Significance
Selective oxidation of aliphatic C–H bonds in alkylphenols is important for both structural derivatization and biological degradation of these fundamental chemicals. However, significant problems are persistently associated with the chemical methods for this oxofunctionalization. In this study, we developed a unique chemomimetic biocatalytic system that is capable of selectively oxidizing p- and m-alkylated phenols in a controllable manner, overcoming the challenges faced by similar chemical oxidation. The structural and bioinformatics analyses of the central P450 biocatalyst CreJ suggest that its substrate flexibility and reaction selectivity could be further leveraged. This novel alkylphenol biooxidation system may hold great potential for application in pharmaceutical, biomanufacturing, and environmental industries once upscaled systems can be further developed in the future.
Keywords: alkylphenol, selective oxidation, chemomimetic biocatalysis, cytochrome P450 enzymes, Corynebacterium glutamicum
Abstract
Selective oxidation of aliphatic C–H bonds in alkylphenols serves significant roles not only in generation of functionalized intermediates that can be used to synthesize diverse downstream chemical products, but also in biological degradation of these environmentally hazardous compounds. Chemo-, regio-, and stereoselectivity; controllability; and environmental impact represent the major challenges for chemical oxidation of alkylphenols. Here, we report the development of a unique chemomimetic biocatalytic system originated from the Gram-positive bacterium Corynebacterium glutamicum. The system consisting of CreHI (for installation of a phosphate directing/anchoring group), CreJEF/CreG/CreC (for oxidation of alkylphenols), and CreD (for directing/anchoring group offloading) is able to selectively oxidize the aliphatic C–H bonds of p- and m-alkylated phenols in a controllable manner. Moreover, the crystal structures of the central P450 biocatalyst CreJ in complex with two representative substrates provide significant structural insights into its substrate flexibility and reaction selectivity.
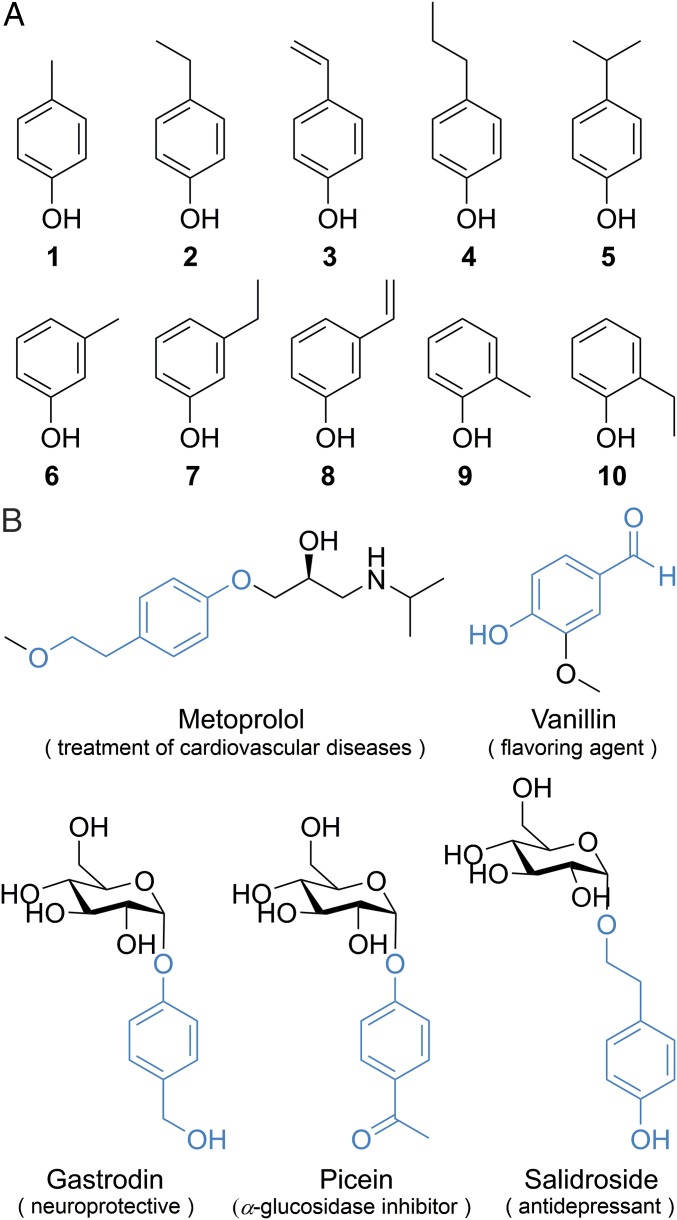
Alkylphenols, p-, m-, and o-alkylated phenols, e.g., 1–10, (Fig. 1A) are among the most important synthetic precursors for manufacturing a vast variety of chemical products including detergents, polymers, lubricants, antioxidants, emulsifiers, pesticides, and pharmaceuticals (1). Alkylphenols are also priority environmental pollutants because they are toxic, xenoestrogenic, or carcinogenic to wildlife and humans (2, 3). The selective oxidation of the aliphatic C–H bonds in alkylphenols serves significant roles not only in generation of functionalized intermediates for synthesizing more downstream products (4), but also in biological degradation of these environmentally hazardous compounds (5). In particular, some oxidized alkylphenols are direct structural motifs in drugs (e.g., metoprolol) (6) and bioactive ingredients of medicinal plants (Fig. 1B).
Fig. 1.
Alkylphenol structures. (A) Representative alkylphenols 1–10. (B) The drug and medicinal plants derived bioactive ingredients that are structurally related to oxidized alkylphenols. The oxidized alkylphenol motifs are highlighted in blue.
Chemically, these oxidative transformations have been practiced for a long time by using superoxides, organocatalyst, high valence transition metals, or strong oxidants such as nitric acid, chromic acid, and potassium permanganate (4, 7, 8). However, a number of problems are persistently associated with the chemical oxidation of aliphatic C–H bonds in alkylphenols: (i) the requirement of protection/deprotection of the phenolic hydroxyl group under reaction conditions that are distinct from those for oxidation complicates the synthetic process; (ii) it is difficult to delicately control the extent of oxidations (i.e., alcohol vs. aldehyde/ketone vs. carboxylic acid); (iii) the oxidation of an aromatic C–H bond may sometimes occur when using strong oxidants; (iv) the regio- and stereoselective oxidation of an unactivated sp3 C–H bond (in alkylphenols) remains a central challenge despite recent advances in combined use of specialized directing groups with transition metal catalysts (9–11) and in biomimetic supramolecular assemblies (12–14); and (v) chemical oxidants could be associated with significant environmental concerns (2, 3). Given these issues, oxidative enzymes with inherent catalytic selectivity and reduced environmental impact may be developed into alternative catalysts for selective oxidation of organic compounds including alkylphenols (15–18).
To date, a majority of efforts have focused on engineering of P450 monooxygenases, which are believed to be the most versatile biocatalysts in nature (19), using either directed evolution and/or rational design (20–22). Recently, a new strategy of substrate engineering has been explored (23, 24), wherein anchoring/directing groups are chemically linked to nonsubstrate compounds. Initial studies demonstrated that the addition of these anchoring groups facilitates enzyme–substrate interactions and enables selective oxidation of C–H bonds in previously inaccessible substrates. Using the P450 monooxygenase PikC as a model system (25, 26), chemically diverse substrates tethered to synthetic anchoring groups containing the N,N-dimethylamine moiety were hydroxylated in regio- or stereoselective manners (27–29). However, these approaches are not completely biological because the generation, installation, and offloading of anchoring groups still rely on chemical synthesis.
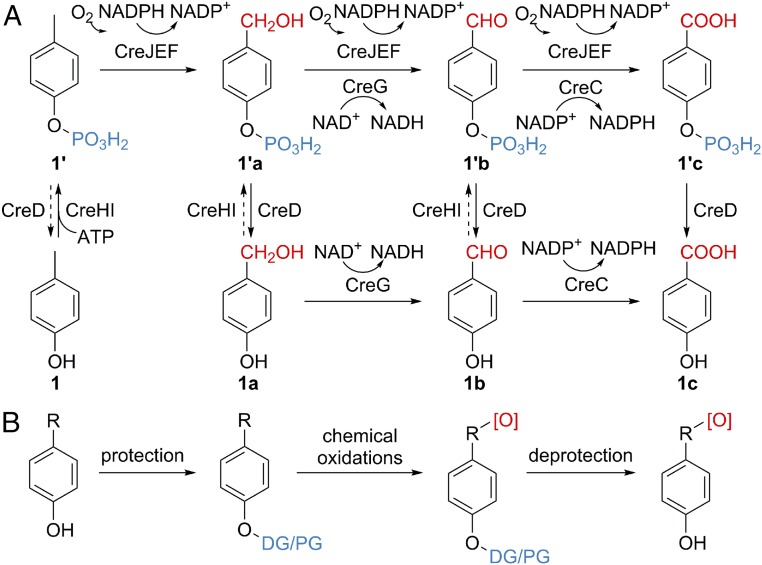
Recently, our laboratories have identified a p-cresol, 1, biodegradation pathway (Fig. 2A) in the Gram-positive bacterium Corynebacterium glutamicum (30). Initially, 1 is phosphorylated to 1′ by a novel 4-methylphenyl phosphate synthase, CreHI, with consumption of ATP. Next, a unique P450 monooxygenase CreJ, when partnered with the native redox partner proteins including ferredoxin CreF and ferredoxin reductase CreE, specifically recognizes phosphorylated intermediates (1′, 1′a, and 1′b) and successively oxidizes the aromatic methyl group into carboxylic acid (1′c) via alcohol (1′a) and aldehyde (1′b) intermediates. Finally, the phosphohydrolase CreD is responsible for removal of the phosphate group to afford 1a–1c. In addition, the alcohol dehydrogenase CreG and the aldehyde dehydrogenase CreC are recruited to reinforce the oxidative transformations of both phosphorylated (1′a and 1′b) and nonphosphorylated (1a and 1b) intermediates.
Fig. 2.
The chemomimetic biooxidation pathway and the classical chemical oxidation route. (A) The chemomimetic p-cresol biodegradation pathway in C. glutamicum. Phosphate anchoring groups are shown in blue. (B) The classical chemical oxidation route. DG, directing group; PG, protecting group. Oxidative functional groups are highlighted in red.
It is striking that this unique catabolic pathway, featuring protection/direction, oxidation, and deprotection steps, mimics the process of classical chemical oxidations (Fig. 2B). To the best of our knowledge, it is rare, if not unprecedented, that a natural biooxidation pathway contains the final deprotection step. As such, this pathway represents a good initial starting point for the development of an entirely biocatalytic system that can achieve selective oxidation of aliphatic C–H bonds in a spectrum of alkylphenols. To achieve this ambitious goal, we first solved the X-ray crystal structure of the central P450 biocatalyst CreJ in complex with its substrate 1′. With understanding of the structural basis for substrate recognition of this P450 enzyme, we next successfully developed a biocatalytic system comprising CreHI, CreJEF/CreG/CreC, and CreD, which is able to selectively oxidize a suite of p- and m-alkylated phenols. Further elucidation of the cocrystal structure of CreJ and p-ethylphenyl phosphate (2′) revealed the molecular mechanism for the regio- and stereoselectivity of CreJ. In sum, the high selectivity, controllable extent of oxidation, generality, and the “green” nature of this alkylphenol biooxidation strategy may find potential application in pharmaceutical, biomanufacturing, and environmental industries upon further development of upscaled systems in the future.
Results
Structural Basis for the Substrate Recognition of CreJ.
The P450 enzyme CreJ (also named CYP288A2 by Nelson et al.) (31) catalyzes the successive oxidations from 1′→1′a→1′b→1′c in the presence of redox partners CreE and CreF, and the electron donor NADPH (30). Although different oxidative modifications of the distal methyl group are well tolerated by CreJ, the tethered phosphate group is required for the activity of this P450 enzyme because the nonphosphorylated substrate cannot be recognized (30). To understand this unique substrate recognition mechanism, the X-ray crystal structure of CreJ in complex with 1′ was solved at 2.0 Å (PDB ID code: 5GWE).
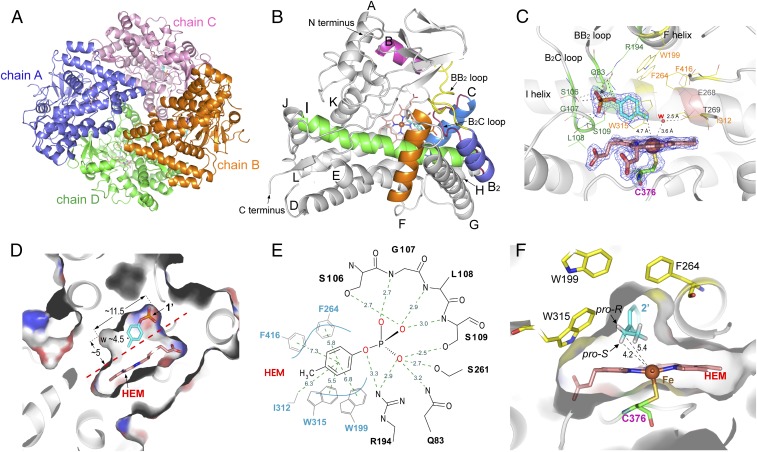
In the cocrystal structure of CreJ and 1′, four molecules that are essentially identical (except some slight variations in flexible loops) in an asymmetric unit (Fig. 3A) were observed (data collection and refinement statistics are available in SI Appendix, Table S1). Each monomer of CreJ adopts a typical triangular P450 fold with a long I helix (Fig. 3B), where the highly conserved acid/alcohol (E268/T269) residues reside (Fig. 3C). Mechanistically, these two amino acids are involved in the required protonation steps to generate the highly reactive heme iron(III)-peroxy (compound 0) and iron(IV)-oxo species (compound I) during a P450 catalytic cycle (SI Appendix, Fig. S1). The absolutely conserved proximal heme thiolate ligand C376 is located at a loop before the L helix (Fig. 3C) (32).
Fig. 3.
Structural analysis of CreJ. (A) An crystal asymmetric unit consisting of four molecules of CreJ. The molecules were traced from T29 to A433 except for chain B, which has gaps due to unresolved electron density between T133 and E141. (B) Monomeric structure of CreJ. (C) Analysis of the amino acid residues in the active site of CreJ. The 2Fo–Fc densities for 1′, heme, and C376 are shown as mesh at 2.0-σ level, and the heme iron is shown as Van der Waals sphere. (D) Cross-sectional view of the catalytic cavity containing the heme group (HEM) and the substrate (1′). (E) Schematic diagram of the interactions between CreJ and 1′ in the active site. Dashed lines denote the electrostatic and hydrophobic interactions with the distance between involved atoms shown in angstroms. (F) Cross-sectional view of the CreJ in complex with 2′. The ethyl group of 2′ enters a cavity, by which the pro-S hydrogen would point to the heme iron and be oxidized to S-2′a during the P450 catalysis.
The substrate binding pocket of CreJ is formed with the I helix as the wall and is covered by the BB2 loop, the B2C loop and the C terminus of F helix (Fig. 3 B and C). These regions (FG helices and BC loop) are generally flexible in P450 enzymes and readily reposition in response to substrate binding (33, 34). The structure adopts a closed conformation in which the putative substrate channel is blocked by the BB2 and B2C loops. The closed cavity that accommodates 1′ has a volume of ∼260 Å3 (11.5 Å × 4.5 Å × 5.0 Å), representing a relatively small substrate-binding pocket (Fig. 3D). The substrate 1′ is anchored and orientated mainly through a complex network of hydrogen bonds that exist between the phosphate moiety and a number of protein backbone amides, as well as hydroxyl and amine groups of amino acid side chains from the BB2/B2C loops and F/I helices (Fig. 3E and SI Appendix, Fig. S2A). Specifically, the 106SGLS109 motif of the B2C loop contributes the majority of hydrogen bond contacts with the phosphate oxygen atoms of the substrate, suggesting its crucial role for substrate anchoring. The positively charged guanidine of R194 from the F helix likely forms a strong electrostatic interaction with the negatively charged phosphate group. In addition, S261 from the I helix and Q83 from the BB2 loop also form hydrogen bonds with 1′.
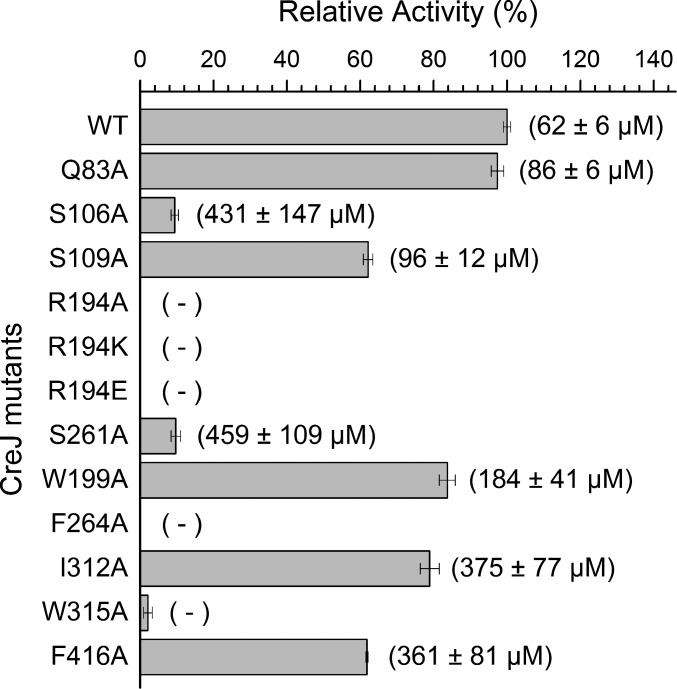
To evaluate whether these active site residues are essential for CreJ catalysis, a series of mutants were constructed, expressed, and purified (SI Appendix, Fig. S3A). The mutant enzymes were then evaluated for their ability, relative to wild-type CreJ, to oxidize substrate 1′ (Fig. 4). Remarkably, the replacement of R194 by either an alanine (R194A), a lysine (R194K), or a glutamine (R194E) residue completely abolished the enzyme activity (Fig. 4). Consistently, no substrate binding could be detected based on the low-to-high spin iron spectral shift (35) of the three R194 mutants (SI Appendix, Figs. S4–S6), whereas the dissociation constant (KD) of wild-type CreJ toward 1′ was determined to be 62 ± 6 μM (SI Appendix, Fig. S7). These results clearly indicate the necessity of the electrostatic interaction between the guanidyl base and the phosphate group for productive substrate binding. The mutants S106A and S261A retained marginal activity (9.4% and 9.7%, respectively) and showed significantly reduced substrate binding affinities (KD = 431 ± 147 μM and 459 ± 109 μM, respectively) compared with wild-type CreJ (Fig. 4 and SI Appendix, Figs. S8 and S9), suggesting that they are also critical for the productive substrate–protein interactions. In contrast, residues Q83 and S109 appear to be less important, as their substitution with alanine had unsubstantial effect on both substrate binding and enzyme activity (Fig. 4 and SI Appendix, Figs. S10 and S11).
Fig. 4.
The relative enzymatic activity and dissociation constant (KD) of CreJ mutants. A total of 1 μM of CreJ was used to transform 1 mM substrate 1′ for 20 min. KD values are shown in brackets. “−” denotes that no valid KD could be deduced. Substrate binding curves used for determining KD are shown in SI Appendix.
Overall, the phosphate moiety of the substrate appears to be gripped by CreJ, whereas the nonpolar portion of 1′ is recognized predominantly by hydrophobic interactions between the benzyl moiety and its surrounding residues including W199, F264, I312, W315, and F416 (Fig. 3E and SI Appendix, Fig. S2B). These residues constitute a hydrophobic pocket that extends beyond the heme, which serves to position the methyl group to be oxidized close to the heme iron reactive center. To examine the function of these hydrophobic residues, five mutants including W199A, F264A, I312A, W315A, and F416A were prepared (SI Appendix, Fig. S3B) and their catalytic activities toward substrate 1′ were evaluated (Fig. 4). As expected, alanine substitution of W199, I312, or F416 displayed unsubstantially negative effects on CreJ activity (Fig. 4 and SI Appendix, Figs. S12–S14). By contrast, the mutation on F264 or W315 abolished the detectable substrate binding (no valid KD could be deduced, SI Appendix, Figs. S15 and S16), accompanied by no or extremely low enzymatic activity (Fig. 4). As shown in Fig. 3E, F264 and W315 are closer to the aromatic ring of 1′ than other hydrophobic residues. More importantly, F264 likely plays an important role in maintaining the proper conformation of R194 via Van der Waals forces, and W315 forms water-mediated H bonds with S109 and heme, contributing to the proper conformation of the B2C loop (SI Appendix, Fig. S2B).
The distance between the methyl carbon atom of 1′ and the heme ion was measured to be 4.7 Å, which is well within the reactive range seen for other P450s (27, 36). Notably, there is small additional space around the methyl group in the catalytic cavity, which is partially occupied by a water molecule. Upon displacement of this water molecule, this space should be sufficient to accommodate oxidized intermediates, including the alcohol 1′a and the aldehyde 1′b that can undergo further oxidation to generate the final product 1′c. Furthermore, this additional space within the catalytic cavity could be leveraged to broaden the substrate specificity of CreJ.
Selective Oxidation of Diverse Alkylphenols by the Chemomimetic Biocatalytic System Composed of CreHI/CreJEF/CreGC/CreD.
The cocrystal structure of CreJ in complex with 1′ suggests that this P450 enzyme might be capable of accepting a broader range of phosphorylated alkylphenols as substrates. If this is true, CreJ could be used to selectively oxidize aliphatic C–H bonds in diverse alkylphenols. However, CreJ’s requirement of a phosphate anchoring group would limit the application of this strategy. To overcome this limitation, we envisioned that CreHI would be able to phosphorylate variant alkylphenols, thereby providing potential properly decorated substrates for CreJ oxidation. Furthermore, if CreD could dephosphorylate diverse oxidized products produced by CreJ, a useful multienzyme system could be established as an alternate to chemical oxidations.
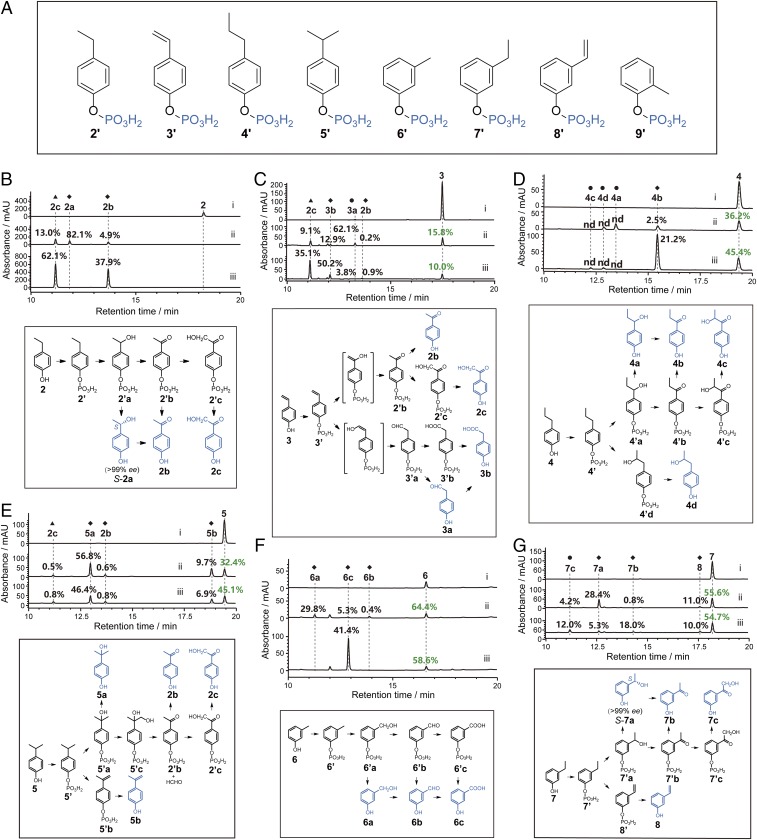
To test these hypotheses, we selected 9 representative alkylphenols, 2–10 (Fig. 1A), namely, p-ethylphenol, 2; p-vinylphenol, 3; p-propylphenol, 4; p-isopropylphenol, 5; m-cresol, 6; m-ethylphenol, 7; m-vinylphenol, 8; o-cresol, 9; and o-ethylphenol, 10 to examine the activity of CreHI, CreJEF, and CreD. As expected, all of these alkylphenols, with the exception of 10, were recognized by CreHI and transformed into corresponding phosphorylated products 2′–9′ (Fig. 5A and SI Appendix, Table S2 and Figs. S17–S25), thus demonstrating the broad substrate specificity of CreHI. The inability of CreHI to phosphorylate 10 might be due to the steric interference of substrate binding or phosphate transfer by the adjacent ethyl group. To evaluate the activity of CreJEF toward 2′–9′, we elected to sequentially add CreHI (with necessary cofactors, including Mg2+, Mn2+, and ATP) and CreJEF (with NADPH) into each reaction mixtures containing an individual alkylphenol substrate. This strategy was pursued due to the difficulty in purifying highly hydrophilic and unstable phosphorylated intermediates. In principle, we reasoned that the phosphorylated intermediates (2′–9′) generated by CreHI catalysis, without isolation and purification, would be recognized by CreJ and converted into the corresponding oxidized phosphoryl products. In these multienzyme reactions, significant substrate consumption and/or new product generation was detected when 2′–7′ were substrates (SI Appendix, Table S2 and Figs. S17–S22). By contrast, m-vinylphenyl phosphate (8′) and o-methylphenyl phosphate (9′) were not oxidized. It is possible that the difficulty in oxidizing compound 9′ is due to the methyl group at ortho position pointing away from the P450 heme-iron reactive center. Surprisingly, CreJ oxidizes 7′ rather than its structural analog 8′. We hypothesize that this might be the result of the intrinsic difference between the sp3 and sp2 C–H bonds.
Fig. 5.
Chemomimetic oxidation of alkylphenols. (A) The tested phosphorylated alkylphenols. (B–G) HPLC analyses (Upper) and putative mechanisms (Lower) of multienzyme reactions using p-ethylphenol (B), p-vinylphenol (C), p-propylphenol (D), p-isopropylphenol (E), m-cresol (F), and m-ethylphenol (G) as substrates. Trace i, negative controls; trace ii, reactions sequentially catalyzed by CreHI/CreJEF (10 μM of each enzyme, 30 °C, 2 h) and CreD (10 μM, 30 °C, 2 h); and trace iii, reactions sequentially catalyzed by CreHI/CreJEF/CreG/CreC (10 μM of each enzyme, 30 °C, 2 h) and CreD (10 μM, 30 °C, 2 h). The initial concentration of substrate was 1 mM in all reactions. The percentage numbers in black indicate the yield (conversion ratio) of a specific product. The percentage numbers in green represent the ratio of unreacted substrates. Due to distinct extinction coefficients, the peak intensity of compounds do not necessarily reflect relative amounts shown in percentage. ◆ indicates that the product was structurally assigned based on HRMS analysis and retention time comparison with the corresponding authentic standard. ▲ indicates that the product was identified by NMR and HRMS. ● indicates that the product was structurally deduced from HRMS and putative mechanism. nd, the specific percentage yield is not determined. Details for product structural assignments are shown in SI Appendix.
To establish the “chemomimetic” alkylphenol oxidation pathway, we carried out a series of one-pot enzymatic reactions using 2–7 as individual substrates, into which CreHI/CreJEF (alternatively with supplementation of CreG/CreC as helper enzymes), and CreD were sequentially added. In this way, the ability of CreD to hydrolyze diverse phosphorylated compounds was readily evaluated. More importantly, the hydrolysis of phosphoryl esters would enable the structural assignments of oxidized products, by which the efficiency and selectivity of oxidation could be ascertained.
When 2 was used as the substrate, CreJ mainly (as reflected by the 82.1% yield of 2a, which was quantified based on HPLC peak area integration) hydroxylates the C–H bond at the benzylic position of 2′ (Fig. 5B). Chiral HPLC analysis, using a manually prepared bis(3-chlorophenylcarbamate)-(cyclohexylamide) chiral column (37), showed only a single enantiomer 2a [>99% enantiomeric excess (ee)] was produced, thus demonstrating the achievement of absolute stereoselectivity during catalysis. Retention time comparison of the product to the enantiomerically pure authentic standard established the S configuration at the benzylic chiral center of 2a (SI Appendix, Fig. S26A). Based on the detection of 2b (4.9% yield) and 2c (13.0% yield), the successive oxidation steps 2′a→2′b→2′c can be readily deduced (Fig. 5B).
To elucidate the structural basis for the 2S stereoselectivity, the crystal structure of CreJ in complex with 2′ was solved at 1.7 Å (PDB ID code: 5XJN). This time, only one P450 molecule existed in one asymmetric unit. The CreJ molecules in CreJ-2′ and CreJ-1′ have ∼0.17 Å rmsd, indicating the essentially identical CreJ structures in both complexes (SI Appendix, Fig. S27A). The phosphate and benzyl moieties in both substrates are almost identically positioned, and only slight conformational changes were observed for the peripheral residues surrounding the aliphatic group of substrates. The terminal methyl group of 2′ in the CreJ-2′ structure occupies the partial cavity space of a water molecule in the CreJ-1′ structure, and the water molecule was not observed in CreJ-2′ (SI Appendix, Fig. S27B).
In the substrate binding pocket of the 5XJN structure (Fig. 3F), the ethyl group of 2′ enters a cavity that is shaped by the heme group and a number of hydrophobic residues, including F264, I312, F416, and T269, in which the pro-S hydrogen atom (4.2 Å) is closer to the heme-iron reactive center than the pro-R hydrogen atom (5.4 Å) of the methylene group in 2′. Although the C–Hpro-S bond somehow points away from the heme iron, the oxygen atom in the genuine ferryl-oxo catalytic species (compound I, SI Appendix, Fig. S1) would likely push away the terminal methyl group, thus counterclockwise rotating the C–C bond that connects the aromatic ring and the ethyl group, to project the pro-S hydrogen atom toward the heme iron. Notably, there is indeed a cavity near F264 that could accommodate the repelled methyl group. These analyses, together with the fact that the benzylic C–H bond is much more reactive than a primary C–H bond, can well explain why S-2′a is exclusively formed. The keto group of 2′b likely results from the second benzylic C–H hydroxylation, which is presumably followed by a spontaneous dehydration. Interestingly, once the benzylic position is fully oxidized, CreJ is able to hydroxylate the primary C–H bond with much higher bond dissociation energy.
Sterically, 3 is highly analogous to 2. However, CreJ oxidized 3 in a pattern that is distinct from 2. Four products including 2b (0.2% yield), 2c (9.1% yield), 3a (62.1% yield), and 3b (12.9% yield) were generated in reactions using 3 as the substrate (Fig. 5C). It appears that both 2b and 2c were derived from 2′b upon the hydroxylation of the benzylic position of 3′. The major product 3a could be derived from the hydroxylation of the terminal sp2 C–H bond in the vinyl group of 3 (compared with the hydroxylation of the benzylic methylene in 2), and followed by isomerization (to 3′a) and hydrolysis (to 3a). We observed that compound 3′a was further oxidized to 3′b, whose subsequent hydrolysis gave rise to 3b. The different hydroxylation site selectivity of CreJ toward 3 and 2 may be due to the presence of the vinyl group in 3. Its double bond could form π–π or electrostatic interactions with neighboring aromatic residues, by which the sp2 C–H bond might adopt a conformation favoring the terminal hydroxylation.
According to the measurement of the substrate binding pocket of CreJ (Fig. 3D), 4′ seems to be too large to be accommodated by this P450 enzyme. However, the detection of multiple oxidative products (4a–d, Fig. 5D) during the transformation of 4 suggests that a catalytic cavity expansion in CreJ could be induced. Notably, the absolute configuration of 4a, 4c, and 4d were not identified due to lack of authentic standards. The low conversion indicates that the size of 4′ might have reached the space limit of the catalytic cavity. When 5 was used as a substrate, in addition to the direct hydroxylation product 5a, three unexpected products including 5b, 2b, and 2c were observed (Fig. 5E). Mechanistically, 5b could result from a direct desaturation of the isopropyl group. A similar mechanism was previously proposed from the oxidation process of ethyl carbamate by CYP2E1 (38). The products 2b and 2c might be derived from an oxidative cleavage event. Putatively, 5′a could be oxidized in one of the methyl groups to generate a diol intermediate (5′c), which could then lose a single carbon unit by the C–C bond cleavage during further oxidation, generating 2′b and a molecule of formaldehyde. This mechanism is not uncommon in the reactions of cholesterol side chain cleavage (39, 40). For example, the transformation from cholesterol to pregnenolone depends on the cleavage of C20,C22 carbon–carbon bond catalyzed by CYP11A1 (41).
Aside from p-alkylphenols, the two m-alkylphenols 6 and 7 were also transformed by CreHI/CreJEF/CreD to different products 6a–c, and 7a–c plus 8, respectively, albeit with lower observed conversion ratios (Fig. 5 F and G). Like S-2a, S-7a turned out to be the only enantiomer (>99% ee) (SI Appendix, Fig. S26B), demonstrating the perfect stereoselectivity of CreJ against 7. The oxidation patterns of 6 and 7 are similar to those of 1 and 2, respectively. The only exception is the production of 8, which is likely attributed to the direct desaturation of the ethyl group.
We reasoned that the addition of CreG (alcohol dehydrogenase) and CreC (aldehyde dehydrogenase) could alter the ratios of oxidative products, provided that the intermediate alcohols or aldehydes could be accepted by CreG or CreC as substrates, respectively (Fig. 5 B–G). For example, in the presence of CreHI and CreJEF, the alcohol 6a, the aldehyde 6b, and the carboxylic acid 6c were all detected (Fig. 5F, trace ii), whereas the addition of CreG and CreC led to the most oxidized compound 6c as the only detectable product (Fig. 5F, trace iii). The improved oxidation could be due to not only the catalytic activity of CreG (for both 6a→6b and 6a′→6b′) and CreC (for both 6b→6c and 6b′→6c′), but also the NAD(P)H recycling effects (Fig. 2A). By contrast, the addition of CreG and CreC showed no positive impact on oxidative reactions of 5 because none of the intermediates or products could be recognized by CreG or CreC (Fig. 5E).
Controlled Oxidation of p-Ethylphenol, 2.
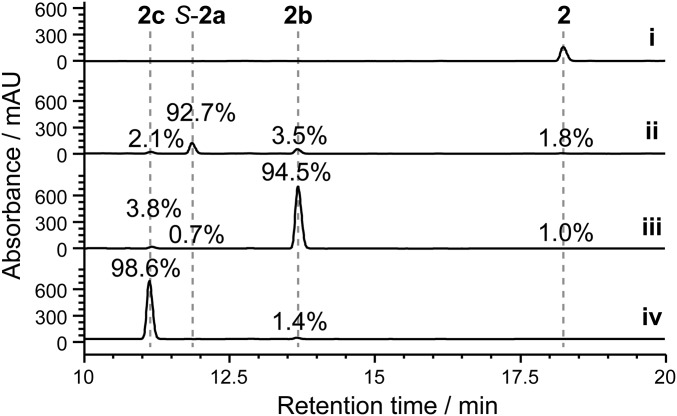
It is challenging to precisely control the extent of oxidation of the aliphatic C–H bonds in an alkylphenol by chemical methods (4). In the majority of our multienzyme reactions (Fig. 5 B–G), a mixture of products was observed. Although the product diversity showcases the ability of this unique biocatalytic system to generate various oxidized products, a single oxidized product is most often preferred in practical application. Accordingly, we chose the substrate 2 as an example to demonstrate that oxidation levels can be deliberately and delicately controlled by careful management of the multienzyme oxidation system.
In the sequential reactions mediated by CreHI/CreJEF and CreD (Fig. 5B, trace ii), the enantiomer S-2a accounted for 82.1% of total products. To further increase this percentage, we simply lowered the concentration of NADPH from 3 mM to 2 mM at the oxidation stage, by which further oxidation of S-2a to 2b and 2c was stalled due to the limited reducing equivalents, thus leading to a 92.7% yield of S-2a (Fig. 6, trace ii). To improve the production of p-acetyl phenol (2b), CreG (10 μM) together with its cofactor NAD+ (2 mM) were added to the reaction mixture upon completion of sequential catalysis by CreHI/CreJEF and CreD. As a result, 94.5% of total detected products were 2b (Fig. 6, trace iii). Finally, oxidation of 2 to 2c was driven to near completion by titration of fresh CreJEF and NADPH in the reaction comediated by CreHI/CreJEF/CreG. Remarkably, final hydrolysis by CreD, produced a 98.6% yield of 2c (Fig. 6, trace iv).
Fig. 6.
Controlled oxidation of p-ethylphenol, 2. Trace i, negative control; trace ii, the reaction was sequentially catalyzed by CreHI/CreJEF (10 μM of each enzyme, 2 mM NADPH, 30 °C, 2 h) and CreD (10 μM, 30 °C, 2 h); trace iii, the reaction mixture of trace ii was supplemented with CreG (10 μM, 30 °C, 2 h) and 2 mM NAD+; trace iv, the reaction was first comediated by CreHI/CreJEF/CreG (10 μM of each enzyme, 30 °C, 2 h), followed by adding an additional batch of CreJEF (10 μM of each enzyme, 30 °C, 2 h) and NADPH (3 mM), and finally catalyzed by CreD (10 μM, 30 °C, 2 h). Due to distinct extinction coefficients, the peak intensity of compounds do not necessarily reflect relative amounts shown in percentage.
Discussion
In this study, the recently discovered p-cresol biodegradation pathway from C. glutamicum (30) comediated by CreHI (for anchoring/directing group installation), CreJEF (for selective oxidation), CreG and CreC (as auxiliary catalysts), and CreD (for anchoring/directing group offloading) was used to construct genuine biosynthetic routes for production of various oxidized alkylphenols. By harnessing the broad substrate specificity and combined activities of CreHI, CreJ, and CreD, selective oxidation of the aliphatic C–H bonds in a representative group of p- and m-alkylphenols was successfully achieved. In particular, this biocatalytic system showed perfect stereoselectivity when hydroxylating p-ethylphenol, 2, and m-ethylphenol, 7 (Fig. 5 B and G), which is highly challenging for chemical oxidation. However, this biocatalytic system was not able to oxidize o-alkylphenols. Interestingly, recent work has demonstrated that P450 BM3 can be engineered to hydroxylate alkylbenzenes to corresponding alkylphenols (42, 43). Should these biocatalysts be coupled to the current alkylphenol oxidation system, an extended biochemical route from alkylbenzenes to oxidized alkylphenols could be created.
To gain more general application of this multienzymatic oxidation system, the substrate specificity of the central catalyst P450 CreJ requires further broadening. Aside from the hydrophilic portion of the substrate binding pocket that serves to tightly anchor the phosphate group, the hydrophobic cavity occupied by the alkyl benzyl moiety is relatively small. This limited space apparently prevents CreJ from accepting bulkier substrates. Thus, protein engineering based on rational design and combinatorial saturation mutagenesis could be used in the future to achieve cavity expansion/reshaping of CreJ, as has been done for P450 BM3 (16, 44).
The phosphate anchoring motif in CreJ is reminiscent of the carboxylate-mediated substrate–enzyme interactions observed in the fatty acid hydroxylase P450BSβ (45) and the P450 fatty acid decarboxylase OleTJE (46). Unlike the phosphate docking site remote from the heme iron in CreJ, the salt bridge for substrate anchoring of the latter two P450 peroxygenases is formed between the substrate carboxylate and a conserved arginine residue at a location proximate to the heme iron. Interestingly, Watanabe and colleagues revealed that nonnative substrates could be recognized by P450BSβ using a free decoy molecule, such as short chain fatty acids (47). This inspired us to test whether the free phosphate group could enable the binding of the free alkylphenol 1. However, no oxidation of 1 by CreJEF was detected, even in the presence of 100 mM inorganic phosphate, which indicates that the covalently bound phosphate group is required for productive substrate binding.
Notably, the hydrophobic pocket of CreJ is mainly shaped by a number of aromatic amino acids, including W199, F264, W315, and F416. It could be speculated that some of these aromatic residues might be involved in π–π stacking interactions with the benzene ring of substrate. If these specific interactions are essential for productive substrate binding, this strategy would be limited to the selective oxidation of alkylphenols. To evaluate this possibility, we preliminarily assessed the reactivity of nonaromatic substrates 1-hexanol phosphate (11′) and 4-methyl cyclohexanol phosphate (12′) with CreJ. It is worth noting that these two phosphorylated compounds were prepared chemically (SI Appendix for synthetic details) because neither compound could be enzymatically prepared by CreHI from 1-hexanol, 11, and 4-methyl cyclohexanol, 12 (SI Appendix, Fig. S28). According to LC/high-resolution mass spectrometry (HRMS) analysis, 11′ and 12′ were both oxidized to corresponding monohydroxylated products, respectively (SI Appendix, Figs. S29 and S30). Although the structures of products have yet to be determined, the substantial conversion of 11′ and 12′ indicate that the chemomimetic oxidation strategy developed in this work could be further extended toward the selective oxidation of aliphatic alcohols, which is currently ongoing in our laboratories.
Compared with other alkyl aromatics biooxidation pathways in nature (5, 48), such as xylene monooxygenase (49–51) and p-cresol methylhydroxylase (52), this unique chemomimetic system features a greater generality because the key P450 enzyme CreJ adopts the phosphate anchoring strategy for substrate recognition and binding. Protein BLAST searches reveal a large number of CreJ analogs (>40% sequence identity) from different genera of microorganisms. Sequence alignment of these homologous proteins demonstrates that the residues involved in phosphate anchoring and hydrophobic interactions with substrates are highly conserved (SI Appendix, Fig. S31), suggesting a new family of P450 enzymes (SI Appendix, Fig. S32). Interestingly, a majority of these P450 genes are clustered with groups of genes analogous to creHI, creEF, as well as creD (SI Appendix, Fig. S33). The widespread creJ-like P450 genes and cre-like gene clusters suggest that similar oxidation systems might be broadly used by diverse microorganisms, from which analogous oxidative biocatalysts with different substrate preference, improved oxidation selectivity, and higher catalytic efficiency could be identified.
Methods
Crystallization, Data Collection, and Structure Determination.
Before crystallization, the protein was diluted to 0.3 mM by mixing with 10 mM Tris⋅HCl (pH 8.0), supplemented with 12 mM substrate 1′ or 2′. Crystallization conditions were determined using commercial high throughput screening kits purchased from Hampton Research and a nanoliter drop-setting Mosquito robot (TTP Labtech) operating with 96-well plates. The crystallization conditions amenable to crystal generations were further optimized in 24-well crystallization plates. The crystals of CreJ in complex with 1′ used for X-ray data collection were obtained in hanging drops containing 0.2 M ammonium sulfate, 0.1 M sodium cacodylate trihydrate, pH 6.4, and 27% PEG8000, whereas the crystals of CreJ in complex with 2′ were obtained in 0.2 M sodium iodide and 20% PEG3350. The data were collected at the Shanghai Synchrotron Radiation Facility, Beamline BL17U (53), in a 100-K nitrogen stream. Data indexing, integration, and scaling were conducted using MOSFLM (54). The CreJ structure was determined by molecular replacement using CCP4 program suite (55) and the PDB file 4RM4 (56) as a search model. The structure was automatically built by ARP/wARP in the CCP4i program package (57). Refinement of the structure was performed using the programs COOT (58) and PHENIX (59). The final model was evaluated using PROCHECK (60). All molecular graphics were created using PyMOL v1.8 (Schrödinger).
Enzymatic Assays.
Unless specified otherwise, all enzymatic assays were carried out in 100 μL of 50 mM Tris·HCl buffer (pH 8.0) at 30 °C for 2 h, and the final concentration of each enzyme was 10 μM. Methanol (3× volume) was added to quench reactions. Precipitated proteins were removed by centrifugation at 20,000 × g for 10 min and the supernatants were used as LC-MS samples. For reactions catalyzed by CreHI, the reaction mixture contained 1 mM substrate, 20 mM MgCl2, 2 mM MnCl2, 2 mM ATP, and purified CreH and CreI. For CreJEF activity toward phosphorylated compounds (2′–9′), the CreHI reactions were used to generate the phosphorylated substrates. Next, CreJ, CreE, and CreF together with 3 mM NADPH were added into the individual post-CreHI reaction mixture.
For one-pot chemomimetic alkylphenol oxidation reactions, the reaction mixtures contained CreHI, CreJEF, and the two optional enzymes CreG (with 2 mM NAD+) and CreC (with 2 mM NADP+), 1 mM substrate 2–7, and necessary cofactors, including 20 mM MgCl2, 2 mM MnCl2, 2 mM ATP, and 3 mM NADPH. After incubation at 30 °C for 2 h, CreD was added into each of the one-pot mixtures, followed by another 2-h incubation at 30 °C.
For controlled oxidation of 2, the reactions were performed as follows: To obtain the predominant product S-2a, the reaction mixture containing 10 μM CreHI, 10 μM CreJEF, 20 mM MgCl2, 2 mM MnCl2, 2 mM ATP, and 2 mM NADPH was first incubated at 30 °C for 2 h. Following this initial incubation, CreD (10 μM) was added into the mixture and allowed to incubate for an additional 2 h. To produce 2b as the major product, CreG (10 μM) with its cofactor NAD+ (2 mM) was added into the above post-CreD reaction mixture, followed by another 2-h incubation. To predominantly produce 2c, the reaction mixture containing 10 μM CreHI, 10 μM CreJEF, 10 μM CreG, 20 mM MgCl2, 2 mM MnCl2, 2 mM ATP, 2 mM NAD+, and 3 mM NADPH was first incubated at 30 °C for 2 h. Subsequently, an additional batch of CreJEF (final concentration of 20 μM, including the first addition) and NADPH (final concentration of 6 mM, including the first addition) were added into the above mixture, followed by another 2-h incubation. Finally, CreD (10 μM) was added into the mixture and followed by incubation at 30 °C for 2 h.
Analytical Methods.
Samples were analyzed on an Agilent 1260 infinity HPLC system (Agilent Technologies) with a photodiode array detector. Reaction mixtures were separated on a ZORBAX SB-C18 column (Agilent Technologies) by using a linear mobile phase gradient ranging from 2% (vol/vol) acetonitrile in 0.1% (vol/vol) TFA aqueous solution to 80% (vol/vol) acetonitrile in 0.1% (vol/vol) TFA aqueous solution over 25 min. The flow rate was set to 1 mL/min and the injection volume was 20 μL. The detection wavelength was set to 275 nm. Substrate consumption and product formation were quantified by HPLC peak area integration using corresponding authentic compounds as standards. Product structural assignments were performed by comparison of a detected compound with the corresponding authentic standard regarding retention time and coinjection confirmation, HRMS, and/or NMR. HRMS data were recorded in the negative and/or positive ionization mode on a MaXis Impact UHR-qTOF System (Bruker).
Supplementary Material
Acknowledgments
We thank the staff of BL17U Beamline at the Shanghai Synchrotron Radiation Facility and Dr. Shilong Fan (Tsinghua University) for assistance during X-ray diffraction data collection and Dr. Jeffrey D. Kittendorf (Alluvium Biosciences) for helpful discussion and proofreading of the manuscript. This work was supported by the National Natural Science Foundation of China under Grant NSFC 31422002, Shandong Provincial Natural Science Foundation Grant JQ201407, the Key Frontier Project of the Chinese Academy of Sciences, QYZDB-SSW-SMC042 (to S.L.), the 973 Project from Ministry of Science and Technology Grant 2012CB7211-04 (to S.-J.L.), and Grant NSFC 31270784 from the National Natural Science Foundation of China (to Y.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5GWE and 5XJN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702317114/-/DCSupplemental.
References
- 1.Fiege H, et al. 2000. Phenol Derivatives. Ullmann's Encyclopedia of Industrial Chemistry (Wiley, Hoboken, NJ)
- 2.Bergé A, et al. Meta-analysis of environmental contamination by alkylphenols. Environ Sci Pollut Res Int. 2012;19:3798–3819. doi: 10.1007/s11356-012-1094-7. [DOI] [PubMed] [Google Scholar]
- 3.Ying GG, Williams B, Kookana R. Environmental fate of alkylphenols and alkylphenol ethoxylates: A review. Environ Int. 2002;28:215–226. doi: 10.1016/s0160-4120(02)00017-x. [DOI] [PubMed] [Google Scholar]
- 4.Hudlicky M. Oxidations in Organic Chemistry. American Chemical Society; Washington, DC: 1990. [Google Scholar]
- 5.Shen XH, Zhou NY, Liu SJ. Degradation and assimilation of aromatic compounds by Corynebacterium glutamicum: Another potential for applications for this bacterium? Appl Microbiol Biotechnol. 2012;95:77–89. doi: 10.1007/s00253-012-4139-4. [DOI] [PubMed] [Google Scholar]
- 6.Briasoulis A, Palla M, Afonso L. Meta-analysis of the effects of carvedilol versus metoprolol on all-cause mortality and hospitalizations in patients with heart failure. Am J Cardiol. 2015;115:1111–1115. doi: 10.1016/j.amjcard.2015.01.545. [DOI] [PubMed] [Google Scholar]
- 7.Kesavan L, et al. Solvent-free oxidation of primary carbon-hydrogen bonds in toluene using Au-Pd alloy nanoparticles. Science. 2011;331:195–199. doi: 10.1126/science.1198458. [DOI] [PubMed] [Google Scholar]
- 8.Newhouse T, Baran PS. If C-H bonds could talk: Selective C-H bond oxidation. Angew Chem Int Ed Engl. 2011;50:3362–3374. doi: 10.1002/anie.201006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang FL, Hong K, Li TJ, Park H, Yu JQ. Organic chemistry. Functionalization of C(sp3)-H bonds using a transient directing group. Science. 2016;351:252–256. doi: 10.1126/science.aad7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen MS, White MC. Combined effects on selectivity in Fe-catalyzed methylene oxidation. Science. 2010;327:566–571. doi: 10.1126/science.1183602. [DOI] [PubMed] [Google Scholar]
- 11.Desai LV, Hull KL, Sanford MS. Palladium-catalyzed oxygenation of unactivated sp3 C-H bonds. J Am Chem Soc. 2004;126:9542–9543. doi: 10.1021/ja046831c. [DOI] [PubMed] [Google Scholar]
- 12.Que L, Jr, Tolman WB. Biologically inspired oxidation catalysis. Nature. 2008;455:333–340. doi: 10.1038/nature07371. [DOI] [PubMed] [Google Scholar]
- 13.Das S, Incarvito CD, Crabtree RH, Brudvig GW. Molecular recognition in the selective oxygenation of saturated C-H bonds by a dimanganese catalyst. Science. 2006;312:1941–1943. doi: 10.1126/science.1127899. [DOI] [PubMed] [Google Scholar]
- 14.Das S, Brudvig GW, Crabtree RH. High turnover remote catalytic oxygenation of alkyl groups: How steric exclusion of unbound substrate contributes to high molecular recognition selectivity. J Am Chem Soc. 2008;130:1628–1637. doi: 10.1021/ja076039m. [DOI] [PubMed] [Google Scholar]
- 15.Roiban GD, Agudo R, Reetz MT. Cytochrome P450 catalyzed oxidative hydroxylation of achiral organic compounds with simultaneous creation of two chirality centers in a single C-H activation step. Angew Chem Int Ed Engl. 2014;53:8659–8663. doi: 10.1002/anie.201310892. [DOI] [PubMed] [Google Scholar]
- 16.Kille S, Zilly FE, Acevedo JP, Reetz MT. Regio- and stereoselectivity of P450-catalysed hydroxylation of steroids controlled by laboratory evolution. Nat Chem. 2011;3:738–743. doi: 10.1038/nchem.1113. [DOI] [PubMed] [Google Scholar]
- 17.Kolev JN, Zaengle JM, Ravikumar R, Fasan R. Enhancing the efficiency and regioselectivity of P450 oxidation catalysts by unnatural amino acid mutagenesis. ChemBioChem. 2014;15:1001–1010. doi: 10.1002/cbic.201400060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang K, Shafer BM, Demars MD, 2nd, Stern HA, Fasan R. Controlled oxidation of remote sp3 C–H bonds in artemisinin via P450 catalysts with fine-tuned regio- and stereoselectivity. J Am Chem Soc. 2012;134:18695–18704. doi: 10.1021/ja3073462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coon MJ. Cytochrome P450: Nature’s most versatile biological catalyst. Annu Rev Pharmacol Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- 20.Behrendorff JB, Huang W, Gillam EM. Directed evolution of cytochrome P450 enzymes for biocatalysis: Exploiting the catalytic versatility of enzymes with relaxed substrate specificity. Biochem J. 2015;467:1–15. doi: 10.1042/BJ20141493. [DOI] [PubMed] [Google Scholar]
- 21.McIntosh JA, Farwell CC, Arnold FH. Expanding P450 catalytic reaction space through evolution and engineering. Curr Opin Chem Biol. 2014;19:126–134. doi: 10.1016/j.cbpa.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fasan R. Tuning P450 enzymes as oxidation catalysts. ACS Catal. 2012;2:647–666. [Google Scholar]
- 23.Polic V, Auclair K. Controlling substrate specificity and product regio- and stereo-selectivities of P450 enzymes without mutagenesis. Bioorg Med Chem. 2014;22:5547–5554. doi: 10.1016/j.bmc.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen AT, May EM, Auclair K. Predictable stereoselective and chemoselective hydroxylations and epoxidations with P450 3A4. J Am Chem Soc. 2011;133:7853–7858. doi: 10.1021/ja200551y. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Ouellet H, Sherman DH, Podust LM. Analysis of transient and catalytic desosamine-binding pockets in cytochrome P-450 PikC from Streptomyces venezuelae. J Biol Chem. 2009;284:5723–5730. doi: 10.1074/jbc.M807592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J Am Chem Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, et al. Selective oxidation of carbolide C-H bonds by an engineered macrolide P450 mono-oxygenase. Proc Natl Acad Sci USA. 2009;106:18463–18468. doi: 10.1073/pnas.0907203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negretti S, et al. Directing group-controlled regioselectivity in an enzymatic C-H bond oxygenation. J Am Chem Soc. 2014;136:4901–4904. doi: 10.1021/ja5016052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayan AR, et al. Enzymatic hydroxylation of an unactivated methylene C-H bond guided by molecular dynamics simulations. Nat Chem. 2015;7:653–660. doi: 10.1038/nchem.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du L, et al. Characterization of a unique pathway for 4-cresol catabolism initiated by phosphorylation in Corynebacterium glutamicum. J Biol Chem. 2016;291:6583–6594. doi: 10.1074/jbc.M115.695320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th Ed Springer; New York: 2015. [Google Scholar]
- 33.Yano JK, et al. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J Biol Chem. 2000;275:31086–31092. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 34.Sherman DH, et al. The structural basis for substrate anchoring, active site selectivity, and product formation by P450 PikC from Streptomyces venezuelae. J Biol Chem. 2006;281:26289–26297. doi: 10.1074/jbc.M605478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenkman JB, Remmer H, Estabrook RW. Spectral studies of drug interaction with hepatic microsomal cytochrome. Mol Pharmacol. 1967;3:113–123. [PubMed] [Google Scholar]
- 36.Schlichting I, et al. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 37.Bai Z, et al. 2014. Chinese Patent CN104311700-A.
- 38.Guengerich FP, Kim DH. Enzymatic oxidation of ethyl carbamate to vinyl carbamate and its role as an intermediate in the formation of 1,N6-ethenoadenosine. Chem Res Toxicol. 1991;4:413–421. doi: 10.1021/tx00022a003. [DOI] [PubMed] [Google Scholar]
- 39.Guengerich FP, Sohl CD, Chowdhury G. Multi-step oxidations catalyzed by cytochrome P450 enzymes: Processive vs. distributive kinetics and the issue of carbonyl oxidation in chemical mechanisms. Arch Biochem Biophys. 2011;507:126–134. doi: 10.1016/j.abb.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ačimovič J, et al. Cytochrome P450 metabolism of the post-lanosterol intermediates explains enigmas of cholesterol synthesis. Sci Rep. 2016;6:28462. doi: 10.1038/srep28462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimoto FK, Jung IJ, Goyal S, Gonzalez E, Guengerich FP. Isotope-labeling studies support the electrophilic compound I iron active species, FeO(3+), for the carbon-carbon bond cleavage reaction of the cholesterol side-chain cleavage enzyme, cytochrome P450 11A1. J Am Chem Soc. 2016;138:12124–12141. doi: 10.1021/jacs.6b04437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoji O, Kunimatsu T, Kawakami N, Watanabe Y. Highly selective hydroxylation of benzene to phenol by wild-type cytochrome P450BM3 assisted by decoy molecules. Angew Chem Int Ed Engl. 2013;52:6606–6610. doi: 10.1002/anie.201300282. [DOI] [PubMed] [Google Scholar]
- 43.Dennig A, Marienhagen J, Ruff AJ, Guddat L, Schwaneberg U. Directed evolution of P450 BM3 into a p-xylene hydroxylase. ChemCatChem. 2012;4:771–773. [Google Scholar]
- 44.Dennig A, Lülsdorf N, Liu H, Schwaneberg U. Regioselektive o-Hydroxylierung monosubstituierter Benzole mit P450 BM3. Angew Chem Int Ed. 2013;125:8617–8620. [Google Scholar]
- 45.Lee DS, et al. Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis. Crystallographic, spectroscopic, and mutational studies. J Biol Chem. 2003;278:9761–9767. doi: 10.1074/jbc.M211575200. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, et al. Hydrogen peroxide-independent production of α-alkenes by OleTJE P450 fatty acid decarboxylase. Biotechnol Biofuels. 2014;7:28. doi: 10.1186/1754-6834-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shoji O, et al. Hydrogen peroxide dependent monooxygenations by tricking the substrate recognition of cytochrome P450BSbeta. Angew Chem Int Ed Engl. 2007;46:3656–3659. doi: 10.1002/anie.200700068. [DOI] [PubMed] [Google Scholar]
- 48.Muhr E, et al. Enzymes of anaerobic ethylbenzene and p-ethylphenol catabolism in ‘Aromatoleum aromaticum’: Differentiation and differential induction. Arch Microbiol. 2015;197:1051–1062. doi: 10.1007/s00203-015-1142-z. [DOI] [PubMed] [Google Scholar]
- 49.Meyer D, Witholt B, Schmid A. Suitability of recombinant Escherichia coli and Pseudomonas putida strains for selective biotransformation of m-nitrotoluene by xylene monooxygenase. Appl Environ Microbiol. 2005;71:6624–6632. doi: 10.1128/AEM.71.11.6624-6632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bühler B, Witholt B, Hauer B, Schmid A. Characterization and application of xylene monooxygenase for multistep biocatalysis. Appl Environ Microbiol. 2002;68:560–568. doi: 10.1128/AEM.68.2.560-568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bühler B, Schmid A, Hauer B, Witholt B. Xylene monooxygenase catalyzes the multistep oxygenation of toluene and pseudocumene to corresponding alcohols, aldehydes, and acids in Escherichia coli JM101. J Biol Chem. 2000;275:10085–10092. doi: 10.1074/jbc.275.14.10085. [DOI] [PubMed] [Google Scholar]
- 52.Cunane LM, et al. Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: Gated substrate entry and proton relays support the proposed catalytic mechanism. J Mol Biol. 2000;295:357–374. doi: 10.1006/jmbi.1999.3290. [DOI] [PubMed] [Google Scholar]
- 53.Qi-Sheng W, et al. The macromolecular crystallography beamline of SSRF. Nucl Sci Tech. 2015;26:010102. [Google Scholar]
- 54.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang A, et al. The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis. Mol Biosyst. 2015;11:869–881. doi: 10.1039/c4mb00665h. [DOI] [PubMed] [Google Scholar]
- 57.Cohen SX, et al. ARP/wARP and molecular replacement: The next generation. Acta Crystallogr D Biol Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.