Significance
This cortical connectivity study shows that neuroplastic reorganization in blind children involves key multisensory integration regions. Using two complementary functional connectivity graph theory analyses, multimodal integration areas exhibited increased connectivity in blind children compared with matched controls; within-group analyses also demonstrated that functional connectivity changes increased with age. Evidence is provided that the spatial cortical expression of genes implicated in neuroplasticity underlies the reorganizational capabilities of the multimodal cortex.
Keywords: blindness, children, neuroplasticity, functional connectivity, CREB family
Abstract
Sensory deprivation reorganizes neurocircuits in the human brain. The biological basis of such neuroplastic adaptations remains elusive. In this study, we applied two complementary graph theory-based functional connectivity analyses, one to evaluate whole-brain functional connectivity relationships and the second to specifically delineate distributed network connectivity profiles downstream of primary sensory cortices, to investigate neural reorganization in blind children compared with sighted controls. We also examined the relationship between connectivity changes and neuroplasticity-related gene expression profiles in the cerebral cortex. We observed that multisensory integration areas exhibited enhanced functional connectivity in blind children and that this reorganization was spatially associated with the transcription levels of specific members of the cAMP Response Element Binding protein gene family. Using systems-level analyses, this study advances our understanding of human neuroplasticity and its genetic underpinnings following sensory deprivation.
Neuroplasticity is an intrinsic ability of the brain to modify and rewire itself following experiences (1). The study of neuroplastic reorganization in blind individuals offers a model through which neuroadaptive processes can be identified. For instance, cross-modal neuroplasticity is a mechanism by which blind individuals recruit visual-related cortices to process sensory information from other perceptual modalities (2–5). In addition to occipital regions, parietal and frontal multimodal integration regions of blind adults are capable of functional connectivity reorganization (6). These brain areas are part of a multimodal integration network that acts as a bridge integrating multisensory functions across cortical regions (7). Our group has previously characterized the hierarchical structure and central position of the multimodal integration network in adult blind subjects (6). Although this finding advances our understanding of how information from primary unimodal cortices is adaptively integrated into higher-order associative areas, it remains unclear if neuroplastic changes within multimodal integration areas also occur in blind children. Furthermore, in addition to clarifying the sites of prominent neuroplastic changes, the biological mechanisms through which neuroplastic alterations occur have yet to be fully elucidated in blind individuals.
Concurrent advances in cellular and molecular biology and neuroimaging provide a unique opportunity to explore the interlinked relationships between genes and neural circuits (8). A neuroimaging endophenotype is informative because it is more closely related to the genetic end product than clinical or behavioral phenotypes (9). Thus, examining properties of systems-level brain organization and cortical gene expression has great potential to expand our understanding of neuroplastic mechanisms, particularly if specific gene expression patterns predispose certain cortical regions to plastic adaptation. In this study, we first applied network-based analyses to investigate whole-brain connectivity profiles in blind children (Fig. S1). Due to widespread adaptation of the cerebral cortex throughout early life (10), the study of blind children serves as an ideal framework through which to investigate systems-level neuroplastic mechanisms. Given that brain organization relies on neural communication within a complex network of connected regions (11), we used graph theory to investigate the connectivity profiles associated with blindness, placing special emphasis on age-dependent changes. Second, we investigated the topological relationship between cortical network reorganization in blind children and gene expression profiles from the Allen Human Brain Atlas, specifically targeting genes implicated in brain plasticity (12, 13) [i.e., cAMP Response Element Binding protein (CREB) family genes]. Here we aimed to delineate the topological distribution of the genes associated to neuroplastic changes in blind children. We hypothesized that blind children, compared with sighted controls, would exhibit increased transformation of multisensory integration area connectivity profiles and that these connectivity changes would correspond to the cortical transcription profiles of genes implicated in neuroplasticity and learning.
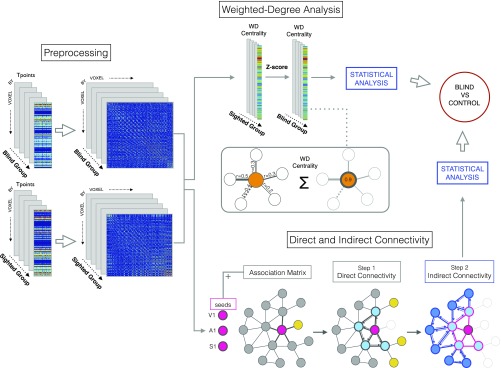
Fig. S1.
Image processing. The time series obtained from BOLD signal preprocessing was used to create a voxel by voxel functional connectivity matrix for each of the two groups (blind children and sighted controls). These matrices served as the inputs for resting-state functional connectivity and graph theory analyses. Next, the weighted degree of each voxel was computed and saved as the value representing the connectivity strength of each voxel (i.e., the sum of the correlations of each node with its neighbors) for the blind and control cohorts. A Z-score was then computed to normalize values and allow for group-level comparisons. Based on the same connectivity matrices, direct and indirect connectivity analyses were performed using three seeds as the starting point, localized in the primary auditory (A1), somatosensory (S1), and visual (V1) cortices. These regions are highlighted in the bottom right portion in dark pink. Direct connectivity with neighboring nodes (black links) was calculated using a graph theory approach, in which only direct connections from a node to connected neighboring nodes are considered [the nodes that were not connected to each other (shown in yellow) were later discarded]. For indirect connectivity analysis, we applied the same principle using direct connectivity nodes as seeds (for visualization purposes, note that direct connections are represented as pink triangles and light blue nodes, indirect connections are represented as blue triangles and dark blue nodes, and the discarded links are faded). Then, between-group statistical analyses were applied.
Results
Connectivity Reorganization in the Cerebral Cortex of Blind Children.
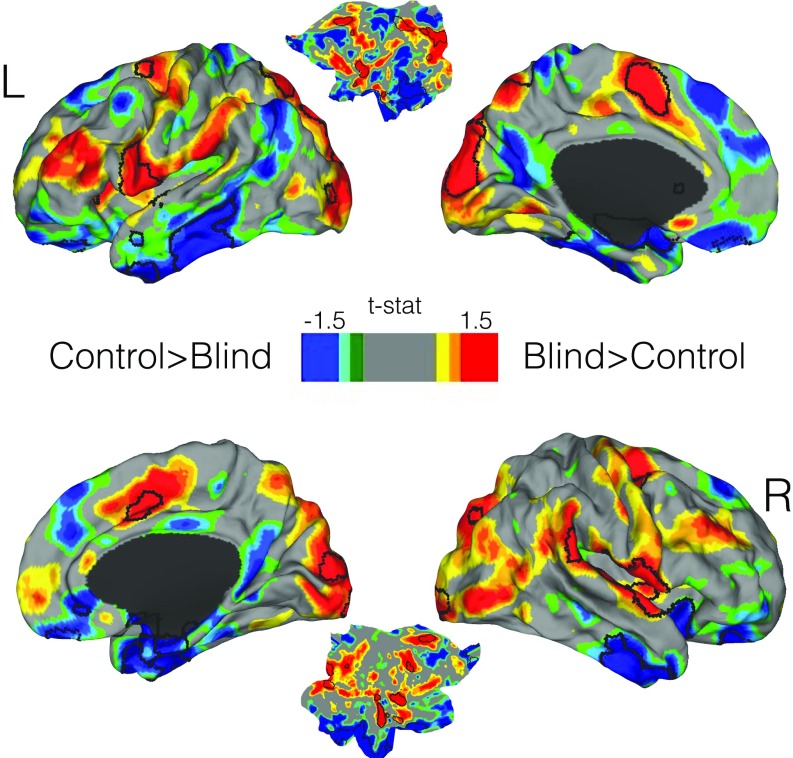
First, the whole-brain voxel-level comparison of weighted-degree maps showed that blind children compared with controls exhibited increased bilateral connectivity in extrastriate occipital cortices, middle cingulate gyri/supplementary motor area (SMA), anterior insula (AI), ventral premotor regions (vPM), and the pars opercularis of the inferior frontal gyri, with the majority of these regions belonging to the multimodal integration network. Also, notable regions that showed only trend-level functional connectivity increases in blind subjects included the parietal operculum [or operculum parietale (OP)] and dorsolateral prefrontal cortex (DLPFC) (Fig. 1 and Fig. S2). Controls compared with blind subjects showed increased bilateral functional connectivity in the anterior frontal poles, inferior temporal gyri, and superior frontal gyri.
Fig. 1.
Connectivity reorganization in blind children compared with their matched controls. A comparison of whole-brain weighted-degree functional connectivity is displayed. A cortical map based on t values is shown in which areas with greater weighted-degree values in blind children are displayed in red and those with greater weighted-degree values in sighted controls are displayed in blue. Statistically significant areas corrected for multiple comparisons are outlined by black lines, and all other areas represent only trend-level findings.
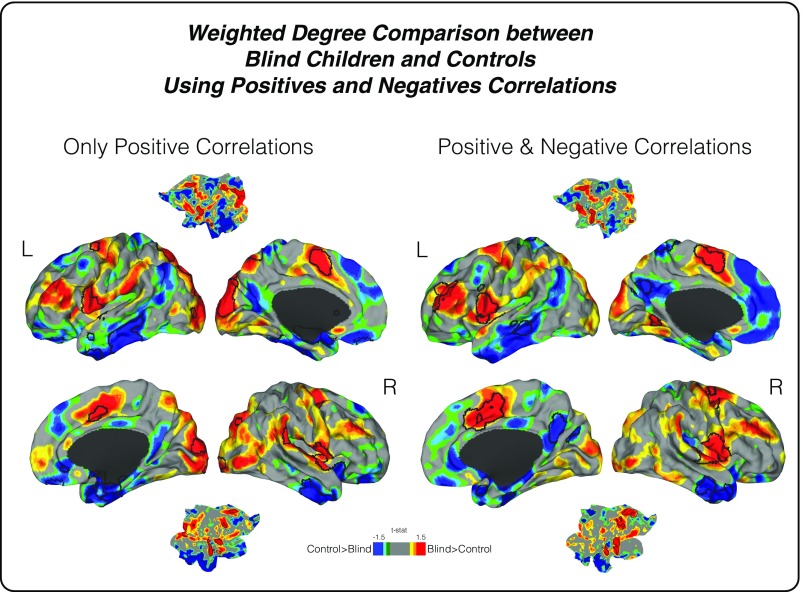
Fig. S2.
Connectivity reorganization in blind children. To compare two different methods for weighted-degree analysis, whole brain connectivity approaches (i) using only positive correlations (illustrated on the Left) and (ii) using both positive and negative correlations (illustrated on the Right) were performed. An uncorrected two-color scale map based on t values is displayed in which red areas have greater weighted-degree values in blind subjects and blue areas show higher connectivity in sighted controls. The black lines represent statistically significant regions. As noted, both analyses had similar results.
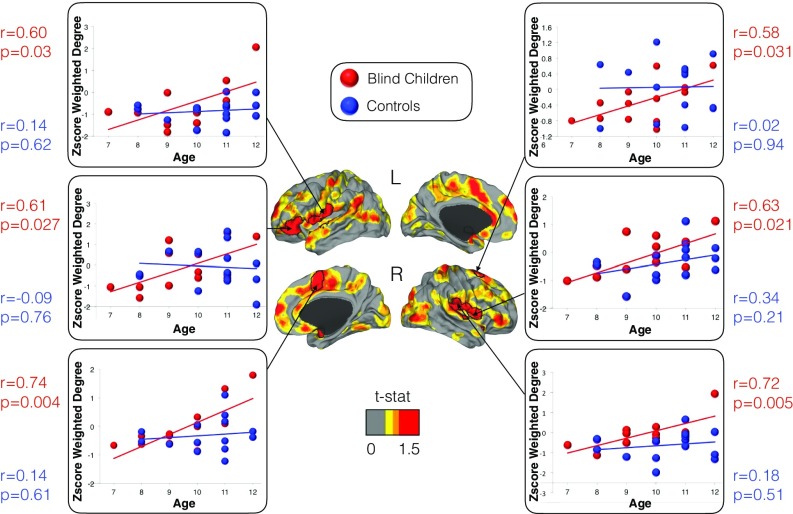
In a second analysis, a whole-brain voxel-level correlation investigation between weighted-degree maps and age in blind individuals showed statistically significant positive associations in bilateral OP, particularly OP4 and OP1; vPM; right SMA; and left inferior frontal gyrus pars triangularis and orbitalis (Fig. 2). Across all of these regions, control subjects showed no statistically significant associations between functional connectivity and age. Thus, in blind children only, the older an individual, the greater the magnitude of functional connectivity observed in multimodal integration regions.
Fig. 2.
The effect of age on the connectivity pattern of blind children. Connectivity changes in multimodal integration regions were positively correlated with age in blind children. A color scale map based on t values is shown with red areas representing the effect of age (i.e., regions with increased connectivity with age) and black lines delineating statistically significant regions corrected for multiple comparisons. In the scatter plots, the connectivity at each significant region is displayed for each subject, with dots for blind children shown in red and those of controls shown in blue. The correlation coefficients and P values are presented next to each scatter plot, again in red for blind children and blue for controls.
Specific Patterns of Connectivity Reorganization in Sensory Processing Streams.
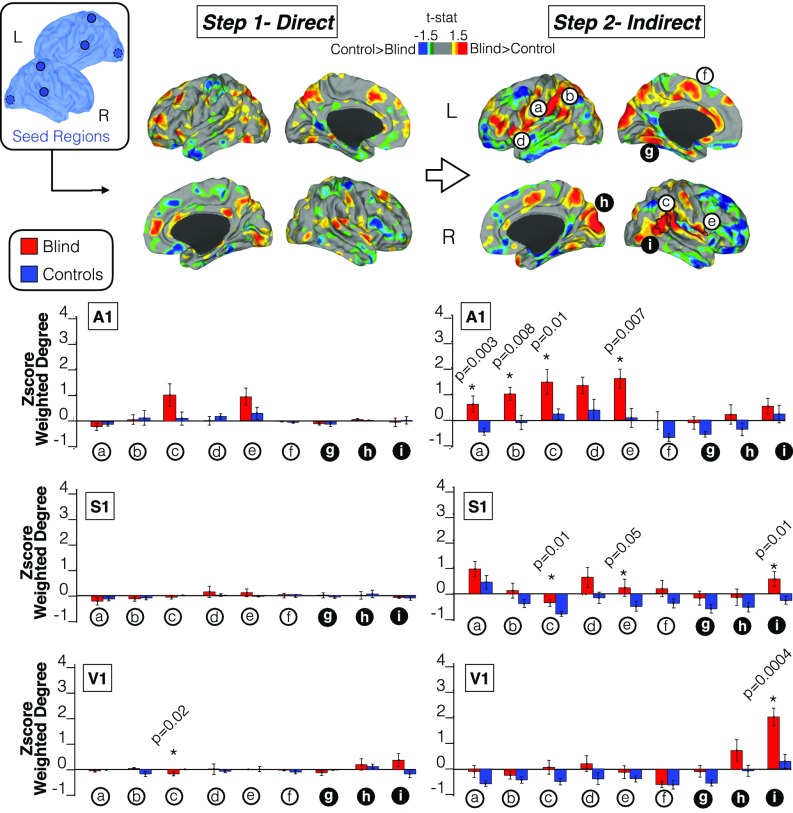
To fully delineate the connectivity reorganization occurring in blind children, we characterized the stepwise connectivity profiles of three major sensory modalities, namely, the visual (V1), auditory (A1), and somatosensory (S1) systems (Fig. 3). We investigated the differences between blind children and their matched controls using direct and indirect connectivity maps. Combined A1, S1, and V1 cortical maps, as well as sensory-specific connectivity profiles, are shown in Fig. 3 (additional cortical maps for each separate seed can be found in Figs. S3–S5).
Fig. 3.
Differences in the direct and indirect connectivity profiles between blind children and controls. The first step measures the direct connectivity of three seeds located in the primary auditory, somatosensory, and visual areas. The second step, representing indirect connectivity, measures indirect connections from the original seed in step 1. A cortical map based on t values is displayed in which areas of greater weighted-degree values are shown in red for blind children and blue for sighted controls. Please note that the areas surrounded by a black line in the cortical maps represent statistically significant regions that survived correction for multiple comparisons. Column charts underneath direct and indirect steps represent the normalized weighted-degree in blind children and controls for multimodal regions (white circle) and unimodal regions (black circle). The P value in the columns indicates the difference between the two cohorts (please note that the different colors, either red or blue, represent the areas where the connectivity is higher between the seeds and unimodal or multimodal areas, in blind children or sighted controls, respectively). Asterisk indicates statistically significant group differences. a = left OP, b = left TPJ, c = right TPJ, d = right AI, e = left AI, f = left SMA, g = right fusiform gyrus, h = right cuneus, and i = right lateral occipital cortex.
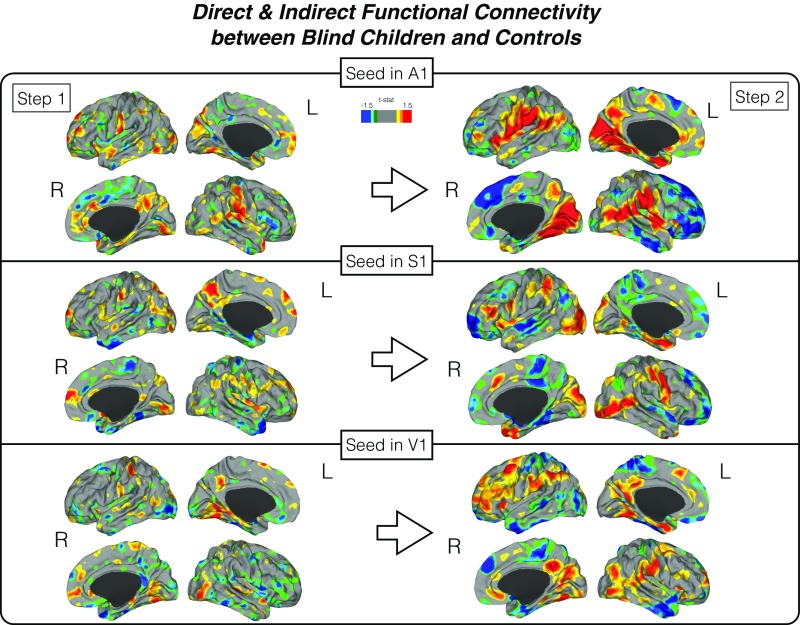
Fig. S3.
Direct and indirect functional connectivity. Differences in the direct (step 1) and indirect (step 2) connectivity between blind children and controls. The first step measures the direct connectivity from seeds located in the primary auditory, somatosensory, and visual areas. Indirect connectivity (step 2) measures indirect connections from the original seed in step 1. A two-color scale map based on t values is displayed in which areas with greater weighted-degree values are shown in red for blind children and blue for sighted controls.
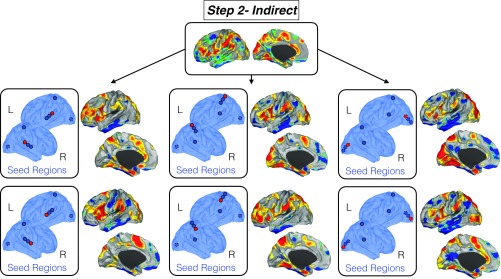
Fig. S5.
Replication of indirect functional connectivity using different primary seed locations. To check the stability of our results, we used different seed locations within the modal cortices in the indirect connectivity analysis. Top shows the map of indirect connectivity differences between blind children and controls reported in the main text. Middle and Bottom report the same map but using different primary seed locations (shown in red dots). Independently of the location of the seeds within the modal cortices, we found analogous findings.
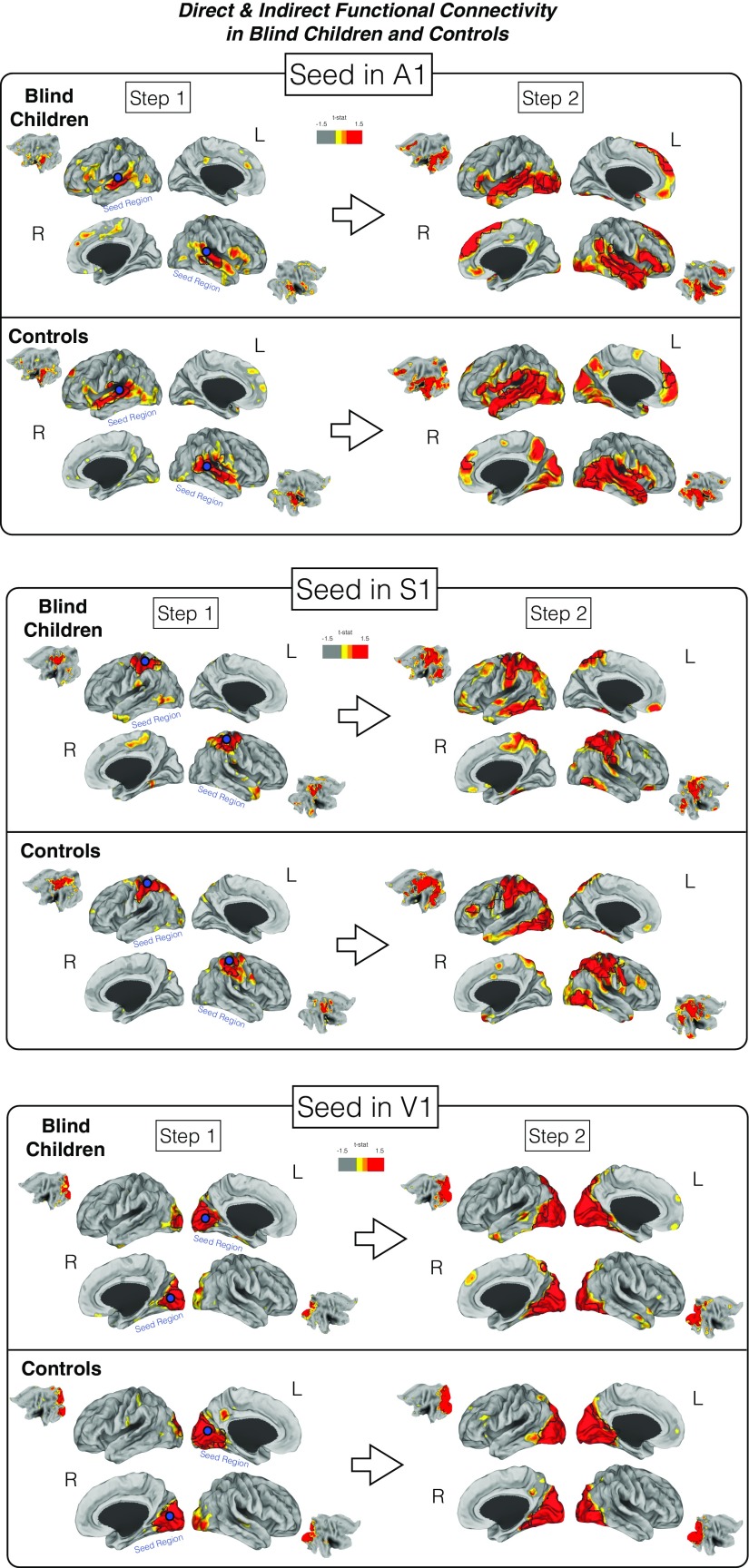
Fig. S4.
Differences in the direct (step 1) and indirect (step 2) connectivity between blind children and controls in relation to a given seed. The first step measures the direct connectivity from selected seeds located in either the primary auditory, somatosensory, or visual areas of either blind or control groups. Please note that seeds originating the analysis are represented as blue circles. Indirect connectivity (step 2) measures indirect connections from the original seed in step 1. The map based on a one-sample t test displays in red areas the greater weighted-degree values. Areas surrounded by a black line represent statistically significant regions that survived multiple comparisons correction.
There were no statistically significant differences between blind children and controls in the direct connectivity maps of primary seeds (Fig. 3, top left cortical maps; see cortical maps for each separate seed in Fig. S3). In contrast, we found that sensory systems in blind children compared with controls demonstrated statistically significant increases in connectivity beyond their direct connections. That is, sensory regions increased connectivity toward multisensory/multimodal integration brain areas including the right temporoparietal junction (TPJ) and anterior insula (Fig. 3, top right cortical maps). Trend level functional connectivity increases were also observed in fusiform gyri; vPM; dorsal/lateral occipital cortices; and dorsal anterior cingulate cortex, SMA, left insula, left TPJ, and OP1. Controls compared with blind subjects showed increased bilateral indirect or step 2 functional connectivity in the anterior frontal poles, inferior temporal gyri, and superior frontal gyri.
When we characterized the specific weight of each modality separate from the overall pattern of connectivity reorganization, the auditory and somatosensory systems drove the majority of connectivity changes, mainly involving connectivity toward multimodal integration regions. Conversely, the visual system was largely associated with adaptations within the dorsal and lateral occipital lobe (Fig. 3, column graphs) but not with multimodal areas or regions outside the occipital lobe. Moreover, auditory and somatosensory cortices reached significant portions of the visual cortex via indirect functional connectivity (Fig. 3 and Fig. S3).
Cortical Relationships of Brain Circuit–Gene Expression.
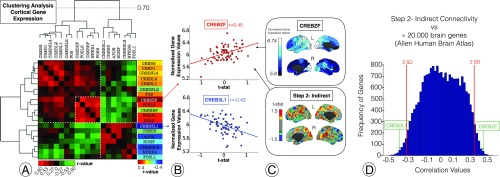
In Fig. 4, the analysis of the relationship between neuroplasticity-related genes transcription levels and brain connectivity changes in blindness is displayed. First, a gene spatial coexpression matrix based on the cortical similarities of gene transcription levels of the Allen Human Brain Atlas is shown (Fig. 4A). The hierarchical clustering analysis of the brain coexpression matrix revealed two distinctive groups of neuroplasticity-related genes (Fig. 4A; association matrix and dendrogram). When we correlated brain gene expression profiles with the connectivity changes characterized in blind subjects (Fig. 4A, right column; Fig. 4B; and Fig. 4C, bottom cortical map, or cortical map of blind subjects compared with sighted controls in the indirect connectivity), the topological distribution of sensory system connectivity reorganization showed a notable positive correlation with the expression patterns of specific members of the CREB family such as the CREBZF gene (Fig. 4B, top scatterplot, r = 0.45, P = 0.000142; Fig. 4C, top cortical map, or raw cortical map of CREBZF gene expression). Conversely, the CREB3L1 gene (Fig. 4B, bottom scatterplot; r = −0.43, P = 0.000271) exhibited a highly significant negative correlation with blind-related connectivity changes. Moreover, CREBZF and CRE3L1 are located above and below two SDs in the overall distribution of similarities between the map of functional connectivity changes in blindness and the cortical expression of ∼20,000 protein-coding genes of the human transcriptome. (For more information about other genes located above and below two standard deviations in the Fig. 4D histogram, please see Dataset S1 and Tables S3 and S4.)
Fig. 4.
Brain plasticity–-gene expression relationships in blindness. (A) An analysis of the relationship between transcription levels of neuroplasticity-related genes from a canonical human brain atlas. A coexpression matrix is shown based on the spatial similarity of transcription levels between genes. The green and red colors represent similarity in the expression between gene’s cortical expression (green for positive correlation and red for negative). (B) An analysis of the relationship between gene expression profiles and main connectivity changes in blind individuals. Note that we correlated the cortical expression profiles of genes and the functional connectivity maps after converting voxel-level connectivity profiles to the 68 Desikan–Killian regions by averaging the intensity values. The x axis represents the blind-sighted step 2 connectivity difference (from Fig. 3). The y axis represents the gene expression score. (C) The spatial distribution of the CREBZF gene (Top) and functional connectivity changes in the indirect connectivity (Bottom) represented in cortical maps. (D) A histogram is displayed presenting the location of CREBZF and CREB3L1 among all topological similarities between ∼20,000 genes and the map of indirect connectivity changes in blind subjects.
Table S3.
GO enrichment analysis of genes positively correlated in Fig. 4D
| GO enrichment annotation | Fold enrichment | P value |
| GO molecular function complete | ||
| ATP-dependent RNA helicase activity | 5.64 | 1.62E-02 |
| RNA binding | 1.80 | 4.12E-04 |
| Ion binding | 1.29 | 7.36E-03 |
| Protein binding | 1.21 | 1.33E-04 |
| GO biological function complete | ||
| tRNA metabolic process | 3.92 | 3.20E-03 |
| mRNA processing | 2.58 | 6.83E-03 |
| DNA metabolic process | 2.29 | 4.11E-04 |
| Chromosome organization | 2.00 | 4.28E-03 |
| Cellular nitrogen compound biosynthetic process | 1.44 | 1.75E-02 |
| Macromolecule modification | 1.44 | 4.97E-02 |
| Cellular macromolecule biosynthetic process | 1.43 | 1.79E-02 |
| Cellular protein metabolic process | 1.42 | 4.21E-02 |
| GO cellular component complete | ||
| Aminoacyl-tRNA synthetase multienzyme complex | 27.46 | 2.10E-02 |
| Nuclear chromosome, telomeric region | 4.13 | 3.09E-02 |
| Nuclear body | 2.23 | 7.82E-04 |
| Catalytic complex | 2.01 | 1.05E-05 |
| Mitochondrion | 1.72 | 9.15E-04 |
| Cytosol | 1.41 | 8.29E-05 |
| Reactome pathways | ||
| Gene expression | 1.74 | 7.40E-04 |
Table S4.
GO enrichment analysis of the genes negatively correlated in Fig. 4D
| GO enrichment annotation | Fold enrichment | P value |
| GO molecular function complete | ||
| System development | 1.51 | 9.58E-03 |
| GO cellular function complete | ||
| Cell periphery | 1.4 | 5.39E-03 |
| Reactome pathways | ||
| Metallothioneins bind metals | 28.60 | 1.70E-04 |
Discussion
The ability of the human brain to reorganize itself following the loss of particular functions (i.e., sight) and to develop new adaptive capabilities has intrigued the scientific community for decades. In this study, early neuroplastic changes in neural circuits were investigated by characterizing the reorganization of functional connections in a sample of blind children. We observed that multimodal integration regions associated with primary cortical systems demonstrated a high degree of connectivity reorganization and that the magnitude of this reorganization was correlated to neuroplasticity-related gene expression in the human brain.
Neuroplasticity in Multimodal Areas of Blind Children.
This study shows that changes in functional connections play a central role in cortical adaptations of blind children. Specifically, multimodal integration areas including the TPJ, OP, vPM, SMA, middle cingulate cortex, and AI displayed enhanced connections with unimodal sensory regions. These findings suggest that compared with age-matched controls, blind children have a greater tendency to establish long-range connections to regions mediating multimodal integration at an early point in their development. This reorganization may occur potentially as a compensatory mechanism to enable subjects to use information from other sensory modalities. Given the substantial neuroplastic reorganization in the multimodal integration regions of blind children and our prior findings in adults with congenital and acquired blindness (6), cross-modal neuroplasticity may involve not only unimodal-to-unimodal alterations but also potentially connectivity changes within multimodal and higher-order associative brain areas.
Historically, the visual cortex was considered to be exclusively devoted to visual processes, but there is now evidence that activity within the human visual cortex plays an active role in multisensory processes (14, 15). In recent years, the occipital cortex, as well as adjacent ventral areas such as the fusiform gyrus (16, 17), have been the main regions of interest in the study of cross-modal phenomena in blindness. Multiple studies have shown that the occipital cortex takes over multiple sensory abilities traditionally belonging to other unimodal cortices, such as auditory (18, 19), tactile (20), or higher cognitive functions, including language processing (21). However, in light of our results, neuroplastic reorganization in blindness is centralized—in network and graph theory sense—beyond the visual cortex or its direct functional connections. Thus, neural plasticity is driven by multimodal connectivity integration, in areas that seem essentially involved in the catalysis of cross-modal adaptation. For instance, our findings integrate well with previous discoveries of cross-modal integration in congenitally blind individuals (22, 23), in which increased connectivity between auditory or somatosensory cortex and specific areas of the occipital lobe is observed. Indeed, we describe similar connectivity increases but at the indirect connectivity level, meaning that a significant amount of cross-modal interactions between visual cortex and other primary systems is likely determined by multimodal relay stations. Furthermore, we theorize that functional connectivity decreases between the occipital lobe and other primary systems in blind adults (14, 15) may relate to impaired connections between primary visual areas and the multimodal integration network and that—potentially as an adaptive phenomenon—increased connectivity between somatosensory or auditory cortices and multimodal areas develop (both phenomena seen in our findings).
Gogtay et al. (10) demonstrated in 4- to 21-y-old subjects that association cortices mature only after lower-order visual and sensory cortices and that phylogenetically older brain areas mature earlier than newer areas. The present functional connectivity study adds to our understanding of the functional connectivity profiles that emerge over the course of early life in blind children. In the blind cohort, connectivity adaptations within multimodal integration areas increased as a function of age (Fig. 2). Notably, this finding suggests that older blind children approximate the connectivity profiles of blind adults (6). Collignon et al. (24) recently demonstrated that transient early-life visual deprivation is associated with long-lasting large-scale cortical reorganizational changes. Although this finding implicates sensory deprivation in the first year of life as facilitating cross-modal plasticity, the within-group analyses of our study suggest that this network reorganization continues to emerge throughout early life in children with sustained blindness.
Neuroplasticity and Genes.
The molecular mechanisms through which neuroplasticity reorganizes the human brain remain unknown. To bridge this gap between connectivity reorganization and the biological basis of neuroplasticity in blindness, we analyzed the spatial distribution of several a priori-selected genes, involved in plasticity, in the human cerebral cortex. The relationship between synchronous blood oxygen level-dependent (BOLD) signal in the brain and the molecular mechanisms underlying it has been explored previously including work by Wang et al. (25), where it was found that a specific group of genes correlated with the resting state activity in the default mode network. Richiardi et al. (26) found evidence that resting state activity correlated with the activity of genes linked to ion channel activity and synaptic function. Thus far, to our knowledge no study has investigated the relationship between neuroplasticity (as measured by functional connectivity) and gene expression. Blindness in children offers a unique opportunity to begin to study this question. The expression of CREB (13) family genes, which are involved in learning and memory, exhibited robust positive spatial associations with the functional connectivity changes of blind children. CREB is a cellular transcription factor that binds to the cAMP response element CRE, increasing the transcription of the somatostatin gene (27). The activation of CREB-dependent gene expression is a crucial step in the molecular cascade that mediates the formation of long-lasting memories. This view is based on both correlative evidence and functional assays that demonstrate, through loss- and gain-of-function experiments, the effect of CREB manipulation on memory performance. Mechanistically, the role of CREB in memory formation is thought to be a consequence of its participation in long-lasting forms of synaptic plasticity including long-term potentiation, as well as modulation of neuronal excitability (12). More broadly, CREB is a key component in diverse physiological processes, including nervous system development and cell survival.
In the developing brain, CREB regulates neuronal plasticity, in part, through structural neural remodeling (28). Accumulating evidence has shown that dendritic spines appear and disappear on a daily basis and that the rate of turnover is increased in response to sensory experience (29, 30), novelty (31), and memory formation (32–34). An emerging model suggests that learning is supported through a balance of spine formation and elimination (35). Within the visual system, it has been shown that CREB expression during the development of the superior colliculus correlates with that of the retinocollicular pathway in mice (36) and cats (37). Moreover, CREB changes in V1 occur after visual deprivation in monkeys (38). Our findings build upon these observations and suggest that CREB-related gene expression drives cortical neuroplastic reorganization in multimodal and higher-order associative brain areas. The magnitude of CREBZF expression was positively associated with increased functional connectivity in blind children. CREBZF activates transcription factors p53 and ATF4 and suppresses the activity of a number of other transcription factors including nuclear receptors containing proteins such as CREB3 (39) (note the inverse associations between these two genes in Fig. 4A). Thus, sensory deprivation in blind children may modulate the expression of different types of CREB-family genes to promote adaptive network-specific neuroplastic changes.
A potential limitation of this study is the comparison of cortical connectivity maps from a child cohort with adult gene expression profiles from the Allen Human Brain Atlas. Currently, the Allen Human Brain Atlas is the only source of high-resolution gene expression data available for neuroanatomical comparisons. Future studies using an analogous dataset of child brain gene expression profiles should seek to replicate the findings of our study. In addition, BOLD resting-state functional MRI (rs-fMRI) measures intrinsic activity correlations between brain regions that seem to be sufficiently constrained by anatomy to reveal informative estimates of connectivity properties (40, 41). However, it is important to emphasize that fcMRI can reflect monosynaptic and polysynaptic connectivity; thus, it should not be considered a direct measure of tract anatomical connectivity. Last, future studies will be needed to investigate if the magnitude of functional connectivity alterations in multimodal integration regions is predictive of adaptive cross-modal processing.
Conclusions
Our study shows that neuroplastic reorganization in blind children involves key multisensory integration regions. We demonstrate significant relationships between the expression of neuroplasticity-related genes and enhanced connectivity in blind children. These findings present opportunities to understand neuroplastic changes in the human brain and may improve future therapeutic treatments for sensory deprivation.
Methods
Participants.
We recruited 17 blind children (8 girls and 9 boys; mean age = 9.4 y, SD = 1.4; range = 7–12 y) and 18 sighted matched controls (10 girls and 8 boys; mean age = 9.9 y, SD = 1.7; range = 8–12 y). Following subject exclusion due to excessive head motion (see exclusion parameters below), the final cohorts for analysis included 13 blind children (4 girls and 9 boys; mean age = 9.6 y, SD = 1.3) and 15 sighted controls (9 girls and 6 boys; mean age = 10.3 y, SD = 1.4). All children had a similar scholastic performance, educational level, and cultural background. Causes of blindness are described in Table S1. Inclusion criteria for the participants were (i) age between 7 and 12 y, (ii) actively participating in school, and (iii) normal scholastic performance. Exclusion criteria were the presence of (i) another sensorial deficit other than blindness, (ii) comorbid neuropsychiatric conditions, and (iii) history of obstetric trauma with cerebral hypoxia. The research protocol was approved by the Ethical Committee of the Hospital Clínico Universitario San Carlos (Madrid) and was in full compliance with the Declaration of Helsinki.
Table S1.
Demographic and clinical characteristics of blind children cohort
| ID | Age, y | Blindness | Light Perception | Handedness | Causes of Blindness |
| B01 | 8 | 6 y old | Yes | Right | Pigmentary retinal dystrophy |
| B02 | 11 | Birth | Yes | Right | Hereditary retinal dystrophy |
| B03 | 8 | Birth | Yes | Right | Pigmentary retinal dystrophy |
| B04 | 9 | Birth | Yes | Right | Optic disk coloboma |
| B05 | 7 | Birth | No | Right | Hereditary retinal dystrophy |
| B06 | 10 | Birth | Yes | Right | Partial optic atrophy |
| B07 | 9 | Birth | Yes | Right | Hereditary optic atrophy |
| B08 | 9 | Birth | Yes | Right | Pigmentary retinal dystrophy |
| B09 | 10 | Birth | No | Right | Anophthalmia |
| B10 | 10 | 1 y old | Yes | Right | Optic atrophy |
| B11 | 12 | 3 y old | Yes | Right | Optic atrophy |
| B12 | 11 | Birth | Yes | Right | Anophthalmia |
| B13 | 10 | Birth | No | Right | Bilateral hypoplasia of the optic nerves (intrauterine varicella-zoster virus infection) |
Each school was provided with written information about the experiment. After principals and teachers approved the research protocol, a formal presentation was organized for all enrolled students, and parents and teachers were provided with detailed information (verbal and written) about the nature and purpose of the research study. Interested parents provided written consent following individual information sessions. Children were also allowed to ask questions in the context of group and individual sessions, and all participants provided assent.
Data Acquisition and Image Preprocessing.
Data acquisition and image preprocessing details are provided in Supporting Information.
Image Postprocessing.
Weighted-degree analysis.
We used weighted-degree analysis to compare group-level functional connectivity profiles between blind and control children and to analyze within-group network correlations with age. Weighted-degree analysis provided the first approximation toward the topological characterization of functional connectivity adaptations in blind children. To obtain the functional connectivity matrices to perform a whole-brain graph analysis, the following steps were applied to the functional connectivity (fcMRI) data at the individual subject level. First, the Pearson’s r correlation coefficient was computed between pairs of voxels across the whole brain using the time course of low-frequency BOLD fluctuations. We then used two complementary strategies for our data: For the first strategy, we applied a false discovery rate correction [Benjamini and Hochberg (42)] at the q = 0.001 threshold level to correct for false positives in the association matrices. This method, which considers only positive correlations, introduces a customized r threshold in each individual that allows for the elimination of network links with low temporal correlation, which are likely to be attributable to noise. For the second strategy, we used both positive and negative correlations without introducing any cutoffs in the association matrices (Fig. S2). Because there were no major differences between strategies, we report the first method in the main text (3). Subsequently, we applied a Fisher transformation to the Pearson’s r correlation coefficient for variance stabilization (4). To evaluate the connectivity strength of each node, we calculated the weighted degree (W; see Eqs. 1 and 2) of each subject by summing all Fisher-transformed correlation values for the nodes in the association matrix (Fig. S2) (5). Finally, we applied a Z-score normalization to the individual weighted-degree maps to gain topological interpretability across the sample.
| [1] |
where
| [2] |
n is the number of nodes in the graph network, and c is the correlation matrix used to calculate the weighted degree.
To compute the weighted-degree differences between blind children and their matched controls, a two-sample t test was applied using age and mean frame displacement as nuisance covariates (41) (Fig. 1). To correct for multiple comparisons, a clusterwise correction was applied. Monte Carlo simulation (3dClustSim, Analysis of Functional Neuroimages, afni.nimh.nih.gov) was performed with 10,000 iterations to estimate the probability of false positive clusters with a P value <0.05 in the analysis. Only results beyond this threshold were considered. To investigate age as a covariate of interest related to neuroplastic changes in blind children, a general linear model was used to quantify its influence (β). Then the t statistic of the mean effect of age in blind children and its P value were computed. Finally, a clusterwise correction accounting for multiple comparisons was applied to detect statistically significant regions correcting for type I errors. Areas surviving multiple comparisons correction (displayed as outlined by black lines in the cortical maps) were selected to visualize the association between age and individual Z-score weighted-degree values (Fig. 2).
Direct and indirect connectivity of sensory systems.
To further characterize the specific networks implicated in sensory and multisensory connectivity adaptations in blind children, we used a second graph theory-based approach, stepwise functional connectivity analysis (7). Given that plastic changes are expected to occur in multiple relay stations across perceptual systems, we used a method that detects both direct and indirect voxel-level functional couplings for a given sensory region. First, we selected three seeds corresponding to unimodal A1, S1, and V1 primary cortices [see Table S2 for Montreal Neurological Institute (MNI) coordinates based on Sepulcre et al. (7), and see Fig. S5 for alternative seeds along the primary cortices]. Then, we identified the position of the seed voxels in the association connectivity matrices of each individual. We subsequently detected and summed the weight of all of the links that were directly connected to each seed region, or a combination of seeds, taking into account only closely connected neighboring nodes showing connectivity with at least one other directly connected node (Fig. S1 and Eq. 3). We then detected and summed all possible weights of indirect connections starting from directly connected nodes (Fig. S1 and Eq. 4). Thus, for this study, we adapted the original stepwise connectivity approach by removing connections that were not directly related to the neighborhood of the sensory seed region within a distance of two links (Fig. S1). Finally, we normalized the final direct and indirect connectivity maps of the sensory modalities using a Z-score approach similar to the weighted-degree approach. To highlight common connectivity effects, combined maps of A1, S1, and V1 from direct and indirect connectivity analysis are presented in Fig. 3. We also extracted the connectivity profiles of the most significant regions within the direct and indirect connectivity maps to better visualize the amount of connections taking place at the regional level. The same regions chosen from indirect connectivity maps were also evaluated in direct connectivity maps to visualize changes in functional connectivity patterns (column charts in Fig. 3).
Table S2.
MNI coordinates for the seeds for the stepwise connectivity (direct and indirect connectivity) analysis
| Region name | MNI coordinates |
| Left primary visual cortex (V1) | −14, −78, 8 |
| Right primary visual cortex (V1) | 10, −78, 8 |
| Left primary auditory cortex (A1) | −54, −14, 8 |
| Right primary auditory cortex (A1) | 58, −14, 8 |
| Left primary somatosensory cortex (S1) | −42, −29, 65 |
| Right primary somatosensory cortex (S1) | 38, −29, 65 |
All reported MNI coordinates correspond to the most lateral, anterior, and superior voxel of the seed.
For the direct connectivity (DC) approach, we applied the following:
| [3] |
A whole-brain voxel-level analysis was computed, where r is each one of the voxels of the brain, c is the connectivity matrix, is the position of the voxel of the selected seed (A1, S1, or V1) inside the matrix, s represents the seed selected where the stepwise analysis begins, and S is the total number of seeds.
For the indirect connectivity (IC) approach, we applied the following:
| [4] |
Multimodal integration network.
Base on Sepulcre et al. (7) and Driver et al. (43), our use of the term “multimodal integration network” includes the DLPFC [Brodmann area (BA) 46], frontal eye fields (BA6), AI, vPM cortex (BA6op), OP1/OP2 (BA40), OP3/OP4 (BA40/43), TPJ (BA39), SMA (BA6), dorsal anterior cingulate cortex (BA 24–32), superior parietal cortex (BA7), and lateral occipital cortex (BA19).
Relationship between gene expression and increased brain connectivity in blind subjects.
We used the Allen Human Brain Atlas, which provides whole-brain genome-wide expression patterns for six human subjects (44), to investigate whether genetic transcription profiles underlie the ability of some brain areas to display adaptive changes after sensory deprivation. In particular, we used a previously generated surface anatomical transformation of the transcription profiles of 20,737 protein-coding genes, based on 58,692 measurements of gene expression in 3,702 brain samples obtained from the 6 adult human subjects of the Allen Human Brain Atlas (45). This anatomical transformation, based on the 68 cortical regions of the Desikan–Killian atlas (46) covering the entire cortex, used individual vectors of the median expression of genes across prespecified cortical brain regions. In a priori analysis, we first selected genes previously involved in brain plasticity (12, 13) (i.e., CREB family genes; list of genes is displayed in Fig. 4). Second, we computed the cortical spatial similarity among them using the magnitude of cortical expression profiles throughout the Desikan–Killian areas and a Pearson correlation approach. Third, we performed a hierarchical clustering analysis of all spatial associations of the CREB family genes to identify clustered genes with related cortical patterns. Fourth, we investigated the spatial cortical similarity between these CREB family genes and the map of significant functional connectivity changes observed in blind subjects. We used again Pearson coefficients to correlate the cortical expression profiles of genes and the functional connectivity map after its conversion from voxel-level to 68 Desikan–Killian regions, using the average intensity. Finally, we calculated the spatial similarity between the significant functional connectivity map and all 20,737 genes transcription cortical maps of the Allen Human Brain Atlas (histogram in Fig. 4D). Therefore, we built a null distribution to validate the associations between the CREB gene family cortical expression and the observed connectivity changes in the blind group. We considered 2 SDs (above and below) as the statistically significant cutoff.
Network and cortical visualization.
Cortical data were visualized on the brain surface using the population-average landmark- and surface-based surface of CARET software (47, 48). Surface images were displayed using a color scale based on t values in Figs. 1–4. Regions surviving multiple comparisons testing were outlined by a black line in the cortical maps. The rest of the image represents the uncorrected values allowing for the inspection of trend connectivity.
SI Methods
Data Acquisition.
Blind children and their controls were scanned on a 3-T (ACHIEVA 3.0T TX; Philips) magnetic resonance imaging (MRI) scanner using an eight-channel phased-array head coil. High-resolution 3D T1-weighted magnetization-prepared rapid acquisition gradient echo (T1W 3D TFE SENSE) images were acquired for anatomic reference (TR = 7.6 ms, TE = 3.5 ms, flip angle = 7 µ, 1.0-mm isotropic voxels). Functional data were acquired using an echo planar imaging pulse sequence sensitive to BOLD contrast (TR = 3,000 ms, TE = 30 ms, flip angle = 90 µ, 3.0-mm isotropic voxels, sequence length= 6 min 12 s, 124 time points). In all cases, head motion was restricted using a pillow and several foam pads, and earplugs were used to attenuate scanner noise. During the functional runs, participants were asked to stay awake and to remain as still as possible. Parents were allowed inside the scanner room to help participants remain still and comfortable.
Image Preprocessing.
rs-fMRI datasets were processed using the MATLAB toolbox, Data Processing Assistant for Resting State (49), based on certain functions of Statistical Parametric Mapping 12 (www.fil.ion.ucl.ac.uk/spm/) software. rs-fMRI preprocessing included (i) removing the first four volumes of each run to allow for T1 equilibration effects; (ii) performing slice-timing correction; (iii) realigning the functional volumes for each subject using a six-parameter (rigid body) linear transformation to correct for head movement within runs; (iv) coregistering individual structural images (T1-weighted images) to the mean functional image; (v) segmenting T1-weighted images into gray matter, white matter, and cerebrospinal fluid; (vi) removing sources of spurious variance from the data through linear regression including 24 motion-related covariates derived from volume-realignment parameters(50, 51), the signal averaged from the deep cerebral white matter and lateral ventricles [global signal regression was not included as a nuisance covariate to prevent the artifactual introduction of negative correlations (52)], and the polynomial quadratic trend; (vii) applying spatial normalization to MNI space (3 mm3) using the parameters from the segmentation of each participant’s high-resolution anatomical acquisition; (viii) conducting spatial smoothing with an isotropic Gaussian kernel of 6-mm FWHM; (ix) applying temporal band-pass filtering (0.01–0.08 Hz) to reduce the effect of low-frequency drift and high-frequency noise (40, 53); and finally, (x) applying downsampling to 8 mm to our data to keep network relationships at the voxel level and to avoid transformation of the images into regions of interest. Motion was quantified using realignment parameters obtained during image preprocessing, which included three translation and three rotation estimates. Subjects with mean global movement greater than 1.5 mm were excluded from further analyses. Scrubbing of the time points with excess head motion (frame displacement > 0.5 mm) was performed through interpolation using an algorithm previously reported in Jenkinson et al. (54). Furthermore, subjects with more than 3 min of discarded time points (time points with a frame displacement > 0.5 mm) were eliminated (55). In both blind and control cohorts, the amount of time points per run discarded was small (less than 5 volumes). All subjects had the same total number of MRI volumes. As an additional corrective measure, global head motion was entered as a nuisance variable in all analyses performed in this study (56).
Gene Ontology Enrichment Analysis.
We ran a Gene Ontology (GO) Enrichment analysis with all of the genes located above and below 2 SD (Tables S3 and S4), and we observed that our genetic profiles mainly relate to transcriptional functions and system development.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health (2T32EB013180-06 to L.O.-T. and K23EB019023 to J.S.), Fundación Mutua Madrileña (4131220 to T.O.), and the Bizkaia Talent and European Commission (AYD-000-289 to I.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.A.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619121114/-/DCSupplemental.
References
- 1.Murray MM, Matusz PJ, Amedi A. Neuroplasticity: Unexpected consequences of early blindness. Curr Biol. 2015;25:R998–R1001. doi: 10.1016/j.cub.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 2.Poirier C, et al. Auditory motion perception activates visual motion areas in early blind subjects. Neuroimage. 2006;31:279–285. doi: 10.1016/j.neuroimage.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 3.Saenz M, Lewis LB, Huth AG, Fine I, Koch C. Visual motion area MT+/V5 responds to auditory motion in human sight-recovery subjects. J Neurosci. 2008;28:5141–5148. doi: 10.1523/JNEUROSCI.0803-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collignon O, et al. Impact of blindness onset on the functional organization and the connectivity of the occipital cortex. Brain. 2013;136:2769–2783. doi: 10.1093/brain/awt176. [DOI] [PubMed] [Google Scholar]
- 5.Striem-Amit E, Amedi A. Visual cortex extrastriate body-selective area activation in congenitally blind people “seeing” by using sounds. Curr Biol. 2014;24:687–692. doi: 10.1016/j.cub.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Ortiz-Terán L, et al. Brain plasticity in blind subjects centralizes beyond the modal cortices. Front Syst Neurosci. 2016;10:61. doi: 10.3389/fnsys.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepulcre J, Sabuncu MR, Yeo TB, Liu H, Johnson KA. Stepwise connectivity of the modal cortex reveals the multimodal organization of the human brain. J Neurosci. 2012;32:10649–10661. doi: 10.1523/JNEUROSCI.0759-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krienen FM, Yeo BTT, Ge T, Buckner RL, Sherwood CC. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc Natl Acad Sci USA. 2016;113:E469–E478. doi: 10.1073/pnas.1510903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer-Lindenberg A. The future of fMRI and genetics research. Neuroimage. 2012;62:1286–1292. doi: 10.1016/j.neuroimage.2011.10.063. [DOI] [PubMed] [Google Scholar]
- 10.Gogtay N, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci USA. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sporns O. Network attributes for segregation and integration in the human brain. Curr Opin Neurobiol. 2013;23:162–71. doi: 10.1016/j.conb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Benito E, Barco A. CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matusz PJ, Retsa C, Murray MM. The context-contingent nature of cross-modal activations of the visual cortex. Neuroimage. 2016;125:996–1004. doi: 10.1016/j.neuroimage.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Tal Z, Geva R, Amedi A. The origins of metamodality in visual object area LO: Bodily topographical biases and increased functional connectivity to S1. Neuroimage. 2016;127:363–375. doi: 10.1016/j.neuroimage.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siuda-Krzywicka K, et al. Massive cortical reorganization in sighted braille readers. Elife. 2016;5:1–26. doi: 10.7554/eLife.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hölig C, Föcker J, Best A, Röder B, Büchel C. Crossmodal plasticity in the fusiform gyrus of late blind individuals during voice recognition. Neuroimage. 2014;103:374–382. doi: 10.1016/j.neuroimage.2014.09.050. [DOI] [PubMed] [Google Scholar]
- 18.Amedi A, et al. Shape conveyed by visual-to-auditory sensory substitution activates the lateral occipital complex. Nat Neurosci. 2007;10:687–689. doi: 10.1038/nn1912. [DOI] [PubMed] [Google Scholar]
- 19.Striem-Amit E, Cohen L, Dehaene S, Amedi A. Reading with sounds: sensory substitution selectively activates the visual word form area in the blind. Neuron. 2012;76:640–652. doi: 10.1016/j.neuron.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Amedi A, Jacobson G, Hendler T, Malach R, Zohary E. Convergence of visual and tactile shape processing in the human lateral occipital complex. Cereb Cortex. 2002;12:1202–1212. doi: 10.1093/cercor/12.11.1202. [DOI] [PubMed] [Google Scholar]
- 21.Bedny M, Pascual-Leone A, Dodell-Feder D, Fedorenko E, Saxe R. Language processing in the occipital cortex of congenitally blind adults. Proc Natl Acad Sci USA. 2011;108:4429–4434. doi: 10.1073/pnas.1014818108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinge C, Eippert F, Röder B, Büchel C. Corticocortical connections mediate primary visual cortex responses to auditory stimulation in the blind. J Neurosci. 2010;30:12798–12805. doi: 10.1523/JNEUROSCI.2384-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawellek DJ, et al. Altered intrinsic neuronal interactions in the visual cortex of the blind. J Neurosci. 2013;33:17072–17080. doi: 10.1523/JNEUROSCI.1625-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collignon O, et al. Long-lasting crossmodal cortical reorganization triggered by brief postnatal visual deprivation. Curr Biol. 2015;25:2379–2383. doi: 10.1016/j.cub.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 25.Wang GZ, et al. Correspondence between resting-state activity and brain gene expression. Neuron. 2015;88:659–666. doi: 10.1016/j.neuron.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richiardi J, et al. IMAGEN Consortium Brain networks. Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348:1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 28.Ortega-Martínez S. A new perspective on the role of the CREB family of transcription factors in memory consolidation via adult hippocampal neurogenesis. Front Mol Neurosci. 2015;8:46. doi: 10.3389/fnmol.2015.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trachtenberg JT, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 30.Holtmaat AJGD, et al. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Kitanishi T, Ikegaya Y, Matsuki N, Yamada MK. Experience-dependent, rapid structural changes in hippocampal pyramidal cell spines. Cereb Cortex. 2009;19:2572–2578. doi: 10.1093/cercor/bhp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu T, et al. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran I, Laplante I, Lacaille J-C. CREB-dependent transcriptional control and quantal changes in persistent long-term potentiation in hippocampal interneurons. J Neurosci. 2012;32:6335–6350. doi: 10.1523/JNEUROSCI.5463-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 2010;33:121–129. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Vierci G, et al. Creb is modulated in the mouse superior colliculus in developmental and experimentally-induced models of plasticity. Int J Dev Neurosci. 2013;31:46–52. doi: 10.1016/j.ijdevneu.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Yu L, Rowland BA, Stein BE. Initiating the development of multisensory integration by manipulating sensory experience. J Neurosci. 2010;30:4904–4913. doi: 10.1523/JNEUROSCI.5575-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lalonde J, Chaudhuri A. Dynamic changes in CREB phosphorylation and neuroadaptive gene expression in area V1 of adult monkeys after monocular enucleation. Mol Cell Neurosci. 2007;35:24–37. doi: 10.1016/j.mcn.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Zhang R, et al. Zhangfei/CREB-ZF—A potential regulator of the unfolded protein response. PLoS One. 2013;8:e77256. doi: 10.1371/journal.pone.0077256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 41.Van Dijk KR, et al. Intrinsic functional connectivity as a tool for human connectomics: Theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 43.Driver J, Noesselt T. Multisensory interplay reveals crossmodal influences on ‘sensory-specific’ brain regions, neural responses, and judgments. Neuron. 2008;57:11–23. doi: 10.1016/j.neuron.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.French L, Paus T. A FreeSurfer view of the cortical transcriptome generated from the Allen Human Brain Atlas. Front Neurosci. 2015;9:323. doi: 10.3389/fnins.2015.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 47.Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28:635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- 48.Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 49.Chao-Gan Y, Yu-Feng Z. DPARSF: A MATLAB toolbox for “pipeline” data analysis of resting-state fMRI. Front Syst Neurosci. 2010;4:13. doi: 10.3389/fnsys.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friston KJ, Williams S, Howard R, Frackowiak RS, Turner R. Movement-related effects in fMRI time-series. Magn Reson Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- 51.Satterthwaite TD, et al. Impact of in-scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: Are anti-correlated networks introduced? Neuroimage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowe MJ, Mock BJ, Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. Neuroimage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- 54.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 55.Yan CG, et al. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satterthwaite TD, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.