Significance
In a quest for antibiotics that may display durable clinical lifetimes, analogs of the glycopeptide antibiotics, including vancomycin, have been designed that not only directly overcome the molecular basis of existing vancomycin resistance but also contain two added peripheral modifications that endow them with two additional independent mechanisms of actions not found in the parent antibiotics. It is shown that such peripherally and binding pocket-modified vancomycin analogs display little propensity for acquired resistance by vancomycin-resistant Enterococci and that both their antimicrobial potency and durability against such challenges follow trends (three > two > one mechanisms of action) that are now predictable.
Keywords: vancomycin, antibiotics, resistant bacterial infections
Abstract
Subsequent to binding pocket modifications designed to provide dual d-Ala-d-Ala/d-Ala-d-Lac binding that directly overcome the molecular basis of vancomycin resistance, peripheral structural changes have been explored to improve antimicrobial potency and provide additional synergistic mechanisms of action. A C-terminal peripheral modification, introducing a quaternary ammonium salt, is reported and was found to provide a binding pocket-modified vancomycin analog with a second mechanism of action that is independent of d-Ala-d-Ala/d-Ala-d-Lac binding. This modification, which induces cell wall permeability and is complementary to the glycopeptide inhibition of cell wall synthesis, was found to provide improvements in antimicrobial potency (200-fold) against vancomycin-resistant Enterococci (VRE). Furthermore, it is shown that this type of C-terminal modification may be combined with a second peripheral (4-chlorobiphenyl)methyl (CBP) addition to the vancomycin disaccharide to provide even more potent antimicrobial agents [VRE minimum inhibitory concentration (MIC) = 0.01–0.005 μg/mL] with activity that can be attributed to three independent and synergistic mechanisms of action, only one of which requires d-Ala-d-Ala/d-Ala-d-Lac binding. Finally, it is shown that such peripherally and binding pocket-modified vancomycin analogs display little propensity for acquired resistance by VRE and that their durability against such challenges as well as their antimicrobial potency follow now predictable trends (three > two > one mechanisms of action). Such antibiotics are expected to display durable antimicrobial activity not prone to rapidly acquired clinical resistance.
Recent years have seen a welcomed refocus on the need for new antibiotics to address the persistent threat of bacterial resistance (1–3). A number of actions have been advanced to address the challenges posed by bacterial resistance now emerging faster than new treatment options. These actions include providing new financial incentives to counter the declining economic interests in developing new antibiotics (4), revamping regulatory criteria for new drug approvals (5), improving the rate of diagnostic characterization of infecting organisms, enhancing nationwide resistance surveillance, encouraging work targeting mechanisms of resistance, and identifying new therapeutic targets for antibiotic discovery (6, 7). The suggested actions also champion antibiotic stewardship (8). Although sounding attractive, the effort to restrict antibiotic use seems counter to their importance, introduces guilt into even their most legitimate of uses, challenges the prevailing practices of initial empirical best guess therapy and prophylaxis deployment, and produces additional disincentives to antibiotic development. Although such initiatives highlight the pressing need for renewed antibiotic discovery and the fundamental importance of antibiotics in modern medicine (9), it has done little to define new approaches that directly address the underlying problem of evolutionarily driven and acquired resistance. The mechanisms of resistance are ancient and increasingly accumulating in pathogenic bacteria, which have now assimilated large elements of this bacterial resistome (10, 11). An additional and perhaps even more important question to ask is if new antibiotics can now be designed that overcome the forces of evolution and selection responsible for bacterial resistance, that are less prone or even impervious to resistance development, that avoid many of the common mechanisms of resistance, and that are more durable than ever before. As an alternative to championing the restricted use of antibiotics or conceding that bacteria will always outsmart us, can durable antibiotics be developed that are capable of continued or even more widespread use? Herein, we describe one such effort to create durable antibiotics by deliberate design that may directly counter such evolutionary forces. We identified the glycopeptide antibiotics as an antibiotic class already endowed with features that avoid many mechanisms of resistance. After introduction of designed structural changes that directly overcome the molecular basis of their only prevalent mechanism of resistance, we have explored the incorporation of peripheral structural modifications in the molecules that provide them with additional and now multiple synergistic mechanisms of action, thereby not only increasing their potency but also, creating prototype durable antibiotics.
In recent disclosures, we have discussed attributes of the glycopeptide antibiotics (12, 13) that have contributed to their sustained effectiveness in the clinic (14). Vancomycin (15), teicoplanin (16), and three recently approved semisynthetic derivatives, oritavancin (17), dalbavancin (18), and telavancin (19), are widely used to treat refractory bacterial infections, including methicillin-resistant Staphylococcus aureus (MRSA) (20). Vancomycin (1) (Fig. 1) (21) was disclosed in 1956 (15) and introduced into the clinic in 1958. After nearly 60 y of clinical use and even with the past use of glycopeptide antibiotics for agricultural livestock (avoparcin), resistant pathogens have only slowly emerged, and vancomycin remains an integral and increasingly important antibiotic today. Clinical resistance was initially observed with vancomycin-resistant Enterococci (VRE; 1987) that was detected only after 30 y of clinical use (22) but now, also includes vancomycin-resistant S. aureus (VRSA; 2002) (23). Treatment options for the latter are limited and presently include antibiotics known to rapidly evoke resistance (24, 25). As a result, these latter antibiotics have been designated as reserve antibiotics to be deployed sparingly to preserve their effectiveness as drugs of last resort against intractable infections. Just as significantly, some VRE organisms, like MRSA, have also reached a stage where they are now resistant to most other common antibiotic classes (26). As a result and because they are already vancomycin-resistant, the CDC has now placed VRE on its serious threat list (27). Most recently, the WHO has released, for the first time, a list of drug-resistant bacteria that pose the greatest threat to human health for which new antibiotics are desperately needed. Both VRE (fourth) and VRSA (fifth) appear on this ranked list (28).
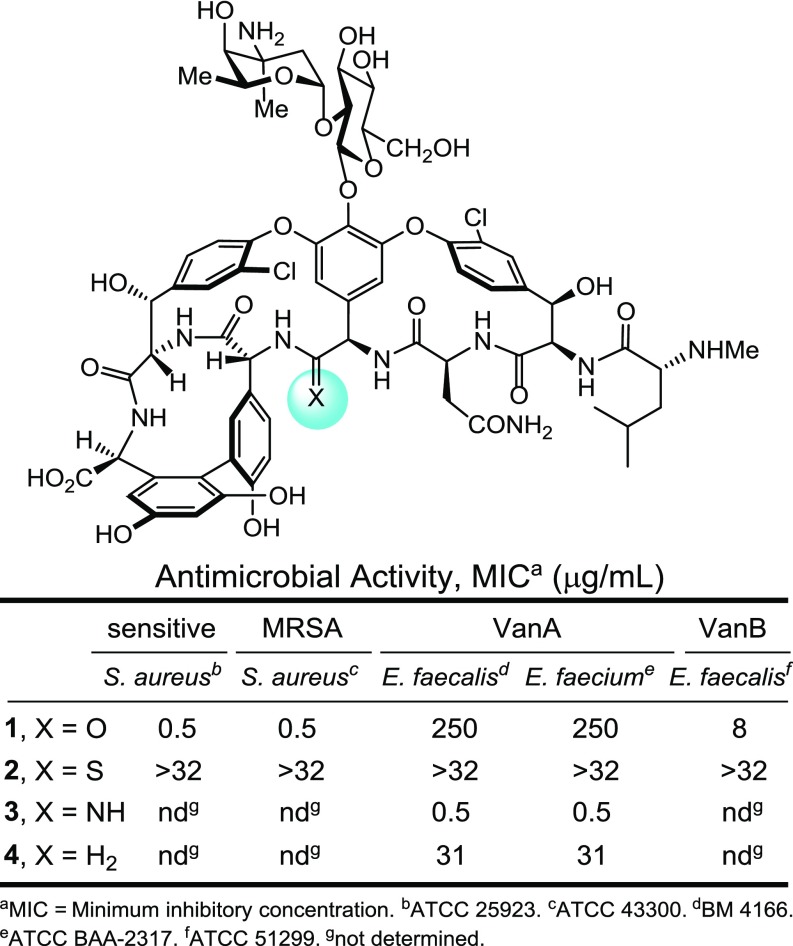
Fig. 1.
Vancomycin (1) and designed binding pocket-modified analogs.
The primary biological target for vancomycin and the glycopeptide antibiotics is bacterial cell wall precursors containing d-Ala-d-Ala, binding to which results in inhibition of cell wall maturation (29). This target is unique to bacteria and contributes to the selectivity of the antibiotic class for bacteria vs. their mammalian hosts. It is also an atypical biological target, being a substrate for an enzymatic reaction and a precursor to a structural component of the bacterial cell wall. It is not a protein or nucleic acid target subject to changes by a single genetic mutation that can result in resistance. The primary mechanism of action of vancomycin involves sequestration of this substrate (d-Ala-d-Ala) for a late-stage enzyme-catalyzed (transpeptidase) reaction used for cell wall cross-linking (13). Thus, the nature of the target (d-Ala-d-Ala) and the antibiotic mechanism of action (sequestration of an enzyme substrate) are difficult for the organism to overcome by a single genetic alteration. Vancomycin is also thought to inhibit the preceding step in the cell wall biosynthesis, the transglycosylase-catalyzed incorporation of lipid intermediate II into the polysaccharide backbone of the bacterial cell wall. In the case of vancomycin, this inhibition also requires d-Ala-d-Ala binding (30–33). However, it is not yet clear whether this occurs through direct binding of the vancomycin disaccharide to the enzyme active site, because cell wall binding contributes to its localization, or whether this occurs by indirect enzyme inhibition. Because there may be two or more mechanisms of action, including those yet unknown or with a role that is not yet fully appreciated (34), full bacterial resistance requires unlikely simultaneous changes that impact each. Further contributing to the durability of vancomycin is the site of action at the bacterial cell wall surface. Cell wall penetration or import is not needed, and this feature allows vancomycin to avoid the common resistance mechanisms mediated by expression levels of proteins involved in transport, efflux, and metabolic deactivation by cytosolic enzymes (35). Finally, it has been suggested that there are genetic features that presently make the glycopeptide antibiotics less susceptible to vertical vs. horizontal gene transfer of resistance (36). Regardless of the origins, it is most revealing that the primary mechanism of clinical resistance to vancomycin (VanA and VanB phenotypes) was transferred to pathogenic bacteria from nonpathogenic organisms that produce vancomycin and use this inducible resistance mechanism to protect themselves during vancomycin production (37). Thus, pathogenic bacteria themselves have not yet evolved effective resistance mechanisms to the glycopeptide antibiotics, even after nearly 60 y of widespread use (38). This latter observation has suggested to us that solutions to VanA and VanB resistance alone may provide antibiotics with durable clinical lifetimes.
It is an intricate mechanism of resistance in which synthesis of the bacterial cell wall precursors continues with installation of the pendant N-terminal d-Ala-d-Ala. Resistant bacteria, like the producer organisms, sense the presence of the antibiotic (39). Through use of a two-component cell surface receptor sensing and intracellular signaling system, the organisms initiate a late-stage remodeling of their peptidoglycan termini from d-Ala-d-Ala to d-Ala-d-Lac to avoid the action of the antibiotic (40). The binding affinity of vancomycin for the altered ligand is reduced 1,000-fold (41, 42), resulting in a corresponding 1,000-fold loss in antimicrobial activity. In a series of studies, we reported the first vancomycin analogs that contain changes at a key single-atom site in its target binding pocket (residue 4 carbonyl O → S, NH, H2), the latter two of which were designed to directly address this underlying molecular basis of resistance to vancomycin (Fig. 1) (43–50). These rationally designed binding pocket modifications reinstated binding to the altered target d-Ala-d-Lac and maintained binding affinity for the unaltered target d-Ala-d-Ala. Such dual target binding compounds were found to reinstate antimicrobial activity against vancomycin-resistant organisms that inducibly or constitutively use d-Ala-d-Lac peptidoglycan precursors and remain active against vancomycin-sensitive bacteria. Moreover, the in vitro antimicrobial potencies of such compounds correlated directly with their absolute dual binding affinities for model target ligands.
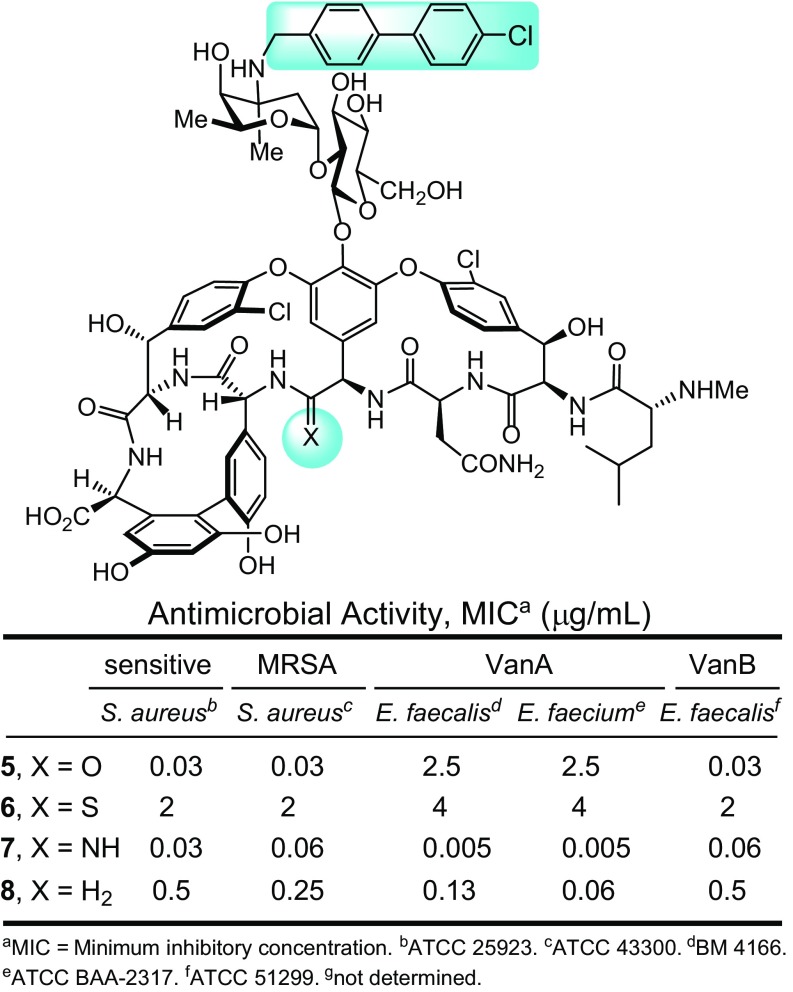
We subsequently found that peripheral functionalization of the binding pocket modified vancomycin analogs, introducing the oritavancin (4-chlorobiphenyl)methyl (CBP) group to the pendant disaccharide and known to enhance antimicrobial potency (17), producing more potent antibiotics (Fig. 2) (47, 48). These analogs exhibited a remarkable spectrum of antimicrobial activity [vancomycin-sensitive S. aureus (VSSA), MRSA, and VanA and VanB VRE] with further improved (ca. 100-fold) and impressive potencies against both vancomycin-sensitive and -resistant bacteria [minimum inhibitory concentration (MICs) = 0.06–0.005 and 0.5–0.06 μg/mL for 7 and 8, respectively]. With the benefit of the examination of the residue 4 thioamide 6, which is incapable of binding either d-Ala-d-Ala or d-Ala-d-Lac (MICs = 2–4 μg/mL), we were able to infer that the activity of such CBP-modified analogs is derived from two synergistic mechanisms of action, only one of which is dependent on d-Ala-d-Ala/d-Ala-d-Lac binding (48).
Fig. 2.
Pocket-modified vancomycins that contain an additional peripheral CBP modification to the pendant disaccharide.
Herein and along with studies that further clarify this second mechanism of action, an alternative peripheral modification that endows the pocket-modified vancomycin analogs with another different second mechanism of action is reported. This modification also provides similarly impressive improvements in antimicrobial potencies against vancomycin-resistant bacteria (VRE). Furthermore, we show that the two peripheral modifications may be combined with the pocket-modified vancomycins to provide even more potent antimicrobial agents with activity that can be attributed to three independent and synergistic mechanisms of action, only one of which requires d-Ala-d-Ala/d-Ala-d-Lac binding. Finally, it is shown that such peripherally and binding pocket-modified vancomycins display little propensity for acquired resistance through serial exposure of VRE and that their durability against such challenges as well as their potency follow trends (three > two > one mechanisms of action) that are shown now to be predictable. Such antibiotics are expected to display even more durable antimicrobial activity than vancomycin or its semisynthetic analogs.
Results and Discussion
These studies were conducted with the methylene pocket-modified vancomycin analog 4 ([Ψ[CH2NH]Tpg4]vancomycin) (48), presently the most readily available of our synthetic analogs prepared by total synthesis (50). Because it also exhibits the more modest dual d-Ala-d-Ala/d-Ala-d-Lac binding affinity and antimicrobial activity against vancomycin-resistant organisms of the two pocket-modified vancomycin analogs (4 vs. 3) (Fig. 1), the impact of alternative or multiple peripheral modifications was anticipated to be most easily quantitated. The alternative peripheral modification examined was C-terminal amide functionalization with incorporation of either a basic amine capable of protonation or a quaternary ammonium salt. Such modifications have been shown to provide improved antimicrobial activity against vancomycin-resistant organisms and were found to act by disrupting bacterial cell wall membrane integrity, increasing cell permeability, and inducing membrane depolarization (51). Although inspired by the nonselective membrane disruption induced by quaternary ammonium salts, the studies herein provide one such modification that exhibits only a subset of such effects (membrane permeability) and acts by a more specific mechanism not resulting in cell lysis. It is a behavior not observed with the naturally occurring glycopeptide antibiotics or their more typical analogs, but the mechanism is one that may contribute to the activity of the semisynthetic drugs dalbavancin and telavancin (52). In vancomycin-resistant organisms, such modifications do not directly contribute to inhibition of cell wall biosynthesis, do not improve d-Ala-d-Lac binding needed to express such effects, and act independent of mechanisms derived from transpeptidase or transglycosylase inhibition.
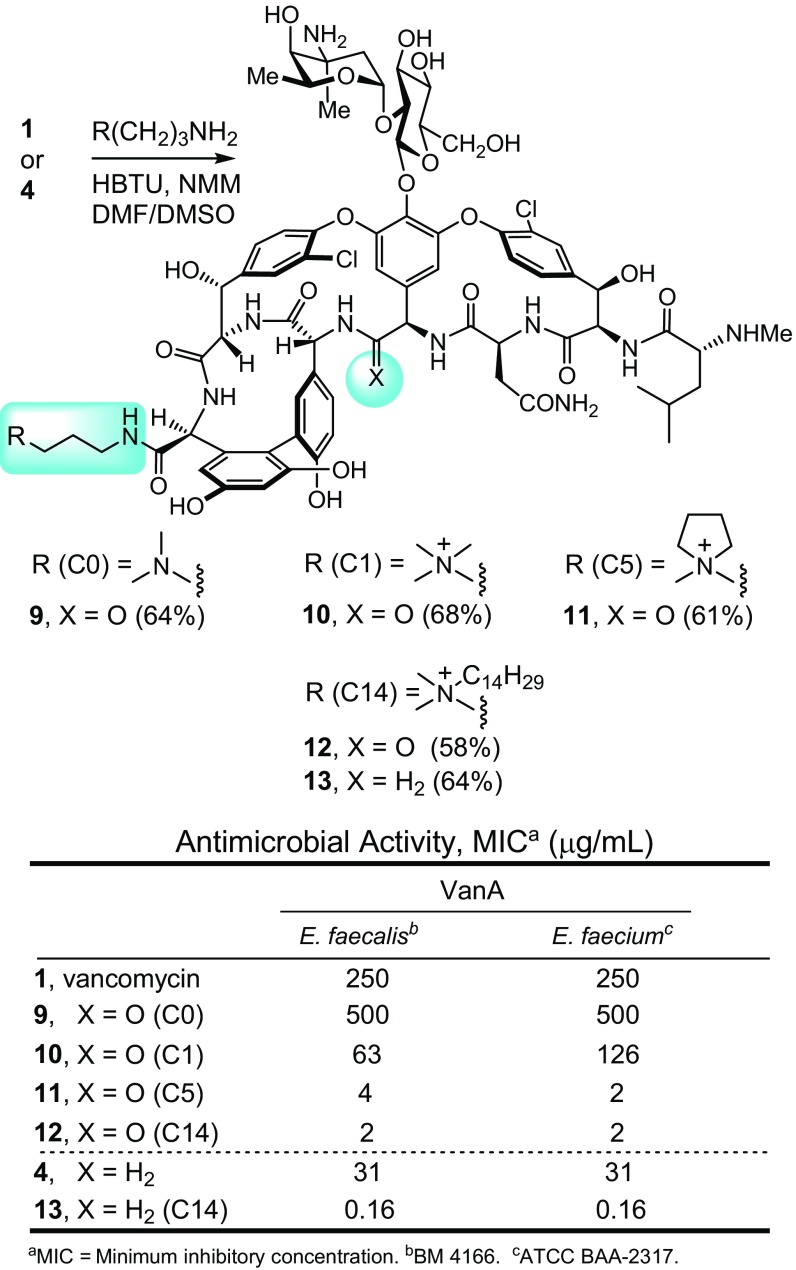
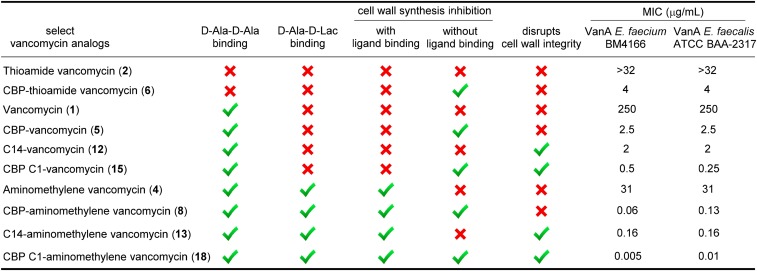
A select series of such vancomycin analogs was first prepared, including 10 and 11 not previously examined. For simplicity, they are referred to as C0 (9) (51), C1 (10), cyclic C5 (11), and C14 (12) (51), denoting the terminal tertiary dimethylamine (9; C0) or the quaternary ammonium salts bearing a methyl (10; C1), C5 cyclic (11; C5), or tetradecyl (12; C14) substituent (Fig. 3). Based on their activity in antimicrobial assays against VanA VRE, the most potent C-terminal modification found in 12 (C14) was incorporated into the analogous C14 derivative 13 of the pocket-modified vancomycin analog 4. This modification was accomplished in a single step from the fully functionalized vancomycin 1 or 4 without need for intermediate protection by coupling the C-terminal carboxylic acid with the corresponding functionalized amines under conditions modified from those previously described (53). These C-terminal changes had a progressively pronounced impact on activity against the VanA vancomycin-resistant organisms (VanA VRE), where the quaternary ammonium salts incrementally increased activity up to 100-fold (12 ≥ 11 > 10 > 9, 1) (Fig. 3). Notably, all of the quaternary ammonium salts (C1, cyclic C5, and C14) improved activity. For the pocket-modified vancomycin analog, the most potent of these modifications produced a 200-fold increase in potency against VanA VRE, reducing the MIC value from the modest activity of 31 μg/mL for 4 to 0.16 μg/mL for 13. This compound is >10-fold more potent than its comparison vancomycin derivative 12 and >1,000-fold more potent than vancomycin itself. Thus, an additional second peripheral modification of a pocket-modified vancomycin analog substantially increased antimicrobial activity against the most stringent of the vancomycin-resistant phenotypes (VanA VRE). As shown below, this improvement arises through an independent second mechanism of action involving induced membrane permeability. This second synergistic mechanism of action incorporated into 4 is different from that observed with the peripheral CBP modification found in 8.
Fig. 3.
Peripheral C-terminal modifications of vancomycin and the binding pocket-modified vancomycin analog 4.
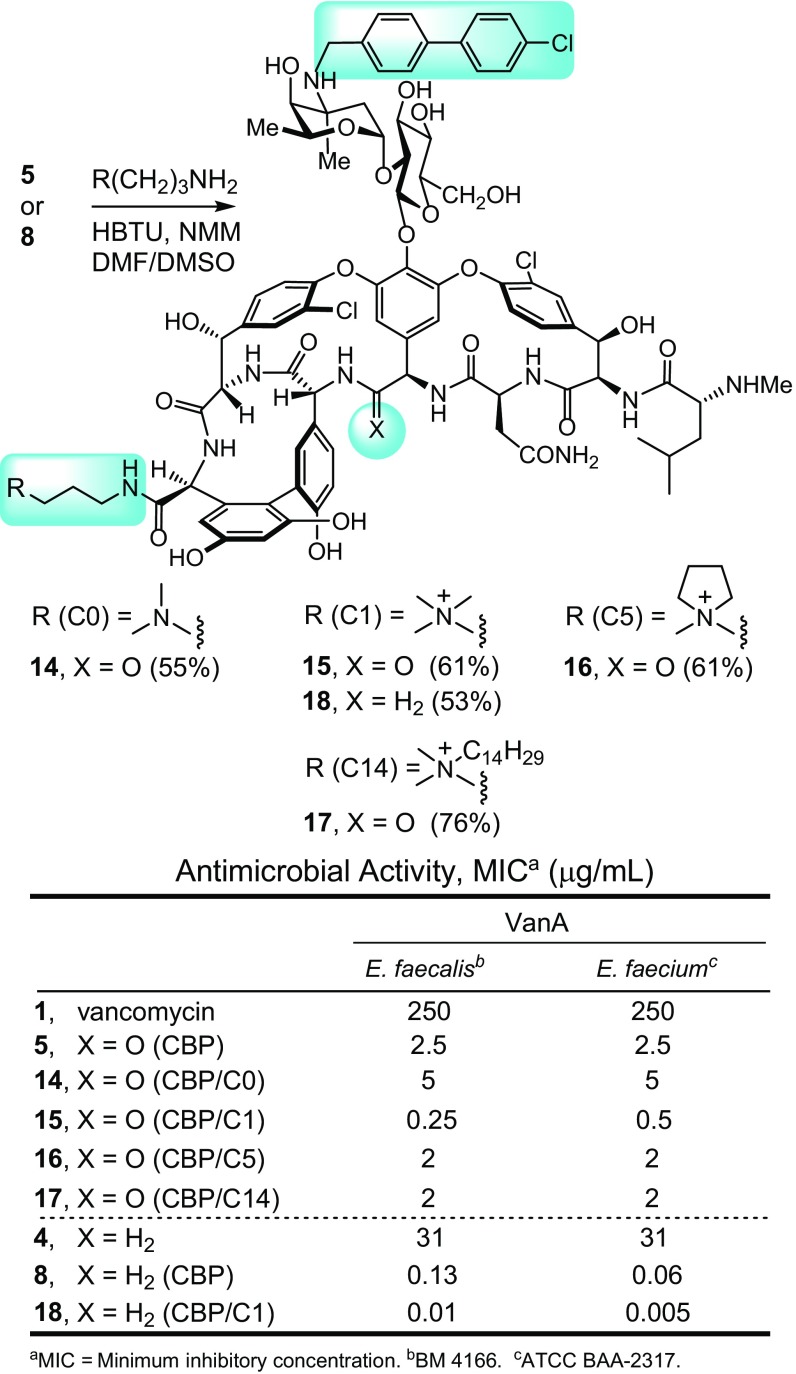
More significantly, the impact of combining the two different peripheral modifications was examined. This combination was explored first with CBP–vancomycin (5), coupling its C-terminal carboxylic acid with the same series of functionalized amines that contain the tertiary dimethylamine (14; C0) or C1 (15), cyclic C5 (16), and C14 (17) quaternary ammonium salts (Fig. 4). Based on their assessment in VanA VRE antimicrobial assays, the most effective C-terminal modification found in 15 (C1) was incorporated into the analogous C1 derivative 18 of the pocket-modified CBP–vancomycin analog 8. Notably, their preparation also required a single-amide bond coupling reaction and was conducted without the need for protected intermediates. This second peripheral modification of CBP–vancomycin did not display the same trends observed with vancomycin itself, with most not altering the antimicrobial activity of CBP–vancomycin against VanA VRE (Fig. 4). The exception was 15, containing the C1 quaternary ammonium salt, which alone displayed a 10-fold increase in activity against VanA VRE, exhibiting exceptional activity for a compound incapable of binding d-Ala-d-Lac. In fact, it represents a compound devoid of the original glycopeptide antibiotic mechanism of action in the resistant organisms but possesses two other effective mechanisms of action independent of d-Ala-d-Ala/d-Ala-d-Lac binding. The impact of these modifications improved the activity beyond what either does alone and as shown below, results from inhibition of bacterial cell wall biosynthesis by direct transglycosylase inhibition caused by the CBP modification and through induced membrane permeability by the C1 quaternary ammonium salt. Although unanticipated, the most effective C-terminal modification is now C1 for the CBP derivatives. Subsequent studies show clearly that it alone imparts membrane permeability not found with derivatives lacking this particular C-terminal modification. For the pocket-modified CBP–vancomycin analog capable of dual d-Ala-d-Ala/d-Ala-d-Lac binding, this second additional peripheral modification with 18 produced a >10-fold increase in potency against VanA VRE relative to 8, lowering the MIC value for 18 to 0.01–0.005 μg/mL (Fig. 4). This vancomycin analog is >10-fold more potent than the CBP derivative 8, >1,000-fold more potent than the pocket analog 4, and a stunning >10,000-fold more potent than vancomycin itself. It is also >25- to 100-fold more potent than its comparison C1/CBP–vancomycin derivative 15 and >250-fold more active than either CBP–vancomycin (5) or C14–vancomycin (12).
Fig. 4.
Combined CBP and C-terminal peripheral modifications to vancomycin and the binding pocket-modified vancomycin analog 4.
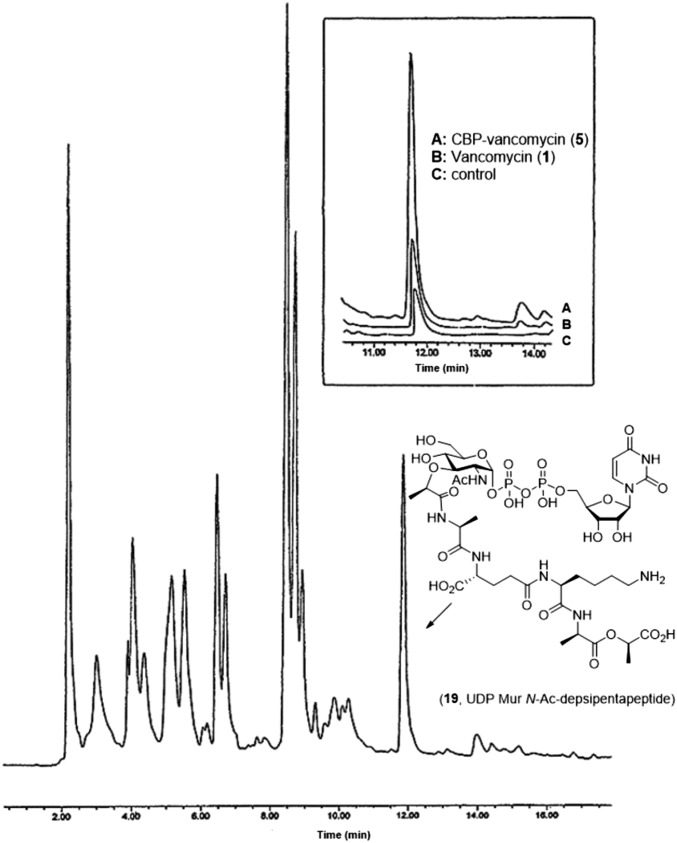
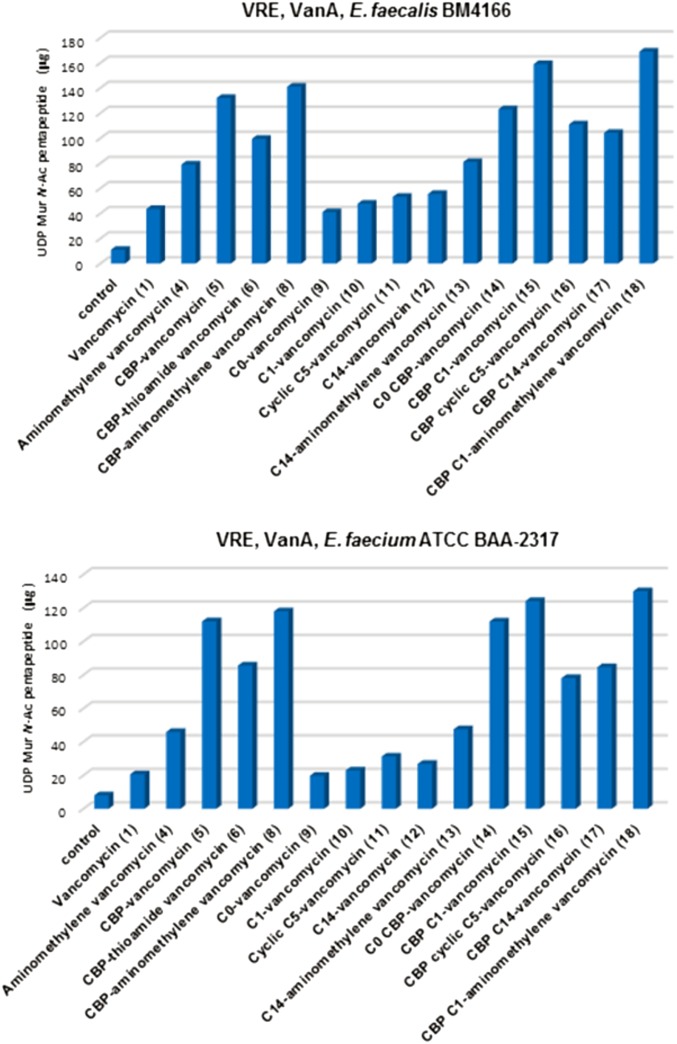
To clarify the contributing mechanisms responsible for the antimicrobial activity in vancomycin-resistant organisms (VanA VRE), the key analogs were examined in a range of assays, two of which defined the origin of their effects. One assay was used to establish inhibition of bacterial cell wall synthesis, and the second measured induced membrane permeability. The inhibition of bacterial cell wall synthesis was established in an assay that quantitates the accumulation of the peptidoglycan precursor UDP-N-acetyl-muramyl-depsipentapeptide (UDPMurNAc-pp; 19) resulting from inhibited cell wall incorporation after antibiotic treatment. This assessment was accomplished by following a protocol that uses vancomycin-resistant Enterococcus faecium with tetracycline pretreatment (30 min at 37 °C) (30). This procedure was reduced in scale and modified to enlist reverse-phase HPLC separation, UV detection, and calibration curve determination of the amount of isolated 19, permitting use with limited sample sizes (Fig. 5). A larger-scale isolation and full characterization of 19 (SI Appendix) provided the material needed to establish a linear calibration curve (SI Appendix). The test compounds were incubated with tetracycline-pretreated VanA VRE for 30 min (37 °C) before the cultured bacteria were collected by centrifugation, washed, and resuspended in pH 7.2 buffer (5 mM Hepes:5 mM glucose; 1:1). The bacterial suspension was heated at 100 °C for 15 min to release cytosolic 19. The entire supernatant was analyzed by semipreparative reverse-phase HPLC with a single injection, and the amount of 19 was quantitated with use of the calibration curve. The results of the evaluation of 5–18 in VanA vancomycin-resistant Enterococcus faecalis (BM 4166) and E. faecium (ATCC BAA-2317) are presented in Fig. 6 and represent the same strains used in the antimicrobial assays.
Fig. 5.
Reverse-phase HPLC analysis of peptidoglycan precursor UDPMurNAc-pp (19) in VanA VRE (E. faecium ATCC BAA-2317).
Fig. 6.
Inhibition of bacterial cell synthesis in VanA VRE. Quantitated accumulation of the peptidoglycan precursor UDPMurNAc-pp (19) in cytosol by vancomycin analogs (35 μg/mL).
The effect of the compounds on cell membrane permeability was examined by measuring cytoplasmic membrane uptake of the fluorescent probe propidium iodide in the same VanA vancomycin-resistant E. faecalis (BM 4166) (SI Appendix, Fig. S1) and E. faecium (ATCC BAA-2317) strains. This fluorescent probe only enters cells with permeabilized cell membranes and is detected by the emission of fluorescence on intracellular nucleic acid binding (54). Fresh midlog-phase VanA VRE in pH 7.2 buffer at 25 °C was preincubated with propidium iodide (10 μM) for 5 min before the test compounds were added and monitored for the fluorescence emission at 617 nm (excitation at 535 nm) over time, both before (5 min) and after (15 min) compound addition. Rapid and sustained increases in fluorescence intensity are observed immediately on addition of test compounds that induce bacterial cell membrane permeability.
Assessments of the compounds, identifying their contributing mechanisms of action in vancomycin-resistant VRE, were conducted with the two assays. The results are discussed below in sets defining first the role of the pocket modification found in 4 and then, the subsequent impact of the peripheral CBP modification to the vancomycin disaccharide found in 5–8. This summary is followed by the discussion of the results from the examination of the peripheral quaternary ammonium salt modifications found in 9–13, including their effects on both unmodified and pocket-modified vancomycin analogs. Finally, the effects of the two combined peripheral modifications in 14–18 are detailed.
Consistent with its inactivity and like the thioamide 2, vancomycin (1) does not effectively inhibit bacterial cell wall synthesis in VanA VRE and does not result in the significant accumulation of 19 in the assay (Fig. 6). In contrast, the pocket-modified analog 4 designed for dual d-Ala-d-Ala/d-Ala-d-Lac binding inhibits bacterial cell wall biosynthesis, resulting in the buildup of the precursor 19 in the assay at levels consistent with its relative model ligand binding affinities and antimicrobial activity. As anticipated, none of these compounds significantly impact membrane integrity, and none result in cytoplasmic membrane permeability as measured by propidium uptake (SI Appendix, Fig. S2). Thus, the antimicrobial activity of 4 correlates directly with its expected impact on bacterial cell wall biosynthesis, binding d-Ala-d-Lac and inhibiting cell wall maturation. Incorporation of the peripheral CBP modification in 5 (ineffective binding to d-Ala-d-Lac) and 6 (ineffective binding to either d-Ala-d-Ala or d-Ala-d-Lac) produced analogs with good activity against VanA VRE that was found to correlate with their ability to inhibit cell wall synthesis of VanA VRE, resulting in the accumulation of 19 in the assay (Fig. 6). Neither compound impact membrane permeability (SI Appendix, Fig. S2). Compounds 5 and 6 exhibit nearly equivalent antimicrobial activity against VanA VRE, and both inhibit cell wall biosynthesis to a similar extent, but 6 is incapable of binding either d-Ala-d-Ala or d-Ala-d-Lac. Thus, this inhibition of bacterial cell wall synthesis is not derived from inhibition of transpeptidase-catalyzed cross-linking derived from d-Ala-d-Ala/d-Ala-d-Lac binding but rather, likely arises from direct inhibition of transglycosylase by the peripherally modified disaccharide. Previous studies of Kahne and coworkers (32, 33) and others (30, 31, 55) have shown such direct inhibition of transglycosylase by 5 and related CBP-bearing analogs. Finally, the potent pocket-modified vancomycin analog 8, containing the peripheral CBP modification, inhibits cell wall synthesis more effectively than 4, lacking the CBP modification and more potently than either 5 or 6, lacking a productive pocket modification. This behavior is the result of the combined effects of the two independent mechanisms of action, both of which impact cell wall biosynthesis but only one of which depends on d-Ala-d-Ala/d-Ala-d-Lac binding. We have interpreted these observations to represent inhibition of both transpeptidase-catalyzed cross-linking, requiring d-Ala-d-Ala/d-Ala-d-Lac binding, and the transglycosylase-catalyzed cell wall incorporation of Lipid II presumably by a direct enzyme interaction that does not require d-Ala-d-Ala/d-Ala-d-Lac binding.
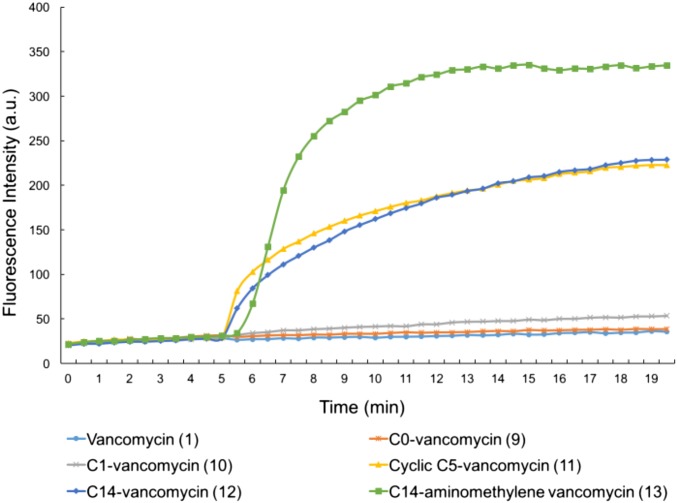
The examination of the analogs that contain the peripheral C-terminal amides with quaternary ammonium salt modifications (9–13) was similarly revealing and clear. Despite the progressive increase in antimicrobial activity observed against VanA VRE with 9–12, little or no change in their ability to inhibit bacterial cell wall synthesis was observed, and they remained, like vancomycin itself, essentially inactive in this assay (Fig. 6). By contrast, the two vancomycin derivatives 11 and 12 that were active against VanA VRE produced pronounced, rapid cell membrane permeability immediately on their addition, whereas the inactive (9) and less active (10) variants did not when examined at 10 μM (Fig. 7). The less active compound 10 exhibited this induced permeability when examined at a higher concentration (100 μM) (SI Appendix, Fig. S3). Here, the antimicrobial activity against VanA VRE can be attributed to a mechanism independent of cell wall biosynthesis and independent of d-Ala-d-Ala/d-Ala-d-Lac binding. The antimicrobial activity correlates with disruption of the cell wall integrity as measured by its increased permeability. The incorporation of the most potent of these peripheral C-terminal modifications into the pocket-modified analog 4 with 13 (C14) further enhanced antimicrobial activity against VanA VRE 200-fold (Fig. 3). This modification did not improve, diminish, or alter the inhibition of cell wall biosynthesis, where 4 and 13 were found to be equally active (Fig. 6). However, it did provide an analog that, unlike 4, produced pronounced cell membrane permeability immediately on addition (Fig. 7). Thus, compound 13 represents a pocket-modified vancomycin analog that displays potent and further improved activity against VanA VRE derived from two independent and synergic mechanisms of action. One mechanism relies on the dual d-Ala-d-Ala/d-Ala-d-Lac binding like 4 and results in effective cell wall synthesis inhibition. The second mechanism is independent of this ligand binding property and derived from induced cell wall permeability. The two combined vancomycin modifications and the accompanying two synergistic mechanisms of action provide a vancomycin analog >1,000-fold more active than vancomycin against the most stringent vancomycin-resistant organisms, VanA VRE, displaying superb in vitro MICs (0.16 μg/mL). It represents now the second such example, complementing the observations made with 8 but with a different second mechanism of action introduced by a second alternative peripheral modification and generalizing the opportunities provided by such design principles.
Fig. 7.
Examination of cell wall permeability induced by compounds 9−13 (10 μM added at 5 min) in VanA VRE (E. faecium ATCC BAA-2317).
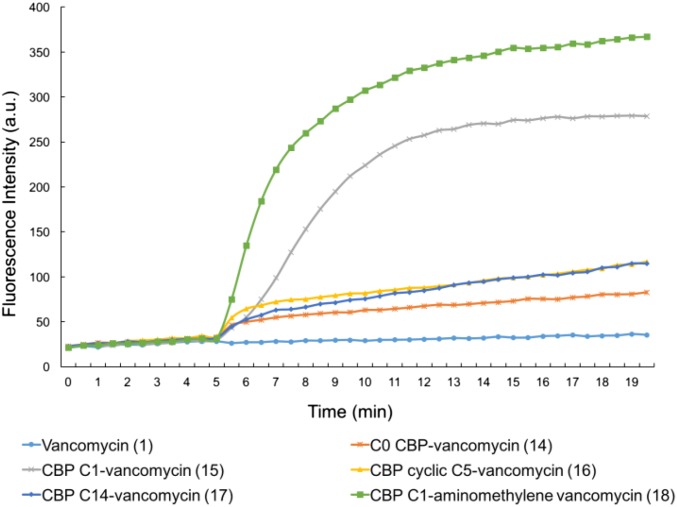
The results of the examination of the analogs that incorporate the two peripheral modifications (14–17) and their combination with the pocket-modified vancomycin analog in 18 were even more revealing. In addition to showing that this may be successfully achieved, they highlight that it is not necessarily the most effective individual variants of the two peripheral modifications that combine to produce the desired effects but rather, that it is a combination that allows expression of the two independent mechanisms. As expected based on the CBP modification, 14–17 inhibit VanA VRE bacterial cell wall synthesis, and their relative activities are reflected in their functional activity in the antimicrobial assays (Fig. 6). The C14 and cyclic C5 quaternary ammonium salts actually diminish the inhibition of cell wall synthesis relative to CBP–vancomycin itself, and C0 was equally active, whereas C1 may have improved activity slightly (activity: 15 > 14 = 5 > 16 and 17). Even more revealing, their examination in the cell wall permeability assay indicates that only C1 combined with the CBP modification induced a pronounced, rapid, and potent cell membrane permeability (Fig. 8). By contrast, the combination of the peripheral CBP modification with the C14 and C5 quaternary ammonium salts was not productive, providing compounds that fail to express the activity. Compound 18, which incorporates the redesigned pocket modification for dual d-Ala-d-Ala/d-Ala-d-Lac binding (blocks cell wall synthesis by ligand binding, including inhibition of transpeptidase-catalyzed cross-linking), the CBP disaccharide modification (blocks cell wall synthesis by direct transglycosylase inhibition without d-Ala-d-Ala/d-Ala-d-Lac binding), and the C1 quaternary ammonium salt C-terminal modification (induces membrane permeability), exhibited the most potent inhibition of cell wall synthesis in the assay of all compounds assessed (Fig. 6) as well as the most pronounced and potent induced cell membrane permeability of all compounds examined (Fig. 8). This behavior indicates that all three mechanisms of action are productively contributing to the expression of the antimicrobial activity of 18 against VanA VRE, resulting in its potent VanA VRE antimicrobial activity (MIC = 0.01–0.005 μg/mL). This compound represents an analog of vancomycin deliberately designed to overcome vancomycin resistance, which incorporates three structural modifications that impart three independent mechanisms of action. Only one mechanism depends on reengineered dual d-Ala-d-Ala/d-Ala-d-Lac ligand binding, and each of three mechanisms contributes to the expression of the antimicrobial activity.
Fig. 8.
Examination of cell wall permeability induced by compounds 14−18 (10 μM added at 5 min) in VanA VRE (E. faecium ATCC BAA-2317).
These latter comparisons provided a direct correlation of the results of the permeability assay with the functional expression of antimicrobial activity where only C1 effectively expresses the functional behaviors. For us, this observation provided compelling evidence that the assay is an accurate readout of the correlated functional behaviors and that the underlying mechanistic interpretation of induced cell permeability is similarly accurate. In addition, it is remarkable that this effect is specific for C1 when combined with the CBP modification, suggesting that the mechanism responsible for induction of membrane permeability may involve specific interactions within the bacterial cell wall.
The results are summarized in Fig. 9 for the key analogs alongside their antimicrobial activity. Within this series, CBP–vancomycin (5) is representative of the potency and characteristics of the clinically approved semisynthetic vancomycin analogs. For VanA VRE, the progression through the series from 1 to 4 to 8 or 13 and finally, to 18 represents vancomycin analogs with zero (1), one (4), two (8 and 13), and three (18) distinct and synergistic mechanisms of action that progressively provide increasingly potent antibacterial activity. This progressive improvement culminates in 18 (MICs = 0.005–0.01 μg/mL), with activity 25,000- to 50,000-fold more potent than vancomycin against VanA VRE. Notably, compound 18 is also 250- to 500-fold more potent than CBP–vancomycin (5), which is representative of the semisynthetic vancomycin analogs presently used in the clinic. Of special note, each structural modification and mechanism of action independently expresses its functional activity at the level of 2–30 μg/mL (1−15 μM) in both the antimicrobial and mechanistic assays, but each provides synergistic improvements in the functional antimicrobial activity when combined.
Fig. 9.
Summary of mechanisms of action of key vancomycin analogs and their individual and cumulative effect on VanA VRE antimicrobial activity.
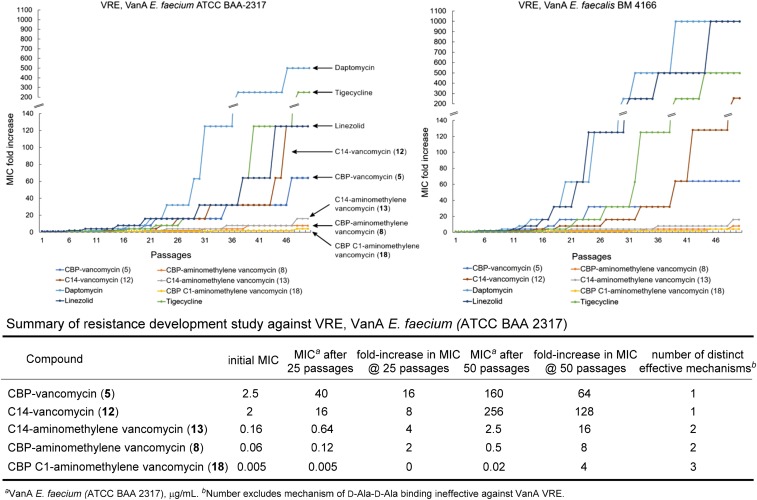
The discussion above focused on the identification of the contributing mechanisms of action and the demonstration that each independently improves antimicrobial activity potency. However, an additional and even more important feature of the expression of multiple independent mechanisms of action is its impact on the rate at which bacterial resistance may emerge. As a result, CBP–vancomycin (5; one mechanism of action), the peripherally C14-modified vancomycin analog (12; one mechanism of action), the peripherally CBP-modified pocket analog 8 (two mechanisms of action), the peripherally C14-modified pocket analog 13 (two mechanisms of action), and the pocket analog 18 that contains the two complementary peripheral C1 and CBP modifications (three mechanisms of action) were examined for their susceptibility to acquired resistance on sublethal (0.5× MIC) serial exposure to the same two VRE bacterial strains, monitoring MICs daily (Fig. 10). Distinct from most related studies enlisting MRSA or other vancomycin-sensitive bacterial strains, this study was conducted with the most stringent (VanA vs. VanB) vancomycin-resistant strains for which the mechanism of action associated with d-Ala-d-Ala binding is no longer effective. Consequently, it is not counted among the number of effective mechanisms of action imbedded in the compound structure. Notably, the antibiotic susceptibility profiles of the VanA VREs used herein indicate that they are resistant to a number of additional classes of antibiotics and on the verge of being classified as multidrug-resistant VRE, indicating that they have already assimilated a number of common resistance mechanisms (SI Appendix). Because the changes for 18 were so small throughout a typical 25-d study, the examination was extended to 50 d. These studies revealed that resistance to 5 and 12 emerged most rapidly and was pronounced (one mechanism). Changes in the potency of both 8 and 13 were much slower and more muted (two mechanisms; only two- to fourfold after 25 passages; 8 slower than 13). No change in susceptibility to 18 was observed after 25 d (three mechanisms), and little change in susceptibility to 18 was observed even after 50 daily passages (only two- to fourfold change in the MICs of 0.01 and 0.005 μg/mL in the two strains). Moreover, the magnitude of the changes in the MICs for the compounds acting by two or more mechanisms is sufficiently small to indicate that none experience a full loss of one of the contributing mechanisms. As such, each mechanism is rendered more robust when combined with structural modifications that provide one or two additional mechanisms of action. Thus, the durability of the antimicrobial activity in such challenges and the effectiveness of each individual mechanism of action as well as the compound potency were found to follow now predictable trends (three > two > one mechanisms of action). Most striking, resistance to daptomycin, linezolid, and tigecycline, each of which is now a frontline single-target antibiotic, arises much faster and is much more pronounced, highlighting the exceptional durability of the antimicrobial activity detailed for 8, 13, and especially 18. Finally, within the series examined, CBP–vancomycin (5) is representative of the expected behavior of the clinically approved semisynthetic vancomycin analogs.
Fig. 10.
Resistance acquisition on serial passaging of VanA VRE in the presence of 0.5× MIC levels of compound. One of two replicate experiments.
The key compounds in the series were examined for in vitro toxicity that might result from the combined mechanisms of action, especially the introduction of structural modifications (quaternary ammonium salt) that might impact host as well as bacterial cell wall integrity. The compounds were examined for RBC hemolytic activity resulting from membrane lysis. No compound in the series, including 18, exhibited any hemolytic activity, even at concentrations >1,000-fold above their MICs (SI Appendix, Fig. S7). In addition, the mammalian cell toxicity of 5, 12, 13, 15, and 18 was established in cell growth inhibition assays against two mammalian cell lines, NIH/3T3 (ATCC CRL-1658; mouse embryonic fibroblast) and HepG2 (ATCC HB-8065; human liver cancer cell line). No growth inhibition (cytotoxic activity) was observed up to 100 μM, the highest dose tested. They were also found to be inactive (>100 μM; highest dose tested) against HCT116 (human colon cancer cell line). Finally, no correlation in activity with cLogP was found for the series of compounds studied, and none of the compounds (5, 12, 13, 15, 17, and 18) exhibited aggregation, higher-order complex formation, or micelle formation at concentrations up to 100 μM in PBS buffer (25 °C), indicating that such effects are not playing a role in the expression of the activity of the compounds. Compound 18 also failed to produce bacterial cell membrane depolarization in the same VanA vancomycin-resistant E. faecium (ATCC BAA-2317) as measured by fluorescence of a released membrane imbedded dye (DiSC35,3,3′-dipropylthiadicarbocyanine iodide) (SI Appendix, Fig. S4) (56). Because the C1 introduction is such a small and seemingly benign structural modification and because it induces membrane permeability without membrane depolarization or cell wall lysis, it suggests a more specific mechanism of action. The mechanism by which 15 and 18 exert their effects on membrane permeability is currently under investigation.
Conclusions
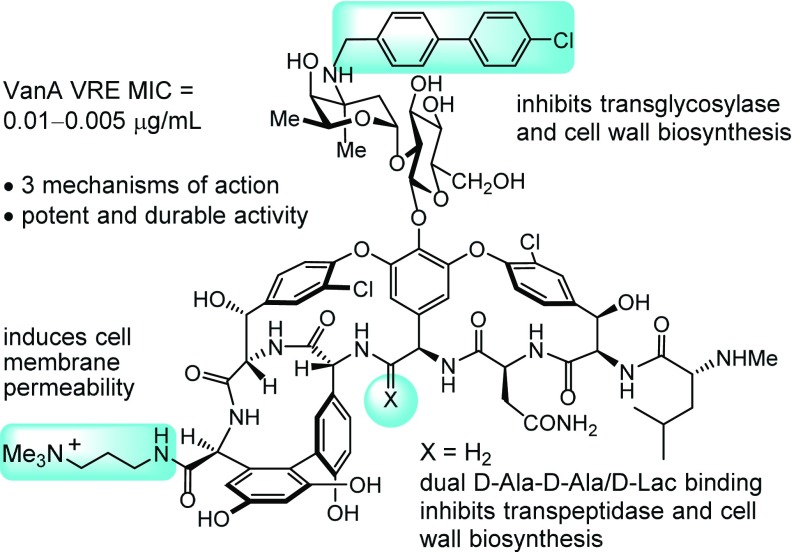
Several programs have disclosed the development of antibiotic products that act by two mechanisms of action. These efforts have included the optimization of a single pharmacophore to independently bind two related targets (e.g., fluoroquinolones targeting both bacterial DNA gyrase and topoisomerase IV); the design of hybrids of two antimicrobial pharmacophores, including the covalent linkage of two antibiotics; the use of combinations of single-target antimicrobials to overcome or avoid resistance (e.g., combination drug treatment of multidrug-resistant TB); and the design of antibiotics that display additional drug target binding contacts to enhance the robustness of target engagement and decrease resistance susceptibility (57). Herein, we described a complementary approach, perhaps a subset of one of these approaches, to design durable antibiotics endowed with multiple synergistic mechanisms of action. This effort has provided prototype antibiotics with three independent mechanisms of action targeting VRE, for which vancomycin is ineffective. Because VRE is already vancomycin-resistant and because many have already reached a point where they are no longer susceptible to most other antibiotic classes, the CDC recently placed VRE on its serious threat list, and the WHO placed it fourth on its list of drug-resistant bacteria that pose the greatest threat to human health. The glycopeptide antibiotics constitute an antibiotic class already endowed with features that avoid many mechanisms of resistance (14). With an understanding of the molecular basis of bacterial resistance to the glycopeptide antibiotics, binding pocket modifications designed for dual ligand binding reinstated binding to the altered target d-Ala-d-Lac and maintained binding affinity for the unaltered target d-Ala-d-Ala. These modifications were found to reinstate antimicrobial activity against vancomycin-resistant organisms that use the altered d-Ala-d-Lac peptidoglycan precursor targets and remain active against vancomycin-sensitive bacteria that use only d-Ala-d-Ala precursors. There is reason to expect that these solutions to VanA and VanB VRE resistance alone may provide antibiotics with durable clinical lifetimes, perhaps approaching those of vancomycin itself (>50 y). Subsequent to these studies, we have explored the peripheral structural changes in the molecules that provide them with additional and now multiple synergistic mechanisms of action. Complementary to our initial disclosure with a carbohydrate CBP modification that produced a 100-fold increase in antimicrobial activity (48), a second peripheral modification at the C terminus of the pocket-modified analogs was detailed herein that enhances antimicrobial activity (200-fold) against VanA VRE by another additional mechanism of action (induced membrane permeability). These two peripheral modifications and their synergistic mechanisms of action were then combined with the pocket modification to provide a vancomycin analog endowed with three independent mechanisms of action, only one of which is dependent on d-Ala-d-Ala/d-Ala-d-Lac binding. This combination not only further increased the antimicrobial potency against VanA VRE (>6,000-fold) but also, reduced the susceptibility to resistance. Thus, the durability of the antimicrobial activity in a resistance challenge and the robustness of each individual mechanism of action as well as the compound potency were shown to follow now predictable trends (three > two > one mechanisms of action). Most striking, resistance to the frontline antibiotics daptomycin, linezolid, and tigecycline, some of which are regarded as durable by today’s standards, was found to arise much faster and more pronounced in the same resistance challenge, highlighting the exceptional durability of the antimicrobial activity detailed for 8, 13, and especially 18 (Fig. 11).
Fig. 11.
Structure of 18, summary of activity, and mechanisms of action.
An important question that these results raise is presently what to do with conventional semisynthetic vancomycin analogs active against VanA VRE that incorporate a single peripheral modification and act by a single mechanism of action that is independent of d-Ala-d-Lac binding [e.g., CBP–vancomycin (5), oritavancin, and C14–vancomycin (12)]. Should their use be encouraged for VRE but at the risk of raising resistance to this otherwise effective approach for other challenging bacterial infections (e.g., MRSA)? The answer would seem to be to encourage their use for challenging vancomycin-sensitive bacterial infections (e.g., VSSA and MRSA), where they are not only more potent than vancomycin but also benefit from two independent mechanisms of action. Clinical resistance or loss in sensitivity to either mechanism would likely be slow to emerge and slower than for vancomycin itself. However, their use against vancomycin-resistant bacteria (e.g., VRE and VRSA), where they are less potent and where only a single and less durable mechanism of action remains operative, likely would more rapidly raise resistance, not only compromising its future use but also, potentially transferring that resistance to other organisms (e.g., MRSA).
The approach used herein, which we suggest represents a case of durable antibiotic discovery by design, relied on the total synthesis of the candidate antibiotics (58–60) to obtain the previously inaccessible compounds. Although not highlighted in the preceding discussion, the total synthesis of the starting pocket-modified aglycon(s) (26 steps) (48), enzymatic installation of the disaccharide (2 steps) (58), and subsequent addition of the two peripheral modifications (2 steps) represent remarkable accomplishments in their own right. Finally, the work herein was conducted with the aminomethylene analog of vancomycin, in which the residue 4 amide carbonyl was removed. A more potent pocket-modified vancomycin analog is the residue 4 amidine (3 vs. 4), which exhibits antimicrobial activity against both vancomycin-resistant and -sensitive bacteria equipotent with the activity that vancomycin displays against vancomycin-sensitive bacteria. Incorporation of such peripheral changes on 3 or 7, providing all three independent mechanisms of action, would be expected to further improve on the already stunning potency of 18 (ca. 30-fold) while displaying the outstanding durability of 18.
Materials and Methods
Synthesis of Vancomycin Analogs.
Full details of the synthesis, purification, and characterization of all compounds reported herein, including copies of the 1H NMR of all tested compounds, are provided in SI Appendix. All reagents were obtained from commercial sources unless noted otherwise.
Bacterial Cell Growth Inhibition Assays.
Full details of the bacterial cell growth inhibition assays with vancomycin-resistant E. faecalis (VanA VRE; BM4166), E. faecium (VanA VRE; ATCC BAA-2317), and vancomycin-resistant E. faecalis (VanB VRE; strain ATCC 51299) are provided in SI Appendix. Compounds were tested in duplicate (n = 2−18 times) at serially diluted concentrations, and the MIC values reported represent the average of 4−36 determinations.
Cell Wall Biosynthesis Inhibition Assay.
Full details of the cell wall biosynthesis assay are provided in SI Appendix. Values reported in Fig. 6 are the average of four measurements (SD ± 10%).
Bacterial Cell Wall Permeability Assay.
Full details of the permeability assay are provided in SI Appendix. Results shown in Figs. 7–9 are one of two to four replicate experiments, each performed at the same time.
Resistance Development Study.
Full details of the study are provided in SI Appendix. Results shown in Fig. 10 are one of two replicate experiments.
SI Appendix.
Full experimental details and copies of 1H NMR spectra are provided. The supplementary data associated with this article can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. A. Mehta [The Scripps Research Institute (TSRI)] for guidance on conducting the cell wall biosynthesis assay and the resistance development assay, Dr. G. Morgan (TSRI) for guidance on the membrane permeability assay, I. Ahmad (TSRI) for guidance on the membrane depolarization assay, Dr. A. Craney (TSRI) for bacterial culturing guidance, and A. Radakovic for conducting the cell growth inhibition assays. We acknowledge financial support from NIH Postdoctoral Fellowship F32 GM114948 (to N.A.I.) and NIH Grant CA041101 (to D.L.B.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 6656.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704125114/-/DCSupplemental.
References
- 1.WHO . Antimicrobial Resistance. Global Report on Surveillance 2014. WHO; Geneva: 2014. [Google Scholar]
- 2.Centers for Disease Control and Prevention . Antibiotic Resistance Threats in the United States. Centers for Disease Control and Prevention; Atlanta: 2013. [Google Scholar]
- 3.Laxminarayan R. Antibiotic resistance: The unfolding crisis. In: Laxminarayan R, Malani A, Howard D, Smith DL, editors. Extending the Cure, Policy Responses to the Growing Treat of Antibiotic Resistance. Resources for the Future; Washington, DC: 2007. pp. 25–37. [Google Scholar]
- 4.Mullard A. Momentum builds around new antibiotic business models. Nat Rev Drug Discov. 2014;13:711–713. doi: 10.1038/nrd4455. [DOI] [PubMed] [Google Scholar]
- 5.Sinha MS, Kesselheim AS. Regulatory incentives for antibiotic drug development: A review of recent proposals. Bioorg Med Chem. 2016;24:6446–6451. doi: 10.1016/j.bmc.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 6.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 7.Wright GD. Solving the antibiotic crisis. ACS Infect Dis. 2015;1:80–84. doi: 10.1021/id500052s. [DOI] [PubMed] [Google Scholar]
- 8.Laxminarayan R. Antibiotic effectiveness: Balancing conservation against innovation. Science. 2014;345:1299–1301. doi: 10.1126/science.1254163. [DOI] [PubMed] [Google Scholar]
- 9.Hamad B. The antibiotics market. Nat Rev Drug Discov. 2010;9:675–676. doi: 10.1038/nrd3267. [DOI] [PubMed] [Google Scholar]
- 10.Wright GD, Poinar H. Antibiotic resistance is ancient: Implications for drug discovery. Trends Microbiol. 2012;20:157–159. doi: 10.1016/j.tim.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 11.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan R, editor. Glycopeptide Antibiotics. Marcel Dekker; New York: 1994. [Google Scholar]
- 13.Kahne D, Leimkuhler C, Lu W, Walsh C. Glycopeptide and lipoglycopeptide antibiotics. Chem Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 14.James RC, Pierce JG, Okano A, Xie J, Boger DL. Redesign of glycopeptide antibiotics: Back to the future. ACS Chem Biol. 2012;7:797–804. doi: 10.1021/cb300007j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick MH, McGuire JM, Pittenger GE, Pittenger RC, Stark WM. Vancomycin, a new antibiotic. I. Chemical and biologic properties. Antibiot Annu. 1955–1956;3:606–611. [PubMed] [Google Scholar]
- 16.Parenti F, Beretta G, Berti M, Arioli V. Teichomycins, new antibiotics from Actinoplanes teichomyceticus Nov. Sp. I. Description of the producer strain, fermentation studies and biological properties. J Antibiot (Tokyo) 1978;31:276–283. doi: 10.7164/antibiotics.31.276. [DOI] [PubMed] [Google Scholar]
- 17.Markham A. Oritavancin: First global approval. Drugs. 2014;74:1823–1828. doi: 10.1007/s40265-014-0295-4. [DOI] [PubMed] [Google Scholar]
- 18.Anderson VR, Keating GM. Dalbavancin. Drugs. 2008;68:639–648. doi: 10.2165/00003495-200868050-00006. [DOI] [PubMed] [Google Scholar]
- 19.Corey GR, Stryjewski ME, Weyenberg W, Yasothan U, Kirkpatrick P. Telavancin. Nat Rev Drug Discov. 2009;8:929–930. doi: 10.1038/nrd3051. [DOI] [PubMed] [Google Scholar]
- 20.Zhanel GG, et al. New lipoglycopeptides: A comparative review of dalbavancin, oritavancin and telavancin. Drugs. 2010;70:859–886. doi: 10.2165/11534440-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 21.Harris CM, Kopecka H, Harris TM. Vancomycin: Structure and transformation to CDP-1. J Am Chem Soc. 1983;105:6915–6922. [Google Scholar]
- 22.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 23.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 24.Brickner SJ, Barbachyn MR, Hutchinson DK, Manninen PR. Linezolid (ZYVOX), the first member of a completely new class of antibacterial agents for treatment of serious gram-positive infections. J Med Chem. 2008;51:1981–1990. doi: 10.1021/jm800038g. [DOI] [PubMed] [Google Scholar]
- 25.Baltz RH, Miao V, Wrigley SK. Natural products to drugs: Daptomycin and related lipopeptide antibiotics. Nat Prod Rep. 2005;22:717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- 26.Arias CA, Murray BE. The rise of the Enterococcus: Beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Centers for Disease Control and Prevention (2017) Available at https://www.cdc.gov/drugresistance/biggest_threats.html. Accessed May 7, 2017.
- 28.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 29.Perkins HR. Vancomycin and related antibiotics. Pharmacol Ther. 1982;16:181–197. doi: 10.1016/0163-7258(82)90053-5. [DOI] [PubMed] [Google Scholar]
- 30.Allen NE, Hobbs JN, Jr, Nicas TI. Inhibition of peptidoglycan biosynthesis in vancomycin-susceptible and -resistant bacteria by a semisynthetic glycopeptide antibiotic. Antimicrob Agents Chemother. 1996;40:2356–2362. doi: 10.1128/aac.40.10.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldman RC, Baizman ER, Longley CB, Branstrom AA. Chlorobiphenyl-desleucyl-vancomycin inhibits the transglycosylation process required for peptidoglycan synthesis in bacteria in the absence of dipeptide binding. FEMS Microbiol Lett. 2000;183:209–214. doi: 10.1111/j.1574-6968.2000.tb08959.x. [DOI] [PubMed] [Google Scholar]
- 32.Ge M, et al. Vancomycin derivatives that inhibit peptidoglycan biosynthesis without binding D-Ala-D-Ala. Science. 1999;284:507–511. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, et al. Vancomycin analogues active against vanA-resistant strains inhibit bacterial transglycosylase without binding substrate. Proc Natl Acad Sci USA. 2003;100:5658–5663. doi: 10.1073/pnas.0931492100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016;537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright GD. Molecular mechanisms of antibiotic resistance. Chem Commun (Camb) 2011;47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- 36.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 37.Marshall CG, Lessard IA, Park I, Wright GD. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob Agents Chemother. 1998;42:2215–2220. doi: 10.1128/aac.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 39.Hong HJ, Hutchings MI, Buttner MJ. Biotechnology and Biological Sciences Research Council, UK Vancomycin resistance VanS/VanR two-component systems. Adv Exp Med Biol. 2008;631:200–213. doi: 10.1007/978-0-387-78885-2_14. [DOI] [PubMed] [Google Scholar]
- 40.Bugg TDH, et al. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: Biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 41.Walsh CT. Vancomycin resistance: Decoding the molecular logic. Science. 1993;261:308–309. doi: 10.1126/science.8392747. [DOI] [PubMed] [Google Scholar]
- 42.McComas CC, Crowley BM, Boger DL. Partitioning the loss in vancomycin binding affinity for D-Ala-D-Lac into lost H-bond and repulsive lone pair contributions. J Am Chem Soc. 2003;125:9314–9315. doi: 10.1021/ja035901x. [DOI] [PubMed] [Google Scholar]
- 43.Crowley BM, Boger DL. Total synthesis and evaluation of [Ψ[CH2NH]Tpg4]vancomycin aglycon: Reengineering vancomycin for dual D-Ala-D-Ala and D-Ala-D-Lac binding. J Am Chem Soc. 2006;128:2885–2892. doi: 10.1021/ja0572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie J, Pierce JG, James RC, Okano A, Boger DL. A redesigned vancomycin engineered for dual D-Ala-D-ala And D-Ala-D-Lac binding exhibits potent antimicrobial activity against vancomycin-resistant bacteria. J Am Chem Soc. 2011;133:13946–13949. doi: 10.1021/ja207142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie J, et al. Total synthesis of [Ψ[C(═S)NH]Tpg4]vancomycin aglycon, [Ψ[C(═NH)NH]Tpg4]vancomycin aglycon, and related key compounds: Reengineering vancomycin for dual D-Ala-D-Ala and D-Ala-D-Lac binding. J Am Chem Soc. 2012;134:1284–1297. doi: 10.1021/ja209937s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okano A, James RC, Pierce JG, Xie J, Boger DL. Silver(I)-promoted conversion of thioamides to amidines: Divergent synthesis of a key series of vancomycin aglycon residue 4 amidines that clarify binding behavior to model ligands. J Am Chem Soc. 2012;134:8790–8793. doi: 10.1021/ja302808p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okano A, Nakayama A, Schammel AW, Boger DL. Total synthesis of [Ψ[C(═NH)NH]Tpg(4)]vancomycin and its (4-chlorobiphenyl)methyl derivative: Impact of peripheral modifications on vancomycin analogues redesigned for dual D-Ala-D-Ala and D-Ala-D-Lac binding. J Am Chem Soc. 2014;136:13522–13525. doi: 10.1021/ja507009a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okano A, et al. Total syntheses and initial evaluation of [Ψ[C(═S)NH]Tpg4]vancomycin, [Ψ[C(═NH)NH]Tpg4]vancomycin, [Ψ[CH2NH]Tpg4]vancomycin, and their (4-chlorobiphenyl)methyl derivatives: Synergistic binding pocket and peripheral modifications for the glycopeptide antibiotics. J Am Chem Soc. 2015;137:3693–3704. doi: 10.1021/jacs.5b01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boger DL. Vancomycin, teicoplanin, and ramoplanin: Synthetic and mechanistic studies. Med Res Rev. 2001;21:356–381. doi: 10.1002/med.1014. [DOI] [PubMed] [Google Scholar]
- 50.Okano A, Isley NA, Boger DL. Total syntheses of vancomycin related glycopeptide antibiotics and key analogues. Chem Rev. April 24, 2017 doi: 10.1021/acs.chemrev.6b00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yarlagadda V, Akkapeddi P, Manjunath GB, Haldar J. Membrane active vancomycin analogues: A strategy to combat bacterial resistance. J Med Chem. 2014;57:4558–4568. doi: 10.1021/jm500270w. [DOI] [PubMed] [Google Scholar]
- 52.Higgins DL, et al. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49:1127–1134. doi: 10.1128/AAC.49.3.1127-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundram UN, Griffin JH. General and efficient method for the solution- and solid-phase synthesis of vancomycin carboxamide derivatives. J Org Chem. 1995;60:1102–1103. [Google Scholar]
- 54.Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R. LIVE/DEAD BacLight: Application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods. 1999;37:77–86. doi: 10.1016/s0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 55.Allen NE, Nicas TI. Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiol Rev. 2003;26:511–532. doi: 10.1111/j.1574-6976.2003.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 56.Te Winkel JD, Gray DA, Seistrup KH, Hamoen LW, Strahl H. Analysis of antimicrobial-triggered membrane depolarization using voltage sensitive dyes. Front Cell Develop Biol. 2016;4:29. doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Discov. 2007;6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 58.Nakayama A, et al. Enzymatic glycosylation of vancomycin aglycon: Completion of a total synthesis of vancomycin and N- and C-terminus substituent effects of the aglycon substrate. Org Lett. 2014;16:3572–3575. doi: 10.1021/ol501568t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boger DL, et al. Total synthesis of the vancomycin aglycon. J Am Chem Soc. 1999;121:10004–10011. [Google Scholar]
- 60.Walker S, et al. Chemistry and biology of ramoplanin: A lipoglycodepsipeptide with potent antibiotic activity. Chem Rev. 2005;105:449–476. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.