both miR-194 mimic and PEITC treatments decreased BMP1 expression (Fig 4C).
Figure 4. BMP1 is a direct target of miR-194.
The BMP1 3′-UTR was amplified from cDNA library and inserted into pGL4.15 vector. The luciferase reporter assay was performed using the recombined construct with PEITC, miR-194 mimic and miR-194 inhibitor treatment. A) sequence of miR-194 and the predicted complementary pairing between miR-194 and human BMP1 3′-UTR (2470–2491); B) relative fold change of luciferase activities tested on the BMP1 3′-UTR reporter genes; C) relative BMP1 mRNA levels after treatment of PETC, miR-194 mimic, and miR-194 inhibitor.
3.5 BMP1 inhibition decreases cellular MMP levels
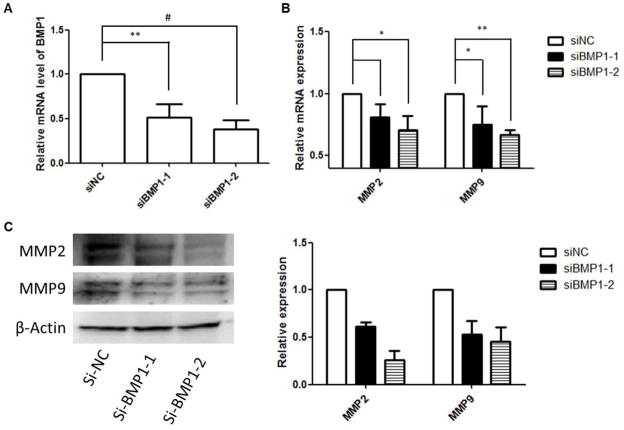
Having demonstrated that BMP1 is a potential target of miR-194, we then tested whether BMP1 was involved in the miR-194-mediated down-regulation of MMP2 and MMP9. We carried out gene silencing by using BMP1 siRNA, and the knock down efficacy was confirmed by qPCR (Fig 5A). After transfection, the cellular MMP2 and MMP9 expression levels were measured by qPCR and western blotting. We found that BMP1 knockdown attenuates MMP2 and MMP9 expression in PC3 cells at both the mRNA and protein levels (Fig 5B & C). Transfection of a negative control siRNA that has no homology to any known mammalian genes was performed in parallel of BMP1 siRNA transfection.
Figure 5. Downregulation of BMP1 mediates the decreased expression of MMP2 and MMP9.
After transfection of BMP1 siRNA, the mRNA and protein levels of MMP2 and MMP9 were determined using qPCR and Western blotting. (A) validation of BMP1 siRNA silencing efficiency by qPCR; B) the relative mRNA levels of MMP2 and MMP9; (B) representative immunoblotting bands of MMP2 and MMP9 protein levels. The relative abundance of each target protein was calculated by normalizing the band intensity to β-actin using ImageJ. The bar chart in the right panel presents the mean ± SEM of three independent experiments.
4. Discussion
Numerous epidemiologic studies have shown the protective role of glucosinolates against cancer, including PC [20, 29, 30]. PEITC is one of the most widely investigated glucosinolate-derived isothiocyanates from cruciferous vegetables, exerting its chemopreventive effects by modulating multiple relevant processes, such as induction of phase II antioxidative/cytoprotective enzymes, inhibition of a prolonged inflammatory response, and regulation of various cellular signaling pathways, including apoptosis, proliferation, angiogenesis, epithelial-mesenchymal transition (EMT), and cancer stem cell self-renewal [31]. In addition, growing evidence has suggested that epigenetic mechanisms, particularly those mediated by miRNAs, are implicated in the chemopreventive effects of PEITC. Consistently with results from previous reports, in the present study, we found that PEITC can decreases MMP2/9 levels and subsequently inhibits PCa cell invasion. Interestingly, we also found that PEITC treatment significantly induces cellular miR-194 expression. Elevated miR-194 levels may account for the attenuated cell invasiveness, suggesting a new mechanism by which PEITC inhibits PCa cell invasion.
miRNA represents a “fine tuning” level of epigenetic regulation that can modulate a wide diversity of biological processes. As such, miRNAs are sensitive to changes in the cellular environment, such as pathological progression and/or chemical interventions. A previous study by Izzotti et al. has shown that the miRNA networks modulated by PEITC are involved in a variety of functions that play roles in protecting environmental cigarette smoke ECS-induced pulmonary carcinogenesis [32]. In this study, we identified the miRNAs that were most affected by PEITC treatment in PCa LNCaP cells; miR-194 and miR-659 were induced by PEITC, whereas miR-17, miR-18a, miR-20a, miR-106a/b, miR-301a were down-regulated. The transcriptional regulation of series of miRNAs is known to occur in a polycistronic manner; thus, alteration of one member in the cluster often coincides with alterations of other members in the cluster [33]. The miR-17-92 cluster, also known as oncomiR-1 (oncogenic miRNA), is among the best-characterized miRNA clusters [34]; it comprises miR-17, miR-18, miR-19a, miR-20a, miR-19b-1 and miR-92a-1. The miR-17-92 cluster was initially found in the 13q31-q32 amplicon in malignant lymphoma [35], which plays an important role in cell cycle regulation, proliferation, apoptosis and other pivotal processes. miR-17-92 overexpression has been widely found in a variety of malignances, including B-cell lymphoma [36], acute myeloid leukemia (AML) [37], hepatocellular carcinoma [38], and breast [39], colon [40], lung [41], and pancreatic [42] cancers. Previous reports have demonstrated that PEITC decreases miR-17 and miR-20a expression, resulting in decreased Myc expression in a reactive oxygen species (ROS)-dependent manner [43]. Correspondingly, our result shows that PEITC decreases the expression of several members of the miR-17-92 cluster, including miR-17, miR-18a, and miR-20a, in PCa cell lines. In addition, elevated miR-106 levels have been detected in prostate tumors with high Gleason scores [44], whereas the oncogenic potential of miR-106 has been identified in human T-cell leukemia [45] and colorectal cancer cells [46]. Interestingly, we also found that PEITC decreases miR-106a/b levels in PCa LNCaP cells. In summary, the evidence mentioned above indicates that miRNA regulation is part of a mechanism that contributes to the anticancer effect of PEITC. An unexpected observation is that PEITC decreases levels of miR-26a/b, which have long been considered as anti-oncomiRs [47, 48]. However, the reduced miR-26a/b levels do not override the anti-invasive activity of PEITC in our study.
miR-194 is at the top of the list of our miRNA profiling candidates. Recent studies have shown that miR-194 acts as a tumor suppressor in liver [49], gastric [24], and non-small cell lung cancers [50]. It is unsurprising that the expression of tumor suppressive miR-194 can be induced by a chemopreventive agent such as PEITC. In the present study, we were interested in the role of miR-194 in PCa cell invasion and metastasis. A Matrigel Transwell assay was performed to investigate the relationship between miR-194 expression and the invasive growth of PCa cells. The number of the PC3 cells that invaded through the membrane was reduced by 55% after ectopic expression of miR-194. In contrast, when an miR-194 inhibitor was applied in the presence of PEITC, the invasiveness of the cells was enhanced. This result suggests that miR-194 at least partially contributes to the anti-invasive effect of PEITC.
Numerous potential targets of miR-194 have been identified in a variety of cells and tissues. In multiple myeloma, miR-194 targets MDM2 expression, which in turn enhances the therapeutic activity of MDM2 inhibitors by increasing their p53-activating effects [51]. It has also been reported that miR-194 overexpression inhibits the proliferation, migration, and invasion of osteosarcoma cells by targeting CDH2 (N-cadherin) and insulin-like growth factor receptor 1 (IGF1R) [52]. Moreover, high levels of miR-194 have been observed in hepatic epithelial cells, which suppress the invasion and migration of mesenchymal-like cancer cells by down-regulating several genes involved in the EMT [49]. Correspondingly, miR-194 represses the expression of the oncogene polycomb complex protein-1 (BMI-1), which accounts for cell invasion and EMT in endometrial cancer cells [53]. In this study, we found that miR-194 has direct complementary pairing with the 3′-UTR of BMP1 mRNA. Previous high-throughput screens have identified BMP1 RNA sequences as among the most up-regulated transcripts in human tumor endothelium associated with angiogenesis [54]. BMP1 reportedly activates transforming growth factor beta-1 (TGFβ1) signaling via cleavage of latent TGFβ-binding protein (LTBP1) [55]; TGFβ activation, along with subsequent roles played by MMPs [56], is important in the tissue remodeling associated with morphogenesis and to cancer metastasis. Thus, BMP1 suppression by miR-194 reduces TGFβ activity, resulting in decreased MMP2 and MMP9 expression, thereby attenuating the invasive capacity of PCa cells. Our present study, together with the aforementioned findings, suggests that miR-194 could modulate multiple steps in the invasion and metastasis cascade.
Like other RNA molecules, miRNAs are transcribed from DNA as immature primary miRNAs (pri-miRNAs) [57] and then are processed by Drosha into precursor miRNAs (pre-miRNAs), which are approximately 70 nt long [58]. Finally, the pre-miRNA hairpins are transported to the cytoplasm and cleaved by Dicer to form mature miRNAs [59]. Based on this biogenesis diagram of miRNA, it should be noted that the initial transcription of miRNAs can be regulated by transcriptional factors. Recent studies have indicated that hepatocyte nuclear factor 1α (HNF-1a) [60] and p53 [51] can bind to the promoter elements of the gene encoding pri-miR-194, thereby inducing the cellular levels of miR-194. The precise mechanisms by which miR-194 expression is regulated in PCa require further exploration. However, it is known that PEITC induces p53 protein expression and p53-dependent transactivation [61]. Thus, PEITC may potentially stimulate transactivation of the miR-194 gene in a p53-dependent manner.
In conclusion, we demonstrated that miR-194 up-regulation helps PEITC suppress PCa cell invasion, suggesting a new mechanism by which PEITC modulates PCa metastasis. As shown above, PEITC may affect a wide range of miRNAs, and each miRNA may have multiple mRNA targets; therefore, PEITC may have a broader functional impact on PCa cells at the miRNA level. Investigating these effects of PEITC will be of interest in future studies.
Acknowledgments
This work is supported in part by R01-CA118947 and R01-AT007065-01 awarded to Dr. Ah-Ng Tony Kong from the National Institutes of Health (NIH), USA. The authors thank all the members in Dr. Ah-Ng Tony Kong’s lab for their helpful discussion of this work.
References
- 1.Scott J, Kuhn P, Anderson AR. Unifying metastasis--integrating intravasation, circulation and end-organ colonization. Nature reviews Cancer. 2012;12:445–446. doi: 10.1038/nrc3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature reviews Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 3.Garzon R, Pichiorri F, Palumbo T, Visentini M, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer research. 2011;71:5950–5954. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sevli S, Uzumcu A, Solak M, Ittmann M, Ozen M. The function of microRNAs, small but potent molecules, in human prostate cancer. Prostate cancer and prostatic diseases. 2010;13:208–217. doi: 10.1038/pcan.2010.21. [DOI] [PubMed] [Google Scholar]
- 6.Saini S, Majid S, Dahiya R. Diet, microRNAs and prostate cancer. Pharmaceutical research. 2010;27:1014–1026. doi: 10.1007/s11095-010-0086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33:135–147. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 8.Bonci D, Coppola V, Musumeci M, Addario A, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nature medicine. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 9.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 10.Kong D, Heath E, Chen W, Cher M, et al. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. American journal of translational research. 2012;4:14–23. [PMC free article] [PubMed] [Google Scholar]
- 11.Kong D, Li Y, Wang Z, Banerjee S, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer research. 2010;70:8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi XB, Xue L, Ma AH, Tepper CG, et al. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. The Prostate. 2011;71:538–549. doi: 10.1002/pros.21270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 15.Kolonel LN, Hankin JH, Whittemore AS, Wu AH, et al. Vegetables, fruits, legumes and prostate cancer: a multiethnic case-control study. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2000;9:795–804. [PubMed] [Google Scholar]
- 16.Abdull Razis AF, Noor NM. Cruciferous vegetables: dietary phytochemicals for cancer prevention. Asian Pacific journal of cancer prevention : APJCP. 2013;14:1565–1570. doi: 10.7314/apjcp.2013.14.3.1565. [DOI] [PubMed] [Google Scholar]
- 17.Stoewsand GS. Bioactive organosulfur phytochemicals in Brassica oleracea vegetables--a review. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 1995;33:537–543. doi: 10.1016/0278-6915(95)00017-v. [DOI] [PubMed] [Google Scholar]
- 18.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. The AAPS journal. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao D, Lew KL, Zeng Y, Xiao H, et al. Phenethyl isothiocyanate-induced apoptosis in PC-3 human prostate cancer cells is mediated by reactive oxygen species-dependent disruption of the mitochondrial membrane potential. Carcinogenesis. 2006;27:2223–2234. doi: 10.1093/carcin/bgl087. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Khor TO, Shu L, Su ZY, et al. Plants vs. cancer: a review on natural phytochemicals in preventing and treating cancers and their druggability. Anti-cancer agents in medicinal chemistry. 2012;12:1281–1305. doi: 10.2174/187152012803833026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang LG, Beklemisheva A, Liu XM, Ferrari AC, et al. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Molecular carcinogenesis. 2007;46:24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Chakravarty S, Dey M. Phenethylisothiocyanate alters site- and promoter-specific histone tail modifications in cancer cells. PloS one. 2013;8:e64535. doi: 10.1371/journal.pone.0064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu C, Gong AY, Chen D, Solelo Leon D, et al. Phenethyl isothiocyanate inhibits androgen receptor-regulated transcriptional activity in prostate cancer cells through suppressing PCAF. Molecular nutrition & food research. 2013;57:1825–1833. doi: 10.1002/mnfr.201200810. [DOI] [PubMed] [Google Scholar]
- 24.Song Y, Zhao F, Wang Z, Liu Z, et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Annals of surgical oncology. 2012;19(Suppl 3):S509–517. doi: 10.1245/s10434-011-1999-2. [DOI] [PubMed] [Google Scholar]
- 25.Gullu IH, Kurdoglu M, Akalin I. The relation of gelatinase (MMP-2 and -9) expression with distant site metastasis and tumour aggressiveness in colorectal cancer. British journal of cancer. 2000;82:249. doi: 10.1054/bjoc.1999.0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krek A, Grun D, Poy MN, Wolf R, et al. Combinatorial microRNA target predictions. Nature genetics. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 29.Jiao D, Eklind KI, Choi CI, Desai DH, et al. Structure-activity relationships of isothiocyanates as mechanism-based inhibitors of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis in A/J mice. Cancer research. 1994;54:4327–4333. [PubMed] [Google Scholar]
- 30.Stoner GD, Morse MA. Isothiocyanates as inhibitors of esophageal cancer. Advances in experimental medicine and biology. 1996;401:13–23. doi: 10.1007/978-1-4613-0399-2_2. [DOI] [PubMed] [Google Scholar]
- 31.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochemical pharmacology. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izzotti A, Larghero P, Cartiglia C, Longobardi M, et al. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010;31:894–901. doi: 10.1093/carcin/bgq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karius T, Schnekenburger M, Dicato M, Diederich M. MicroRNAs in cancer management and their modulation by dietary agents. Biochemical pharmacology. 2012;83:1591–1601. doi: 10.1016/j.bcp.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell death and differentiation. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ota A, Tagawa H, Karnan S, Tsuzuki S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer research. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 36.He L, Thomson JM, Hemann MT, Hernando-Monge E, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Lu J, Sun M, Mi S, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, et al. Elevated expression of the miR-17- 92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. The American journal of pathology. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Z, Willmarth NE, Zhou J, Katiyar S, et al. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dews M, Homayouni A, Yu D, Murphy D, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nature genetics. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osada H, Takahashi T. let-7 and miR-17-92: small-sized major players in lung cancer development. Cancer science. 2011;102:9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 42.Szafranska AE, Davison TS, John J, Cannon T, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 43.Jutooru I, Guthrie AS, Chadalapaka G, Pathi S, et al. Mechanism of action of phenethylisothiocyanate and other reactive oxygen species-inducing anticancer agents. Molecular and cellular biology. 2014;34:2382–2395. doi: 10.1128/MCB.01602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang H, Studach L, Hullinger RL, Xie J, Andrisani OM. Down-regulation of RE-1 silencing transcription factor (REST) in advanced prostate cancer by hypoxia-induced miR-106b~25. Experimental cell research. 2014;320:188–199. doi: 10.1016/j.yexcr.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer research. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 46.Zhang GJ, Li JS, Zhou H, Xiao HX, et al. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. Journal of experimental & clinical cancer research : CR. 2015;34:73. doi: 10.1186/s13046-015-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu J, He ML, Wang L, Chen Y, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer research. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 48.Zhu Y, Lu Y, Zhang Q, Liu JJ, et al. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic acids research. 2012;40:4615–4625. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng Z, Fu X, Chen X, Zeng S, et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–2157. doi: 10.1002/hep.23915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, Liu T, Fang O, Leach LJ, et al. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1) Oncogene. 2014;33:1506–1514. doi: 10.1038/onc.2013.108. [DOI] [PubMed] [Google Scholar]
- 51.Pichiorri F, Suh SS, Rocci A, De Luca L, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer cell. 2010;18:367–381. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Han K, Zhao T, Chen X, Bian N, et al. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. International journal of oncology. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong P, Kaneuchi M, Watari H, Hamada J, et al. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Molecular cancer. 2011;10:99. doi: 10.1186/1476-4598-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St Croix B, Rago C, Velculescu V, Traverso G, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 55.Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. The Journal of cell biology. 2006;175:111–120. doi: 10.1083/jcb.200606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sehgal I, Thompson TC. Novel regulation of type IV collagenase (matrix metalloproteinase-9 and -2) activities by transforming growth factor-beta1 in human prostate cancer cell lines. Molecular biology of the cell. 1999;10:407–416. doi: 10.1091/mbc.10.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y, Kim M, Han J, Yeom KH, et al. MicroRNA genes are transcribed by RNA polymerase II. The EMBO journal. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 59.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 60.Krutzfeldt J, Rosch N, Hausser J, Manoharan M, et al. MicroRNA-194 is a target of transcription factor 1 (Tcf1, HNF1alpha) in adult liver and controls expression of frizzled-6. Hepatology. 2012;55:98–107. doi: 10.1002/hep.24658. [DOI] [PubMed] [Google Scholar]
- 61.Huang C, Ma WY, Li J, Hecht SS, Dong Z. Essential role of p53 in phenethyl isothiocyanate-induced apoptosis. Cancer research. 1998;58:4102–4106. [PubMed] [Google Scholar]