Significance
Interactions between RNA and protein molecules are critical for many cellular processes. Bacterial cells rely on RNA–protein interactions to regulate gene expression in response to an ever-changing environment. To understand such regulation, it is key to identify the processes controlled by RNA-binding proteins. In this study, we have taken a RNA ligand-centered approach to chart the physiological processes controlled by a class of RNA-binding proteins harboring the highly conserved cold-shock domain. This approach revealed cold-shock proteins CspC and CspE to be critical for the stress response and virulence in the enterobacterial pathogen Salmonella enterica serovar Typhimurium, emphasizing RNA-binding proteins as major players in bacterial infection.
Keywords: RNA-binding protein, cold-shock protein, Salmonella, bacterial pathogenesis, stress response
Abstract
The functions of many bacterial RNA-binding proteins remain obscure because of a lack of knowledge of their cellular ligands. Although well-studied cold-shock protein A (CspA) family members are induced and function at low temperature, others are highly expressed in infection-relevant conditions. Here, we have profiled transcripts bound in vivo by the CspA family members of Salmonella enterica serovar Typhimurium to link the constitutively expressed CspC and CspE proteins with virulence pathways. Phenotypic assays in vitro demonstrated a crucial role for these proteins in membrane stress, motility, and biofilm formation. Moreover, double deletion of cspC and cspE fully attenuates Salmonella in systemic mouse infection. In other words, the RNA ligand-centric approach taken here overcomes a problematic molecular redundancy of CspC and CspE that likely explains why these proteins have evaded selection in previous virulence factor screens in animals. Our results highlight RNA-binding proteins as regulators of pathogenicity and potential targets of antimicrobial therapy. They also suggest that globally acting RNA-binding proteins are more common in bacteria than currently appreciated.
The myriad of coding and noncoding RNAs in a cell generally do not act in isolation but rapidly associate with RNA-binding proteins (RBPs) to execute their functions. Recent methods relying on the global capture of polyadenylated transcripts in eukaryotes have dramatically expanded our knowledge of RBP activity. These approaches have revealed many previously unsuspected RBPs, suggesting a much wider scope of posttranscriptional control based on thousands of new RBP–mRNA interactions (1, 2). Nevertheless, the functions of many of these newly discovered RBPs remain unclear.
In contrast to the situation in eukaryotes, there is a paucity of knowledge about RBPs in prokaryotes. This lack of knowledge is compounded further by the lack of a poly(A) tail on functional transcripts, which precludes similar global discovery studies of bacterial RBP networks. Extensive profiling of cellular targets of the small RNA (sRNA) chaperone Hfq and the translational repressor CsrA (3–7) have shown that large posttranscriptional networks also exist in bacteria. In addition, the ProQ protein has recently been identified as a previously overlooked global RBP in Salmonella enterica (8). As new methods are developed to identify RPBs in bacteria globally, it is essential to be able to analyze their functions systematically to understand their cellular and physiological roles (9).
Cold-shock proteins (CSPs) constitute the largest nonribosomal RBP family in Gram-negative bacteria, including the model species Escherichia coli and Salmonella enterica. Their conserved nucleic acid-binding cold-shock domain makes them members of a widespread RBP superfamily that includes the well-investigated eukaryotic Y-box proteins (10, 11). Of the nine and six CSP paralogs present in E. coli and S. enterica, respectively, CspA is the best characterized in terms of function (12–14). CspA is an RNA chaperone that accumulates during growth at low temperatures and modulates both the transcription and translation of target genes required for bacterial survival in these conditions (15, 16). Intriguingly, other family members, such as CspC and CspE, are not induced in response to cold shock but are highly expressed at higher temperatures. Although their cellular abundance (17) and high sequence conservation (18) suggest important functions in bacteria, the physiological roles of these other CSPs remain to be fully understood. Is their function to safeguard the bacteria until CspA is expressed upon a drop in environmental temperature, or do they serve temperature-independent functions as suggested by earlier work in E. coli (19, 20)? Given their shared RNA-binding domain, do all CSPs target the same transcripts, or is there functional specialization?
Here, we report a comparative analysis of the RNA targets of all CSPs in the facultative intracellular bacterial pathogen S. enterica serovar Typhimurium (henceforth “Salmonella”), discovering an essential cold-shock–independent function of CspC and CspE in bacterial virulence. We show that the activities of these two RBPs affect a fifth of the Salmonella transcriptome, including many genes involved in the stress response and virulence.
Importantly, our approach prioritizes an association of in vivo RNA targets with cellular pathways over direct phenotypic assessment, and we show how this strategy overcomes a problematic molecular redundancy of CspC and CspE that likely explains why these proteins have evaded selection in previous virulence factor screens in animals (21). Our results with the CSP family demonstrate that RNA ligand-centric approaches are a viable route in the challenging quest to unravel RBP networks in bacteria.
Results and Discussion
RNA Ligand Profiles Suggest Functional Specialization of Salmonella CSPs.
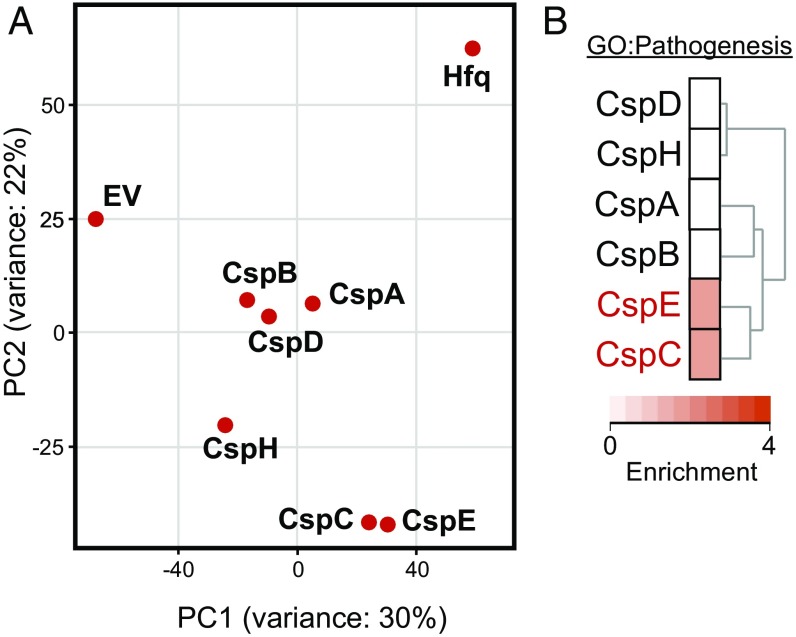
To understand the physiological roles of the CSP family members in Salmonella, we applied an RNA ligand-centric approach to reveal the cellular processes in which they are involved. The screening was performed with bacteria grown at 37 °C to early stationary phase, a condition in which host cell invasion genes are expressed (22, 23). Building on our previous successful approach to define RNA targets of Hfq in vivo (3), we compared RNA ligand profiles among the six Salmonella CSPs derived from RNA immunoprecipitation and high-throughput sequencing (RIP-seq) by principle component analysis (PCA) and functional enrichment analysis (Fig. 1 and SI Appendix, Fig. S1 and Dataset S1). The PCA revealed two clusters formed by CspA/CspB/CspD and by CspC/CspE, respectively, indicating that the proteins within each cluster bind similar sets of RNA ligands (Fig. 2A). In contrast, neither CspH nor Hfq (the latter used as a positive control for RNA pull-down) clustered with any of the other proteins. Interestingly, clustering in the PCA also reflects the phylogenetic relationship of CSP amino acid sequences from E. coli, with two major evolutionary branches separating CspH from the rest of the CSPs and with the latter branch being further divided into clades that separate CspC/CspE from CspA/CspB (18). A similar grouping was observed by applying Pearson correlation and fold-change cluster analysis (SI Appendix, Fig. S1).
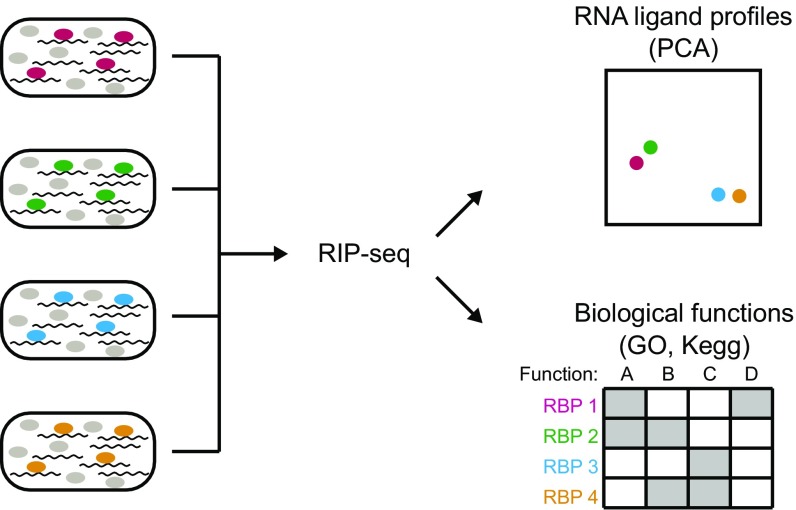
Fig. 1.
RNA ligand profiling as a means to predict RBP function. Shown is the schematic representation of the combination of RIP-seq, PCA, and functional enrichment analysis to uncover similarities in RNA ligand profiles and associated functional pathways of different RBPs.
Fig. 2.
Global RNA ligand profiling of Salmonella CSPs. (A) PCA plot based on the transcripts identified by RIP-seq for each indicated protein or the control. EV, empty vector. (B) Enrichment of the GO term “Pathogenesis” in the transcripts identified by RIP-seq for each Salmonella CSP. The dendrogram on the right displays the clustering of the functional profiles.
To begin to understand the biological functions of the six Salmonella CSPs, the cellular processes associated with the RNA ligands bound by each family member were explored by an enrichment analysis using gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations (24). We observed that the transcripts associated with the CSPs were generally enriched for GO terms of general processes such as metabolism and translation; however, each CSP also showed transcript enrichment with different specific processes (SI Appendix, Fig. S2). Intriguingly, CspC and CspE, but none of the other family members, were associated with stress responses and pathogenicity (Fig. 2B and SI Appendix, Fig. S2).
To determine the impact of these proteins on Salmonella gene expression, we performed an RNA-sequencing (RNA-seq) experiment comparing a strain deleted for both cspC and cspE (∆cspCE) to the isogenic WT strain. Together, these two proteins significantly impact gene expression, with >1,000 genes found to be differentially expressed between the two strains (590 down-regulated and 536 up-regulated genes upon cspC/cspE deletion). Differentially expressed transcripts included both mRNAs and sRNAs but not housekeeping tRNAs (SI Appendix, Fig. S3 and Dataset S2). Consistent with the RIP-seq results, GO term and KEGG pathway enrichment analyses of the differentially expressed genes also suggested that CspC and CspE are involved in the response to various cellular stresses and virulence (SI Appendix, Fig. S3).
Validation of RNA Binding and Regulation by CspC and CspE.
The RIP-seq analysis indicated that CspC and CspE bind a large number of cellular transcripts (Dataset S1). To validate these predictions, we purified the proteins and selected 10 transcripts that are enriched in the RIP-seq data, down-regulated in ∆cspCE strain, and known to be involved in the pathways enriched in our global data analysis and two nonenriched transcripts for in vitro gel shift analysis. In these assays, CspC and CspE bound the enriched transcripts with dissociation constants ranging from 0.3 to 1.8 μM (SI Appendix, Fig. S4A). This result is in good agreement with a previous study, which reported dissociation constants of 2.5 μM (CspC) and 0.5 μM (CspE) for systematic evolution of ligands by exponential enrichment (SELEX)-enriched RNAs (25), suggesting that the transcripts assayed here indeed are high-affinity RNA ligands. The nonenriched transcripts were not bound by either CspC or CspE even at the highest concentration tested (7.5 μM) (SI Appendix, Fig. S4B). These results indicate that the RIP-seq analysis faithfully identified high-affinity cellular RNA ligands of CspC and CspE.
Many mRNAs identified in the RIP-seq analysis also showed altered expression in the absence of CspC and CspE (Datasets S1 and S2), suggesting that these proteins may impact the cellular fate of their RNA ligands. One such ligand, specifically enriched in CspC and CspE coimmunoprecipitation and down-regulated in the ∆cspCE mutant compared with the WT strain, is the ecnB mRNA encoding a bacteriolytic lipoprotein suggested to mediate adaptation to starvation (26). Northern blot analysis verified that ecnB mRNA steady-state levels were reduced in the ∆cspCE mutant and could be restored to above WT levels upon complementing CspC or CspE from plasmids (SI Appendix, Fig. S5A). In gel shift assays, CspC and CspE bound 5′ radiolabeled ecnB mRNA with high affinity, and these interactions could be outcompeted by adding an excess of unlabeled ecnB mRNA (SI Appendix, Fig. S5 B and C).
To test whether the reduced level of ecnB mRNA in the ∆cspCE mutant was caused by altered RNA stability, we performed rifampicin run-out experiments. The deletion of cspC and cspE each reduced the ecnB mRNA half-life fourfold, whereas overexpression of either CspC or CspE resulted in hyperstabilization (SI Appendix, Fig. S5D). In Salmonella, RNase E is the major ribonuclease responsible for turnover of mRNAs. Recent genome-wide mapping of RNase E cleavage sites identified several sites within the ecnB mRNA (27), suggesting transcript protection from cleavage as a potential mechanism of CspC and CspE action. Combining the ∆cspCE mutant with the rne701 allele expressing a truncated RNase E variant abolished the ∆cspCE-dependent reduction of ecnB mRNA levels, suggesting that RNase E is responsible for the decreased stability of ecnB mRNA in the absence of CspC and CspE (SI Appendix, Fig. S5E). Indeed, inactivation of RNase E activity using a temperature-sensitive mutant also abolished the down-regulation of ecnB mRNA in the ∆cspCE strain (SI Appendix, Fig. S5F). In line with the in vivo data described above, we observed in an in vitro assay that ecnB mRNA was readily degraded when incubated with purified RNase E; however, RNase E-dependent degradation was fully inhibited when the ecnB mRNA was preincubated with CspC or CspE (SI Appendix, Fig. S5G). Taken together, this in-depth analysis of the ecnB mRNA reveals one model for how CspC and CspE RNA-binding activity affects gene expression, namely by protecting their ligands from being targeted by RNase E, thereby increasing transcript stability.
CspC and CspE Affect Gene Expression During Host Cell Infection.
A number of our findings indicate that CspC and CspE are global regulators of gene expression involved in the Salmonella stress response and virulence. These findings include the large number of RNA ligands that coimmunoprecipitate with CspC and CspE, the pervasive changes in global gene expression upon their deletion (affecting ca. 20% of the transcriptome), their high cellular levels compared with other abundant RBPs (SI Appendix, Fig. S6), and the functional enrichment analysis (Fig. 2). To confirm the relevance of these changes for infection, we used Dual RNA-seq to compare gene-expression patterns in WT and ∆cspCE strains of Salmonella replicating inside human (HeLa) cells (28). The results (SI Appendix, Fig. S7 and Dataset S3) recapitulated the major contributions of CspC and CspE to the Salmonella stress response and virulence, and this time-course approach also revealed the dynamic effects of CspC and CspE on gene expression during infection. Before infection, motility and flagellar pathways were dysregulated in the ∆cspCE mutant, whereas after 16 h of infection major virulence factors involved in Salmonella pathogenesis, particularly the PhoP/Q regulon, were down-regulated, highlighting the potential contribution of CspC and CspE to both invasion and survival in the host. Unexpectedly, this dataset for Salmonella growing inside HeLa cells also revealed progressive induction of other members of the CSP family, namely CspA, CspB, and CspD in the ∆cspCE strain, compared with WT (SI Appendix, Fig. S7). Therefore, our global data support a previously predicted cooperative behavior of CSP proteins during cold-shock stress in E. coli, where deletion of cspA, cspB, and cspG renders cspE expression cold-inducible (29, 30).
CspC and CspE Mediate Resistance to Oxidative and Membrane Stress.
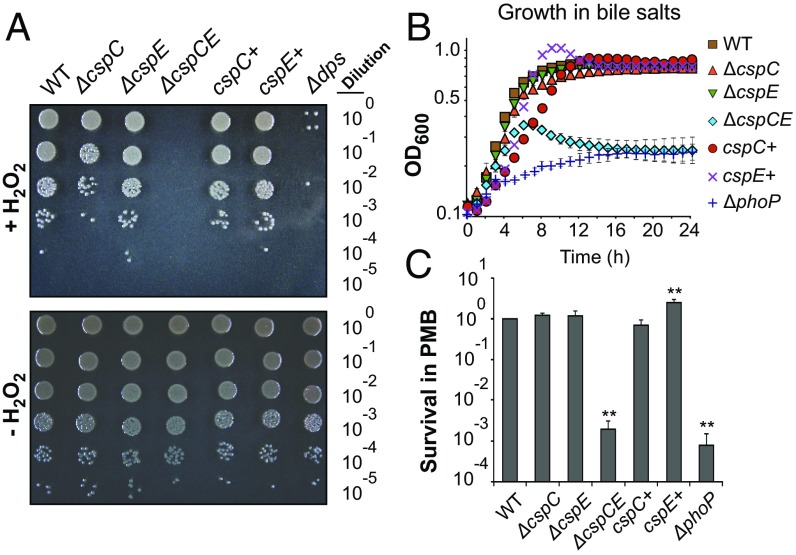
Although the GO-term enrichment analysis indicated that CspC and CspE are involved in the stress response and virulence, the specific cellular pathways under their control remained unclear. To this end, we used the RNA ligands of CspC and CspE as a guide to identify affected biological processes using targeted phenotypic assays. For instance, analysis of the RIP-seq and RNA-seq data revealed enrichment and/or differential expression of several transcripts associated with oxidative stress protection and iron-dependent hydrogen peroxide killing [e.g., nrfA (SL1344_4213) (31), dps (SL1344_0806) (32), and uspA (SL1344_3556) (33)]. Northern blot analysis confirmed that full expression of uspA and dps requires the expression of either CspC or CspE (SI Appendix, Fig. S8A), in agreement with previous studies in E. coli (20, 33, 34). In addition, in vitro binding assays showed a direct and specific interaction between uspA or dps mRNA and each of the two proteins (SI Appendix, Fig. S8 B and C). When Salmonella was challenged with hydrogen peroxide, the ∆cspCE strain showed a strongly reduced survival similar to that of the positive control strain ∆dps (35), indicating an important contribution of CspC and CspE to oxidative stress tolerance (Fig. 3A).
Fig. 3.
CspC and CspE contribute to stress resistance. (A) Growth of the indicated Salmonella strains on LB agar plates after a 2-h challenge with H2O2 (2 mM final concentration) (Upper) or after mock treatment (Lower). The ∆dps strain was used as a positive control for sensitivity to oxidative stress. (B) Growth of Salmonella in liquid LB medium supplemented with 3% bile salts as determined by measuring the OD at 600 nm. (C) Survival of Salmonella grown in LB medium after a 1-h challenge with polymyxin B (4 µg/mL final concentration) as determined by counting cfus (**P < 0.001). In B and C, the ∆phoP strain was used as a positive control for sensitivity to membrane stress. Data points refer to the mean of three independent experiments and error bars indicate SEs.
Several transcripts previously linked to membrane stress, including resistance to bile salts and antimicrobial peptides, coimmunoprecipitated with CspC and CspE and were down-regulated in the ∆cspCE compared with the WT strain (SI Appendix, Table S1 and Dataset S2). To test if CspC and CspE impact membrane stress, we monitored bacterial survival in the presence of bile salts (Fig. 3B) or the antimicrobial peptide polymyxin B (Fig. 3C), respectively. A ∆phoP strain was included as a positive control for bile and polymyxin B sensitivity (36, 37). Although growth in LB medium alone was not affected by the absence of CspC and/or CspE (SI Appendix, Fig. S9A), the ∆cspCE double mutant displayed a strong growth defect after exposure to either of these two membrane-damaging compounds, indicating that CspC and CspE have an important role in membrane stress tolerance. Notably, the impaired growth of the ∆cspCE strain in bile salts was not caused by toxicity: The cells remained viable and could be recultivated after removal of the stressor (SI Appendix, Fig. S9 B and C).
Most importantly, single deletions of either cspC or cspE resulted in only minor, if any, changes in growth during oxidative and membrane stress conditions (Fig. 3), whereas complementing the ∆cspCE mutant with either of the two proteins in trans restored resistance to WT levels (Fig. 3). This result strongly suggests that CspC and CspE act in a redundant fashion under the tested virulence-related stress conditions, consistent with the high similarity of their RNA ligand profiles (Fig. 2A).
CspC and CspE Are Required for Motility and Biofilm Formation.
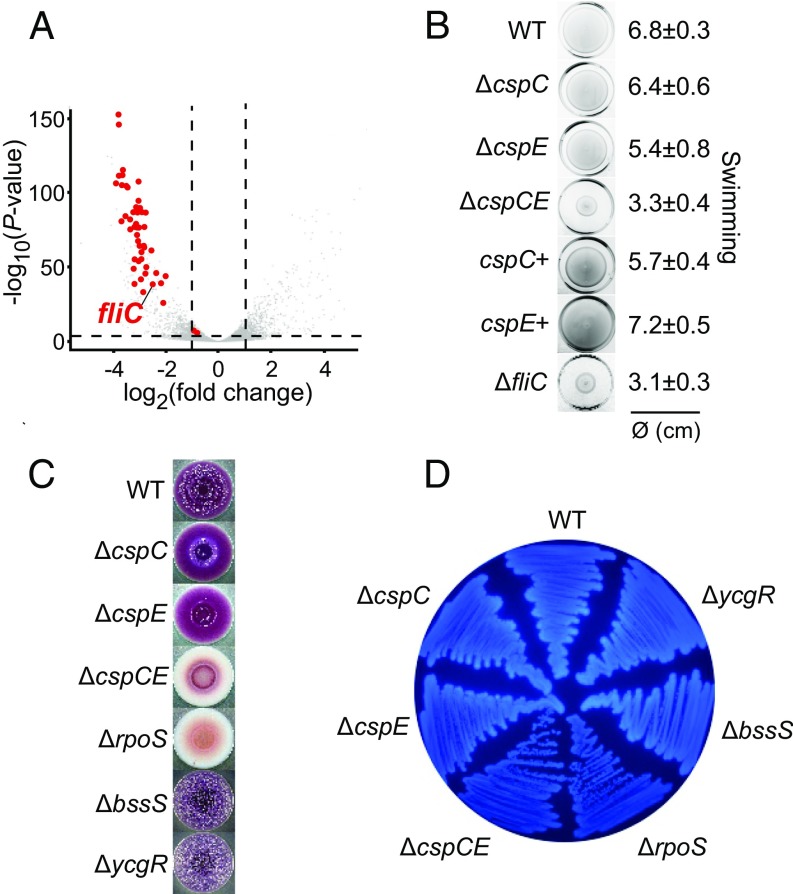
The RNA-seq experiments also suggested that CspC and CspE control the expression of genes involved in bacterial motility, because almost all genes related to motility and chemotaxis were down-regulated upon deletion of cspC and cspE (Fig. 4A and SI Appendix, Table S2). Among the most strongly repressed motility-related genes was fliC, which encodes flagellin, the structural protein of the flagellum and a pathogen-associated molecular pattern (PAMP) used by the host to recognize bacteria and trigger an immune response (38). Upon double deletion of cspC and cspE, fliC expression was reduced both at the mRNA and protein level (SI Appendix, Fig. S10 A and B), indicating that CspC and CspE are critical for fliC expression and thus for the formation of flagella. Consistent with this requirement, the ∆cspCE strain was strongly impaired in both swimming and swarming ability compared with WT bacteria (Fig. 4B and SI Appendix, Fig. S10C). Salmonella also uses flagella to mediate contact with host cells. Accordingly, the ∆cspCE strain phenocopied the reduced invasion of the ∆fliC mutant (SI Appendix, Fig. S10D) and showed reduced adherence (SI Appendix, Fig. S10E) in the HeLa infection model. Because the invasion assay was performed after infection synchronization, the observed phenotype may be attributed partly to the down-regulation of flagellin (39, 40) but also could reflect a flagellin-independent reduction of host cell adhesion in the ∆cspCE strain (SI Appendix, Fig. S10 D and E).
Fig. 4.
CspC and CspE are critical for motility and biofilm formation. (A) Global changes in gene expression between WT and ∆cspCE Salmonella as inferred from RNA-seq (LB medium, OD600 of 2.0). Motility-related genes are shown as red dots, and all other genes are indicated by gray dots. The gene encoding the major flagellin, fliC, is labeled. Dashed lines denote the thresholds set for regulation, namely a fold change >2 and a P value <0.05. (B) The indicated Salmonella strains were stabbed into 0.3% soft agar plates and incubated for 6 h at 37 °C. Numbers on the right indicate the average and SD of the swimming zone diameter. The ∆fliC strain was used as a positive control for impaired motility. (C) Ten-microliter overnight cultures of each indicated Salmonella strain were spotted onto an LB agar plate supplemented with Congo red and Coomassie brilliant blue and were incubated 8 d at 28 °C. (D) The indicated strains were grown on LB agar supplemented with Calcofluor during 48 h at 28 °C to monitor cellulose production. In C and D the ∆rpoS strain was included as a positive control for biofilm formation deficiency, and the ∆bssS and ∆ycgR strains were positive controls for enhanced biofilm formation.
CspC and CspE RNA ligand profiles also suggested that these proteins bind and regulate genes involved in biofilm formation (SI Appendix, Table S2 and Dataset S1). Salmonella forms biofilms on gallstones during gallbladder colonization to resist high bile concentrations (41). Because the ∆cspCE strain was highly sensitive to bile (Fig. 3B), we assessed whether CspC and CspE also affected Salmonella biofilm formation, using crystal violet staining to visualize the attachment of Salmonella to plastic surfaces. This assay indicated impaired biofilm formation in the ∆cspCE mutant (SI Appendix, Fig. S10F). Repeating the assay with cholesterol, which is commonly used to mimic gallstones (42), also showed reduced biofilm formation in the absence of CspC and CspE (SI Appendix, Fig. S10G). Moreover, formation of the red, dry, and rough (rdar) morphotype on Congo red plates, which requires the production of extracellular matrix (ECM) components such as curli fibers and cellulose, was virtually abolished in ∆cspCE (Fig. 4C). Finally, deletion of cspC and cspE also reduced bacterial binding to the fluorescent dye calcofluor, an indicator of cellulose production (Fig. 4D).
The down-regulation of both motility and biofilm formation in the ∆cspCE strain may seem contradictory, because these two opposite lifestyles are inversely regulated (43). However, there are other regulators that control both these processes; for example, the E. coli sRNAs OmrA and OmrB inhibit expression of the master regulators of both biofilm and motility (44, 45). Interestingly, Salmonella adherence and biofilm formation on cholesterol are mediated by the flagellin protein FliC (46), suggesting that the observed reduction in biofilm formation on cholesterol-coated surfaces in the ∆cspCE strain also may be connected to the down-regulation of FliC expression (Fig. 4A and SI Appendix, Fig. S10 A and B).
Deletion of cspC and cspE Renders Salmonella Avirulent.
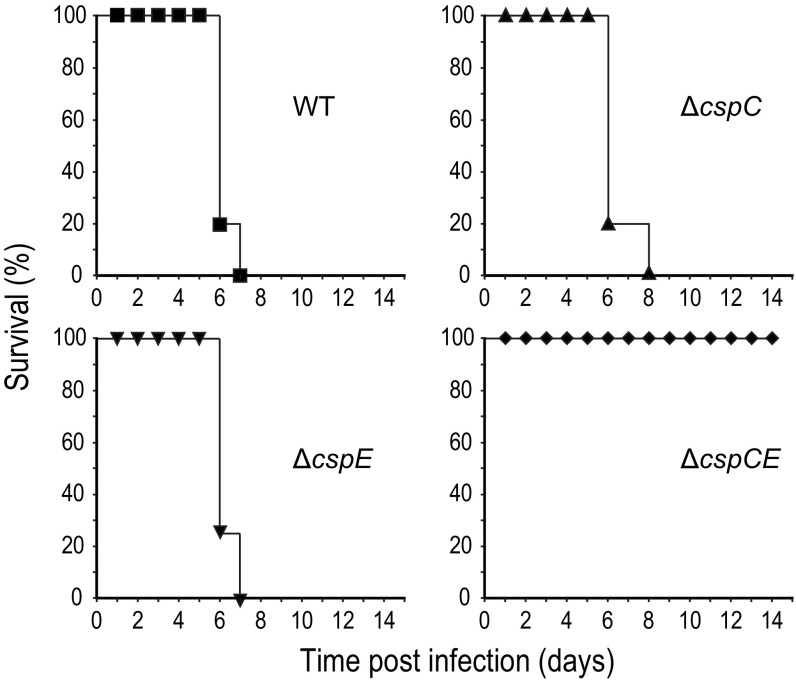
We demonstrated the role of CspC and CspE in a range of physiological processes related to infection. Within the host, Salmonella is exposed to oxidative stress through the production of reactive oxygen (ROS) and reactive nitrogen species (RNS) produced by cells of the innate immune system (47). Bile salts that disrupt the bacterial cell envelope are present in the body compartments where infecting Salmonella resides, such as the gallbladder and the small intestine (48). Although Salmonella notably depends on the flagellum during host cell invasion (49), biofilm formation is a well-known bacterial defense mechanism during the colonization of the gallbladder (42). Because the deletion of cspC and cspE strongly affected all the above-mentioned processes and resulted in the down-regulation of many virulence-related genes, including the PhoP/Q regulon (SI Appendix, Figs. S7 and S11 and Tables S3 and S4), we investigated if CspC and CspE were required for Salmonella pathogenicity in vivo. To this end, survival of female BALB/c mice systemically infected with Salmonella was monitored over time. Mice infected with WT or ∆cspC and ∆cspE single-mutant strains survived between 6 and 8 d post infection (dpi), whereas none of the mice infected with the ∆cspCE double-deletion strain showed any signs of illness over 14 d of infection (Fig. 5). In other words, the sum of their individual activities in diverse virulence-associated pathways makes CspC and CspE essential for Salmonella pathogenicity.
Fig. 5.
CspC and CspE are required for Salmonella virulence in mice. The graphs show the survival of female BALB/c mice after i.p. infection with 103 cfus of the indicated Salmonella strains. Five mice were monitored for each Salmonella strain.
Conclusions
Although a few bacterial RBPs are known to play important roles in pathogenicity (50, 51), the physiological impact of most bacterial RBPs remains unknown. Our ligand-centered functional profiling proved to be a successful route to uncovering important physiological roles of CspC and CspE. Analysis of functional pathways and biological processes enriched among RNAs detected by RIP-seq indicated that CspC and CspE bind to a similar set of RNA ligands (Fig. 2A). Transcriptome analysis of the ∆cspCE mutant, in vitro and in the context of an infection (SI Appendix, Figs. S3 and S7), revealed differential expression of many genes previously linked to the stress response and virulence, including members of regulons known to be critical for Salmonella pathogenicity, such as PhoPQ (SI Appendix, Fig. S11 and Table S4). Several mRNAs in the PhoPQ regulon, including phoP mRNA itself, are bound with high affinity by CspC and CspE (SI Appendix, Fig. S4A), suggesting that CspC/E act directly on both phoP and downstream mRNAs in the same regulon. Possibly, targeting both the master regulator and a specific set of downstream genes may enable discoordinate regulation within a regulon. Interestingly, we found that some pathways regulated by CspC and CspE, e.g., the PhoP/Q-related and motility genes, are differentially temporally regulated during infection. The transcriptomic data during infection reveal that flagella genes are mainly down-regulated at the beginning of infection (during invasion), whereas PhoP/Q-dependent genes, including virulence-related genes, are down-regulated only later during the infection time course (during full replication). Phenotypic assays strongly indicated that CspC and CspE are crucial for stress resistance, motility, and biofilm formation (Figs. 3 and 4). Each of these phenotypes has previously been linked to Salmonella virulence. Consequently, Salmonella lacking both proteins became avirulent during systemic mouse infection (Fig. 5).
In most of our experiments, the absence of both CspC and CspE resulted in strong phenotypes, whereas single-gene deletions had little or no effect, suggesting redundant functions for these two proteins. This notion is consistent with a recent study in E. coli showing functional redundancy for CspC and CspE in regulating the rpoS mRNA (52). Note, however, that, in contrast to E. coli (19, 53), we do not observe CspC/E-dependent changes in rpoS expression in Salmonella or strongly RpoS-dependent transcripts such as the sRNA SdsR (Dataset S2) (54, 55), suggesting important differences in CSP-mediated regulation between these closely related species. Functional redundancy might further explain why single-transposon insertions in cspC or cspE did not result in fitness defects within different animal models of salmonellosis (21). Interestingly, the gradual induction of other CSP proteins during infection in ∆cspCE indicates some level of feedback and functional compensation in the CSP network, although clearly not enough to rescue the strain under the conditions tested. This observation raises a number of interesting questions: How is this feedback mediated? Are there conditions under which these proteins can still complement one another, despite their deep evolutionary divergence? What forces have shaped the functional diversification of CSP proteins?
Another important open question is the nature of the regulatory events underlying the gene-expression changes observed for the ∆cspCE mutant. Our analysis of one mRNA ligand, the ecnB mRNA, indicates that CspC and CspE are able to protect their ligands against RNase E activity and stabilize transcripts. Future work should address the relative importance of this newly described mechanism compared with previously described roles for CSPs as chaperones facilitating RNA melting (56) or promoting antitermination (15). This issue could be addressed by establishing in vivo UV crosslinking (4) for CspC/E to determine direct binding sites at a global level; these data then could be integrated with global profiling of transcript decay data obtained in rifampicin run-out experiments (57) and with available information on RNase E cleavage sites (27) to understand the degree to which this mechanism accounts for the large number of gene-expression changes in the absence of CspC and CspE.
Taken together, the findings presented in this study emphasize the emergence of RBPs as key posttranscriptional regulators of bacterial physiology and pathogenesis and demonstrate that global RNA ligand profiling is a rapid approach to linking RBP activity to phenotypes. The expanding number of bacterial RBPs that impact bacterial virulence suggests that this group of proteins should be considered as potential targets for antimicrobial therapy.
Materials and Methods
Bacterial Strains and Growth Conditions.
The bacterial strains and plasmids used in this study are listed in SI Appendix, Tables S5 and S6, respectively. For all experiments, single colonies were grown overnight at 37 °C (or 28 °C when required) in LB medium (5 g/L of yeast extract, 5 g/L of NaCl, and 10 g/L of Tryptone/Peptone ex casein; Roth), diluted 1:100 in fresh medium, and further grown to the desired density. When appropriate, the medium was supplemented with 3% bile salts (Sigma-Aldrich), 0.2% l-Arabinose (Roth), 50 μg/mL kanamycin, 30 μg/mL chloramphenicol, or 100 μg/mL ampicillin.
Protein Purifications, in Vitro Transcription, and Gel Mobility Shift Assay.
Recombinant C-terminal Strep-tagged Salmonella CspC and CspE were overexpressed and purified from E. coli BL21 (DE3) using Strep-Tactin Sepharose (IBA GmbH, no. 2-1202-001) (see SI Appendix, Materials and Methods for details). In vitro transcription and gel mobility shift assays, including competition and nuclease assays, were performed as described in SI Appendix, Materials and Methods using conditions used in previous studies of CSPs (15).
Polymyxin B Assay.
The polymyxin B (Sigma-Aldrich) survival assay was described previously (58); see SI Appendix, Materials and Methods for details.
Oxidative Stress Assay.
Bacterial resistance to hydrogen peroxide (Roth) was measured as described previously (59) with slight modifications; see SI Appendix, Materials and Methods for details.
Motility Assay.
Swimming assays were performed on 0.3% agar plates as described in ref. 60 with slight modifications; see SI Appendix, Materials and Methods for details. Swarming assays were performed on 0.5% agar plates.
Biofilm Assays.
Rapid attachment biofilm assays were performed in 1:20 Tryptic Soy Broth (TSB) medium (Sigma-Aldrich) in the presence or absence of cholesterol (BD Biosciences) as described in ref. 61; see SI Appendix, Materials and Methods for details. The rdar morphotype or cellulose production was determined qualitatively after bacterial growth at 28 °C on LB agar without NaCl supplemented with 40 µg/mL Congo red (Merck) and 20 µg/mL Coomassie Brilliant Blue (Roth) or 50 µg/mL Calcofluor (Sigma-Aldrich), respectively; see SI Appendix, Materials and Methods for details.
Bacterial Infection Assays.
In vitro Salmonella infection assays were performed as previously described (62, 63); see SI Appendix, Materials and Methods for details.
Mouse Infection.
Female BALB/c mice (Jackson Laboratory) were inoculated i.p. with ∼103 cfu of the different Salmonella strains (n = 5 per group) and observed for morbidity up to 14 dpi.
Global Datasets, RIP-Seq, RNA-Seq, Computational Analyses.
Salmonella-harboring plasmids expressing FLAG-tagged CspA, CspB, CspC, CspD, CspE, CspH, Hfq, or an empty vector (EV) were grown in LB (at 220 rpm, 37 °C; Innova 44, New Brunswick) supplemented with appropriate antibiotics. At an OD600 of 0.2, l-Arabinose was added to a final concentration of 0.2% to induce expression of the tagged proteins. At an OD600 of 2.0, 100 OD of culture were collected by centrifugation (4,800 × g, 40 min, 4 °C) and subjected to RNA coimmunoprecipitation as described previously (3); see SI Appendix, Materials and Methods for details. For the RNA-seq experiments, two [inside HeLa cells, as described previously (28)] or three (in vitro cultures in LB, 37 °C, OD600 of 2.0) replicates of the WT and ∆cspCE strains, respectively, were analyzed.
Supplementary Material
Acknowledgments
We thank B. Plaschke, V. McParland, and T. Achmedov for excellent technical assistance, the Eulalio laboratory for assistance with the in vitro infection model, G. Matera for assistance with the gel-shift assay, U. Römling and Y. Chao for providing bacterial strains, and S. Gorski for editing the manuscript. E.H. is supported by the Wenner-Gren Foundation. L.B. was supported in part by a research fellowship from the Alexander von Humboldt Foundation. The J.S.G. laboratory was sponsored by NIH Grant AI116917. The J.V. laboratory received funds from Deutsche Forschungsgemeinschaft Priority Program SPP1258 (Vo875/3‐2), the Bavarian BioSysNet program, and Federal Ministry of Education and Research (BMBF) eBio programme RNASys.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE91086).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620772114/-/DCSupplemental.
References
- 1.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 3.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmqvist E, et al. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016;35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sittka A, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang A, et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 8.Smirnov A, et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci USA. 2016;113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barquist L, Vogel J. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu Rev Genet. 2015;49:367–394. doi: 10.1146/annurev-genet-112414-054804. [DOI] [PubMed] [Google Scholar]
- 10.Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: Highly versatile regulators of gene expression. Bioessays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 11.Wolffe AP. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 12.Giuliodori AM, et al. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol Cell. 2010;37:21–33. doi: 10.1016/j.molcel.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 13.Gualerzi CO, Giuliodori AM, Pon CL. Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol. 2003;331:527–539. doi: 10.1016/s0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- 14.Jones PG, VanBogelen RA, Neidhardt FC. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 17.Ishihama Y, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: Multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 19.Phadtare S, Inouye M. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J Bacteriol. 2001;183:1205–1214. doi: 10.1128/JB.183.4.1205-1214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phadtare S, Tadigotla V, Shin W-H, Sengupta A, Severinov K. Analysis of Escherichia coli global gene expression profiles in response to overexpression and deletion of CspC and CspE. J Bacteriol. 2006;188:2521–2527. doi: 10.1128/JB.188.7.2521-2527.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri RR, et al. Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet. 2013;9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kröger C, et al. An infection-relevant transcriptomic compendium for Salmonella enterica serovar typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reference Genome Group of the Gene Ontology Consortium The gene ontology’s reference genome project: A unified framework for functional annotation across species. PLoS Comput Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phadtare S, Inouye M. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol Microbiol. 1999;33:1004–1014. doi: 10.1046/j.1365-2958.1999.01541.x. [DOI] [PubMed] [Google Scholar]
- 26.Bishop RE, Leskiw BK, Hodges RS, Kay CM, Weiner JH. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J Mol Biol. 1998;280:583–596. doi: 10.1006/jmbi.1998.1894. [DOI] [PubMed] [Google Scholar]
- 27.Chao Y, et al. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell. 2017;65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westermann AJ, et al. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 29.Phadtare S, Inouye M, Severinov K. The nucleic acid melting activity of Escherichia coli CspE is critical for transcription antitermination and cold acclimation of cells. J Biol Chem. 2002;277:7239–7245. doi: 10.1074/jbc.M111496200. [DOI] [PubMed] [Google Scholar]
- 30.Xia B, Ke H, Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- 31.Kern M, Volz J, Simon J. The oxidative and nitrosative stress defence network of Wolinella succinogenes: Cytochrome c nitrite reductase mediates the stress response to nitrite, nitric oxide, hydroxylamine and hydrogen peroxide. Environ Microbiol. 2011;13:2478–2494. doi: 10.1111/j.1462-2920.2011.02520.x. [DOI] [PubMed] [Google Scholar]
- 32.Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyström T, Neidhardt FC. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol. 1994;11:537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 34.Phadtare S, Inouye M. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol. 2004;186:7007–7014. doi: 10.1128/JB.186.20.7007-7014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almirón M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 36.Groisman EA, Parra-Lopez C, Salcedo M, Lipps CJ, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Velkinburgh JC, Gunn JS. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect Immun. 1999;67:1614–1622. doi: 10.1128/iai.67.4.1614-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi F, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 39.Elhadad D, Desai P, Rahav G, McClelland M, Gal-Mor O. Flagellin is required for host cell invasion and normal Salmonella pathogenicity Island 1 expression by Salmonella enterica serovar Paratyphi A. Infect Immun. 2015;83:3355–3368. doi: 10.1128/IAI.00468-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen JE, et al. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar dublin compared to the broad host range serovar S. typhimurium. BMC Microbiol. 2013;13:67. doi: 10.1186/1471-2180-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prouty AM, Gunn JS. Comparative analysis of Salmonella enterica serovar typhimurium biofilm formation on gallstones and on glass. Infect Immun. 2003;71:7154–7158. doi: 10.1128/IAI.71.12.7154-7158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford RW, et al. Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci USA. 2010;107:4353–4358. doi: 10.1073/pnas.1000862107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Povolotsky TL, Hengge R. ‘Life-style’ control networks in Escherichia coli: Signaling by the second messenger c-di-GMP. J Biotechnol. 2012;160:10–16. doi: 10.1016/j.jbiotec.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 44.De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmqvist E, et al. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J. 2010;29:1840–1850. doi: 10.1038/emboj.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford RW, Reeve KE, Gunn JS. Flagellated but not hyperfimbriated Salmonella enterica serovar typhimurium attaches to and forms biofilms on cholesterol-coated surfaces. J Bacteriol. 2010;192:2981–2990. doi: 10.1128/JB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang FC. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 48.Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 49.Fàbrega A, Vila J. Salmonella enterica serovar typhimurium skills to succeed in the host: Virulence and regulation. Clin Microbiol Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Vakulskas CA, Potts AH, Babitzke P, Ahmer BMM, Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev. 2015;79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shenhar Y, Biran D, Ron EZ. Resistance to environmental stress requires the RNA chaperones CspC and CspE. Environ Microbiol Rep. 2012;4:532–539. doi: 10.1111/j.1758-2229.2012.00358.x. [DOI] [PubMed] [Google Scholar]
- 53.Cohen-Or I, Shenhar Y, Biran D, Ron EZ. CspC regulates rpoS transcript levels and complements hfq deletions. Res Microbiol. 2010;161:694–700. doi: 10.1016/j.resmic.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 54.Fröhlich KS, Haneke K, Papenfort K, Vogel J. The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res. 2016;44:10406–10422. doi: 10.1093/nar/gkw632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker A, Gottesman S. Small RNA regulation of TolC, the outer membrane component of bacterial multidrug transporters. J Bacteriol. 2016;198:1101–1113. doi: 10.1128/JB.00971-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phadtare S, Severinov K. Nucleic acid melting by Escherichia coli CspE. Nucleic Acids Res. 2005;33:5583–5590. doi: 10.1093/nar/gki859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen H, Shiroguchi K, Ge H, Xie XS. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol Syst Biol. 2015;11:781. doi: 10.15252/msb.20145794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Appia-Ayme C, et al. ZraP is a periplasmic molecular chaperone and a repressor of the zinc-responsive two-component regulator ZraSR. Biochem J. 2012;442:85–93. doi: 10.1042/BJ20111639. [DOI] [PubMed] [Google Scholar]
- 59.Farizano JV, Torres MA, Pescaretti MdeLM, Delgado MA. The RcsCDB regulatory system plays a crucial role in the protection of Salmonella enterica serovar typhimurium against oxidative stress. Microbiology. 2014;160:2190–2199. doi: 10.1099/mic.0.081133-0. [DOI] [PubMed] [Google Scholar]
- 60.Forbes SJ, Eschmann M, Mantis NJ. Inhibition of Salmonella enterica serovar typhimurium motility and entry into epithelial cells by a protective antilipopolysaccharide monoclonal immunoglobulin A antibody. Infect Immun. 2008;76:4137–4144. doi: 10.1128/IAI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koopman JA, et al. Inhibition of Salmonella enterica biofilm formation using small-molecule adenosine mimetics. Antimicrob Agents Chemother. 2015;59:76–84. doi: 10.1128/AAC.03407-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azriel S, Goren A, Rahav G, Gal-Mor O. The stringent response regulator DksA is required for Salmonella enterica Serovar typhimurium growth in minimal medium, motility, biofilm formation, and intestinal colonization. Infect Immun. 2015;84:375–384. doi: 10.1128/IAI.01135-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maudet C, et al. Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat Commun. 2014;5:4718. doi: 10.1038/ncomms5718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.