Significance
Synthesis of new RNA and removal of incorrect nucleotides during proofreading by RNA polymerase involve the transfer of two protons. Here, we show that a polymerase component, the trigger loop, does not directly mediate proton transfer during these reactions, as previously proposed. Instead, the trigger loop plays a central role in transcription as a positional catalyst, by orienting the reactants and promoting the polymerase backtracking necessary for proofreading. This positional-catalyst model of trigger-loop function explains its diverse effects on polymerase catalysis and reconciles contradictory reports in the literature. By establishing that the trigger loop is not an acid–base catalyst, our results also guide the search for alternative proton donors and acceptors for reactions in the active site of polymerase.
Keywords: transcription, proofreading, acid–base catalysis, trigger loop, RNA hydrolysis
Abstract
The active site of multisubunit RNA polymerases (RNAPs) is highly conserved from humans to bacteria. This single site catalyzes both nucleotide addition required for RNA transcript synthesis and excision of incorrect nucleotides after misincorporation as a proofreading mechanism. Phosphoryl transfer and proofreading hydrolysis are controlled in part by a dynamic RNAP component called the trigger loop (TL), which cycles between an unfolded loop and an α-helical hairpin [trigger helices (TH)] required for rapid nucleotide addition. The precise roles of the TL/TH in RNA synthesis and hydrolysis remain unclear. An invariant histidine residue has been proposed to function in the TH form as a general acid in RNA synthesis and as a general base in RNA hydrolysis. The effects of conservative, nonionizable substitutions of the TL histidine (or a neighboring TL arginine conserved in bacteria) have not yet been rigorously tested. Here, we report that glutamine substitutions of these residues, which preserve polar interactions but are incapable of acid–base chemistry, had little effect on either phosphoryl transfer or proofreading hydrolysis by Escherichia coli RNAP. The TL substitutions did, however, affect the backtracking of RNAP necessary for proofreading and potentially the reactivity of the backtracked nucleotide. We describe a unifying model for the function of the RNAP TL, which reconciles available data and our results for representative RNAPs. This model explains diverse effects of the TL basic residues on catalysis through their effects on positioning reactants for phosphoryl transfer and easing barriers to transcript backtracking, rather than as acid–base catalysts.
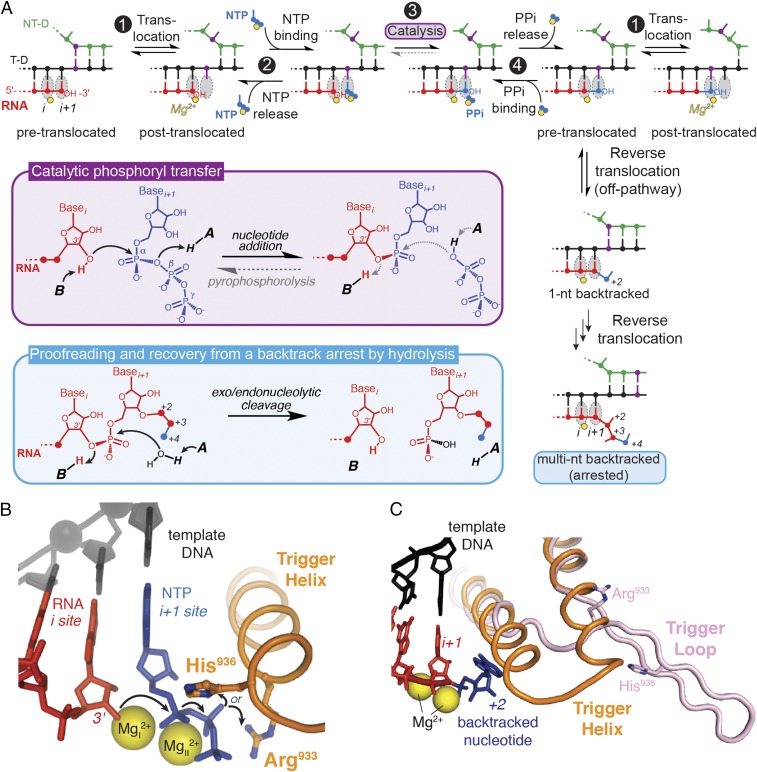
Transcription of DNA-encoded messages into RNA is a complex, highly regulated process performed by multisubunit RNA polymerases (RNAPs) in all free-living organisms. At the heart of transcription lies the nucleotide addition cycle (NAC), which mediates extension of an RNA transcript 1 nt at a time as RNAP reads the DNA template. The NAC is comprised of a repeating sequence of translocation of the nucleic-acid template to align the next position for RNA extension, nucleoside triphosphate (NTP) binding, phosphodiester bond formation, and pyrophosphate release (Fig. 1A, steps 1–4). Once every 104–105 nucleotide, RNAP incorporates an incorrect ribobase (1, 2). This misincorporation event interrupts the NAC and forces the polymerase to reverse-thread (backtrack) the nucleic-acid template to excise the misincorporated nucleotide either with or without the help of an extrinsic factor (Fig. 1A, blue box) (3, 4). Backtracking is the first step required for nucleotide excision, and hence constitutes an essential component of cotranscriptional proofreading by RNAP and of transcriptional fidelity in general, which is vital to the survival of organisms.
Fig. 1.
Activities of RNA polymerase. (A) Nucleotide addition, pyrophosphorolysis, and backtracking events in RNAP active site. RNA synthesis (sequence of events at the top) is accomplished in a NAC consisting of four steps. Step 1: translocation of DNA and RNA to position the 3′-OH of RNA in i site on RNAP for the reaction with an incoming NTP; step 2: NTP binding to i+1 site; step 3: formation of a new phosphodiester bond; and step 4: pyrophosphate release. Template DNA is shown in black, nontemplate DNA in green, RNA in red, incoming NTP in blue, and catalytic magnesium ions in yellow. One position on template/nontemplate DNA is highlighted in purple, to illustrate translocation. Chemical reactions of the reversible phosphoryl transfer (step 3) and hydrolysis of a backtracked RNA are shown in purple and blue boxes, respectively. The flow of electrons in the reactions is marked with arrows. Protons are supplied by the putative general acid (“A”) and base (“B”). (B) Basic residues of the trigger helix as candidates for a general acid–base catalyst (PDB ID code 2O5J). The electron flow in the nucleotide addition reaction is indicated with arrows. (C) Structure of a 1-nt backtracked elongation complex (PDB ID code 4WQS), with the trigger loop in a partially unfolded, “bent” conformation (shown in pink). The folded trigger helix (from PDB ID code 2O5J) is shown for reference in orange.
The events of the NAC are accompanied by dynamic changes in the RNAP architecture, including kinking of the bridge helix and folding of the polymorphous trigger loop (TL) to close the active site of RNAP. Folding of the TL into the α-helical trigger helices (TH), although not absolutely required for catalysis, enhances the rate of nucleotide addition ∼104- to 105-fold in bacteria (5, 6) and significantly in archaeal RNAP, even at suboptimal temperature (7). The TL is also thought to play roles in other aspects of RNAP function, including translocation, selection of a correct NTP, transcriptional pausing, proofreading RNA hydrolysis, and termination (7–11). It appears that the TL evolved as a central controller in the catalytic mechanism of RNAP.
One contribution of the TL/TH to catalysis is hypothesized to be direct participation of its residues in acid–base chemistry during transcription and proofreading (10, 12). The chemical reaction of nucleotide addition by RNAP (step 3 and purple box in Fig. 1A) requires transfer of two protons: deprotonation of the 3′ hydroxyl of the terminal nucleoside monophosphate (NMP) on the RNA transcript and protonation of the departing pyrophosphate of the incoming NTP (13, 14). The identities of the acid and base (“A” and “B” in Fig. 1A, respectively) that participate in the transfer of these protons are currently uncertain. The bacterial TH contains two basic amino acids that are sufficiently close to the reaction center to partake directly in nucleotide addition: the invariant histidine (TL His, β′ H936 in Escherichia coli RNAP) (Fig. 1B), proposed previously to act as a general acid (12, 15); and an arginine (TL Arg, β′ R933 in E. coli RNAP) (Fig. 1B), which is conserved in bacterial RNAPs but varies in polymerases from other organisms (e.g., it is an asparagine in eukaryotic Pol I and Pol II, generally a lysine in Pol III, and a mix of arginine and asparagine in archaeal RNAPs).
In crystal structures of transcribing polymerases, both the TL His (5, 12) and Arg are within the hydrogen-bonding or ion-pairing distance of the incoming NTP phosphates (Fig. 1B), and could thus protonate the departing pyrophosphate during nucleotide addition. Several structurally distinct nucleic acid polymerases in fact appear to use a basic amino acid (lysine) positioned similarly to the TL His as a general acid in nucleotide addition (14), suggesting a potentially conserved mechanism of phosphodiester bond formation across various polymerases. These interactions of the TL His with the β-phosphate oxygens of a matched NTP have been interrogated by molecular dynamics simulations and shown to be significantly stronger when the TL His is protonated than when it is unprotonated (16). Another set of simulations suggested that the TL His aids the exit of pyrophosphate out of the active site following NMP addition, through hydrogen-bonding contacts to pyrophosphate (17). The recently solved crystal structure of an E. coli σs transcription initiation complex indeed places the TL His in coordination with both the pyrophosphate and the phosphate of the 3′-terminal nucleotide of RNA (18). In vivo, several TL His substitutions (Ala, Asn, Asp, and Phe) are lethal to yeast, whereas others (Lys, Arg, Trp, Tyr, Gln, and Leu) cause variable growth defects but are viable (19–22). In bacterial RNAPs, only the effects of a TL His-to-Ala substitution have been studied (9, 10, 23), with inconclusive results. Even less is known about the possible involvement of the TL Arg in catalysis.
Similarly to nucleotide addition, factor-independent excision of nucleotides from a backtracked RNA transcript involves the transfer of two protons, but in the reverse direction (blue box in Fig. 1A). The same protein residues (the TL His or Arg) could catalyze acid–base chemistry of this factor-independent RNA hydrolysis, although steric effects preclude their participation in Gre- or transcription factor IIS (TFIIS)-dependent hydrolysis. A body of inconclusive and sometimes contradictory data on the involvement of the TL in the intrinsic cleavage of RNA by RNAP has been reported. Fouqueau et al. found that a TL deletion (ΔTL) in an archaeal RNAP from Pyrococcus furiosis does not affect transcript hydrolysis (7). Similarly, in E. coli RNAP, neither the ΔTL nor a TL His-to-Ala substitution significantly decreases intrinsic cleavage of a 2-nt backtracked RNA “locked” by the mismatched bases at its 3′ end (8). In contrast, another study found that TL His-to-Ala or ΔTL variants of E. coli and Thermus aquaticus RNAPs are deficient at hydrolyzing both the RNA with a single and multiple 3′ mismatches (10). Uncertainty about the function of the TL in acid–base catalysis is further exacerbated by crystal structures of backtracked elongation complexes (ECs) in which the TL His or Arg is positioned too far from the relevant nucleotides to function as a general base during hydrolysis (Fig. 1C) (24–26).
An issue with studies of the TL His and Arg residues to date is that most rely on Ala substitutions that do not differentiate between polar and acid–base contributions to catalysis. Furthermore, these drastic changes may alter the available space in RNAP active site and thus complicate the interpretation of the results (Table S1 for calculated side-chain volumes). To address these concerns and provide more rigorous tests of function, we compared several E. coli RNAP mutants: H936A, which removes polar and acid–base contributions; H936Q and R933Q, which preserve the potential for polar interactions but not their acid–base capabilities; and a double mutant (R933Q/H936Q). We tested the effect of these TL substitutions on known RNAP activities (nucleotide addition, pyrophosphorolysis, and factor-independent exo- and endonucleolytic hydrolysis). Our results suggest that the TL contributes to RNAP catalysis of these reactions by positioning reactants in the active site, rather than directly donating protons to the reaction.
Table S1.
Calculated volume of relevant amino acid side chains
| Side chain identity | Molar volume (cm3/mol) |
| Histidine | 60.6 |
| Arginine | 73.3 |
| Glutamine | 69.8 |
| Tyrosine | 90.2 |
| Leucine | 63.9 |
| Alanine | 31.9 |
Results
The TL His Is Not Essential for Efficient Transcript Elongation or Pyrophosphorolysis.
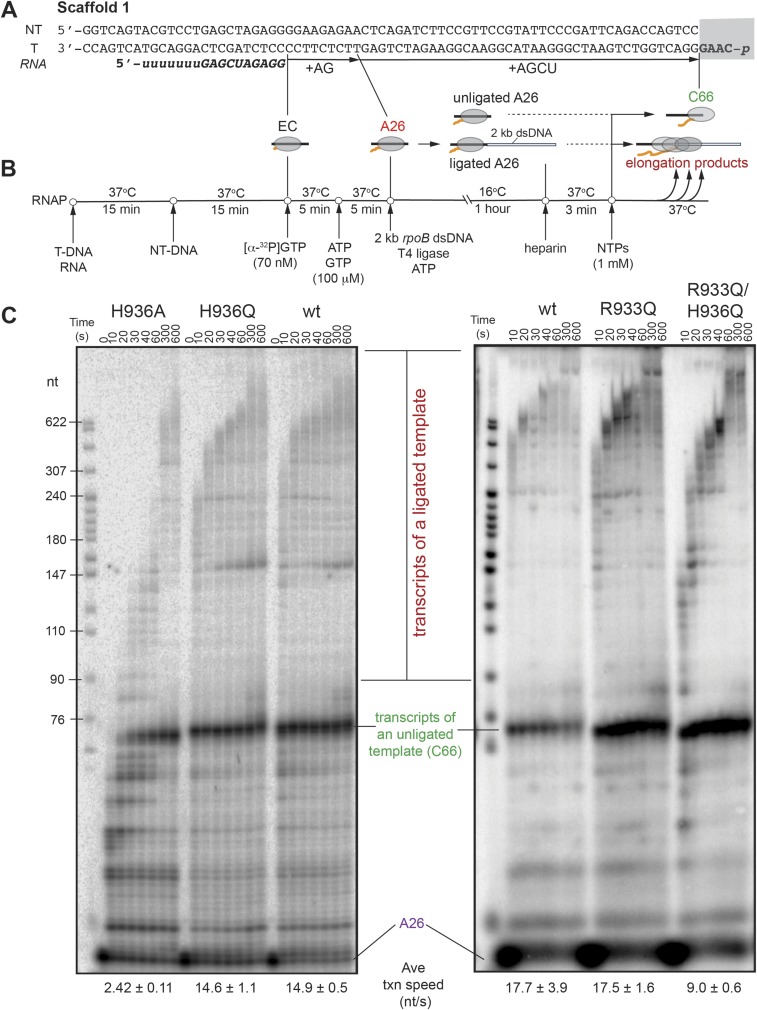
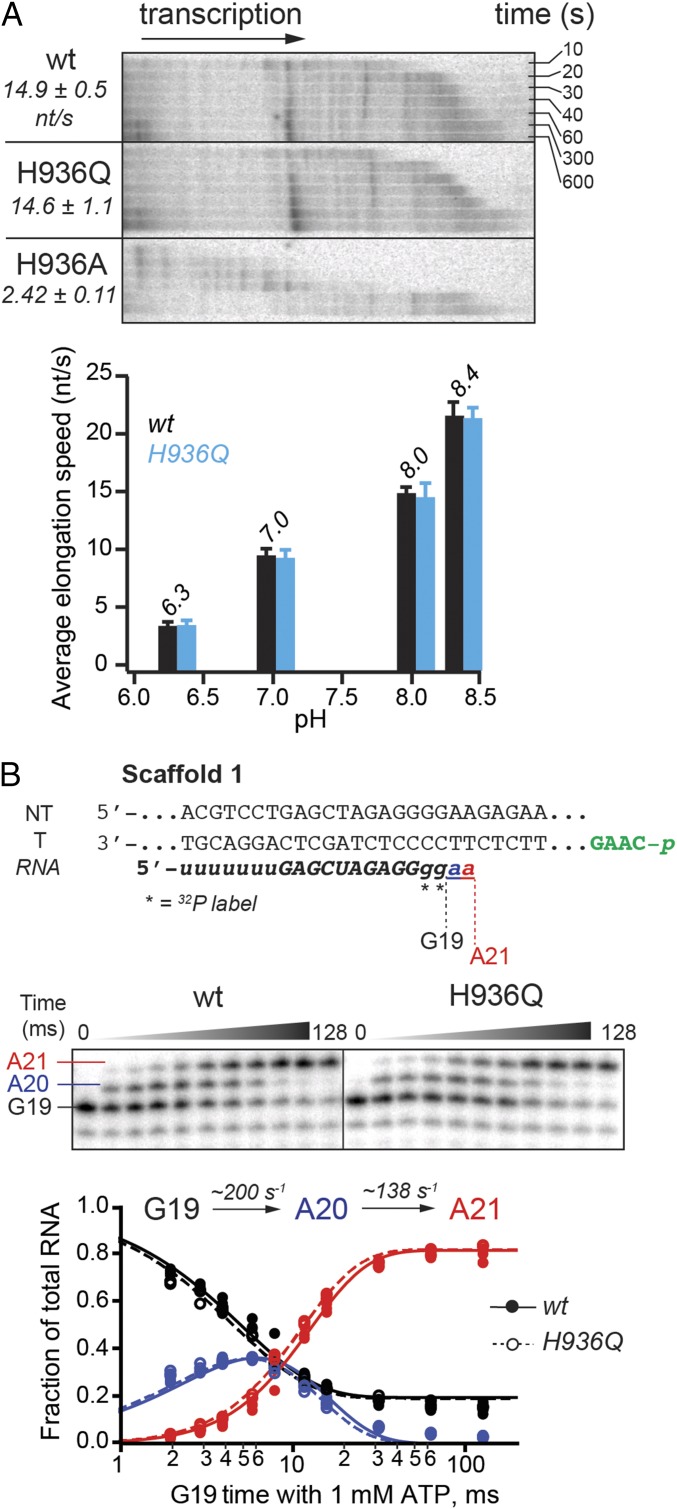
To assess the TL His contribution to physiologically relevant processive NMP addition, we developed an experimental method that uses an ∼2-kb DNA fragment of the naturally transcribed E. coli rpoB gene as a transcription template to allow ample time for manual rate measurements. The assay builds on the previously reported scaffold ligation approach (27, 28) and offers the advantage of bypassing initiation at a promoter, often compromised by mutations in the polymerase, and measuring nucleotide addition rates at physiological NTP concentrations (1 mM) and temperature (37 °C) for E. coli, which are otherwise too fast to measure manually. The ECs were assembled on a short scaffold (∼75 bp) with the polymerase, RNA primer, nontemplate DNA, and a template DNA strand bearing a 5′ phosphate for subsequent ligation (Fig. S1). The ECs were radiolabeled by reaction with [α-32P]GTP, extended to an A26 position by incubation with a subset of NTPs, and then ligated to the 2-kb rpoB DNA fragment. Transcription of the ligated template was initiated by addition of 1 mM NTPs and its progress was monitored at various time points at 37 °C (Materials and Methods and Fig. S1 A and B for additional details). A glutamine (Gln) substitution of the TL His did not affect NMP addition rate across tested buffer pH conditions appropriate for an expected histidine pKa (Fig. 2A) (7, 8). The alanine (Ala) mutant, on the other hand, was approximately sixfold slower at pH 8; this could be because of the loss of hydrogen-bonding interactions with the incoming NTP, compromised TL folding into TH necessary for efficient transcription, or both.
Fig. S1.
Ligated-scaffold transcription assay. (A) Nucleic-acid scaffold 1 used in elongation rate measurements. The starting 17mer RNA is italicized and bolded, with the sequence complementary to the template DNA shown in uppercase letters. Positions of halted 26-nt RNA and 66-nt run-off transcript are marked as A26 and C66, respectively. The 4-nt overhang and 3′ phosphate of the template DNA, necessary for ligation, are boxed. (B) Schematic of the experimental set-up for EC reconstitution, labeling, and ligation to a StyI-treated dsDNA fragment of rpoB (see Materials and Methods for details). RNAP and nucleic acids at various stages are represented as a cartoon. (C) Elongation of ligated A26 complexes by the wild-type and mutant RNAPs at pH 8.0. Indicated are the positions of halted A26 complexes, run-off transcript of unligated ECs (C66), and elongation products of successfully ligated ECs. Average transcription speeds were determined as previously described (41).
Fig. 2.
Nucleotide addition by TL His mutants. (A) Multiround nucleotide addition assay. RNA extension by the RNAPs was monitored using scaffold 1 ligated to a 2-kb double-stranded DNA (dsDNA) fragment of rpoB gene via a phosphorylated overhang (in green on scaffold 1) complementary to a StyI-generated sticky end on the dsDNA (Materials and Methods and Fig. S1). Transcription progress along the ligated scaffold over time was visualized on a denaturing 8% polyacrylamide gel (Upper, time course at pH 8 is shown). Average transcription speeds are reported in nucleotides per second. The wild-type and H936Q RNAPs elongated at approximately the same speed across the tested pH range (Lower, tested pH values are shown above the data bars). (B) Addition of two consecutive nucleotides to an RNA primer. RNA in the starting EC was radiolabeled by incorporation of [α-32P]GMP (G19) and further extended in the reaction with 1 mM ATP, to form A20 and A21 RNAs (Top; see Fig. S1 and Table S2 for complete oligonucleotide sequences). The reactions were quenched with HCl at various time points, and RNA products separated by gel electrophoresis (Middle). Quantified AMP addition data from at least three replicates per time point for the wild-type (closed circles) and the H936Q RNAP (open circles) closely matched. Data from all replicates were globally fitted to a single-intermediate reaction model (solid and dashed curves, respectively), to obtain individual rate constants shown (Bottom).
A caveat with the processive transcription assay described above is the possibility that during the multiround nucleotide addition, postcatalysis steps other than chemistry (e.g., translocation, pyrophosphate release, conformational changes, and pause escape) may limit the overall rate of elongation. To ensure that such kinetic steps did not mask the effect of the H936Q mutation on chemistry, we measured the rates of single-nucleotide addition in the wild-type and H936Q variant (Fig. 2B). ECs were formed on the same unligated nucleic-acid scaffold as above, labeled with [α-32P]GTP, and then reacted with 1 mM ATP in a quench-flow instrument to follow synthesis of the next two phosphodiester bonds (Materials and Methods). No measurable difference was observed between the ability of the wild-type RNAP and that of the H936Q mutant to extend the RNA (Fig. 2B), consistent with the results of the multiround nucleotide addition experiment.
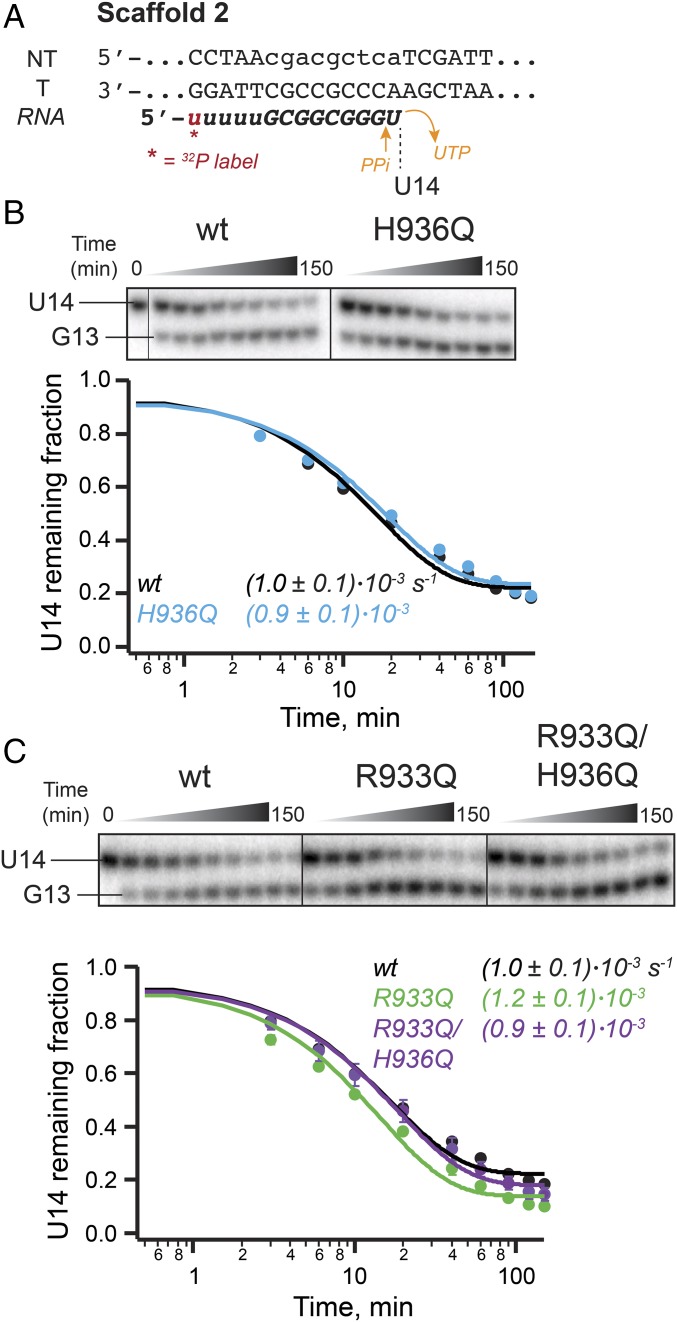
Phosphoryl transfer (step 3 in Fig. 1A) can occur in reverse at high pyrophosphate concentrations, a process termed pyrophosphorolysis, during which the TL His or some general base would be expected to remove a proton from pyrophosphate. To determine if the TL His participates in acid–base catalysis of this reverse reaction, we tested the ability of the H936Q polymerase to perform pyrophosphorolysis. The sequence of the nucleic-acid scaffold was designed such that the EC would favor the pretranslocated register, placing the 3′ ribonucleotides (GU) in a position from which pyrophosphorolysis is possible (Fig. 3A) (29). Indeed, pyrophosphorolysis produced a single RNA cleavage product, 1-nt shorter than the starting RNA (Fig. 3B). As evident from the overlay of the wild-type and H936Q RNAP pyrophosphorolysis kinetic data (Fig. 3B), the mutation did not compromise the pyrophosphorolysis reaction of the polymerase.
Fig. 3.
Pyrophosphorolysis by TL mutants. (A) RNA sequence of the nucleic-acid scaffold 2 was designed to promote reconstitution of RNAP in the pretranslocated register from which pyrophosphorolysis of the terminal UMP is possible. (B) Pyrophosphorolysis by the TL H936Q mutant. RNA cleavage data were fit to a single-exponential function. The wild-type and H936Q RNAPs catalyzed pyrophosphorolysis with similar kinetics. Error bars are smaller than data points. (C) Pyrophosphorolysis by the TL Arg mutants. All pyrophosphorolysis reactions were performed in the presence of apyrase, to degrade the released UTP, thereby favoring forward reaction (i.e., cleavage of the starting RNA). Refer to Table S2 for complete nucleic-acid sequences.
The TL His Does Not Serve as a General Base for Intrinsic Cleavage of RNA, but Facilitates RNAP Backtracking.
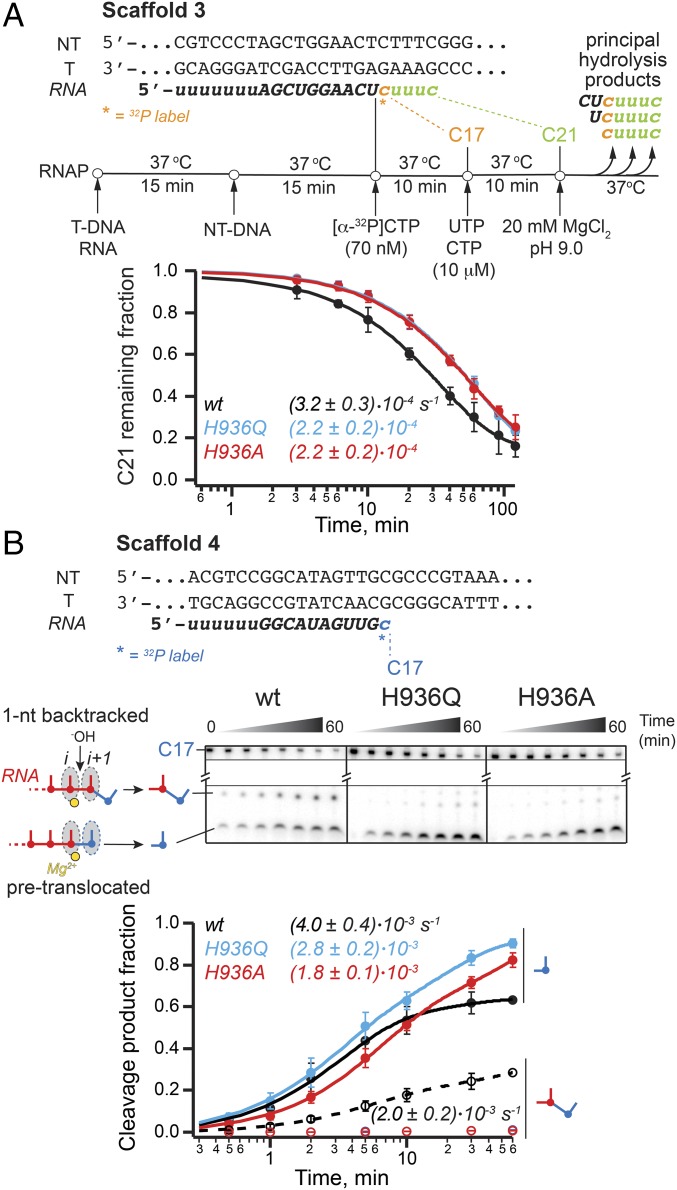
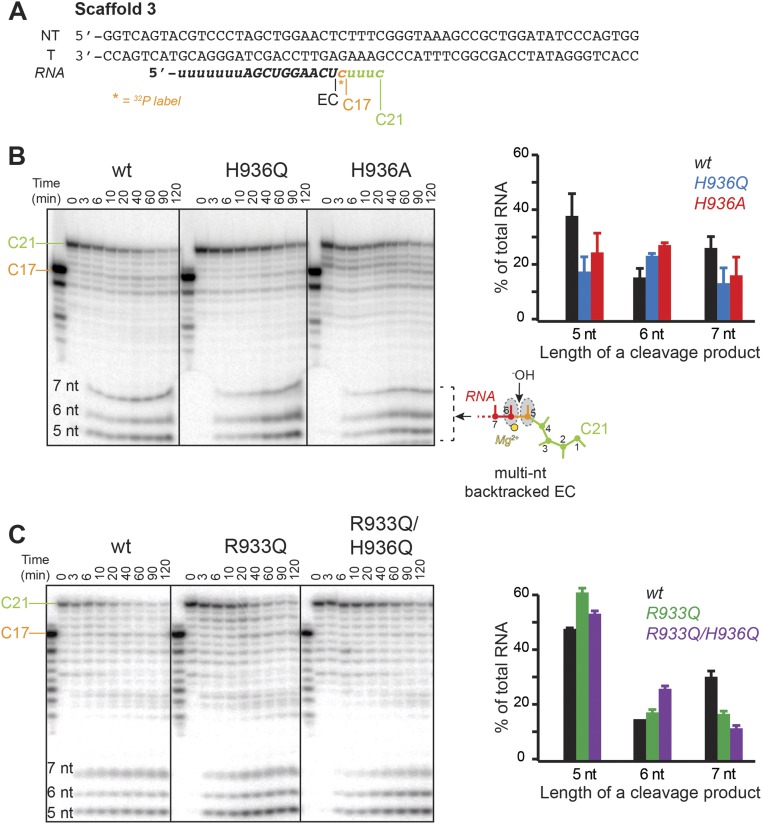
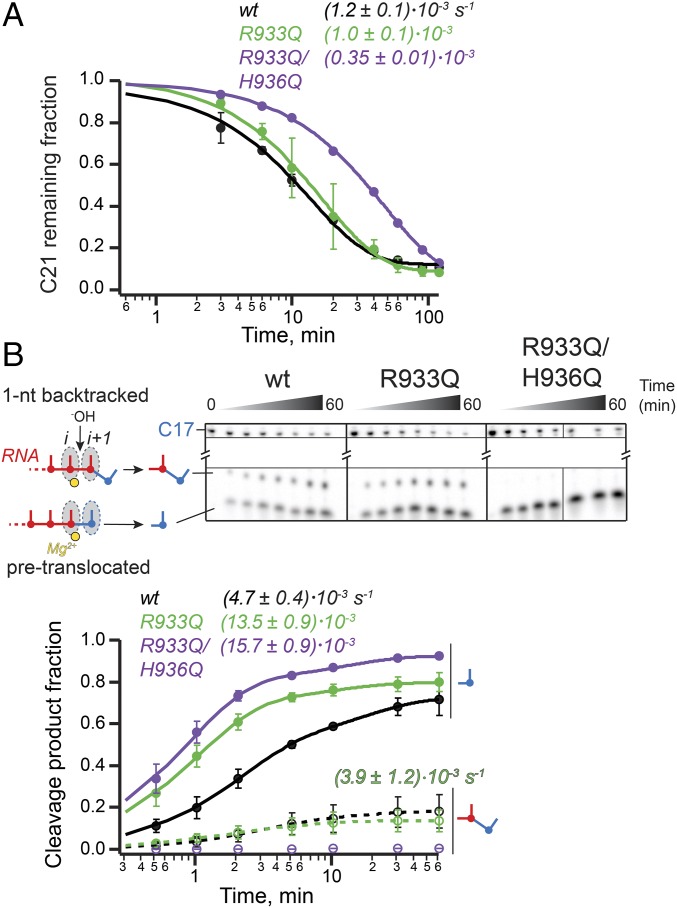
To study the role of the TL in RNA hydrolysis, we used nucleic-acid sequences on which RNAP backtracks following incorporation of cognate nucleotides. Kotlajich et al. identified a pause position on a λPR-bglF template at which E. coli RNAP backtracked by 4–6 nt (30). We took advantage of this natural backtrack pause sequence to assess the effect of the TL His mutations on the cleavage of multinucleotide backtracked RNA. Briefly, ECs were radiolabeled by incorporation of [α-32P]CMP, elongated to the pause site, and allowed to equilibrate among various backtrack registers. The intrinsic cleavage of backtracked RNA was then initiated with 20 mM Mg2+ in pH 9.0 buffer (Materials and Methods and Fig. 4A). Both TL His mutants (Ala and Gln) were capable of hydrolyzing the multinucleotide backtracked RNA, at most twofold slower than the wild-type polymerase (Fig. 4A and Fig. S2B). Although the overall rate of hydrolysis of the starting RNA (C21) was unaffected by the TL His substitutions, the extent of the RNA backtracking changed, as evidenced by the difference in the pattern of cleavage products in the final reactions (Fig. S2B). Specifically, the RNA in the wild-type ECs was backtracked by fewer nucleotides than in the TL His mutants, indicating that the His was able to stabilize the earlier backtracked states more readily than the Gln or Ala.
Fig. 4.
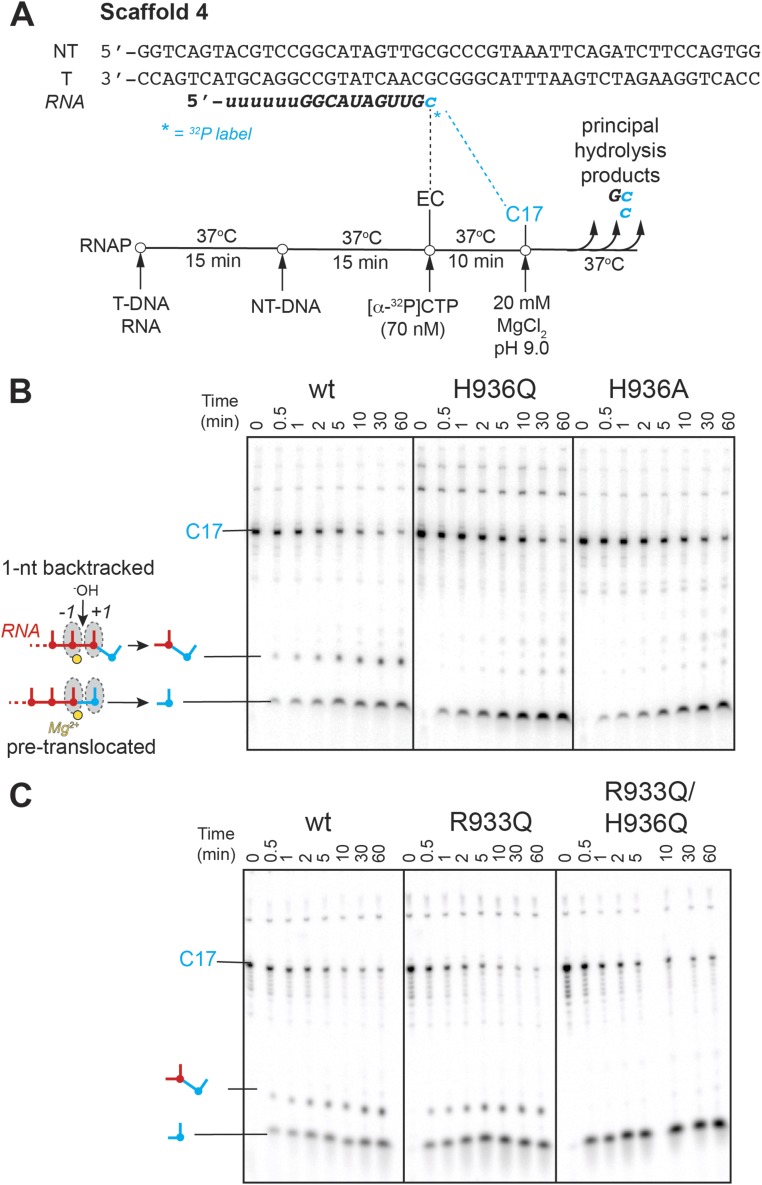
Intrinsic RNA cleavage by TL His mutants. (A) RNA hydrolysis in ECs backtracked by 4–6 nt. The starting RNA in the scaffold 3 was radiolabeled at the 3′-end by incorporation of [α-32P]CMP and extended to the halt position (C21), previously demonstrated to cause RNAP to backtrack by multiple nucleotides (30) (Upper; refer to Table S2 and Fig. S2 for complete sequence of the nucleic acids and Fig. 1A for translocation states). Hydrolysis of the backtracked ECs was initiated with high concentration of Mg2+ in pH 9.0 buffer. Single-exponential kinetics of the C21 RNA disappearance is illustrated in the Lower panel. The TL His substitutions affected hydrolysis only slightly. (B) Exonuclease activity and backtracking. When reconstituted on the shown nucleic-acid scaffold 4 (Upper; see Table S2 for sequences) and walked one position forward to C17, wild-type RNAP partitions between pretranslocated and 1-nt backtracked states, as evidenced by the presence of both the 1- and 2-nt cleavage products in the hydrolysis reactions (Lower and Fig. S3). The TL His mutants preserve the ability to cleave 1 nt off of the RNA, and do so with kinetics similar to the wild-type RNAP, but fail to backtrack, as suggested by the lack of the 2-nt product (open blue and red circles, Lower, and Fig. S3). Thin black lines separate gel portions containing C17 reactant and short cleavage products. See Fig. S3B for an image of the full gel.
Fig. S2.
Intrinsic RNA cleavage by the TL mutants in ECs backtracked by multiple nucleotides. (A) Nucleic-acid scaffold 3 used in cleavage experiments. The starting 16mer RNA was radiolabeled at 3′-end by incorporation of [α-32P]CMP and extended to the halt position (C21), previously demonstrated to backtrack by multiple nucleotides (30). (B) Cleavage of the C21 RNA by TL His mutants. The backtrack positions giving rise to the cleavage products observed on the gel (5–7 nt in length) are numbered on the cartoon of the EC. (Inset) Distribution of major cleavage products in end-point hydrolysis reactions of the wild-type and TL His mutant polymerases. (C) Cleavage of the C21 RNA by TL Arg mutants. (Inset) Populations of hydrolysis products in end-point reactions of the wild-type and TL Arg variants.
To investigate cleavage in the 1-nt backtracked register, we used the recently identified consensus pause sequence (31). At the consensus pause, ECs exist in a dynamic equilibrium between the pretranslocated and 1-nt backtracked states (31, 32). ECs were assembled on a consensus pause scaffold 1-nt upstream from the pause site, advanced to the pause location in a reaction with [α-32P]CTP, and subjected to intrinsic RNA cleavage (Materials and Methods and Fig. S3A). Two radioactive cleavage products were observed in the reactions of the wild-type RNAP, 1 and 2 nt in length, resulting from cleavage of RNA in the pretranslocated (exonucleolytic) and 1-nt backtracked (endonucleolytic) states, respectively (Fig. 4B). The 1-nt product accounted for the majority of the cleaved RNA (60% after 10 min), indicating that at the consensus pause the equilibrium lies toward the pretranslocated EC. The TL His mutants, on the other hand, lacked the 2-nt hydrolysis product, yet still efficiently cleaved 1 nt off of the starting RNA (Fig. 4B). Assuming that 1- and 2-nt cleavage reactions occur by the same mechanism, these observations suggest that the TL His side chain is not required for the intrinsic hydrolysis of RNA, in agreement with the findings described above for the multinucleotide backtracked ECs. Importantly, although the ability of RNAP to cleave RNA was unaffected by the TL His substitutions (as evidenced by the similar rates of the 1-nt cleavage product appearance), the TL His variant polymerases failed to stabilize the 1-nt backtracked state (as evidenced by the lack of the 2-nt product formation with these RNAPs), with implications described further in Discussion, below.
Fig. S3.
Endo- and exonuclease activities of the TL variants. (A) Nucleic-acid scaffold 4 used in cleavage experiments. The scaffold is based on the published consensus pause sequence (31), on which a portion of ECs was shown to backtrack by one nucleotide (32). The 16mer RNA was extended to the pause position by incorporation of [α-32P]CMP, and the resulting C17 ECs allowed to equilibrate between the pretranslocated and 1-nt backtracked registers. RNA hydrolysis was commenced with high pH and Mg2+ concentration. (B) Cleavage of the C17 RNA by TL His mutants. The 1- and 2-nt cleavage products are marked on the gel. (C) Cleavage of the C17 RNA by TL Arg mutants. The translocation state of an EC that gives rise to each cleavage product is shown as a cartoon.
The TL Arg Is Not the Alternative General Acid–Base Catalyst in Bacterial RNAP.
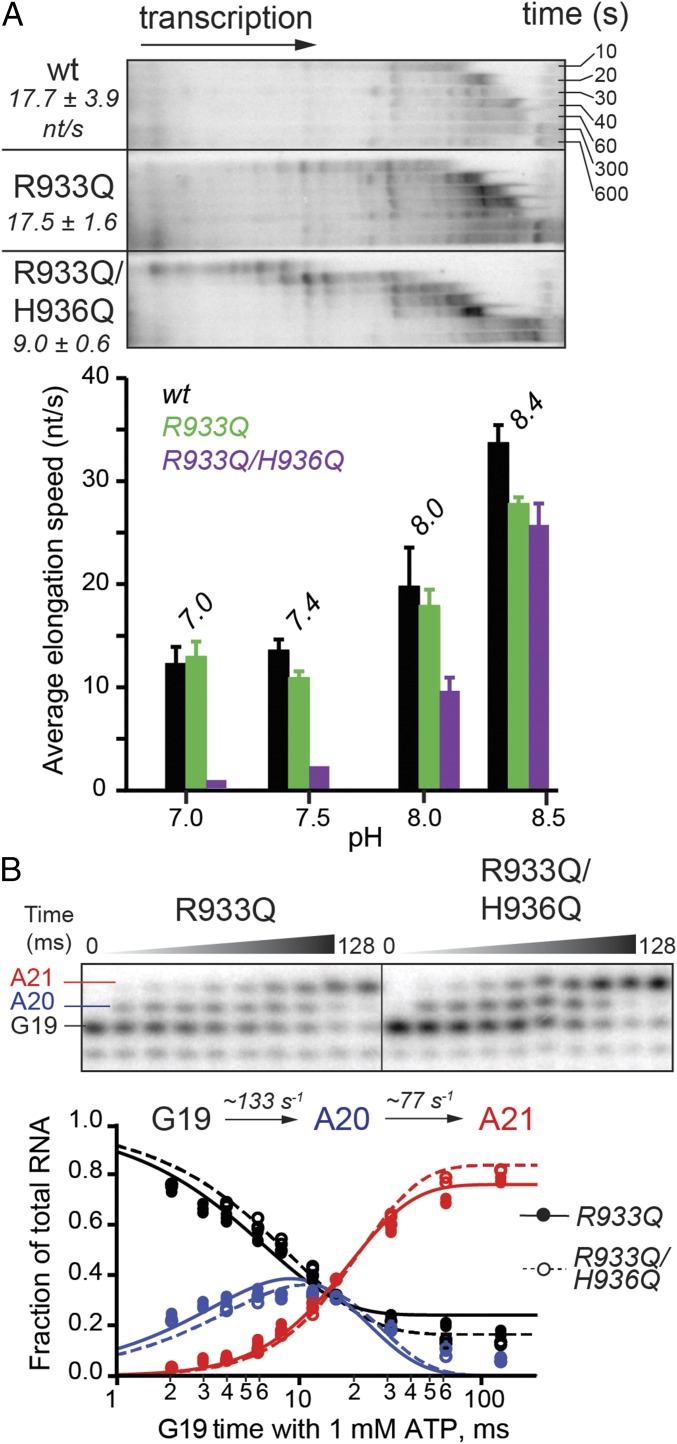
A TL arginine side chain (β′ R933 in E. coli RNAP), located near the TL His (Fig. 1B) and conserved among bacterial RNAPs, could catalyze acid–base chemistry in place of the His, or potentially rescue the H936Q mutant, provided the TL His and Arg have similar pKa values in the RNAP active site. To evaluate the function of the TL Arg, we tested RNAPs with a Gln substitution of R933 alone, or in combination with the H936Q mutation. The single substitution of R933 with Gln left processive elongation rates largely unaffected across the tested range of buffer pH, whereas the double mutant elongated at about half the wild-type rate at pH 8 and even slower at lower pH (Fig. 5A). These effects of the doubly substituted TL at low pH are readily explained by effects on salt bridges involved in TL folding (SI Discussion). However, the difference between R933Q and R933Q/H936Q RNAPs disappeared in a single-nucleotide addition experiment (Fig. 5B), with both mutants somewhat impaired (two-thirds of the wild-type rate for addition of the first AMP, and approaching half of the wild-type rate for the second AMP incorporation). The discrepancy in the effect of the combined mutations on the processive vs. single-nucleotide transcription at pH 8 could potentially be explained by increased sequence-specific pausing of the R933Q/H936Q mutant on a long DNA template (Fig. 5A). Consistent with the lack of difference in single-nucleotide addition rates, pyrophosphorolysis was not compromised in either R933Q or R933Q/H936Q RNAPs (Fig. 3C).
Fig. 5.
Nucleotide addition by TL Arg variants. (A) Multiround nucleotide addition by a point R933Q mutant and a double mutant carrying glutamine substitutions at both the TL Arg and His. Average transcription speeds are reported in nucleotides per second. The pH activity profiles are shown (Lower). (B) Addition of two consecutive nucleotides to an RNA primer in pH 8.0 buffer. A 1.5- and 1.8-fold decrease in the rates of the first and second AMP addition, respectively, was observed relative to the wild-type RNAP (Fig. 2B). The experiments and data analysis were performed in a manner identical to those with the TL His mutants (Fig. 2).
Similarly to the TL His variants, the TL Arg mutants performed both endo- and exonucleolytic cleavage of RNA (Fig. 6 and Figs. S2C and S3C). Unlike the TL His mutants, the R933Q polymerase was fully capable of achieving the 1-nt backtracked state and of hydrolyzing the resulting backtracked RNA (Fig. 6B). This result suggests the TL Arg does not play a role in proofreading by RNAP, in agreement with the lack of strict conservation of an Arg at this position of the TL, and is consistent with a role of the TL His in 1-nt backtracking.
Fig. 6.
Intrinsic RNA cleavage by TL Arg mutants. (A) RNA hydrolysis in ECs backtracked by multiple nucleotides was not reduced by the R933Q substitution. The double mutant hydrolyzed RNA ∼3.4-fold slower. (B) Exonuclease activity and backtracking of RNAP are not compromised by the R933Q mutation. The experiments and data analysis were performed in a manner identical to those with the TL His mutants (Fig. 4). Thin black lines separate gel portions containing C17 reactant and short cleavage products. See Fig. S3C for an image of the full gel.
SI Discussion
Polymerases bearing Gln substitutions at both the TL His and Arg extended RNA with nearly the wild-type kinetics at alkaline pH (8.0 and 8.4). At acidic pH, however, the R933Q/H936Q RNAP showed impaired transcription (Fig. 5A). This defect could indicate that: (i) the TL Arg indeed rescues the TL His-to-Gln mutant in an acid–base reaction, and vice versa; (ii) at least one ion-pair interaction between the TH His or Arg and NTP phosphates is necessary for optimal NTP positioning; or (iii) at least one ion-pair interaction of the His or Arg with other TL residues, or other parts of RNAP, is required for efficient TL folding and unfolding during processive elongation by bacterial RNAP. A requirement for the formation of an ion-pair involving either the His or Arg (models ii and iii) should be most obvious at lower pH, where the majority of the His or Arg side chains, but not Gln, are protonated (i.e., positively charged) and thus able to form charge–charge interactions. Indeed, the defect in processive elongation caused by the His/Arg double mutation was exacerbated by acidic pH (Fig. 5A). Because the difference from wild-type of the multinucleotide addition rate for the R933Q/H936Q and R933Q enzymes disappeared when a single-nucleotide addition was monitored (Fig. 5B), we favor model iii. The folding of the TL into TH indeed involves the formation and breakage of a series of salt bridges, as observed in the simulated TL–TH transition in pol II (46). In particular, a salt bridge between the TL His and an aspartate (Saccharomyces cerevisiae Rpb1 D1359) stabilizes an open conformation of the TL. The simulations suggest that the transition from an open TL to a closed TH occurs in two steps, with a partially folded TL as an intermediate. Existence of the intermediate conformer raised the possibility that full opening of the TL may not be required during processive elongation, and may only take place during pausing or backtracking. This possibility would explain the pronounced effect of the TL His/Arg double substitution on multiround nucleotide addition, which is clearly accompanied by pauses (see transient RNA bands on transcription gels in Fig. S1C), and not on pause-free single-nucleotide addition. Taken together, our findings argue against the TL His or Arg functioning as a general acid or a base in either transcription or factor-independent proofreading and can be readily explained by roles in ionic interactions.
Discussion
To address the uncertainty about the role of the conserved TL His in phosphoryl transfer and hydrolysis, as well as the contribution of the TL generally to intrinsic transcript cleavage, we studied RNAPs in which the TL His and Arg residues were substituted with Gln, which preserved the capacity for polar interactions but not acid–base catalysis. We found that the Gln substitutions have little effect on most activities of RNAP. This observation strongly indicates that neither the TL His nor Arg functions as an acid–base catalyst during either phosphoryl transfer or hydrolysis. The TL His-to-Gln substitution provided additional useful insight into intrinsic RNA cleavage because it inhibited hydrolytic removal of a dinucleotide but not of a single nucleotide from the 3′ end of a consensus-pause RNA. To interpret these findings, we describe below a unified model for TL function as a positional rather than acid–base catalyst. The positional catalyst model of TL function builds on prior descriptions of TL positioning effects and can explain the diverse effects of TL mutants on intrinsic cleavage reported in the literature (7, 8, 10, 33).
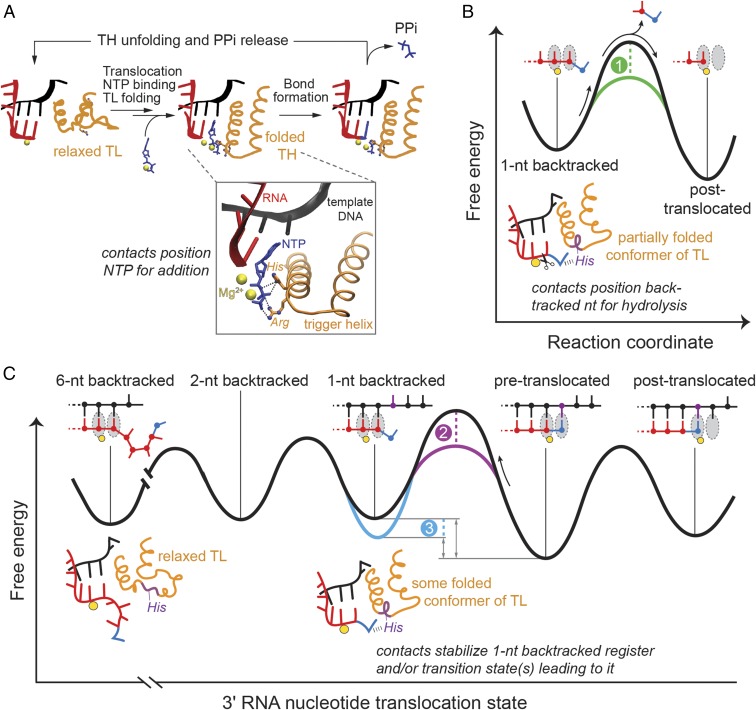
The TL/TH may serve as a positional—rather than an acid–base—catalyst in RNAP in two distinct ways during: (i) phosphoryl transfer and (ii) backtracking and intrinsic RNA hydrolysis. During phosphoryl transfer, the TH His (and Arg in bacteria) makes contacts to the phosphorylated reactants that are indispensable for optimal positioning of the electrophile (α-phosphate of the incoming NTP during addition or the 3′ phosphodiester bond of RNA during pyrophosphorolysis) for the SN2 attack of the nucleophile (the nascent RNA’s 3′ hydroxyl or a pyrophosphate’s oxygen) (Fig. 7A, Inset). These proposed positioning contacts are well documented structurally (5, 12, 18) and are likely both electrostatic and steric in nature: the TH side chains form salt bridges (i.e., hydrogen bonds and charge–charge interactions) with the substrates and push them together to achieve a reactive configuration. Such dual nature of protein–substrate interactions can explain deficient in vitro elongation by the His-to-Ala mutants (Fig. 2A; see also ref. 8), and can also explain the robust in vitro activity of E. coli RNAP mutants (Fig. 2) and viability of yeast carrying a His-to-Gln TL mutation (20). The reactive positioning may potentially even be satisfied solely by steric effects, which could explain the recently reported viability of yeast containing a sterically conservative His-to-Leu substitution in the TL (22) (see calculated side-chain volumes in Table S1). Additionally, these contacts may aid folding of the TL into the TH necessary for catalysis. In contrast, a role for the TL residues in proton transfer to or from pyrophosphate in the phosphoryl transfer reaction is not supported by our finding that the single-turnover reactions are uncompromised by Gln substitutions in the TL. At most, such a role must be dispensable rather than obligate and be easily assumed by other proton donors or acceptors in the active site. This conclusion is consistent with previous negative tests for a role of the TL His in proton-transfer chemistry during nucleotide addition (e.g., ref. 8).
Fig. 7.
Positional catalyst model of TL/TH function. The TL/TH serves as a positional, not an acid–base, catalyst in transcription and proofreading by RNAP. During the bond formation step of the nucleotide addition cycle (A), the TL is folded into the active site, thus allowing the His and Arg to orient the NTP phosphates for the substitution reaction through hydrogen-bonding contacts (Inset, PDB ID code 2O5J). Upon incorporation of an incorrect nucleotide, RNAP reverse threads the nucleic-acid template (backtracks) and the TL assumes a partially folded conformation, such that its invariant His can form ionic and hydrogen-bonding contacts with the backtracked RNA nucleotide (B and C). The TL/TH His could then contribute to proofreading RNA hydrolysis by directly positioning the backtracked ribobase for excision, thereby lowering the activation barrier for the reaction (B, green dashed line). Alternatively, the interactions with the His could stabilize 3′ nucleotide in the backtracked state (C, blue dashed line) or reduce the energy of the transition states/intermediates leading to that state (C, purple dashed line), consequently promoting RNAP passage into the 1-nt backtracked register. The stabilization of the 1-nt backtracked state in turn eases the path of RNAP to subsequent, multinucleotide backtracked registers.
The second way in which the TL functions as a positional catalyst occurs during interruptions to the NAC that lead to backtracking and transcript hydrolysis, which together underlie transcriptional proofreading. The TL can act as a positional catalyst of proofreading RNA hydrolysis in at least three different scenarios. First, the dynamic fluctuations of the TL could shift a backtracked nucleotide into the reactive alignment with water, effectively lowering the activation barrier to hydrolysis (effect 1 in Fig. 7B). This would be analogous to the TL contribution to phosphoryl transfer (Fig. 7A), except that the TL would remain in one or multiple partially folded conformations to avoid steric clash with the backtracked nucleotide (Fig. 1C) (26). A role for the TL His in orienting the backtracked nucleotide for cleavage has been proposed previously (10), although without distinguishing between potential TL His functions in positioning vs. as a general base. Crystal structures reveal that a single backtracked nucleotide, such as would arise after misincorporation, is shifted to a different position for hydrolysis upon binding of the GreA/B or TFIIS cleavage factors (24, 26). Thus, it is plausible that a partially folded TL aids transient occupancy of a similarly shifted state required to achieve the trigonal bipyramidal geometry for cleavage in the absence of GreA/B or TFIIS, even though the TL His cannot reach the backtracked base directly (Fig. 1C) (24, 26).
In the second and third scenarios, the TL could aid entry into or increase occupancy of the initially backtracked state or further backtracked states, either by lowering the barrier to backtracking or by stabilizing a 1-nt backtracked complex (effects 2 and 3, respectively, in Fig. 7C). The polymorphous nature of the TL lends itself to either possibility because the TL could adopt a conformation that positions the TL His, Arg, or other residues to accomplish stabilization of a ground or transition state along the translocation coordinate. Indeed, a molecular dynamics simulation of pyrophosphate release observed transient interactions of the TL His with departing pyrophosphate (17). A similar interaction of the TL His with the 3′-most RNA phosphodiester bond could aid RNA backtracking.
Our results strongly support backtracking assistance by the TL His. We found that Gln substitution for the TL His showed no effect on the rate of intrinsic cleavage of 1 nt from a pretranslocated EC, but eliminated cleavage of 2 nt from the backtracked conformation (Fig. 4B), consistent with the TL His aiding entry into the initial backtracked state (Fig. 7C). Furthermore, the same substitution altered the population distribution of ECs among the 4-, 5-, and 6-nt backtracked states, consistent with preferential stabilization of the earlier, 4-nt backtracked state by the TL His, a preferential stabilization that is lost in the TL His mutants (Fig. S2B). Taken together, these effects of the TL His are most readily explained by the positional catalyst model rather than by direct participation of the TL His in the acid–base chemistry of the cleavage reaction.
The positional catalyst model for the TL function in intrinsic RNA hydrolysis can explain diverse and sometimes seemingly contradictory results published to date. Yuzenkova et al. (10) observed large effects of the TL deletion or Ala substitution of the TL His on RNA cleavage in ECs containing a single mismatched 3′ U or A. These effects could reflect either a role of the TL His in repositioning the mismatched nucleotide into the hydrolysis-competent backtracked state (Fig. 7B) or the TL His favoring the backtracked over the hydrolysis-resistant pretranslocated register (Fig. 7C), because the mismatch could be accommodated in the pretranslocated register by 3′-nt fraying. The short downstream duplex on the scaffolds used in Yuzenkova et al. (10), the downstream end of which lies within the RNAP DNA binding cleft, may perturb the energetic landscape and make the pretranslocated state more accessible to a mismatched 3′ nt. Such TL effects on translocation register bias could be altered by TL His protonation, which may provide an explanation for pH effects on cleavage that depend on the TL His (10), and could also underlie some of the effects of 3′-nt structure on cleavage rates observed on short scaffolds elsewhere (34). In yeast RNAPII, substitution of the TL His with tyrosine (Tyr) stimulates intrinsic transcript cleavage, also in a pH-dependent fashion. Tyr can interact with the RNA bases in multiple modes altered by its deprotonation (planar stacking, edge stacking, hydrogen bonding, and so forth), possibly facilitating positional catalysis by the TL Tyr and explaining the diverse ways this substitution alters transcript cleavage (33). In γ-proteobacteria, the TL contains a large insertion (sequence insertion 3, SI3) (35, 36) that may modulate TL dynamics, thereby affecting positional catalysis of intrinsic RNA cleavage by the TL. Indeed, partial deletion of SI3 or binding of a monoclonal antibody to it inhibited RNA hydrolysis (37), whereas complete removal of SI3 boosted intrinsic cleavage rates of certain mismatched scaffolds (38). Finally, although the TL is generally thought to play minimal role in GreA/B- or TFIIS-mediated transcript cleavage because of steric clash that displaces the TL away from the active site upon cleavage factor binding, we note that TL effects on factor-stimulated transcript hydrolysis are still possible via positional catalysis because the TL could modulate the location of the transcript before binding of the cleavage factors.
Conclusion
The unifying model of the TL as a positional catalyst, rather than an acid–base catalyst, provides a parsimonious explanation for varied effects of TL substitutions on phosphoryl transfer and transcript hydrolysis. Nevertheless, significant questions remain. The participants in proton transfer during phosphoryl transfer and hydrolysis catalyzed by RNAP remain to be identified, and both non-TL RNAP side chains and a metal-ion–activated water molecule are possibilities that remain to be tested rigorously. Furthermore, we cannot exclude the possibility that the roles of TL side chains change in different conditions, are altered upon interaction of transcription factors with the EC, or differ among diverse RNAPs, as demonstrated by the effects of lineage-specific variations in bacterial RNAPs on RNA hydrolysis (38) and the effects of the TL mutations on transcription start site selection in yeast (39). The plasticity of the RNAP active site and the polymorphous nature of the TL afford intriguing opportunities for regulation via positional catalysis that provide fertile ground for future study.
Materials and Methods
Nucleic Acids and Nucleotides.
All RNA and DNA oligonucleotides were acquired from Integrated DNA Technologies and were gel-purified on 8 M urea, 8% (for DNA) or 15% (for RNA) polyacrylamide (PA: 19:1 acryalmide:bisacrylamide) gels in 1.25 mM Na2EDTA and 44 mM Tris-borate, pH 8.3 (TBE). The gel pieces containing desired oligonucleotides were excised and soaked overnight in buffer (10 mM Tris-Cl pH 7.5, 1 mM EDTA, 100 mM NaCl) to extract the nucleic acids. The extracted oligos were then purified using DEAE Sepharose resin (Bio-Rad) and ethanol-precipitated. For the ligated-scaffold transcription assay, a 2.3-kb double-stranded DNA (dsDNA) fragment of an rpoB gene from pRL785 plasmid (Table S2) was ligated to the scaffold downstream of RNAP. The fragment was PCR-amplified using forward primer (#10551) (Table S2), coding for a StyI cleavage site, and reverse primer #10242 (Table S2). The amplified fragment was then spermine-precipitated and digested with restriction enzyme StyI to generate a sticky end for ligation to the A26 EC. The longer, 2.3-kb DNA fragment of the digest was spermine-precipitated and stored in TB (20 mM Tris-acetate, pH 8.0, 40 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT) before use. Unlabeled NTPs were from GE Healthcare Life Sciences. 32P-labeled NTPs were obtained from PerkinElmer.
Table S2.
Strains, plasmids, and oligonucleotides
| Stock no. | Name | Description or sequence | Source or Note |
| Strains | |||
| RL2245 | DH10B (DE3) | Escherichia coli F- endA1 recA1 galE15 galK16 nupG rpsL(StrR) ∆lacX74 araD139 ∆(ara,leu)7697 mcrA ∆(mrr-hsdRMS-mcrBC) ∆ompT774::frt λ(DE3 [lacI, PlacUV5-T7, ind1, sam7, nin5]) | Present work |
| Plasmids | |||
| 2956 | pRM756 | Expresses wild-type E. coli RNAP (α2ββ′ω) with His10 tag on the β′ C terminus | (47) |
| 1663 | pRL663 | Encodes rpoC fragment of E. coli RNAP. Used as PCR template for site-directed mutagenesis. | (48) |
| 1785 | pRL785 | Encodes rpoB fragment of E. coli RNAP. Used as PCR template for preparation of dsDNA for ligated-scaffold transcription experiment. | Derivative of pRL702 (49), with NcoI site directly fused to N terminus of rpoB |
| Oligonucleotides (5′→3′) | |||
| 10160 | T-DNA, scaffold 1 | p-CAAGGGACTGGTCTGAATCGGGAATACGGAACGGAAGATCTG AGTTCTCTTCCCCTCTAGCTCAGGACGTACTGACC | |
| 10161 | NT-DNA, scaffold 1 | GGTCAGTACGTCCTGAGCTAGAGGGGAAGAGAACTCAGATCTTCCGTTCCGTATTCCCGATTCAGACCAGTCC | |
| 6354 | NT-DNA, scaffold 2 | GGTCAGTACGTCCTAACGACGCTCATCGATTCAGACAGG | |
| 6355 | T-DNA, scaffold 2 | CCTGTCTGAATCGAACCCGCCGCTTAGGACGTACTGACC | |
| 9580 | NT-DNA, scaffold 3 | GGTCAGTACGTCCCTAGCTGGAACTCTTTCGGGTAAAGCCGCTGGATATCCCAGTGG | |
| 9581 | T-DNA, scaffold 3 | CCACTGGGATATCCAGCGGCTTTACCCGAAAGAGTTCCAGCTAGGGACGTACTGACC | |
| 8334 | T-DNA, scaffold 4 | CCACTGGAAGATCTGAATTTACGGGCGCAACTATGCCGGACGTACTGACC | |
| 9563 | NT-DNA, scaffold 4 | GGTCAGTACGTCCGGCATAGTTGCGCCCGTAAATTCAGATCTTCCAGTGG | |
| 8855 | RNA, scaffold 1 | UUUUUUUGAGCUAGAGG | |
| 11001 | RNA, scaffold 2 | UUUUUGCGGCGGGU | |
| 9594 | RNA, scaffold 3 | UUUUUUUAGCUGGAACU | |
| 8401 | RNA, scaffold 4 | UUUUUUGGCAUAGUUG | |
| Primers (5′→3′) | |||
| 10242 | Rev (PCR) | TCGGATACGAGGATGGAGTC | |
| 10551 | For (PCR) | CTACCGCCTTGGCGAACCGG | |
| 10746 | R933Q (SDM) | GGGTACACAGCTGACCATGCAGACGTTCCACATCGGTGG | |
| 10747 | H936Q (SDM) | GCTGACCATGCAGACGTTCCAGATCGGTGGTGCGGC | |
Proteins.
T4 DNA ligase, high-fidelity (HF) StyI, SacII, BsmI, Q5 HF DNA polymerase, T4 polynucleotide kinase, and apyrase were obtained from New England Biolabs. Wild-type E. coli RNAP was prepared by isopropyl-β-d-thiogalactopyranoside (IPTG)-induced expression from pRM756 in E. coli strain #2245 (Table S2), and purified by polyethylenimine (PEI) and ammonium sulfate precipitation, anion-exchange FPLC on a GE HisTrap HP 5-mL column, and heparin affinity FPLC on a GE HiTrap Heparin HP 5-mL column, as described in detail previously (6). RNAP variants were prepared by using the QuikChange II Site-Directed Mutagenesis Kit with plasmid pRL663, using the mutagenic primers #10746 and #10767 for the glutamine substitutions at rpoC positions 933 and 936, respectively (Table S2). The mutant rpoC genes were generated in pRL663 and then moved into the pRM756 RNAP overexpression plasmid using restriction enzymes SacII and BsmI. β′ R933Q RNAP, H936Q RNAP, and β′ R933Q/H936Q RNAP were expressed and purified as described above for the wild-type enzyme.
EC Reconstitution and Transcript Labeling.
Full sequences of the nucleic acids used in scaffolds 1–4 are listed in Table S2. To form template DNA (T-DNA)/RNA hybrids of scaffolds 1, 3, and 4, 10 µM T-DNA was annealed to 5 µM RNA in a thermocycler, as described previously (40) in either TB or HB (25 mM Hepes-KOH, pH 8.0, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, 25 µg/mL acetylated BSA), for the transcription and hydrolysis assays, respectively. The scaffold for pyrophosphorolysis assay (scaffold 2 in Fig. 3A) contained 5 µM RNA, 7.5 µM T-DNA, and 9.4 µM nontemplate DNA (NT-DNA), with all three oligonucleotides annealed simultaneously. Each RNAP (1.5 µM) was incubated at 37 °C with the appropriate scaffold (0.5 µM) for 15 min in the corresponding buffer. To complete EC reconstitution on scaffolds 1, 3, and 4 (Table S2), 1.5 µM NT-DNA was added at 37 °C for another 15 min. The ECs (150 nM) with scaffolds 1, 3, and 4 were incubated with either trace [α-32P]GTP or [α-32P]CTP for elongation and hydrolysis experiments, respectively, for 5 min at 37 °C to form either the labeled G19 ECs or the C17 ECs.
Ligated-Scaffold Transcription Assay of Multiround Nucleotide Addition.
Labeled G19 ECs formed on scaffold 1 (Fig. S1A and Table S2) were extended to A26 position by incubation with ATP and GTP (100 µM each) for 5 min at 37 °C in TB, pH 8.0 (Fig. S1B). The 2.3-kb DNA fragment was ligated downstream of the EC by incubation of A26 EC (5 nM) with the 2.3-kb DNA fragment (15 nM), ATP (1 mM), and T4 ligase (25 units/µL) at 16 °C for 1 h in TB (Fig. S1 A and B). Following the ligation reaction, the ligated A26 ECs were incubated with heparin (100 µg/mL) for 3 min at room temperature, to eliminate any nonspecific interactions of ligase with DNA. For transcription experiments at various pH, the ligated A26 ECs were then immobilized on 10 µL Novex “HisTag Isolation & Pulldown” Dynabeads by incubation at room temperature for 10 min. Bead-tethered complexes were washed three times with the buffer of desired pH (40 mM potassium phosphate, pH 7.0 or 7.4, 5 mM MgCl2, 1 mM DTT; or 20 mM Tris-acetate, pH 8.4, 40 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT). The ligated A26 ECs were then elongated in 1 mM NTPs in their corresponding buffers. Samples were withdrawn at 10, 20, 30, and 40 s, and 1, 5, and 10 min and quenched with 2× stop buffer (8 M urea, 30 mM Na2EDTA, 90 mM Tris-borate buffer, and 0.05% each Bromophenol blue and Xylene cyanol). An additional 5-s time point was taken for pH 8.4, where the reactions were generally faster than at more acidic pH. All samples were heated at 95 °C for 2 min and separated by electrophoresis on an 8 M urea, 8% PA gel in TBE, alongside a radiolabeled pBR322 DNA ladder. The gel was then exposed to a PhosphorImager screen, which was scanned with a Typhoon PhosphorImager. The gels were quantified with ImageQuant software (GE Healthcare), and densitometry plots for each sample were created by converting pixels to transcript length with the pBR322 DNA as a reference, via a four-factor polynomial fitting in KaleidaGraph. By summing the product of each transcript length and its signal intensity and dividing the sum by total intensity, the average transcript length was determined (41). Elongation rates were calculated by dividing the average transcript length for each sample by the reaction time. Rates for at least three independent samples were determined.
Single-Nucleotide Addition Assay.
Labeled G19 complexes formed on scaffold 1 (Fig. S1A and Table S2) were prepared as described above. Complete extension of RNA to G19 position was accomplished in a subsequent reaction with unlabeled GTP (100 nM) for 3 min at 37 °C. Radiolabeled G19 complexes were passed through G-50 resin, pre-equilibrated in TB, to remove unincorporated nucleotides. G19 ECs were reacted with 1 mM ATP for 2–128 ms at 37 °C in a quench-flow instrument (KinTek RQF-3). The reactions were quenched with 2 M hydrochloric acid, immediately neutralized to pH 8.0 with unbuffered 3 M Tris base, phenol:chloroform-extracted, and ethanol-precipitated for 1 h at −20 °C. To aid precipitation of the nucleic acids, torula yeast RNA was added to the quenched samples to a final concentration of 125 µg/mL. Pellets of the ethanol-precipitated samples were dried and resuspended in 1× stop buffer (4 M urea, 15 mM Na2EDTA, 45 mM Tris-borate, pH 8.3, 0.025% each Bromophenol blue and Xylene cyanol). Dissolved pellets were heated at 95 °C for 2 min, and radiolabeled RNA species were resolved by electrophoresis on an 8 M urea, 15% PA, TBE gel. Gels were exposed to a PhosphorImager screen, which was scanned using a Typhoon PhosphorImager and quantified with ImageQuant software. The amount of RNA of a given length at each time point was calculated as a fraction of all RNA species in the quenched reaction (i.e., G19, A20, and A21). Quench-flow data were fitted to a single-intermediate kinetic model in Mathematica, using nonlinear least squares regression to extract the individual rates of nucleotide addition.
Pyrophosphorolysis.
Scaffold 2 (Fig. 3A and Table S2) was used to reconstitute ECs for the assay, based on the high propensity of an EC with a 3′-rGrU to occupy the pretranslocated state on the polymerase, as previously tested (29). RNA was 5′-radiolabeled before scaffold preparation, in a reaction with [γ-32P]ATP catalyzed by T4 polynucleotide kinase. ECs were reconstituted in Reconstitution Buffer (RB; 20 mM Tris-acetate, pH 8.0, 20 mM potassium acetate, 0.1 mM EDTA) with 450 nM RNAP and 150 nM scaffold, for 15 min at 37 °C. Assembled ECs were immobilized on magnetic Ni beads for 10 min at room temperature, and repeatedly washed with RB. Pyrophosphorolysis was commenced by resuspension of the beads in RB containing 5 mM magnesium acetate, 0.5 mM sodium pyrophosphate, and 1.5 units/mL apyrase. Apyrase was included in the assay to break down released UTP, and in doing so to push the pyrophosphorolysis reaction forward, as previously described (29). Reaction progress was monitored by withdrawing aliquots at indicated time points, quenching with 2× stop buffer, and visualizing RNA species on a 20% denaturing PAGE. Decay kinetics of the starting RNA was determined by fitting the quantified data to a single-exponential function.
Multinucleotide (5–7 nt) Cleavage Assay.
Labeled C17 ECs formed on scaffold 3 (Fig. S2A and Table S2) were immobilized on 15-µL Ni beads as described above. The beads were washed with HB three times to remove unincorporated [α-32P]CTP and then resuspended in 100 µL HB. The “C17” sample was withdrawn and the remaining buffer was removed. The beads were resuspended in HB prewarmed to 37 °C containing 10 µM each CTP and UTP, and incubated for 10 min to form C21 ECs (Fig. 4A). The beads were then washed with Wash Buffer (WB; same as HB but without Mg2+) six times to remove unincorporated NTPs and any cleaved RNA products, and resuspended in 100 µL WB. At this point the “C21” sample was withdrawn. The remaining buffer was removed, and the beads were resuspended in 100 µL WB and split into two 50 µL samples for replicates. All WB was then removed from each sample. Transcript cleavage was initiated by resuspension of the bead-bound C21 complexes in Cleavage Buffer (CB; 25 mM Tris⋅HCl pH 9.0, 50 mM KCl, 20 mM MgCl2, 1 mM DTT, 5% glycerol, and 25 µg acetylated BSA per milliliter) preheated to 37 °C. Samples were withdrawn at 3, 6, 10, 20, 40, 60, and 120 min and mixed with 2× stop buffer as described above. Samples were heated, separated in an 8 M urea, 20% PA, TBE gel, and scanned as described above. The amount of the starting C21 RNA and cleavage product (5, 6, and 7 nt in length) bands was quantified with ImageQuant software and used to calculate the fraction of each RNA species for each time point. The C21 fractions were plotted vs. time for each polymerase and fitted to a single-exponential decay model using Igor graphing software to obtain rate constants. Time courses from at least three independent cleavage experiments were fitted.
One- vs. Two-Nucleotide Cleavage Assay.
Similarly to the multinucleotide cleavage experiment described above, labeled C17 ECs formed on scaffold 4 (Fig. S3A and Table S2) were immobilized on 15-µL Ni beads. The beads were washed with WB four times to remove unincorporated [α-32P]CTP and resuspended in 105 µL WB, from which the “C17” sample was withdrawn. The C17 EC mixture was split into two 50-µL samples for replicates. Remaining WB was removed and transcript cleavage was initiated by resuspension in 50 µL CB preheated to 37 °C. Samples were withdrawn at 0.5, 1, 2, 5, 10, 30, and 60 min and quenched with 2× stop buffer. Samples were heated, separated in a 25% urea-PAGE gel, and scanned. The amounts of each C17, 2-nt, and 1-nt RNA band were quantified with ImageQuant software and used to obtain rate constants for both the formation of the 1- and 2-nt RNA products, as described above. Time courses from at least three independent cleavage experiments were used in the fitting.
Side-Chain Volume Calculations.
To determine the relative steric bulk of the relevant side chains (Table S1), we calculated their molar volumes with density functional theory (42, 43) using Gaussian 09 (44). Each side chain was truncated at the Cα-side chain bond, with a hydrogen replacing the α-carbon (e.g., the side chain of alanine was calculated as methane), and the geometries were optimized at the B3LYP/6–31+G* level in a polarizable continuum model (45) for water.
Acknowledgments
We thank the members of the R.L. laboratory for fruitful discussions and valuable input throughout the course of this work; and Daniel Roston (University of Wisconsin–Madison) for performing side-chain volume calculations. This work was supported by NIH Grant GM038660 (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702383114/-/DCSupplemental.
References
- 1.Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 2.Imashimizu M, Oshima T, Lubkowska L, Kashlev M. Direct assessment of transcription fidelity by high-resolution RNA sequencing. Nucleic Acids Res. 2013;41:9090–9104. doi: 10.1093/nar/gkt698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 5.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 6.Windgassen TA, et al. Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;42:12707–12721. doi: 10.1093/nar/gku997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouqueau T, Zeller ME, Cheung AC, Cramer P, Thomm M. The RNA polymerase trigger loop functions in all three phases of the transcription cycle. Nucleic Acids Res. 2013;41:7048–7059. doi: 10.1093/nar/gkt433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Yuzenkova Y, Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc Natl Acad Sci USA. 2010;107:10878–10883. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-Nahum G, et al. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro C, et al. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc Natl Acad Sci USA. 2007;104:4267–4272. doi: 10.1073/pnas.0608952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro C, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat Struct Mol Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho ATP, Fernandes PA, Ramos MJ. The catalytic mechanism of RNA polymerase II. J Chem Theory Comput. 2011;7:1177–1188. doi: 10.1021/ct100579w. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, et al. RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc Natl Acad Sci USA. 2010;107:15745–15750. doi: 10.1073/pnas.1009898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Da LT, Wang D, Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc. 2012;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B, Zuo Y, Steitz TA. Structures of E. coli σS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci USA. 2016;113:4051–4056. doi: 10.1073/pnas.1520555113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braberg H, et al. From structure to systems: High-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu C, et al. High-resolution phenotypic landscape of the RNA polymerase II trigger loop. PLoS Genet. 2016;12:e1006321. doi: 10.1371/journal.pgen.1006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuzenkova Y, et al. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 26.Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 2003;371:233–251. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- 28.Palangat M, et al. Efficient reconstitution of transcription elongation complexes for single-molecule studies of eukaryotic RNA polymerase II. Transcription. 2012;3:146–153. doi: 10.4161/trns.20269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hein PP, Palangat M, Landick R. RNA transcript 3′-proximal sequence affects translocation bias of RNA polymerase. Biochemistry. 2011;50:7002–7014. doi: 10.1021/bi200437q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kotlajich MV, et al. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. eLife. 2015;4:e04970. doi: 10.7554/eLife.04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larson MH, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imashimizu M, et al. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Čabart P, Jin H, Li L, Kaplan CD. Activation and reactivation of the RNA polymerase II trigger loop for intrinsic RNA cleavage and catalysis. Transcription. 2014;5:e28869. doi: 10.4161/trns.28869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 35.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: Implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Chlenov M, et al. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase beta’ subunit. J Mol Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 37.Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta’ subunit inhibit transcript cleavage and transcript elongation. J Biol Chem. 1998;273:24912–24920. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]
- 38.Esyunina D, et al. Lineage-specific variations in the trigger loop modulate RNA proofreading by bacterial RNA polymerases. Nucleic Acids Res. 2016;44:1298–1308. doi: 10.1093/nar/gkv1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin H, Kaplan CD. Relationships of RNA polymerase II genetic interactors to transcription start site usage defects and growth in Saccharomyces cerevisiae. G3 (Bethesda) 2014;5:21–33. doi: 10.1534/g3.114.015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 41.Haft RJF, et al. Correcting direct effects of ethanol on translation and transcription machinery confers ethanol tolerance in bacteria. Proc Natl Acad Sci USA. 2014;111:E2576–E2585. doi: 10.1073/pnas.1401853111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becke AD. Density-functional thermochemistry. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 43.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 44.Frisch MJ, et al. Gaussian 09, Revision E.01. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
- 45.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 46.Wang B, Predeus AV, Burton ZF, Feig M. Energetic and structural details of the trigger-loop closing transition in RNA polymerase II. Biophys J. 2013;105:767–775. doi: 10.1016/j.bpj.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell. 2013;50:882–893. doi: 10.1016/j.molcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D, et al. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 49.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]