Significance
Botulinum neurotoxins (BoNTs) are the most potent biological toxins. These proteases function by cleaving SNARE proteins, leading to paralysis and death from respiratory failure. BoNT serotype A (BoNT/A) has the most prolonged symptoms due to an extraordinarily stable catalytic light chain (LCA). BoNT/A intoxication can occur through ingestion (either sporadically or from a common food source), as a consequence of a clinical mishap, or potentially through bioterrorism, which would require mass mechanical ventilation. We report that LCA persistence is due to a particular deubiquitinating enzyme that binds a specific region of LCA and prevents its ubiquitin-dependent proteasomal degradation. These findings represent an essential step toward developing targeted molecular approaches to reducing morbidity and mortality from this toxin.
Keywords: synaptosomal-associated protein 25, synaptic transmission, motoneuron, USP9X, toxin persistence
Abstract
Botulism is characterized by flaccid paralysis, which can be caused by intoxication with any of the seven known serotypes of botulinum neurotoxin (BoNT), all of which disrupt synaptic transmission by endoproteolytic cleavage of SNARE proteins. BoNT serotype A (BoNT/A) has the most prolonged or persistent effects, which can last several months, and exerts its effects by specifically cleaving and inactivating SNAP25. A major factor contributing to the persistence of intoxication is the long half-life of the catalytic light chain, which remains enzymatically active months after entry into cells. Here we report that BoNT/A catalytic light chain binds to, and is a substrate for, the ubiquitin ligase HECTD2. However, the light chain evades proteasomal degradation by the dominant effect of a deubiquitinating enzyme, VCIP135/VCPIP1. This deubiquitinating enzyme binds BoNT/A light chain directly, with the two associating in cells through the C-terminal 77 amino acids of the light chain protease. The development of specific DUB inhibitors, together with inhibitors of BoNT/A proteolytic activity, may be useful for reducing the morbidity and public health costs associated with BoNT/A intoxication and could have potential biodefense implications.
Botulinum neurotoxins (BoNTs) are the most potent natural toxins known (1, 2). At least seven distinct serotypes (A–G) of BoNTs have been identified, all of which block acetylcholine release from the presynaptic terminals of motoneurons at neuromuscular junctions, thereby causing flaccid paralysis (3, 4). BoNTs consist of a catalytic light chain (LC) metalloprotease disulfide linked to a heavy chain (5). The heavy chain binds to specific receptors on the presynaptic terminal and delivers the LC protease into the cell (6). Upon entry, BoNT LC cleaves SNARE proteins and inhibits neurotransmission (7). This property of BoNTs has been exploited in the clinic to treat many neuromuscular disorders. Due to their high potency, BoNTs have been designated as category A biodefense threat agents by the US Centers for Disease Control and Prevention (8). No antidote exists to reverse the symptoms of BoNT intoxication after onset of paralysis.
One salient feature of BoNT intoxication, particularly by serotype A (BoNT/A), which blocks neurotransmission by cleaving SNAP25, is the persistence or long duration of paralysis, lasting 2–6 mo from a single exposure to the toxin (9, 10). Although desirable for therapeutic applications, this extreme persistence presents special challenges for the clinical management of intoxication. Symptomatic treatment, including mechanical ventilation, must be provided for months during the recovery process and could quickly overwhelm the medical care system in an outbreak. The mechanisms for the differential persistence of BoNTs are not fully understood. Several factors may contribute to BoNT persistence, but the relative lifetime of BoNT LC proteases in the presynaptic terminal appears to play a predominant role (7, 10, 11). Cells normally degrade foreign proteins such as bacterial or viral proteins through the ubiquitin-proteasome system (UPS). In this regard, the catalytic LC of BoNT/E (LCE), which also cleaves SNAP25 but causes only transient paralysis, is degraded relatively rapidly as a consequence of ubiquitination by the E3 ubiquitin ligase TRAF2 (12). In contrast, the LC of BoNT/A (LCA) is quite stable (12), which is consistent with the proteolytic activity of LCA remaining detectable in neuronal cultures for months (13, 14). The activity of ubiquitin ligases toward proteins is highly specific; accordingly, despite 35% sequence identity between LCE and LCA, there is no evidence that TRAF2 ubiquitinates the highly stable LCA (12, 15).
Here we report that LCA persists by evading proteasomal degradation because of its association with valosin-containing protein (VCP) complex-interacting protein 135 kDa (VCIP135), a deubiquitinating enzyme (DUB) that is also known as VCPIP1. VCIP135 stabilizes LCA by removing ubiquitin from LCA, thereby limiting its polyubiquitination and degradation by the proteasome. In the absence of VCIP135, LCA is targeted for degradation by the homologous to E6-AP carboxy-terminus protein (HECT) domain ubiquitin ligase HECTD2. The functional antagonism between HECTD2 and VCIP135 determines the lifetime of LCA and hence toxin persistence. Moreover, knockdown of VCIP135 accelerates the recovery of cultured motoneurons from BoNT/A intoxication as assessed by levels of intact SNAP25.
Results
Identification of VCIP135 as a DUB That Stabilizes BoNT/A Catalytic LC.
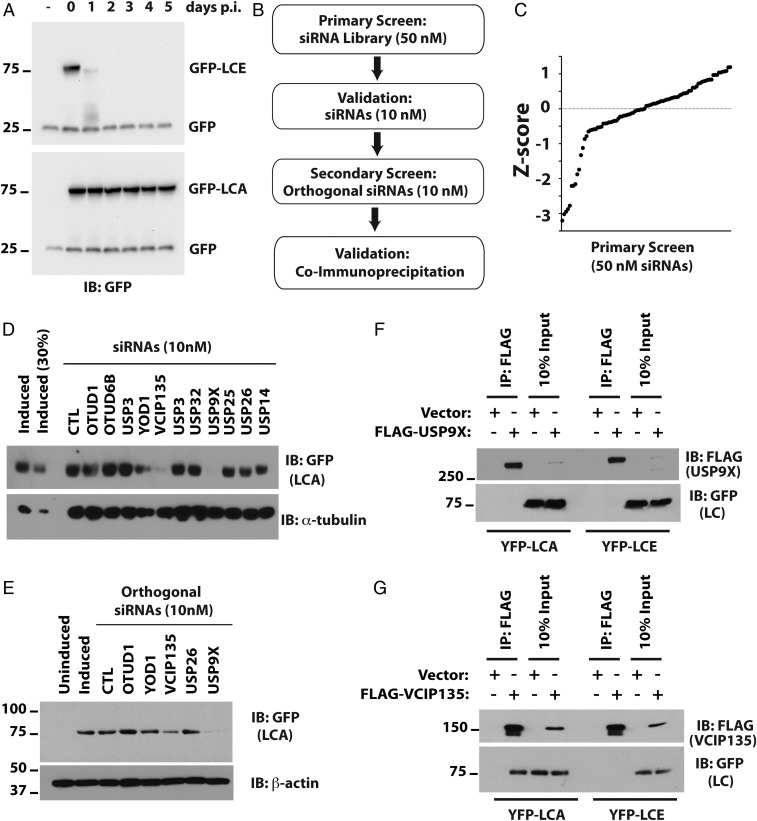
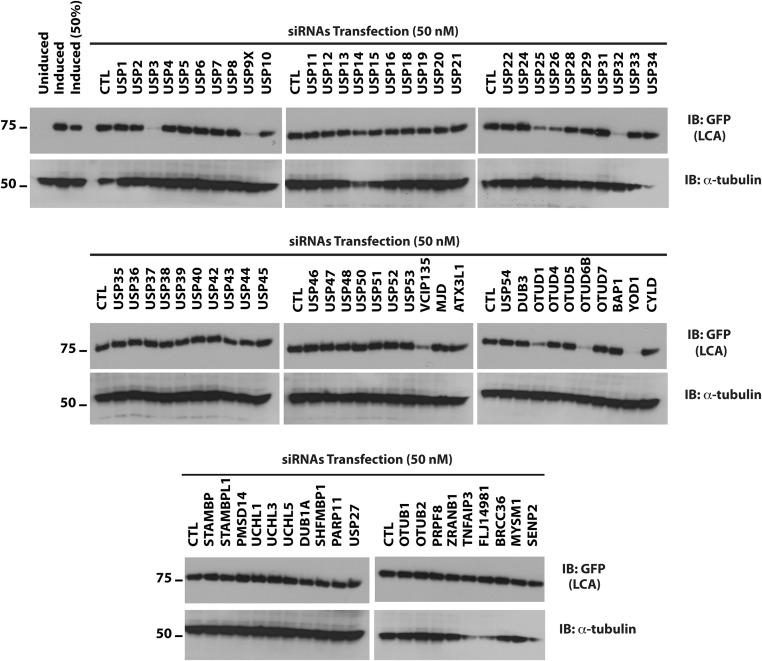
We have described a tractable cell culture model for studying the persistence of LCA (12). In this model, HEK293 cells are induced with doxycycline to transiently express codon-optimized GFP-tagged BoNT LCs. Following removal of doxycycline to turn off gene expression, LCA persists for at least 5 d, whereas LCE is undetectable after 2 d (Fig. 1A). In a previous study, we found that LCA shows detectable, albeit minimal, ubiquitination in the presence of proteasome inhibitor (12). However, given its already long half-life, no significance could be attributed to this finding. We postulate that LCA is shielded from the UPS by interaction with a cellular protein. Ubiquitination is reversible and depends on dynamic interactions between the substrate and its ubiquitin ligase and DUB. To determine whether LCA is stabilized by interacting with a DUB, we used this cell culture model to screen siRNAs targeting DUBs (Fig. 1B). We performed the primary screen using 50 nM siRNAs, a concentration commonly used in such screens (Fig. S1). The z scores for the primary screen are summarized in Fig. 1C. As expected, most of the siRNAs did not substantially affect LCA levels. We focused on 11 genes that resulted in significant reductions in the level of LCA. To minimize potential off-target effects caused by higher concentrations of siRNAs (16–18), we validated the primary screen using a fivefold lower concentration (10 nM) of siRNAs. siRNAs targeting three DUBs—YOD1, USP9X/FAF-X, and VCIP135—substantially reduced the levels of LCA compared with control (CTL) siRNAs (Fig. 1D). We further assessed the effects of targeting these genes using orthogonal siRNAs from a different source (Fig. 1E). The secondary screen confirmed that two DUBs—USP9X and VCIP135—stabilized LCA. USP9X is the human ortholog of Drosophila melanogaster fat-facets, which is a DUB known to affect synaptic organization in Drosophila (19). USP9X has also been implicated in regulating autophagy, which ultimately results in degradation of proteins by lysosomal proteases (20–23). Additionally, it plays roles in other critical processes, including cell proliferation and apoptosis (24–26). Indeed, we observed reduced cell viability when USP9X expression was diminished for 7 d by RNAi. Little is known about the functions of VCIP135, except that it is implicated in the reassembly of the Golgi and the endoplasmic reticulum following mitosis (27–29).
Fig. 1.
Identification of DUBs that stabilize BoNT/A catalytic LC. (A) Cell culture model for persistence of LCA. Flp-in/T-REX/ 293 cells were induced with doxycycline for 16 h to express GFP-LCE and GFP-LCA. The persistence of GFP-LCs was monitored following doxycycline withdrawal (postinduction, p.i.). GFP served as the loading and transfection control. (B) Algorithm for siRNA library screening. (C) The z score for primary screen was computed based on median and SD of the sample (Materials and Methods). (D) Validation of potential hits from primary screen with low concentrations (10 nM) of the same siRNAs compared with control (CTL) siRNAs. The lane labeled “Induced (30%)” contains 30% of the cell lysate compared with other lanes. (E) Secondary screen with low concentrations (10 nM) of orthogonal siRNAs from a different vendor. α-tubulin and β-actin served as loading controls in D and E, respectively. (F and G) Coimmunoprecipitation assays to evaluate interactions of LCA with USP9X (F) or VCIP135 (G).
Fig. S1.
Identification of DUBs that stabilize BoNT/A catalytic LC. Flp-in/T-REX/293 cells were induced with doxycycline for 16 h to express GFP-LCA. On the following day, cells were washed, trypsinized, replated, and allowed to recover overnight. The primary screen was carried out with 50 nM siRNAs and cells harvested 3 d posttransfection. Levels of LCA were monitored by Western blotting.
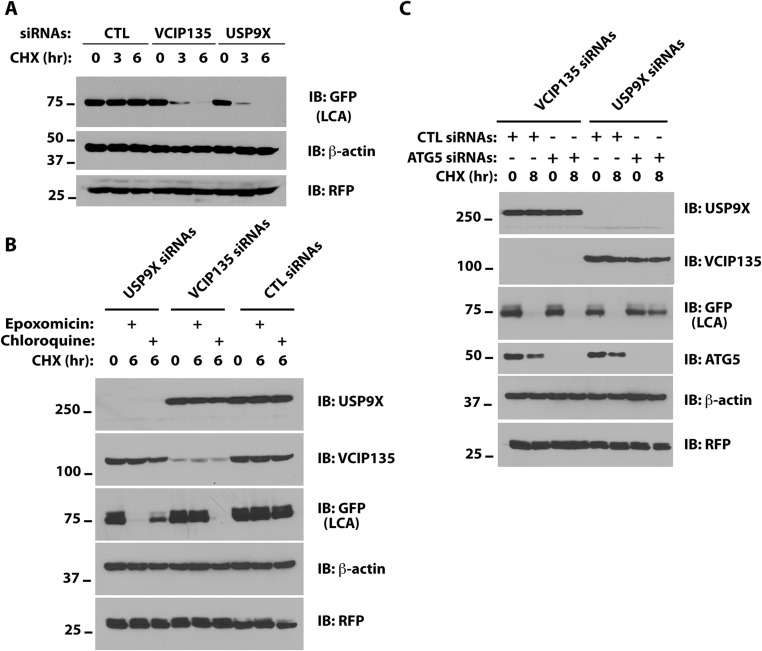
To determine whether depletion of these DUBs reduces the stability of LCA, we used cycloheximide (CHX) to inhibit new protein synthesis and monitored LCA levels. Although depletion of either USP9X or VCIP135 increased the turnover of LCA tagged with YFP, which is recognized by GFP antibody (Fig. S2A), this increased turnover appears to depend on different cellular pathways in USP9X- and VCIP135-depleted cells. Epoxomicin, a proteasome inhibitor, blocked YFP-LCA degradation in VCIP135-depleted cells but not in USP9X-depleted cells (Fig. S2B). In contrast, chloroquine, an inhibitor of lysosomal degradation, partially inhibited the degradation of YFP-LCA in USP9X-depleted cells but had no effect in VCIP135-depleted cells (Fig. S2B). As USP9X has been implicated in regulating autophagy (20–23), we assessed whether this degradative process contributes to the increased turnover of YFP-LCA in USP9X-depleted cells. To address this, we used siRNAs to reduce the level of ATG5, a protein required for autophagy, and evaluated loss of YFP-LCA. In USP9X-depleted cells, concomitant depletion of ATG5 restored the stability of YFP-LCA (Fig. S2C). In contrast, depletion of ATG5 failed to restore stability of YFP-LCA in VCIP135-depleted cells (Fig. S2C).
Fig. S2.
Different pathways for LCA degradation in USP9X- and VCIP135-depleted cells. (A) HEK293 cells were transfected with YFP-LCA, and the indicated siRNAs and degradation of YFP-LCA (recognized by GFP antibody) were assessed by CHX chase and Western blotting. (B) M17 cells transfected with VCIP135 siRNAs and YFP-LCA were treated with the indicated inhibitors with or without CHX for 6 h and YFP-LCA monitored. (C) M17 cells were transfected with YFP-LCA and either VCIP135 or USP9X siRNAs with or without ATG siRNAs and degradation of YFP-LCA assessed. RFP was included as a transfection efficiency control and as a loading control along with β-actin.
We next determined whether these DUBs physically associate with LCA by coimmunoprecipitation. Cells were transfected with YFP-LCA and either USP9X or VCIP135 epitope-tagged with FLAG. DUBs were immunoprecipitated and associated LCA detected by immunoblotting. No interaction of either LCA or LCE with USP9X was observed (Fig. 1F). These results suggest that USP9X does not interact with LCA and that depletion of USP9X may enhance LCA degradation indirectly by increasing autophagy. In contrast, LCA, but not LCE, coimmunoprecipitated with VCIP135, indicating that VCIP135 interacts specifically with LCA (Fig. 1G).
The Catalytic Activity of VCIP135 Is Necessary for Stabilizing LCA.
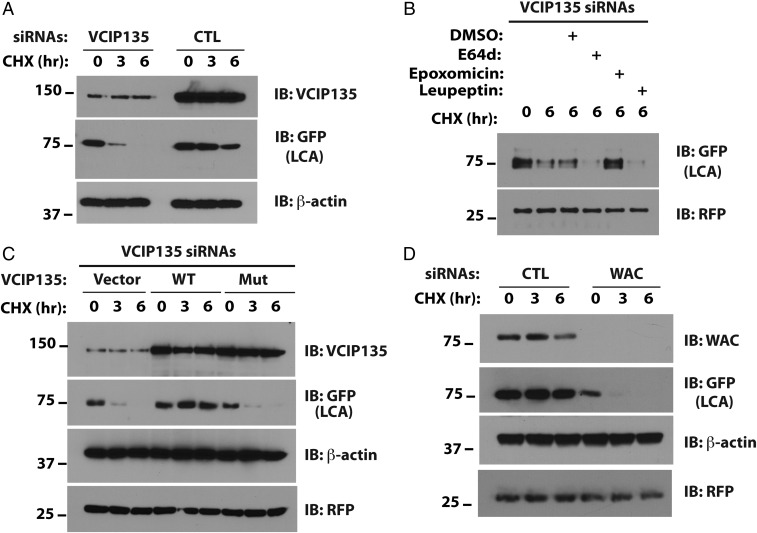
To confirm the effects of VCIP135 on the stability of LCA, we turned to M17, a human neuroblastoma cell line sensitive to BoNT/A (Fig. 2A). Depletion of VCIP135 accelerated LCA degradation. As with nonneural cells, this accelerated degradation was attenuated by epoxomicin, again supporting the idea that LCA is targeted for UPS-mediated degradation in the absence of VCIP135 (Fig. 2B). In contrast, degradation of LCA in VCIP135-depleted cells was not impeded by inhibitors of lysosomal degradation (E64d, leupeptin; Fig. 2B). To determine whether the DUB activity of VCIP135 is required to stabilize LCA, either wild-type (WT) or catalytically inactive mutant VCIP135 (C218A) was re-expressed in VCIP135-depleted cells. Only WT VCIP135 restored LCA stability, suggesting that active deubiquitination by VCIP135 is necessary for LCA stability (Fig. 2C).
Fig. 2.
The catalytic activity of VCIP135 is necessary for stabilizing LCA. (A) M17 cells were transfected with YFP-LCA and the indicated siRNAs and degradation of YFP-LCA (recognized by GFP antibody) assessed by CHX chase and Western blotting. (B) Cells transfected with VCIP135 siRNAs and YFP-LCA were treated with CHX and the indicated inhibitors or vehicle control (DMSO) for 6 h. RFP served as a transfection and loading control. (C) Cells were transfected with VCIP135 siRNAs and subsequently transfected with YFP-LCA and the indicated VCIP135 constructs. (D) Cells were transfected with YFP-LCA and the indicated siRNAs.
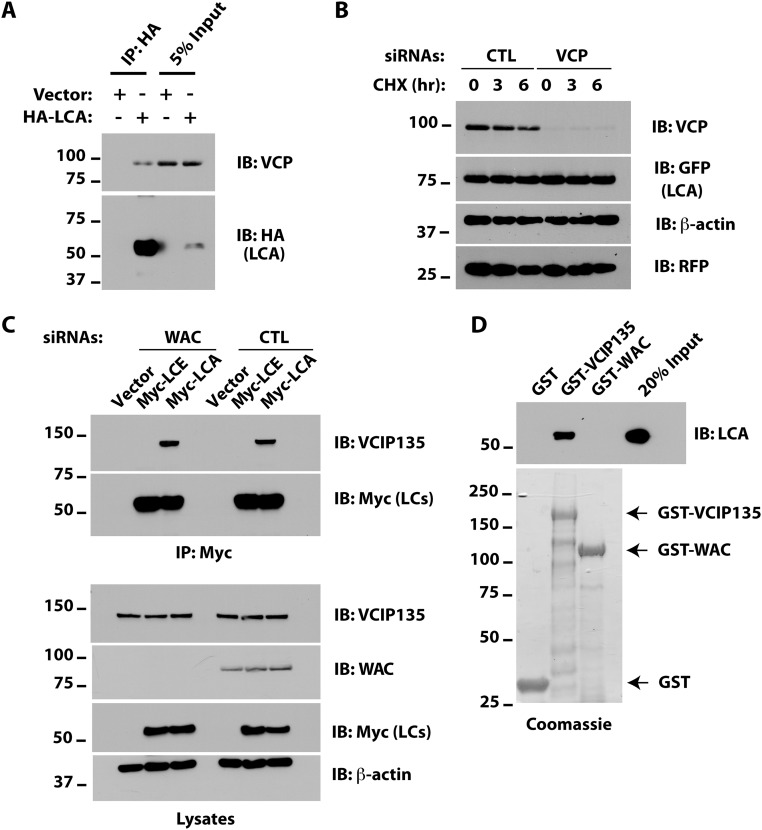
As VCIP135 interacts with VCP/p97, we asked whether LCA also associates with VCP. Although LCA coimmunoprecipitated VCP (Fig. S3A), depletion of the latter had little effect on LCA stability (Fig. S3B), suggesting that VCP is not critical for LCA stability. VCIP135 also associates with the protein WW domain-containing adaptor with coiled-coil (WAC), which stimulates the deubiquitinating activity of VCIP135 (30). WAC is not required for the association of VCIP135 with LCA in cells (Fig. S3C). Consistent with this result, bacterially expressed VCIP135, but not WAC, bound LCA in vitro, indicating a direct interaction between VCIP135 and LCA (Fig. S3D). However, similar to depletion of VCIP135, silencing of WAC enhanced LCA degradation (Fig. 2D), providing additional evidence for the crucial role of the enzymatic activity of VCIP135 in stabilizing LCA.
Fig. S3.
Different pathways for LCA degradation in USP9X- and VCIP135-depleted cells. (A) M17 cells were transfected with HA-tagged LCA. After 48 h, cells were harvested and HA-LCA immunoprecipitates were immunoblotted as indicated. (B) Cells were transfected with YFP-LCA and either CTL or VCP siRNAs. Degradation of YFP-LCA was assessed by CHX chase. (C) Cells were transfected with CTL or WAC siRNAs. After 18 h, cells were transfected with plasmids encoding either Myc-LCA or Myc-LCE. Myc-LC proteins were immunoprecipitated with Myc agarose and samples immunoblotted as indicated. (D) E. coli lysates containing ∼0.2 μg 6×His-LCA and 2 μg of GST fusion proteins were incubated with Glutathione Sepharose 4B for 2 h at 4 °C and LCA binding assessed by immunoblotting. The relative amounts of GST fusion proteins used in the binding assay were assessed by enrichment using Glutathione Sepharose 4B and Coomassie staining and are shown below. Bands beneath the full-length proteins in the Coomassie gel represent degradation products and/or products of incomplete translation.
The C Terminus of LCA Interacts with VCIP135.
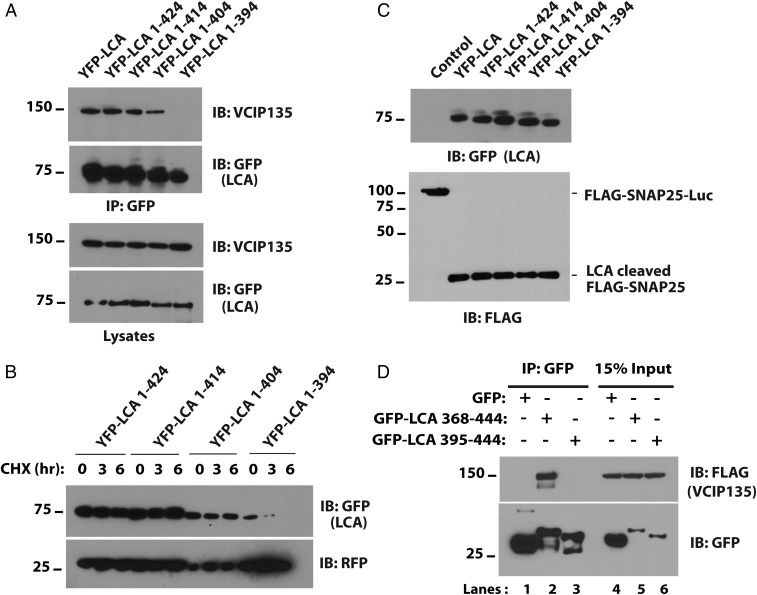
Both long-lived LCA and the relatively short-lived LCE cleave SNAP25 (15). Sequence alignment shows divergence in both their N and C termini. However, the N terminus of LCA is required for its association with SNAP25 and contributes to its potency (31). For this reason, to determine the region of LCA important for association with VCIP135, we focused on the C terminus of LCA. A series of truncation mutants of YFP-LCA were generated and their interaction with VCIP135 assessed. Coimmunoprecipitation with VCIP135 was disrupted when the C-terminal 50 residues of LCA were deleted (YFP-LCA 1–394; Fig. 3A). Consistent with the importance of this interaction, YFP-LCA 1–394 showed a dramatic decrease in stability (Fig. 3B). All of the truncations were active and cleaved a transfected SNAP25 fusion protein, suggesting that they are properly folded (Fig. 3C). These findings imply that the C-terminal 50 residues of LCA are necessary for association with and deubiquitination by VCIP135. Nevertheless, the C-terminal 50 amino acids of LCA fused to GFP (GFP-LCA 395–444) was not sufficient to interact with VCIP135, as assessed by coimmunoprecipitation (Fig. 3D). However, a larger fragment spanning the C-terminal 77 amino acid residues of LCA (GFP-LCA 368–444) was sufficient to coimmunoprecipitate VCIP135 (Fig. 3D, Upper, compare lanes 2 and 3). These results identify a C-terminal region of LCA that both mediates association with VCIP135 and allows for the stabilization of LCA by this DUB.
Fig. 3.
The C terminus of LCA interacts with VCIP135. (A) HEK293 cells were transfected with the indicated C-terminal deletion mutants of YFP-LCA. YFP-LCA mutants were immunoprecipitated with GFP antibody and immunoblotted for VCIP135. (B) HEK293 cells were transfected with deletion mutants as in A, and degradation of YFP-LCAs was assessed. (C) HEK293 cells were transfected with FLAG-SNAP25-luciferase and the indicated YFP-LCAs. After 36 h, the proteolytic activity of the truncation mutants was assessed by evaluating cleavage of FLAG-SNAP25-luciferase using FLAG antibody. (D) HEK293 cells were transfected with the C-terminal 77 or 50 residues of LCA fused to GFP. After 30 h, GFP-LCA fusions were immunoprecipitated with GFP antibody and coimmunoprecipitated VCIP135 assessed.
HECTD2 Promotes the Degradation of LCA.
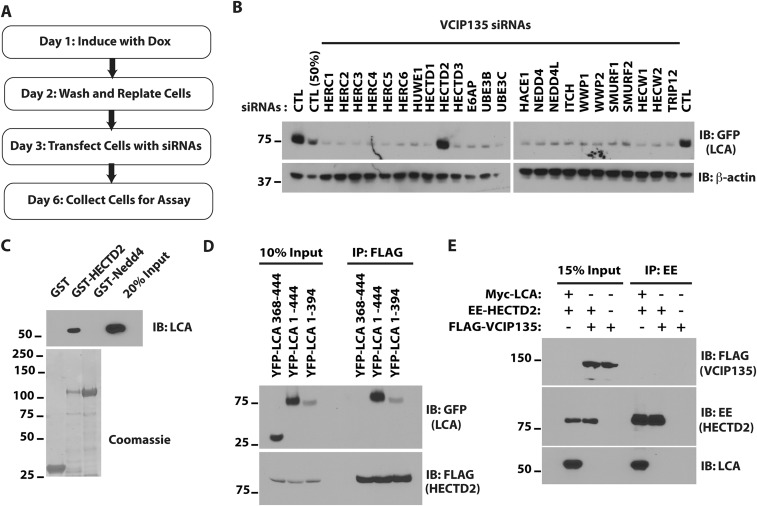
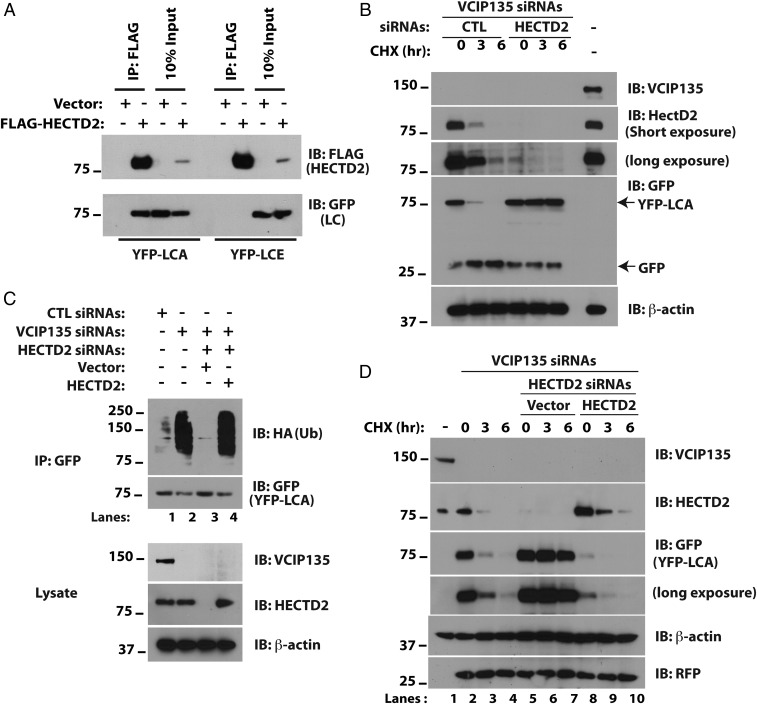
The results presented above support the idea that VCIP135 stabilizes LCA and that loss of expression of this DUB accelerates clearance of LCA via the UPS. This degradation is presumably mediated by a cellular E3 ubiquitin ligase. To identify the E3 involved, we performed a siRNA screen for ubiquitin ligases that result in increased LCA levels in VCIP135-depleted cells. HECTD2 was identified as such an E3 (Fig. S4 A and B). HECTD2 is implicated in regulating PIAS1 (32) and as a candidate susceptibility gene in Alzheimer’s disease and Creutzfeldt–Jakob disease (33, 34). Consistent with the result of the siRNA screen, HECTD2 coimmunoprecipitates LCA but not LCE (Fig. 4A). This interaction is specific and direct, as bacterially expressed HECTD2, but not another HECT E3 Nedd4, bound LCA in vitro (Fig. S4C). More importantly, depletion of HECTD2 restored LCA stability in the absence of VCIP135 (Fig. 4B). In accord with the antagonistic functions of HECTD2 and VCIP135, depletion of VCIP135 led to increased ubiquitination of LCA in cells (Fig. 4C, compare lanes 1 and 2), which was reversed by concomitant depletion of HECTD2 (Fig. 4C, lane 3). Ubiquitination of LCA was restored by re-expression of HECTD2 (Fig. 4C, lane 4), demonstrating that this is not an off-target effect of the HECTD2 siRNAs. Similarly, re-expression of HECTD2 resulted in rapid degradation of LCA in cells depleted of both endogenous VCIP135 and HECTD2 (Fig. 4D, middle two panels, compare lanes 8–10 to 5–7).
Fig. S4.
Identification of a ubiquitin ligase for BoNT/A catalytic LC. (A) Schematic of the screen. (B) Western blots of siRNA screen of HECT domain ubiquitin ligases identifying HECTD2 as a putative ubiquitin ligase for LCA. The lane labeled “CTL (50%)” contains 50% of the cell lysate compared with other lanes. (C) E. coli lysates containing ∼0.2 μg 6×His-LCA and 2 μg of GST fusion proteins were assessed as in S3D. (D) Cells were transfected with FLAG-HECTD2 and full-length YFP-LCA or the indicated deletion mutants. FLAG-HECTD2 was immunoprecipitated with M2 agarose and coimmunoprecipitated YFP proteins assessed. (E) Cells were transfected with the indicated plasmids. Immunoprecipitation for EE-tagged HECTD2 was followed by immunoblotting as indicated.
Fig. 4.
HECTD2 promotes the degradation of LCA. (A) HEK293 cells were transfected with plasmids encoding FLAG-HECTD2 and YFP-LCA as indicated. After 36 h, cells were treated with MG132 for 8 h to inhibit the proteasome and lysed in Triton X-100 buffer. (B) M17 cells depleted of VCIP135 were transfected with YFP-LCA and either CTL or HECTD2 siRNAs. After 48 h, cells were treated with CHX and loss of YFP-LCA assessed. Cotransfected GFP was used to monitor transfection efficiency. (C) M17 cells were transfected with YFP-LCA, HA-ubiquitin, and the indicated siRNAs and plasmids. After 72 h, cells were treated with MG132 for 8 h followed by cell lysis, immunoprecipitation of YFP-LCA, and immunoblotting for HA-ubiquitin. (D) Cells were transfected with the indicated siRNAs and replated for overnight culture. After 16 h, cells were transfected with either vector or plasmid expressing HECTD2 or were not subject to further transfection (lanes 1–4).
Unlike VCIP135, HECTD2 can interact with LCA lacking its C-terminal 50 amino acids (Fig. S4D) and exhibits no binding to the C-terminal 77 amino acids of LCA. This and the absence of a discernable interaction between the DUB and E3 when coexpressed in cells (Fig. S4E) is consistent with the independent and direct binding of each to LCA. These results support an antagonistic relationship between VCIP135 and HECTD2 in determining the stability of LCA, where VCIP135 plays a dominant role leading to the persistence of the catalytic LC.
Depletion of VCIP135 Accelerates Recovery of Full-Length SNAP25 from BoNT Intoxication.
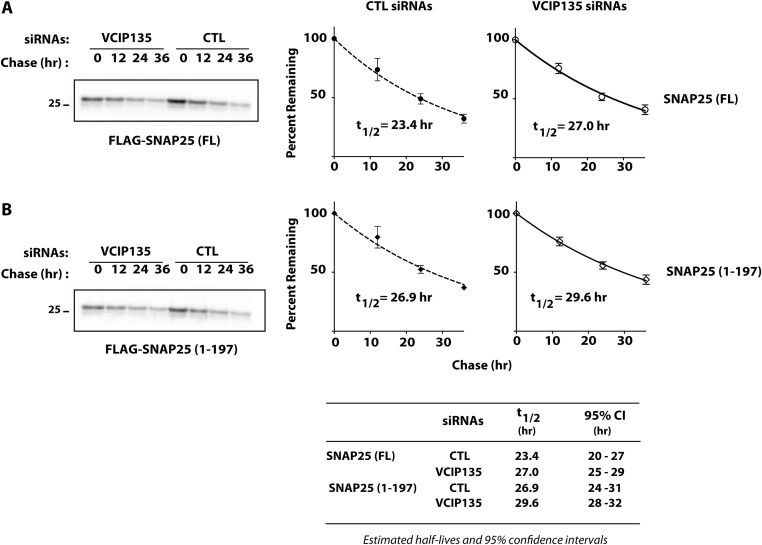
As motoneurons are the primary targets of BoNTs, a critical question is whether inhibition of VCIP135 accelerates the recovery of motoneurons from intoxication by BoNT/A holotoxin. BoNT/A inhibits synaptic transmission by cleaving SNAP25 after residue 197, generating a shorter cleaved SNAP25 (SNAP251–197), which is defective in synaptic vesicle fusion. Restoration of synaptic function following BoNT/A intoxication is dependent upon, and thus correlated with, the recovery of full-length SNAP25 (13, 35). A major factor contributing to the long duration of BoNT/A toxicity is the persistence of proteolytic activity of LCA, which continues to cleave newly synthesized SNAP25 (13, 14, 36). As a result, overexpression of WT SNAP25 is unable to restore exocytosis in BoNT/A intoxicated cells. In contrast, expression of a mutant SNAP25 resistant to BoNT/A cleavage restores exocytosis (37). Depletion of VCIP135 by itself had no significant effect on the measured half-lives of either SNAP25 or SNAP251–197 (Fig. S5 A and B). If VCIP135 is functionally important in the context of BoNT/A intoxication, its loss should accelerate recovery of full-length SNAP25 due to enhanced degradation of LCA. We therefore assessed the effect of VCIP135 depletion on the recovery of full-length SNAP25 in motoneurons treated with BoNT/A holotoxin.
Fig. S5.
Effect of VCIP135 depletion on turnover of SNAP25. M17 cells were transfected with CTL or VCIP135 siRNAs 18 h before transfection with (A) FLAG-SNAP25 or (B) FLAG-SNAP251–197. Turnover of SNAP25 proteins was assessed by pulse-chase metabolic labeling. Representative data shown on the Left and summarized on the Right (mean ± SD; n = 3). The half-lives of full-length SNAP25 and SNAP251–197 were ∼23 and 27 h, respectively, comparable to their reported half-lives of 24 h in cultured neurons (36). Importantly, depletion of VCIP135 had little effect on the measured half-lives for SNAP25 (t1/2 = 27 h) or SNAP251–197 (t1/2 = 30 h); P > 0.99 by Bonferroni’s multiple comparison tests.
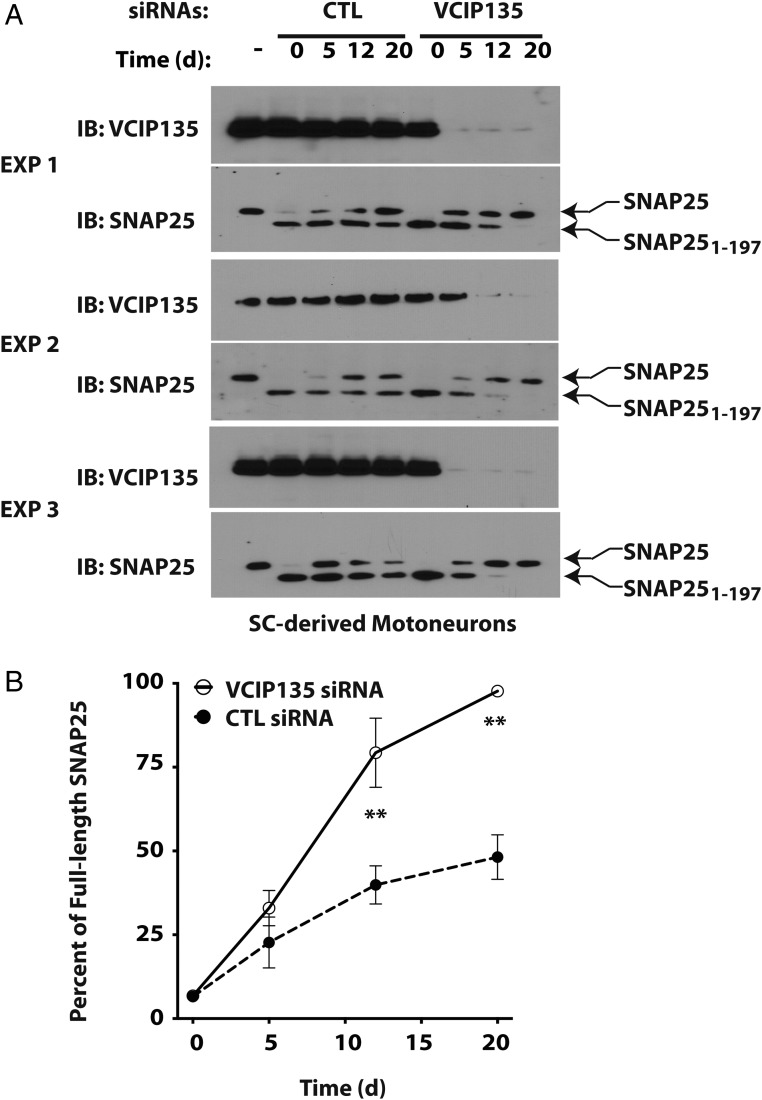
Motoneurons were generated by directed differentiation of mouse embryonic stem cells; these motoneurons show sensitivity to BoNT/A holotoxin comparable to that of cultured primary neurons (38). We next treated the motoneurons with BoNT/A holotoxin, which cleaved endogenous SNAP25 to produce the more rapidly migrating SNAP251–197 (Fig. 5A, Lower panels of each experiment). Cells were then transfected with either CTL or VCIP135 siRNAs, and the rate of recovery of full-length endogenous SNAP25 was monitored (quantified in Fig. 5B). In three separate experiments, depletion of VCIP135 accelerated the recovery of full-length SNAP25 (t1/2 = 6.2 d) compared with CTL siRNAs (t1/2 = 22.1 d). There was some variability in the rate of recovery of intact SNAP25, which could reflect normal biological variability in the rate of SNAP25 synthesis and turnover, transfection efficiency (Fig. 5 legend), or the expression of other modulators.
Fig. 5.
Depletion of VCIP135 accelerates recovery of full-length SNAP25 from BoNT. (A) Motoneurons derived from mouse embryonic stem cells were treated with BoNT/A holotoxin. After 20 h, cells were washed with media to remove residual toxin and transfected with either CTL or VCIP135 siRNAs. Cell lysates were collected at the indicated days postintoxication and levels of full-length and cleaved SNAP25 monitored by Western blotting, which migrated as expected at 25 and 24 kDa, respectively. (B) Semiquantitative analysis of the recovery of full-length SNAP25 as a percentage of total SNAP25. Data were shown as mean SEM (n = 3). **P < 0.01 based on Bonferroni’s multiple comparison tests (Table 1). Note that the initial knockdown of VCIP135 in experiment 2 was ineffective; regardless, the corresponding VCIP135 data were included in this analysis.
Discussion
There is currently no treatment for BoNT intoxication. Although several antibodies have been developed, these are only useful for preventing further uptake of the toxin in infected individuals. A single exposure to BoNT/A toxin can lead to prolonged muscle paralysis, lasting for months. Thus, even with the use of antitoxin antibodies, patients may require respiratory support for several months. The long-lasting effect of BoNT/A presents a major obstacle in the treatment of botulism and constitutes a major unaddressed bioterrorism threat and public health management cost.
In general, cells rapidly eliminate foreign proteins, such as bacterial toxins. LCA stands out for its cellular longevity, leading to persistent toxicity due to ongoing proteolysis of newly synthesized SNAP25. Different mechanisms had been proposed to account for persistence of BoNT/A intoxication (10, 11). We have previously provided a molecular mechanism for the differential persistence of LCA compared with LCE by demonstrating that LCE is recognized by the ubiquitin ligase TRAF2 and rapidly degraded by the UPS (12). In contrast, LCA is stable and there is no evidence that it is a target of TRAF2. Here, we establish how a cellular DUB, VCIP135, plays a dominant role in stabilizing LCA in cells by interacting with the C-terminal region of LCA, deubiquitinating the toxin, and rescuing it from proteasomal degradation. The rapid degradation of LCA in the absence of VCIP135 also allowed us to unmask a role for a ubiquitin ligase, HECTD2, in promoting degradation of LCA, an effect which is normally antagonized by the activity of VCIP135.
An important limitation in studying BoNT is that it is not feasible to monitor the small number of LC molecules (less than 1,000) needed to block neurotransmission (39). This has necessitated the transfection of plasmids encoding BoNT LCs that have been codon-optimized for mammalian expression to study BoNT persistence (12, 40–42). It is natural to ask whether discoveries based on transfection of BoNT LCs can be extended to intoxication with the holotoxin. The finding that SNAP25 recovery is accelerated with loss of VCIP135 in neurons intoxicated with BoNT/A holotoxin provides support for using recombinant BoNT LCs to study the mechanisms of BoNT persistence.
A second DUB, USP9X, also appears to be important for the turnover of LCA. However, we have not observed an interaction between USP9X and LCA. In addition, unlike the case with depletion of VCIP135, the accelerated degradation of LCA in the absence of USP9X depends on autophagy. These findings raise the possibility that loss of USP9X enhances the turnover of LCA due to indirect effects on autophagy. It might be attractive to target USP9X in botulism. Unfortunately, the reduced cell viability observed with loss of USP9X expression limits our ability to determine whether this effect is relevant at the low level of LCA that characterizes BoNT/A intoxication.
Finally, we have not observed toxicity from depletion of VCIP135 in HEK293 cells, neuroblastoma cells, or motoneurons. This suggests that agents targeting VCIP135 activity, or disrupting interactions between VCIP135 and LCA, may be efficacious in combination with inhibitors targeting the protease activity of LCA to accelerate patient recovery following BoNT/A intoxication.
Materials and Methods
Plasmids and Reagents.
Construction of codon-optimized LCA and LCE in pEGFP-C1, pEYFP-C1, pCMV-Tag2C, pcDNA5/TO/Frt, as well as HA-Ubiquitin, FLAG-tagged murine SNAP25 and FLAG-SNAP25-Luciferase reporter plasmid has been described (12). FLAG-SNAP251–197 was generated by PCR with FLAG-SNAP25 as a template and subcloned into pcDNA3.1(+) FLAG vector between BamHI and XhoI. Truncation mutants of LCA were generated by PCR with YFP-LCA as a template and subcloned into pEYFP-C1 vector from XhoI to ApaI. The C-terminal 77- and 50-residue fragments were subcloned into pEGFP-C1 between XhoI and EcoRI sites. For expression in Escherichia coli, 6×His-tagged LCA was generated by PCR and subcloned into pET3a from NdeI to XhoI. FLAG-USP9X was a gift from Vishva Dixit, Genentech, South San Francisco, CA (24); FLAG-HA-VCIP135 (Addgene) was a gift from Wade Harper, Harvard Medical School, Boston, MA (43). Catalytically inactive VCIP135 was generated by mutation of Cys-218 to Ala by site-directed mutagenesis. Murine Nedd4 in PGEX-KG has been described (44). Full-length VCIP135 was generated by PCR and subcloned into pGEX6P-1 from BamHI to SalI. Full-length HECTD2 was generated by PCR using an EST (CloneID 4811444; Open Biosystems) as template and subcloned into pcDNA3.1(+) FLAG, pcDNA3.1(+) Glu–Glu (EE) tag, or pGEX6P-1 from BamHI to XhoI. Full-length WAC was generated by PCR from an EST (CloneID 3347345; Dharmacon) and subcloned into pGEX6P-1 from BamHI to XhoI. All constructs were verified by DNA sequencing. Additional siRNAs targeting murine VCIP135 were purchased from Qiagen; siRNAs targeting ATG5 were from Cell Signaling Technology.
MG132, chloroquine, and Leupeptin were from Sigma-Aldrich; Epoxomicin and E64d were from ApexBio; cOmplete Protease Inhibitor Mixture tablets were from Roche; Protein G Sepharose was from GE Healthcare Life Sciences; rabbit polyclonal GFP antibody was from Clontech; mouse monoclonal antibodies for GFP, RFP, and ubiquitin (P4D1) were from Santa Cruz Biotechnology; rat monoclonal HA-antibody (3F9) was from Roche; mouse monoclonal FLAG antibody (M2), rabbit SNAP25 antibody targeting the N terminus of SNAP25, FLAG antibody (M2)-conjugated agarose, and HA antibody (HA7)-conjugated agarose were from Sigma-Aldrich; rabbit antibodies recognizing ATG5, VCP and Glu–Glu tag, VCIP135, HECTD2, and mouse antibodies for α-tubulin and β-actin were from Cell Signaling; mouse monoclonal antibody against LCA was from BEI Resources; and rabbit polyclonal WAC, VCIP135, and HECTD2 antibodies were from Novus Biologicals.
Tetracycline-Induced Expression of BoNT LCs and siRNA Screening.
To develop a tractable cell culture model for BoNT persistence, Flp-in/T-REX/ 293 cells (Invitrogen) were treated with 2 μg/mL doxycycline for 16 h to induce expression of the GFP-LCs. Cells were washed three times in serum-free media, trypsinized, and replated into tetracycline-free media. Chase began at this time (time = 0). Lysates were collected daily for 5 d and levels of GFP-LCs assessed by Western blotting.
For siRNA screening, we collaborated with Dharmacon to assemble siRNA libraries targeting the ubiquitin pathway. These reagents are now commercially available as The Human Deubiquitinating Enzyme and The Ubiquitin Conjugation Subsets 1–3 (Dharmacon). To screen for DUBs involved in LCA stabilization, we treated Flp-in/T-REX/293 cells with doxycycline to induce the expression of GFP-LCA. On the following day, cells were trypsinized and replated into 24-well plates and allowed to recover for 24 h. They were then transfected with siRNAs (50 nM) and cell lysates collected for Western blot analysis 3 d posttransfection. The z score was calculated using the median and SD (σ):
For validation, the experiment was repeated using 10-nM siRNAs. Positives from these experiments were then assessed using 10 nM of distinct siRNAs from a different supplier (Qiagen).
To screen for a putative ubiquitin ligase for LCA, we again induced expression of GFP-LCA with doxycycline and replated the cells the following day. After allowing cells to recover for 24 h, they were transfected with VCIP135 siRNAs together with siRNAs targeting specific ubiquitin ligases and cell lysates collected 3 d posttransfection.
Cell Culture and Transfection.
Unless indicated otherwise, all reagents were from Invitrogen. HEK293 cells were cultured in DMEM containing 10% FBS, 2 mM glutamine, and 2% penicillin–streptomycin; HEK-Flp-in/T-REX/293 cells in DMEM supplemented with 10% tetracycline-free FBS (Clontech), 2 mM glutamine, 2% penicillin–streptomycin, 100 μg/mL zeocin, and 15 μg/mL blasticidin; and M17 human neuroblastoma cells (ATCC) in DMEM media containing 10% FBS, 0.1 mM β-mercaptoethanol, 2 mM glutamine, 2% penicillin/streptomycin, and 2 mM nonessential amino acids (NEAA). Lipofectamine RNAiMAX was used for transfecting 50 nM siRNA, and HiPerfect HTS (Qiagen) was used when validating effects of knockdowns in screens using 10 nM siRNAs in the screen. In other experiments, we used Lipofectamine 3000 for plasmid transfection and HiPerfect (Qiagen) or Dharmafect 3 (Dharmacon) for siRNA transfection.
Differentiation of Mouse ES Cells into Motoneurons.
Motoneurons were derived by directed differentiation of mouse ES cells (HBG3) in which GFP expression is driven by mouse motoneuron-specific Hb9 promotor, using retinoic acid/Sonic Hedgehog/Purmorphamine-based induction methods as described (38). The HBG3 cell line was kindly provided by Thomas M. Jessell and Hynek Wichterle, Columbia University, New York, NY.
Ubiquitination Assays.
For assessment of ubiquitination, cells were transfected with siRNAs to deplete the endogenous protein(s) of interest. After 24 h, cells were transfected with plasmid encoding YFP-tagged LCA and other plasmids as indicated. After 36 h, cells were treated with proteasome inhibitor (MG132) for 6 h and then lysed in Triton X-100 Buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 10 mM iodoacetamide, 50 μM MG132 complete protease inhibitors) with 2% SDS. Lysates were heated for 2 min to denature proteins and immediately diluted with additional Triton X-100 buffer to a final concentration of 0.1% SDS. After centrifugation at 1,000 × g for 5 min, the clarified lysates were precleared by incubation with Protein G Sepharose at 4 °C for 60 min. YFP-LCA was immunoprecipitated from the precleared lysate by incubation with rabbit polyclonal GFP antibody and Protein G Sepharose at 4 °C for 2 h with constant mixing. Immunoprecipitates were washed with 3 × 20 bed volumes of 1% Triton X-100 buffer and samples resolved by SDS/PAGE followed by immunoblotting with HA antibody.
Coimmunoprecipitation.
Cell lysates in 1% Triton X-100 buffer were clarified and precleared as above and incubated with the indicated antibodies prebound to protein G Sepharose or with antibody-conjugated agarose. After washing extensively with Triton lysis buffer, immunoprecipitated proteins were eluted in 2× sample buffer, resolved by SDS/PAGE, and immunoblotted as indicated.
Metabolic Labeling.
Pulse chase analysis was performed 48 h after transfection. Briefly, cells were starved for 1 h in Met/Cys-free media and labeled for 45 min with 100 μCi/mL [35S]-Met/Cys. After labeling, cells were washed with PBS and cultured in chase media containing 10× unlabeled Met. At the indicated times, cells were washed with PBS and lysed with modified RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate 10 mM iodoacetamide, 50 μM MG132, and cOmplete protease inhibitors). Immunoprecipitation was carried out as described above, after resolution by SDS/PAGE samples were analyzed on a Typhoon FLA 7000 (GE).
CHX Chase.
Cells were transfected with the indicated plasmids. On the following day, they were trypsinized and replated. CHX (50 μg/mL) was added to inhibit protein synthesis 36–48 h after transfection. At the specified times, cells were lysed in modified RIPA buffer and processed for immunoblotting.
BoNT Intoxication and Recovery.
Mouse motoneurons were treated with 1 nΜ BoNT/A holotoxin (Metabiologics) for 16 h to cleave endogenous SNAP25. After extensive washing to remove the remaining toxin, the cells were transfected with CTL or VCIP135 siRNAs. Fresh media was provided every 3 d and transfections repeated every 5 d. Depletion of VCIP135 in motoneurons did not result in cytotoxicity. Cells were lysed in modified RIPA buffer and collected at the indicated times and assessed by immunoblotting for full-length and cleaved SNAP25 as well as VCIP135 by Western blotting.
Statistical Analysis.
Levels of full-length SNAP25 and SNAP251–197 were estimated from immunoblot data using a Typhoon FLA 7000. Data were analyzed with Prism (GraphPad Software). Data for recovery of full-length SNAP25 were analyzed by two-way ANOVA (Time × siRNAs) and Bonferroni’s multiple comparison tests; a P value less than 0.05 was considered significant (Table 1).
Table 1.
Statistical analysis of SNAP25 recovery following BoNT/A intoxication
| Bonferroni’s multiple comparisons test | Mean difference | 95% CI | Adjusted P value |
| VCIP135 siRNAs–CTL siRNAs | |||
| Time = 0 | 0.002504 | −0.2303–0.2353 | >0.9999 |
| Time = 5 | 0.1025 | −0.1303–0.3353 | 0.9331 |
| Time = 12 | 0.3944 | 0.1616–0.6272 | 0.0008** |
| Time = 20 | 0.4951 | 0.2623–0.7279 | <0.0001** |
P < 0.01.
To estimate the half-time for recovery of full-length SNAP25 (t1/2), the data were fitted with a single exponential achieving full recovery. The estimated t1/2 for control cells was 22.1 d with 95% CI 16–37 d; estimated t1/2 for VCIP135-depleted cells was 6.2 d with 95% CI 4.6–9.5 d. In experiment 2, the recovery for VCIP135 siRNA-transfected cells at day 5 was different from others in the group, as VCIP135 depletion from the first round of siRNA transfection was apparently not successful. Nevertheless, we did not exclude this data point but used all of the data in both statistical analysis and estimation of t1/2 for SNAP25 recovery.
Protein Expression and In Vitro Interactions.
Expression of GST and 6×His-tagged proteins were induced in Rossetta E. coli (EMD Millipore) with 0.2 mM IPTG and expression proceeded at 18.5 °C for 20 h. For in vitro binding experiments, lysates containing ∼0.2 μg of 6×His-tagged LCA and 2 μg GST fusion proteins were incubated with Glutathione Sepharose 4B for 2 h at 4 °C. The beads were then collected by centrifugation and washed with 4 × 1 mL of binding buffer (50 mM Tris·HCl, pH 8.0, 150 mM NaCl, and 0.1% Triton X-100) and processed for immunoblotting.
Acknowledgments
We thank Kevin Lorick for assistance with assembling siRNA libraries; Daniel Stringer and Charles Shoemaker (Tufts University) for helpful discussions; Meredith Metzger, Jadranka Loncarek, Stan Lipkowitz, and R. Andrew Byrd for critical reading of the manuscript; and Alayna Stepp, Emma Gergel, Haley Smith, Matthew Villanueva (all supported by WHK internships), Rhyan Maditz, and Sera Choi for technical assistance. This research is supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (Y.C.T., M.Y., L.T., and A.M.W.); National Institute of Allergy and Infectious Diseases Grant R01-AI093467 and Defense Threat Reduction Agency Grant W81XUH-12-2-0031 (to A.K. and G.A.O.); National Institute of Allergy and Infectious Diseases Grant R33-AI101387 (to E.K. and S.B.); and the Defense Threat Reduction Agency/Joint Science and Technology Office (S.B.).
Footnotes
Conflict of interest statement: G.A.O. has a financial interest in Synaptic Research LLC, which is developing therapeutics for BoNT intoxication, including the delivery of designer ubiquitin ligases.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621076114/-/DCSupplemental.
References
- 1.Simpson LL. Identification of the characteristics that underlie botulinum toxin potency: Implications for designing novel drugs. Biochimie. 2000;82:943–953. doi: 10.1016/s0300-9084(00)01169-x. [DOI] [PubMed] [Google Scholar]
- 2.Habermann E, Dreyer F. Clostridial neurotoxins: Handling and action at the cellular and molecular level. Curr Top Microbiol Immunol. 1986;129:93–179. doi: 10.1007/978-3-642-71399-6_2. [DOI] [PubMed] [Google Scholar]
- 3.Johnson EA, Montecucco C. Botulism. Handb Clin Neurol. 2008;91:333–368. doi: 10.1016/S0072-9752(07)01511-4. [DOI] [PubMed] [Google Scholar]
- 4.Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco C, Schiavo G. Structure and function of tetanus and botulinum neurotoxins. Q Rev Biophys. 1995;28:423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- 6.Dolly JO, Black J, Williams RS, Melling J. Acceptors for botulinum neurotoxin reside on motor nerve terminals and mediate its internalization. Nature. 1984;307:457–460. doi: 10.1038/307457a0. [DOI] [PubMed] [Google Scholar]
- 7.Pantano S, Montecucco C. The blockade of the neurotransmitter release apparatus by botulinum neurotoxins. Cell Mol Life Sci. 2014;71:793–811. doi: 10.1007/s00018-013-1380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnon SS, et al. Working Group on Civilian Biodefense Botulinum toxin as a biological weapon: Medical and public health management. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 9.Eleopra R, Tugnoli V, Rossetto O, De Grandis D, Montecucco C. Different time courses of recovery after poisoning with botulinum neurotoxin serotypes A and E in humans. Neurosci Lett. 1998;256:135–138. doi: 10.1016/s0304-3940(98)00775-7. [DOI] [PubMed] [Google Scholar]
- 10.Shoemaker CB, Oyler GA. Persistence of Botulinum neurotoxin inactivation of nerve function. Curr Top Microbiol Immunol. 2013;364:179–196. doi: 10.1007/978-3-642-33570-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossetto O, Pirazzini M, Montecucco C. Botulinum neurotoxins: Genetic, structural and mechanistic insights. Nat Rev Microbiol. 2014;12:535–549. doi: 10.1038/nrmicro3295. [DOI] [PubMed] [Google Scholar]
- 12.Tsai YC, et al. Targeting botulinum neurotoxin persistence by the ubiquitin-proteasome system. Proc Natl Acad Sci USA. 2010;107:16554–16559. doi: 10.1073/pnas.1008302107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller JE, Neale EA, Oyler G, Adler M. Persistence of botulinum neurotoxin action in cultured spinal cord cells. FEBS Lett. 1999;456:137–142. doi: 10.1016/s0014-5793(99)00948-5. [DOI] [PubMed] [Google Scholar]
- 14.Whitemarsh RC, Tepp WH, Johnson EA, Pellett S. Persistence of botulinum neurotoxin a subtypes 1-5 in primary rat spinal cord cells. PLoS One. 2014;9:e90252. doi: 10.1371/journal.pone.0090252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binz T, et al. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. J Biol Chem. 1994;269:1617–1620. [PubMed] [Google Scholar]
- 16.Tsai YC, et al. Differential regulation of HMG-CoA reductase and Insig-1 by enzymes of the ubiquitin-proteasome system. Mol Biol Cell. 2012;23:4484–4494. doi: 10.1091/mbc.E12-08-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semizarov D, et al. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci USA. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 19.DiAntonio A, et al. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 20.Rott R, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci USA. 2011;108:18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasso D, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011;286:8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas RL, Gustafsson AB. MCL1 is critical for mitochondrial function and autophagy in the heart. Autophagy. 2013;9:1902–1903. doi: 10.4161/auto.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germain M, et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011;30:395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwickart M, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 25.Nagai H, et al. Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol Cell. 2009;36:805–818. doi: 10.1016/j.molcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Naik E, et al. Regulation of proximal T cell receptor signaling and tolerance induction by deubiquitinase Usp9X. J Exp Med. 2014;211:1947–1955. doi: 10.1084/jem.20140860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchiyama K, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kano F, et al. NSF/SNAPs and p97/p47/VCIP135 are sequentially required for cell cycle-dependent reformation of the ER network. Genes Cells. 2005;10:989–999. doi: 10.1111/j.1365-2443.2005.00894.x. [DOI] [PubMed] [Google Scholar]
- 30.Totsukawa G, et al. VCIP135 deubiquitinase and its binding protein, WAC, in p97ATPase-mediated membrane fusion. EMBO J. 2011;30:3581–3593. doi: 10.1038/emboj.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S, Barbieri JT. Association of botulinum neurotoxin serotype A light chain with plasma membrane-bound SNAP-25. J Biol Chem. 2011;286:15067–15072. doi: 10.1074/jbc.M111.224493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coon TA, et al. The proinflammatory role of HECTD2 in innate immunity and experimental lung injury. Sci Transl Med. 2015;7:295ra109. doi: 10.1126/scitranslmed.aab3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lloyd SE, et al. HECTD2 is associated with susceptibility to mouse and human prion disease. PLoS Genet. 2009;5:e1000383. doi: 10.1371/journal.pgen.1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd SE, Rossor M, Fox N, Mead S, Collinge J. HECTD2, a candidate susceptibility gene for Alzheimer’s disease on 10q. BMC Med Genet. 2009;10:90. doi: 10.1186/1471-2350-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duregotti E, et al. Snake and spider toxins induce a rapid recovery of function of botulinum neurotoxin paralysed neuromuscular junction. Toxins (Basel) 2015;7:5322–5336. doi: 10.3390/toxins7124887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foran PG, et al. 2003. Evaluation of the therapeutic usefulness of botulinum neurotoxin B, C1, E, and F compared with the long lasting type A. Basis for distinct durations of inhibition of exocytosis in central neurons. in J Biol Chem 278:1363–1371.
- 37.O’Sullivan GA, Mohammed N, Foran PG, Lawrence GW, Oliver Dolly J. Rescue of exocytosis in botulinum toxin A-poisoned chromaffin cells by expression of cleavage-resistant SNAP-25. Identification of the minimal essential C-terminal residues. J Biol Chem. 1999;274:36897–36904. doi: 10.1074/jbc.274.52.36897. [DOI] [PubMed] [Google Scholar]
- 38.Kiris E, et al. Embryonic stem cell-derived motoneurons provide a highly sensitive cell culture model for botulinum neurotoxin studies, with implications for high-throughput drug discovery. Stem Cell Res (Amst) 2011;6:195–205. doi: 10.1016/j.scr.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanig JP, Lamanna C. Toxicity of botulinum toxin: A stoichiometric model for the locus of its extraordinary potency and persistence at the neuromuscular junction. J Theor Biol. 1979;77:107–113. doi: 10.1016/0022-5193(79)90141-3. [DOI] [PubMed] [Google Scholar]
- 40.Fernández-Salas E, Ho H, Garay P, Steward LE, Aoki KR. Is the light chain subcellular localization an important factor in botulinum toxin duration of action? Mov Disord. 2004;19:S23–S34. doi: 10.1002/mds.20006. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-Salas E, et al. Plasma membrane localization signals in the light chain of botulinum neurotoxin. Proc Natl Acad Sci USA. 2004;101:3208–3213. doi: 10.1073/pnas.0400229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo CL, Oyler GA, Shoemaker CB. Accelerated neuronal cell recovery from Botulinum neurotoxin intoxication by targeted ubiquitination. PLoS One. 2011;6:e20352. doi: 10.1371/journal.pone.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatakeyama S, Jensen JP, Weissman AM. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]