Significance
The emergence of novel infectious diseases is often associated with cross-species virus transmission. Virus host range is owed, in large part, to genetic variation of its envelope gene. The illustrative example presented here is based on a structural study of the Rous sarcoma virus (RSV) avian retrovirus passaged twice through rodents. RSV heterotransmission was enabled by spontaneous envelope gene mutations, which allow virus entry even into human and other receptor-free cells. Through three independent biochemical assays, we established that these mutations caused conformational change similar to the one that follows virus–receptor interaction. The newly identified envelope mutations responsible for the broad retrovirus host range contribute to our understanding of retroviruses’ pathogenicity and their ability to cross the transclass barriers.
Keywords: Rous sarcoma virus, retrovirus, virus entry, envelope glycoprotein, receptor-independent entry
Abstract
The extent of virus transmission among individuals and species is generally determined by the presence of specific membrane-embedded virus receptors required for virus entry. Interaction of the viral envelope glycoprotein (Env) with a specific cellular receptor is the first and crucial step in determining host specificity. Using a well-established retroviral model—avian Rous sarcoma virus (RSV)—we analyzed changes in an RSV variant that had repeatedly been able to infect rodents. By envelope gene (env) sequencing, we identified eight mutations that do not match the already described mutations influencing the host range. Two of these mutations—one at the beginning (D32G) of the surface Env subunit (SU) and the other at the end of the fusion peptide region (L378S)—were found to be of critical importance, ensuring transmission to rodent, human, and chicken cells lacking the appropriate receptor. Furthermore, we carried out assays to examine the virus entry mechanism and concluded that these two mutations cause conformational changes in the Env variant and that these changes lead to an activated, or primed, state of Env (normally induced after Env interaction with the receptor). In summary, our results indicate that retroviral host range extension is caused by spontaneous Env activation, which circumvents the need for original cell receptor. This activation is, in turn, caused by mutations in various env regions.
The emergence of novel infectious diseases is often associated with cross-species virus transmission. For example, wildlife viruses have caused such severe human diseases as AIDS, Ebola, severe acute respiratory syndrome (SARS), and influenza. However, the genetic mechanisms determining how viruses cross the species boundary and adapt to new hosts have not been properly understood (1). The ability of a virus to enter the host cell is the first and crucial step in determining host specificity. Viruses causing the above-mentioned diseases possess envelope class I fusion glycoproteins, and their entry into the host cell shares very similar features. There may be a yet unidentified universal mechanism of cross-species transmission.
For decades, avian retrovirus Rous sarcoma virus (RSV) has been a driving force in efforts to understand acutely transforming retroviruses. The establishment of proper cell culture conditions and an in vitro assay of RSV transforming activity have led to a generally accepted description of retrovirus entry, replication, composition, and genetics (2).
RSV belongs to avian sarcoma and leukosis viruses (ASLVs), which are part of the alpharetrovirus genus. RSV and the other ASLVs naturally infect only avian species; however, experimental RSV infection was achieved in hamsters and rats. Studies of RSV transforming activity in mammalian cells enabled the discovery of the tight association that exists between the viral genome and the genome of the transformed cell and corresponds to the RSV provirus state (3) (reviewed in ref. 4). Furthermore, mammalian RSV-transformed cells turned out to be virogenic, which means that they contain the virus genome but do not produce infectious viruses. The nonpermissiveness of mammalian cells can be overcome by cell fusion with permissive chicken cells that provide the necessary cell factors and thus rescue virus production ability (5, 6).
Despite the significant contribution of RSV-transformed mammalian cells to our understanding of RSV life cycle, it is yet to be explained how mammalian cells were directly infected by avian retroviruses, despite lacking any of the known cell receptors required for ASLV cell infection.
ASLV cellular receptors have been well described; they are denoted as tumor virus (Tv) loci and are highly specific for individual virus subgroups (7). The Tva receptor enables the infection of ASLV subgroup A (ASLV-A) (8), Tvc is the receptor for ASLV-C (9), Tvj is the receptor for ASLV-J (10), and various Tvb alleles confer sensitivity to the B, D, and E subgroups (11, 12).
To understand how RSV entered mammalian cells, we had to focus on the structure of the viral Env, which enables the virus to enter the cell (13) and whose alterations are responsible for changes in the virus host range. ASLV mutants with extended host ranges, including mammalian tropism, were observed in experiments aimed at overcoming avian subgroup barriers (14, 15) or at characterizing viruses that had escaped a block produced by HR2-based inhibitor treatment in chicken cells (16). Mutations were found in the host range region 1 (hr1) of the SU Env subunit or in the heptad repeat region 1 (HR1) of the transmembrane (TM) Env subunit.
In this paper, we investigate mutations in the Prague RSV subgroup C (PR-RSV-C) env gene passaged twice through rodents (H20-RSV). We have identified a series of env mutations that do not match the already described mutations extending the host range. Two of these, in particular a mutation located in the fusogenic peptide, facilitate RSV entry into mammalian cells. We show that the envelope glycoprotein of H20-RSV (EnvH20) has changed its conformation, contains reactive thiolate, and is able to bind liposomes even in the absence of receptor priming. We therefore propose that mutations found in the H20-RSV env are responsible for the Env activation that normally follows receptor binding and that they endow the envelope glycoprotein with a conformation that allows the H20-RSV virus to infect cells in the absence of an appropriate receptor.
Results
Mutations in the Mammalian-Passaged RSV env Enable Virus Entry into Hamster Cells.
Because the retroviral Env plays a decisive role in virus entry, we decided to assess the env gene structure of PR-RSV-C passaged twice through rodents and to compare its composition with PR-RSV-C passaged in chicken only. The first rodent passage was done in rats, generating the XC-RSV provirus, whereas the second passage was done in hamsters, yielding the H20-RSV provirus (Fig. 1A).
Fig. 1.
Virus transmission to mammalian cells and envelope glycoprotein alteration. (A) Schema of rat and hamster infection with original PR-RSV-C virus. PR-RSV-C virus was injected into a chicken, the resulting tumor was minced and injected into a rat, which developed a tumor caused by XC-RSV. The virus rescued from XC cells was again multiplied in a chicken and injected into a Syrian hamster, where H20 tumor harboring H20-RSV was obtained (for details see Materials and Methods). (B) Diagram of the envelope glycoprotein domain structure with positions of described mutations responsible for mammalian tropism (black arrows) (14, 16) and found amino acid mutations (red arrows). Host-range regions (hr1/2) and variable regions (vr1/2/3) in the surface subunit (SU) are depicted in gray boxes. The fusion peptide (FP), heptad repeats (HR1/2), and membrane spanning domain (MSD) in the transmembrane subunit (TM) are shown in white boxes. (C) Comparison of gp85 amino acid sequences from original parental virus PR-RSV-C (sequence from GenBank V01197.1) (17) and viruses rescued from RSV-transformed rat (XC) or hamster (H20) cells. h and vr regions in SU are depicted in gray boxes. FP, HR1, HR2, and MSD in TM are shown in white boxes. Their position is depicted according to refs. 18, 19.
To our surprise, the eight amino acid substitutions found in the rodent-passaged H20-RSV did not match the already described mutations responsible for the ASLV extended host range. The latter mutations were observed in the hr1 region of the SU Env subunit (14, 15) and in the HR1 region of the TM Env subunit (16) (Fig. 1B). The next question concerned the origin of these env mutations. Using virus rescued from rat XC cells (XC-RSV), we verified that all of the eight mutations were already present before the second mammalian passage (done in hamsters). Using PCR, we synthesized and sequenced proviral env genes in XC cells and showed that the same mutations were present in the original XC proviruses, matching those in the rescued XC-RSV (Fig. 1C). It should be noted that XC cells contain multiple proviruses (20); however, all of the env sequences we obtained uniformly contained the eight mutations. This observation shows that mutations in env were selected already in the first round of mammalian cell infection and that at least some of these mutations might affect RSV entry into mammalian cells.
To compare the ability of viruses to enter mammalian cells, we measured the amounts of the newly made viral DNA synthesized in the NIL hamster cell line and its derivative NIL-Tvc (equipped with the Tvc receptor) at different intervals after infection with PR-RSV-C and H20-RSV. For both viruses, viral DNA was efficiently synthesized in NIL-Tvc (Fig. 2A). After infection of NIL cells with H20-RSV, we detected approximately one log order less of viral DNA. However, a sharp two-log decrease in viral DNA level was observed when NIL cells were exposed to PR-RSV-C (on day 9 after infection, no viral DNA was detected), indicating that H20-RSV was more efficient in triggering viral DNA synthesis than the original PR-RSV-C. In subsequent experiments, we used RSV-based retroviral vectors, denoted replication-competent ASLV long terminal repeat with a splice acceptor (RCAS) (21). The env gene in the RCAS-EnvC-GFP vector originates from PR-RSV-C, and the env gene in the RCAS-EnvH20-GFP vector originates from H20-RSV (Fig. 2B). As shown in Fig. 2 C and E, this substitution enabled the infection of NIL cells and resulted in a three-order-of-magnitude increase in NIL cell infection over the limit of detection (which represents a background signal from uninfected NIL cells in flow cytometry measurements). Similarly, when we compared the ratio of NIL to NIL-Tvc infection, we observed a significant increase in the case of RCAS-EnvH20-GFP (Fig. 2D).
Fig. 2.
Viruses equipped with EnvH20 are able to infect hamster cells. (A) Amount of newly made env viral DNA in infected cells (NIL or NIL-Tvc) was quantified in different time points postinfection. The results were normalized to chGAPDH expression and are presented relative to the sample of NIL-Tvc infected PR-RSV-C on day 1. (B) Schematic diagram of chimeric reporter vector RCAS-EnvH20-GFP prepared from RCAS-EnvC-GFP and H20-RSV virus. Restriction sites and position of primers used for the env gene substitution are depicted. (C) Infection of NIL and NIL-Tvc cells with RCAS-GFP viruses containing EnvC or EnvH20 was scored by flow cytometry 3 d later. Titers in GFP-transducing units were determined as described in SI Materials and Methods. Error bars show the SD of two independent experiments in parallel. The limit of detection (signal from uninfected NIL cells) is marked with a dashed line. Significant differences are marked by asterisks (***P < 0.001). (D) Relative infectivity of NIL cells infected with RCAS-EnvC-GFP and RCAS-EnvH20-GFP. Infectivity is expressed as the percentage of the viral titer on NIL-Tvc cells. (E) Examples of FACS diagrams showing the percentages of GFP+ cells after infection of NIL cells with RCAS-EnvC-GFP and RCAS-EnvH20-GFP viruses.
To assess the role of individual mutations, we subdivided the H20-RSV env gene into four parts using suitable restriction sites (Fig. 3A). Individual restriction fragments then replaced the corresponding regions of RCAS-EnvC-GFP vector. Either part 1 (encompassing the SU region upstream of the vr1 variability region and containing the D23G mutation) or part 4 (spanning the TM region and containing the L378S, G464S, and L503V mutations) were sufficient to enable entry into NIL cells, with the efficiency of infection being more than one or two orders of magnitude over the limit of detection (Fig. 3 B and C).
Fig. 3.
Mutations in the first part of SU (D23G) and in the fusion peptide (L378S) are responsible for the virus-extended host range. (A) Schematic diagram showing restriction sites (dashed lines) that were used to divide EnvH20 into four parts, which were tested separately as chimeras with EnvC. Positions of the found amino acid mutations are shown with red arrows. (B and D) Infection of hamster cell lines NIL and NIL-Tvc (B), human cell lines HEK293, RPE1-hTERT, and chicken L15 cell line (Tvc−) (D) with RCAS-GFP viruses harboring EnvC-H20 chimeras and RCAS-EnvC-GFP single mutants L378S, G464S, and G464S was scored by flow cytometry 2 (HEK293) or 3 (NIL, RPE1-hTERT, L15, NIL-Tvc) days later. Titers were determined as described in SI Materials and Methods. The limit of detection (signal from uninfected cells) is marked with a dashed line. (C) Relative infectivity of hamster NIL cells with different RCAS-GFP viruses is expressed as the percentage of the viral titer on NIL-Tvc cells. Error bars show the SD of two independent experiments in parallel. Significant differences are marked by asterisks (*P = 0.05–0.01, **P = 0.01–0.001, ***P < 0.001). NS, not significant.
Because part 4 harbors three mutations, we applied site-directed mutagenesis to introduce these mutations into RCAS-EnvC-GFP (Fig. 3A). The mutated viruses were then tested for NIL infectivity (Fig. 3B), with the data presented as a ratio of NIL/NIL-Tvc infection (Fig. 3C). Unequivocally, the L378S mutation in the fusion peptide played a decisive role in the increased NIL cell infectivity.
In summary, we show that H20-RSV mammalian tropism is caused by mutations in Env, particularly D32G or L378S. Both of these mutations can separately facilitate entry into hamster cells.
D32G and L378S Mutations Enable Virus Entry into Human Cells and Receptor-Deficient Chicken Cells.
We extended our observations to human cells, namely, human embryonic kidney cells (HEK293) and human telomerase reverse transcriptase (hTERT)-immortalized retinal pigment epithelial cells (RPE1-hTERT) (Fig. 3D). Surprisingly, HEK293, in contrast to RPE1-hTERT cells, were significantly more sensitive to RCAS-EnvC-GFP infection than NIL cells. However, the highest infection efficiency was observed, in both human cell lines, with the RCAS chimera harboring the H20 env gene. The increased infectivity was clearly mediated by parts 1 and 4 of the H20-RSV env gene. The importance of the L378S mutation in the fusion peptide was confirmed using the aforementioned RCAS vector with this mutation.
Furthermore, we wanted to know whether our mutations that arose in rodents were similarly efficient in infecting chicken cells lacking the corresponding receptor. For subgroup C, to which PR-RSV-C belongs, we used line 15 (L15) chicken cells, in which the tvc receptor gene was inactivated by a mutation introducing a premature stop codon (9). Although the virus with EnvC could not infect L15 cells, the virus with EnvH20 was able to enter these cells with an efficiency of two orders of magnitude higher over the limit of detection (Fig. 3D). The D32G mutation had a very mild effect (construct EnvC-H20Part1), whereas mutation in the fusion peptide (construct EnvC-H20Part4, L378S) was almost as effective as the entire H20 env gene (Fig. 3D).
H20 Envelope Glycoprotein Has an Altered Conformation Similar to a Receptor-Primed Activated State.
For the RCAS-EnvH20-GFP virus, we observed significant loss of infectivity in permissive cells (NIL-Tvc, DF-1) in comparison with the RCAS-EnvC-GFP virus (Fig. 2C and Fig. S1A). Such a drop in infectivity might have been caused by lower virus production or by decreased infectivity of the virus produced. To distinguish between the two options, we tested reverse transcription activity (Fig. S1B) and virus-like particle (VLP) formation (Fig. S1C), observing a very small difference between the two viruses. This finding indicates that the virus production being comparable, the RCAS-EnvH20-GFP virus particles are less infective than RCAS-EnvC-GFP. Because the two viruses differ only in the env gene, these results suggest that EnvH20 is less stable or less efficient in facilitating virus entry.
Fig. S1.
Virus harboring EnvH20 has a lower titer than the virus with original EnvC despite the same RT activity and VLP formation. (A) Infection of NIL-Tvc and DF-1 cells with RCAS-EnvC-GFP and RCAS-EnvH20-GFP viruses was scored by flow cytometry 2 (DF-1 cell) or 3 (NIL-Tvc) days later. Titers were determined as described in Materials and Methods. The results are presented as titers relative to RCAS-EnvC-GFP. (B) RT activity of RCAS-EnvC and RCAS-EnvH20-GFP was measured using the PERT assay described in SI Materials and Methods. The results of the PERT assay are presented relative to RCAS-EnvC-GFP. (C) VLP production of DF-1 cells infected with RCAS-EnvC-GFP or RCAS-EnvH20-GFP was estimated by Western blot analysis. Expression of Gag product p27 in the medium was determined with anti-p27 antibody.
Next, we applied a treatment to explore the stability and conformation of the RCAS-EnvH20-GFP virus. The first approach, which was originally introduced by Bova-Hill et al. (22), was based on thermostability testing. The infectivity of RCAS-EnvH20-GFP substantially decreased with the time of exposure to 44 °C as measured by both flow cytometry and viral DNA quantification (Fig. 4A).
Fig. 4.
EnvH20 has similar features as a receptor-primed Env. (A) The virus with EnvH20 is thermosensitive. Virus stocks of similar titers were incubated at 44 °C for the indicated time. The titers were determined on NIL-Tvc 3 d after infection using flow cytometry. The amount of proviral DNA in infected DF-1 cells was measured 1 d after infection using RT-PCR. The results are presented as the percentage of the original titer (full lines) or the amount of DNA (dashed lines) remaining after heating. (B) The virus with EnvH20 is inactivated by low pH. Purified viruses of similar titers were incubated with low (pH 5) or neutral (pH 7.5) treatment at 37 °C for 30 min before infection of DF-1 cells. The titers were determined by flow cytometry 2 d later. (C) Formation of TM oligomers triggered by increasing temperature. The virus was incubated for 20 min at the indicated temperature. Samples were lysed in Laemmli loading buffer and analyzed by SDS/PAGE without boiling. TM was detected by immunoblotting with an antibody against its C-terminal part. Only the 70-kDa isoform of TM is shown. (D) Formation of TM oligomers at low pH. The virus was incubated for 30 min at the indicated pH at room temperature. Samples were neutralized, lysed in Laemmli loading buffer, and analyzed by SDS/PAGE without boiling. TM was detected by immunoblotting with an antibody against its C-terminal part. (E) The virus with EnvH20 binds liposomes. Viruses of similar titers were incubated with or without liposomes at 37 °C and then the mixture was separated by sucrose gradient centrifugation. Sample fractions were collected and analyzed by Western immunoblotting using anti-p27 antibody. T, top; B, bottom. (F) The virus containing EnvH20 is inhibited with PMB. Purified viruses of similar titers were incubated at 37 °C for 30 min with increasing concentration of PMB. Viruses were diluted in medium and spinoculated on DF-1 cells. The percentage of GFP+ cells was measured by flow cytometry 2 d after infection. Titers below the limit of detection are marked with “<”. Error bars show the SD of the two independent experiments in parallel. Significant differences are marked by asterisks (*P = 0.05–0.01, **P = 0.01–0.001, ***P < 0.001). NS, not significant.
This observation prompted us to test whether EnvH20 is constitutively activated in a way similar to receptor priming. It has previously been shown that a virus activated by a soluble receptor is very unstable at low pH (23). Therefore, we tested RCAS-EnvH20-GFP and discovered that it behaved similarly to a receptor-primed virus and that it was almost completely inactivated by low-pH treatment, in contrast to RCAS-EnvC-GFP, which remained stable at low pH (Fig. 4B).
Next, we used a biochemical assay to test the oligomeric state of the Env TM subunit. It has been determined that the ASLV Env TM is in a metastable conformation. Env TM from purified virions lysed with buffer containing 1% SDS and 5% β-mercaptoethanol migrate in SDS/PAGE as a monomer (approximately 30 kDa) at both neutral and low pH. After receptor priming, Env is refolded, but at neutral pH the conformation detected in SDS/PAGE is still predominantly monomeric. After low pH treatment of receptor-primed Env, additional rearrangement occurs and TM forms SDS-resistant oligomers, which can be detected by SDS/PAGE and migrate at around 70–150 kDa (23, 24). TM oligomerization can also be triggered by heat treatment, and it has been shown that if the Env is in the receptor-primed state, the temperature for TM oligomerization is lower than for unbound Env (24). We analyzed the temperature threshold that triggers TM oligomerization and observed only a small difference between EnvC and EnvH20. Envs of both viruses formed small amounts of TM oligomers even at low temperature (25 °C); these amounts increased with temperature (Fig. 4C). We then compared the ability of EnvC and EnvH20 to form TM oligomers at neutral pH and low pH at room temperature. In both cases, a certain amount of TM oligomers formed at neutral pH; however, EnvH20 TM oligomerization was clearly stronger following low pH treatment than that of EnvC (Fig. 4D). This observation shows that EnvH20 behaves like an Env in the receptor-primed state and has a conformation that enables the triggering of TM oligomerization by low pH.
Moreover, we showed that RCAS-EnvH20-GFP is able to bind liposomes without receptor priming, in contrast to RCAS-EnvC-GFP (Fig. 4E). It has been described that after being activated with a soluble receptor, ASLV Env changes conformation and the fusion peptide is exposed and can be inserted in liposomes composed of phosphatidylcholine and cholesterol. These virus–liposome complexes can be detected by density gradient centrifugation, in which the liposomes float on top of the gradient and virions migrate to its bottom (25). In the case of RCAS-EnvH20-GFP, we observed some virions (detected by virus protein p27) on the top with liposomes; in contrast, all RCAS-EnvC-GFP virions were detected in the bottom fraction (Fig. 4E), showing that EnvH20 can form virus–liposome complexes and supporting the conclusion that EnvH20 behaves similarly to Env in the receptor-primed state.
Finally, we tested whether infection can be inhibited by the thiol-specific alkylating reagent PEG–maleimide–biotin (PMB); the maleimide group reacts specifically with reduced thiols to form stable thioether bonds. It has been shown that receptor priming induces formation of reactive thiolate in SU, and, consequently, that the virus becomes prone to PMB inhibition when incubated with a soluble receptor (26). We show that RCAS-EnvH20-GFP was inhibited by PMB, in contrast to RCAS-EnvC-GFP, whose infectivity was inhibited only partially at the highest PMB concentration (Fig. 4F). Stronger PMB inhibition indicates that EnvH20 has an exposed thiol as seen in the receptor primed state. In summary, all these experiments show that EnvH20 has similar features as a receptor-primed Env.
The Level of Env Activation Correlates with the Efficiency of Receptor-Independent Entry.
To determine how individual mutations contribute to Env activation, we analyzed the sensitivity to pH inactivation and PMB-induced inhibition of RCAS-GFP viruses harboring EnvC and EnvH20 chimeras. We observed that the SU D32G mutant virus that exhibited less efficient entry into mammalian cells was inactivated at low pH and was inhibited by PMB only partially. The virus with a fusion peptide mutation enabling more efficient entry into mammalian cells exhibited more pronounced PMB inhibition and low pH-induced virus inactivation. The most sensitive virus to both the aforementioned inhibitory assays was RCAS-EnvH20-GFP, which contains the entire EnvH20 with both mutations (Fig. 5 A and B). In both assays, we observed that the level of inactivation and inhibition correlated with the ability to enter mammalian cells (Fig. 5 C and D). As both these assays should reveal the degree of Env activation, we conclude that the level of Env activation correlates with the efficiency of receptor-independent entry.
Fig. 5.
The level of Env activation correlates with the efficiency of receptor-independent entry. (A) Virus inactivation by low pH. Purified viruses harboring EnvC-H20 chimeras were incubated with low (pH 5) or neutral (pH 7.5) treatment at 37 °C for 30 min before infection of DF-1 cells. The titers were determined by flow cytometry 2 d later. (C) PMB inhibition of virus infection. Purified viruses harboring EnvC-H20 chimeras were incubated at 37 °C 30 min with increasing concentration of PMB. The titers were determined on DF-1 cells by flow cytometry 2 d after infection. Titers below the limit of detection are marked with “<”. Error bars show the SD of two independent experiments in parallel. (B and D) Correlation between relative NIL infectivity and virus inactivation at low pH (B) or PMB inhibition of virus infection (expressed as residual infectivity after 1 mM PMB) (D).
Mutation in TM Subunit Facilitates Entry into Mammalian Cells Regardless of SU Subgroup.
Because the L378S mutation in TM of EnvH20 was sufficient for virus entry into mammalian cells, we next examined whether TM-H20 could facilitate mammalian cell infection also with an SU subgroup different from SU-C. We prepared RCAS virus with SU-B combined with EnvH20-TM and analyzed its ability to enter human RPE1-hTERT cells. As shown in Fig. 6A, in contrast to the original virus RCASBP-B-GFP (RCAS with Bryan polymerase, Env subgroup B) the virus with TM-H20 and SU-B (EnvB-H20Part4) was able to infect human cells almost as efficiently as the virus with TM-H20 and SU-C (EnvC-H20Part4). To confirm that the ability to infect human cells was, as shown above with other viruses, coupled with Env activation, we tested the sensitivity of EnvB-containing viruses to PMB inhibition. In contrast to RCASBP-B-GFP, the virus with EnvB-H20Part4 was significantly inhibited by PMB, to almost the same extent as the virus with EnvC-H20Part4 (Fig. 6B).
Fig. 6.
Mutation in TM facilitates entry into mammalian cells independent of SU. (A) Relative infectivity of human RPE1-hTERT cells with RCAS-EnvC-GFP, RCASBP-B-GFP, and RCAS-GFP viruses bearing chimeric Envs (SU-C or SU-B with TM from EnvH20) is expressed as the percentage of the viral titer on DF-1 cells. The percentage of GFP+ cells was scored by flow cytometry 2 (DF-1) or 3 (RPE1-hTERT) days after infection. (B) PMB inhibition of virus infection. Purified viruses were incubated at 37 °C for 30 min with increasing concentration of PMB. The titers were determined on DF-1 cells by flow cytometry 2 d after infection. Titers below the limit of detection are marked with “<”. Error bars show the SD of two independent experiments in parallel. Significant differences are marked by asterisks (**P = 0.01–0.001, ***P < 0.001). NS, not significant.
These results show that the TM mutation found in EnvH20 causes Env activation and enables mammalian cell infection even when combined with SU-B. This supports our hypothesis that the extended host range is due to Env activation and not to a new mammalian receptor recognition.
Discussion
To elucidate the mechanisms enabling cross-species virus transmission, we analyzed genetic changes in the RSV avian retrovirus variant that repeatedly infected and transformed mammalian cells. We thus followed the strategy of original virus reconstruction after repeated passaging across a class barrier. According to the classical definition, we dealt with a post hoc ergo propter hoc situation.
It is generally known that the retroviral Env is responsible for virus entry into the cell, enabling, as the first step, its interaction with a specific cell surface receptor (13). We therefore analyzed the env gene structure and identified a total of eight amino acid substitutions in rodent-passaged RSV (H20-RSV). We found the same changes in XC cell-harbored proviruses representing the first mammalian passage. The same pattern of amino acid substitutions remained during the second cycle of rodent cell infection; therefore, we assumed that the amino acid changes identified played a role in the virus ability to infect receptor-deficient cells.
The amino acid substitutions identified in EnvH20 did not match the previously described ASLV mammalian-tropic mutants (14–16, 27, 28), reviewed by Coffin (29). We therefore proceeded to assess the contribution of each individual substitution by testing its effect in the NIL hamster cell line infection. Two of them were found to be of decisive importance, namely, a substitution (D32G) upstream of the vr1 region and another substitution (L378S) at the end of the fusion peptide (Fig. 1). In general, two distant env mutations can exert a joint effect in some env recombinants (30). We found that a greater role was played by the amino acid substitution in the fusion peptide; however, cell lines (especially human) varied as to their sensitivity. For example, the sensitivity of RPE1-hTERT cells followed the pattern observed in NIL, with the following two differences: (i) viruses with mutations enabling an extended host range were even more efficient (the percentage of infected cells was more than one order of magnitude higher than in NIL cells), and (ii) both amino acid substitutions (D32G and L378S) contributed to the RPE1-hTERT infection almost equally. HEK293 cells were found to be sensitive even to the original RCAS-EnvC-GFP (env gene from PR-RSV-C), in contrast to their background-level sensitivity to PR-RSV-B (14). The increased sensitivity of HEK293 cells to subgroup C viruses is reflected by an increased background level in the virus entry efficiency assays. However, the virus with EnvH20 as well as the virus with the two aforementioned mutations retained the highest infection efficiency for HEK293 cells. It should be noted that there are significant differences in sensitivity of individual human cell lines to HIV infection, which have not been fully explained so far (reviewed in ref. 31). Some of these differences can be explained by low-affinity Env interactions with adhesion molecules such as heparin sulfate proteoglycans. These interactions alone do not trigger Env conformational changes required for entry; however, they are important for virus attachment and significantly increase infectivity (32). We can only speculate about the role of the cell membrane lipid bilayer variation among cell lines, but it is already known that various cell membrane components are cooperative in their interaction with viral Gag (33). These interactions might be a source of variation of cell line susceptibility and might play a role in ASLV Env interaction.
ASLV entry has been studied in detail and it has been shown that ASLV Env uses a two-step fusion activation mechanism. Persuasive evidence supports the view that the ASLV Env, specifically its SU part, interacts with a cell receptor at neutral pH, resulting in Env conformational changes, heptad repeat exposure, and fusion peptide insertion in the cell membrane (13, 34–36), reviewed in ref. 37. It has previously been determined that the formation of reactive thiolate on Cys-38 is another essential consequence of receptor priming (26). The second step is dependent on acidic pH (23, 38, 39), which initiates further conformational alterations, six helix bundle (6HB) formation, and permits fusion between the virus and the cell membrane (39).
The existence of a stable receptor-primed intermediate form of ASLV Env is supported by several lines of evidence. For example, ASLV remains stable for many hours in cells that are transiently blocked with lysosomotropic agents increasing endosomal pH (23, 40). Another example is ASLV inhibition with a heptad repeat peptide (called R99), which can block virus infection when added at the cell surface (38, 41). Env conformation is probably paused in a pre-6HB conformation, following the insertion of the fusion peptide into the target membrane and before acidic pH triggering.
Particular events occurring after receptor priming have been described by assays based on the incubation of the virus with a soluble receptor. The receptor-primed virus becomes highly unstable at low pH (23) and its conformation changes. Consequently, the virus can now bind liposomes via the fusion peptide insertion (25, 34) and Cys-38 reactive thiolate is formed (26). Subsequent low pH triggering of the receptor-primed ASLV Env results in the formation of SDS-resistant 6HB oligomers (24). Moreover, it has been shown that the virus primed with a soluble receptor was able to enter receptor-deficient cells (42).
It was tempting to speculate that the virus mutants that were able to enter receptor-deficient mammalian cells had Env glycoprotein conformational changes normally triggered by the Env–receptor interaction. The study of the thermostability of PR-RSV-C and mammalian-tropic Schmidt-Ruppin RSV strain (SR-RSV-D) (22) gave us the first hint. In line with our results, the virus harboring PR-RSV-C Env (EnvC) was thermostable, but the infectivity of the virus with SR-RSV-D Env decreased with time of exposure to 44 °C. In contrast to EnvC, our mutant with EnvH20 behaved like the thermosensitive SR-RSV-D. Next, we tested whether EnvH20 had similar characteristics to a receptor-primed Env. We showed that the virus with EnvH20, in contrast to the virus with EnvC, can be inactivated by low pH and is efficiently inhibited with PMB, which blocks reactive thiolates. Furthermore, EnvH20, in contrast to EnvC, is able to bind liposomes, and its TM subunit oligomerization is triggered by low-pH treatment (Fig. 4). All these observations show that EnvH20 behaves as a receptor-primed Env.
To identify the contribution of particular mutations to Env activation, we tested how efficiently EnvC-H20 chimeras are inactivated by low pH and inhibited by PMB. We determined that the greater their efficiency in mammalian cell infection, the greater their sensitivity to low pH and PMB inhibition (Fig. 5). Therefore, the level of Env activation correlates with the efficiency of receptor-independent entry. Conformational transition from an unbound state of Env to an active (primed) state requires energy to overcome an activation barrier (43). We hypothesize that Env-activating mutations decrease this activation barrier, so the Env can be activated spontaneously, which would explain why we detect only a certain proportion of activated Env and that its amount is dependent on the particular mutations.
Other mammalian-tropic ASLV mutations have been found in hr1 in SU and HR1 in TM. These mutants with extended host range were originally discovered in experiments aimed at overcoming avian subgroup barriers (14, 15) or at characterizing viruses that had escaped a block produced by the R99 peptide (HR2-based inhibitor) treatment (16); their ability to infect mammalian cells was observed as a side effect.
Alterations in the fusion peptide have also been observed in other ASLV mutants. A study (44) selected for ASLV with a decreased pH threshold of membrane fusion activation, revealing that mutations in hr1 and in the fusion peptide region influence the threshold. Its authors suggest that the increased acid requirement exhibited by mutants may be due to their increased stability. In contrast, our study indicates that mutations in the fusion peptide can also destabilize Env. Another mutation in the fusion peptide was identified after selecting for improved replication capacity of an ASLV mutant that lacked critical cysteine residues flanking the fusion peptide. The mutant Env was unable to form stable TM oligomers, and a positively charged mutation in the middle of its fusion peptide partially restored this ability (45).
Together with our present data, the mammalian-tropic ASLV mutations reported so far span the entire env region and target different Env domains (upstream from vr1, hr1, fusion peptide, and HR1). In contrast, ASLV interacts with the corresponding avian receptors mainly through two host range (hr) regions (46). The extent of the env mutations contributing to mammalian tropism does not support the involvement of a specific unknown receptor. This is also corroborated by our finding that the mutations residing in H20-TM can facilitate human cell entry mediated by SU-B, which recognizes an avian receptor different from the one recognized by SU-C (Fig. 6).
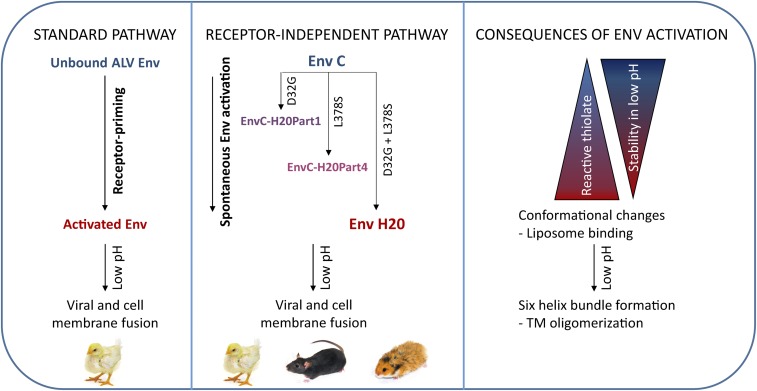
Taken together, we detected two mutations (D32G and L378S) in different regions of Env, which contribute to its activation (Fig. 7). We propose that these mutations change the Env conformation, in such a way that the Env activation barrier is decreased and Env can be spontaneously activated in a way similar to activation caused by receptor binding. We hypothesize that due to the spontaneous Env activation, low-affinity Env interactions with currently unknown molecules commonly present on the cell surface are sufficient to mediate the virus entry. Because the virus circumvents the need for Tvc or another cognate receptor required for Env priming, it is also able to enter the cells of various receptor-deficient species.
Fig. 7.
Proposed model of receptor-independent entry. (Left) Viral Env is normally activated by binding with the receptor. Activated Env takes a conformation enabling further changes that occur at low pH in endosomes and lead to viral and cell membrane fusion. (Middle) Mutations D32G and L378S change EnvC conformation and shift Env close to the active state, which normally follows receptor priming. These viruses are therefore able to be spontaneously activated with efficiency, depending on the mutation type. After low pH exposure, activated Env facilitates viral and cell membrane fusion. (Right) Consequences of Env activation can be observed in different steps. The level of Env activation positively correlates with increased formation of reactive thiolate and decreased stability at low pH. After activation, the conformation is changed: Heptad repeats are exposed and the fusion peptide is inserted into the liposome membrane. After low pH treatment, formation of the six-helix bundle can be determined by TM oligomerization assay.
Mutations leading to host range extension have been described in envelope glycoproteins of other retroviruses (mouse mammary tumor virus, Moloney murine leukemia virus) (47, 48). Although these mutations are located in the SU part of Env and the authors hypothesize that they could be associated with a new host receptor binding, we cannot exclude the possibility that such mutations influence the Env activation too. Lentivirus transmission to new species has been studied mainly in relation to host-encoded restriction factors. Nevertheless, the study of HIV Env mutants responsible for the infection of cells lacking the CD4 virus receptor provided evidence that important mutations were not located only in SU subunit, where subsequently a coreceptor binding site was revealed, but also some mutations were located in TM subunit (gp41) (49). Similarly, mutations in gp41 influence the virus infectivity and fusogenic activity (50). Furthermore, it has recently been shown that HIV Env mutations in gp41 associated with CD4-independent infection promote the exposure of the HR1 peptide on the gp41 and increase the overall sensitivity of HIV to impairments such as exposure to cold and the binding of antibodies and small molecules (51). Moreover, the Env conformation of some CD4-independent SIV strains is similar to constitutively “open” conformation of HIV-1 when it is bound to CD4 (52).
Other viruses with class I fusion proteins have exhibited an extended host range following the mutation of their fusion subunits. In the case of coronaviruses, there are several hints that mutations in S2 (analogous to the retrovirus TM subunit) are associated with the ability to infect receptor-deficient cells. Similar mutations identified in the mouse hepatitis virus (MHV)—at the junction between its fusion peptide (called PP3) and HR1—resulted in an expanded host range, which included hamsters (53). In a way similar to our findings, the substitutions at the PP3–HR1 junction can be coupled with substitution at the beginning of SU. Furthermore, it has been shown that the heptad repeat region was a major target of adaptive evolution in MERS-CoV–related viruses (54).
A broad range of extended host range mutants suggests that such mutants may represent a first step in cross-species transmission. On the one hand, the env mutations described enable the virus to infect receptor-deficient cells; on the other hand, they interfere with virus infectivity by destabilizing Env and, probably, by increasing the accessibility of Env to neutralizing antibodies. Additionally, it has been shown that some ASLV extended host range mutants have enhanced cytopathogenicity (27). Therefore, the infection of mammalian cells by RSV represents a convenient system for the detection of the first events in cross-species transmission evolution due to the fact that RSV is not able to replicate in mammalian cells. We suppose that in a permissive system the virus would quickly adapt to new conditions. ASLV evolved to use different avian receptors and it is possible that such mutants as described here serve as intermediates in the virus adaptation for a new host receptor. Hypothetically, these Env-activating mutations enable the initial infection of a hitherto resistant host. Once the virus achieves good conditions for its replication, the Env can adapt for a new high-affinity receptor and the virus returns back to receptor-dependent Env activation.
Apart from mutations in env, we also analyzed other mutation in H20-RSV. We identified a few changes relative to the PR-RSV-C sequence (GenBank V01197.1) in gag (T221P, A1638G, and A2293G), pol (G3645A and G4947A), src (insertion of AGG in position 7248, A7410G, A7977G, G8087A, and A8226G), and LTR (G918A). None of these mutations influenced H20-RSV infectivity, as the virus rescued from H20 cells replicated and transformed chicken cells efficiently. In this analysis, we did not consider substitutions in H20-RSV that lead to synonymous mutations and amino acid substitutions occurring in other ASLV subgroups.
As mentioned above, the RSV mutants entering mammalian cells do not represent “adapted” viruses replicating in host cells and transmissible among individuals, such as the duck-adapted RSV (55). Although the virus is able to enter the cell and integrate the proviral DNA, its replication is hampered in later stages of infection (5), preventing or highly suppressing virus production. For tumor induction, newborn hamsters had to be inoculated again with tumor cell suspension derived from chicken sarcoma induced by the virus rescued from rat XC cells (56). The live cell application could significantly augment and prolong virus infection. Virus-producing chicken cells can also associate with the host mammalian cells, as proposed by Svoboda and Chyle (57), allowing direct virus transfer from cell to cell via intercellular bridges or exosomes (reviewed in refs. 58–60). Both processes may act either independently or jointly.
In summary, the identification of mammalian-tropic mutants enabled us to explore the molecular mechanisms responsible for virus entry into nonpermissive cells. What complements of the chicken cell genome are required for an increase or establishment of mammalian cell permissiveness to RSV replication is a subject of our ongoing investigation.
Materials and Methods
XC and H20 Cell Line Origin.
XC and H20 are rodent tumor cell lines transformed with the PR-RSV-C strain. The rat XC cell line harbors several provirus copies per genome (20), whereas the hamster H20 cell line possesses only a single provirus (61). Both cell lines produce either no or negligible infectious virus (3, 56). The XC cell line is derived from a tumor that was induced by inoculation of PR-RSV-C–infected chicken tissues into newly born rats (62). The H20 cell line was prepared in a similar way with the difference that chicken sarcoma tissue induced by XC-RSV, which was rescued by transfection from XC cells, was inoculated into newly born Syrian hamsters (56).
TM Oligomerization Assay.
The TM oligomerization assay was performed similarly as previously described (23, 24, 45). The temperature threshold that triggers TM oligomerization was determined by incubating the purified virions in HN buffer [10 mM Hepes, 130 mM NaCl (pH 8.0)] at the indicated temperature for 20 min before lysis with Laemmli loading buffer (1% SDS, 10% glycerol, 0.1 M Tris-Cl, pH 6.8, 0.1% bromophenol blue, 5% β-mercaptoethanol). The pH of HN buffer with virions was adjusted with predetermined volumes of 100 mM Hepes (pH 3.8 or 7.4), and the samples were incubated at room temperature for 30 min. The samples were then neutralized with 1 M Tris (pH 7.6 or 9.5), lysed with Laemmli loading buffer, and analyzed by immunoblotting after SDS/PAGE in 11% polyacrylamide gels with a rabbit antibody specific for the carboxyl terminus of the TM subunit of ASLV Env (1:1,000) (23).
Virus Liposome Binding Assay.
The virus liposome binding assay is a modification of the protocol previously described (39). A total of 105 GFP-transducing units of each virus in 100 µL of cultivation media was incubated with 250 µL of liposomes (5 mM) at 37 °C for 30 min. A total of 350 µL of ice-cold 80% sucrose-PBS was added to the preparation to bring it to 40% sucrose, and 700 µL of this solution was layered on 3.2 mL of 50% sucrose-PBS and overlaid with 1 mL of 30% sucrose-PBS and 100 µL of 5% sucrose-PBS. The samples were ultracentrifuged at 152,000 × g at 4 °C for 2 h. Three 400-µL aliquots followed by one 600-µL aliquot were taken from the air–liquid interphase. The top 400 µL and the last 600 µL (fraction above 50% sucrose, called bottom) of each sample were mixed with 5× Laemmli loading buffer, boiled for 10 min, and separated in 13% SDS/PAGE. The presence of virions was detected by immunoblotting with the anti-ASLV rabbit p27 Antiserum (Charles River Laboratories, 1:1,000).
Statistical Analysis.
We performed Welch t test to determine significance of difference in virus infectivity. In graphs, significant differences are marked by asterisks (*P = 0.05–0.01, **P = 0.01–0.001, ***P < 0.001).
SI Materials and Methods
Cell Cultures.
The chicken fibroblast cell line DF-1, which is free of ASLV-related endogenous (ev) loci (63), was obtained from S. Hughes, National Cancer Institute, Bethesda. L15 cells are primary chicken embryo fibroblasts prepared from 10-d-old embryos of chicken inbred line L15, which harbors a mutation introducing a premature stop codon in Tvc receptor (9). Hamster cell line NIL-2 (herein termed NIL) is a spontaneously transformed cell line derived from Syrian hamster embryo (64). NIL-2 cells expressing Tvc receptor (NIL-Tvc) were derived from NIL-2 and their preparation was described previously (9). HEK293 (human embryonic kidney cells) and RPE1-hTERT (hTERT-immortalized retinal pigment epithelial cells) were purchased from the American Type Culture Collection. DF-1, L15, NIL, and NIL-Tvc cells were grown in 1:1 DMEM:F-12 medium (Life Technologies) supplemented with l-glutamine, 5% calf serum, 1–5% FCS, 1% chicken serum, and 10% tryptose phosphate broth (Life Technologies). HEK293 and RPE1-hTERT cells were grown in DMEM (Sigma) supplemented with 10% FCS and 1× antibiotic–antimycotic solution (Sigma).
Virus Rescue from H20 Cells.
X-irradiated H20 cells were fused with DF-1 by means of PEG as described before (5). Virus production in culture medium was 5–15 focus-forming units (FFU)/mL 4 d after fusion in contrast to untreated, treated, or self-fused H20 cells, which produced no infectious virus. The virus was then replicated in DF-1 cells to ∼105 FFU. The viral particles were concentrated by ultracentrifugation through a 25% sucrose cushion at 32,000 rpm for 2 h in a Beckman SW38 rotor.
DNA Extraction and RSV env Gene Amplification.
The genomic DNA was isolated by phenol–chloroform extraction from the XC, H20, or XC-RSV–infected DF-1 cells. The RSV env gene was amplified using the forward primer PolEnd_fw (5′-TTTGGGTACCCTCTCGAAAAGT) and reverse primer EnvEnd_StuI_rv (5′-ACAGGCCTTTTGCATCTTCCTGTATTCAGTA). The following PCR conditions were used: 98 °C for 30 s, 36 cycles of 98 °C for 15 s, 65 °C for 30 s, and 72 °C for 60 s, and terminal extension at 72 °C for 7 min with Phusion polymerase (NEB). The resulting PCR product of 1,869 bp in length was sequenced from both sides with the primers used in the PCR amplification.
Construction of Virus Reporter Vectors and Virus Propagation.
The original virus bearing PR-RSV-C was expressed from plasmid pPrC, which was obtained from K. Beemon, Johns Hopkins University, Baltimore (65) and amplified in DF-1 cells with a resulting titer ∼105 FFU. The viral particles were concentrated by ultracentrifugation through 25% sucrose cushion at 32,000 rpm for 2 h in a Beckman SW38 rotor. RCAS-EnvC-GFP and RCAS-EnvH20-GFP were prepared by env replacement from retroviral vector RCASBP-C-GFP (66) transducing the green fluorescent protein (GFP) reporter gene. env from H20 cells and from pPrC was amplified using the forward primer PolEnd_fw and reverse primer EnvEnd_StuI_rv by Phusion polymerase with the same conditions as mentioned above. PCR products were adenylated and cloned in pGEM-T Easy plasmid vector (Promega). The resulting pGEM-EnvC and pGEM-EnvH20 as well as vector RCASBP-C-GFP were digested with KpnI and StuI restriction enzymes and the 1,853-bp fragment containing the 3′ end of pol and the entire env gene from RCASBP-C-GFP was replaced with equal-length fragments from pGEM-EnvC or pGEM-EnvH20 (Fig. 2B). Chimeric constructs EnvC-H20Parts1–4 were prepared by replacement of one specific part of EnvC by EnvH20. pGEM-EnvC and pGEM-EnvH20 were digested with NheI and AgeI for EnvH20Part1, AgeI and HindIII for EnvH20Part2, HindIII and Xma for EnvH20Part3, and Xma and StuI for EnvH20Part4. The cleaved fragment from pGEM-EnvC was replaced with the equal-length fragment from pGEM-EnvH20. The resulting chimeric pGEM-EnvC-H20Parts1–4 was digested with KpnI and StuI and inserted into cleaved RCASBP-C-GFP in the same way as described for RCAS-EnvC-GFP. EnvC-L378S, EnvC-G464S, and EnvC-L503V are constructs harboring EnvC with one specific mutation and were prepared by PCR mutagenesis and In-Fusion cloning. Fifty nanograms of pGEM-EnvC was amplified with primers bearing mutation, primers 5′-CGCAAGCCTCAAGAGAAATTGAGAGAC and 5′-TTCTCTTGAGGCTTGCGCAGCT for EnvC-L378S, 5′-TAAGATCAGCGTGGACAGCGACC and 5′-GTCCACGCTGATCTTATTGACATGTTTC for EnvC-G464S, and 5′-ATTGCTAGTGGTGTGCCTGCCTT and GGCACACCACTAGCAATAAAATAACTACAAGCC for EnvC-L503V. The following PCR conditions were used: 98 °C for 30 s, 36 cycles of 98 °C for 15 s, 70 °C for 30 s, and 72 °C for 3 min, and terminal extension at 72 °C for 7 min with Phusion polymerase (NEB). The resulting PCR product (4,900 bp) was cleaved with DpnI to get rid of the original pGEM-EnvC and recircularized by In-Fusion HD Cloning Kit (Clontech) according to the manufacturer’s protocol. pGEM-EnvC with particular mutation was digested with KpnI and StuI and inserted into cleaved RCASBP-C-GFP in the same way as described for RCAS-EnvC-GFP. EnvB-H20Part4 was prepared from RCASBP-B-GFP (66). Part of the env gene encoding the surface subunit of EnvB was amplified with primers 5′-TCGATTTTTGGGTACCCTCTCGGA and 5′-CTTGTGCAGCTGCTACCC from template RCASBP-B-GFP with Phusion polymerase. The resulting PCR product was inserted into pGEM-EnvC-H20Part4, which was cleaved with KpnI–XmaI using In-Fusion HD Cloning Kit. The resulting vector pGEM-EnvB-H20Part4 as well as vector RCAS-EnvC-GFP were cleaved with KpnI and BstBI, and the 1,797-bp fragment from RCAS-EnvC-GFP was discarded and replaced with same-length fragments from pGEM-EnvB-H20Part4. All viruses were propagated in the same way. DF-1 cells were transfected with viral constructs using Lipofectamine 3000 (Thermo Fisher Scientific) according to the producer’s protocol and the viruses were amplified for 2–3 wk. The medium was then collected, centrifuged (3,850 × g, 20 min, 4 °C) and filtered through a 0.45-µm filter. Aliquots were stored at −80 °C.
Quantification of Proviral DNA in Infected Cells.
NIL and NIL-Tvc cells were seeded at a density of 5 × 103 per well in a 96-well plate. Five hours after seeding, the cells were pretreated for 30 min with 50 µL of medium with polybrene (20 µg/mL) and infected with 50 µL of medium containing virus. Before infection, all viral stocks were treated with DNase for 30 min in 37 °C to get rid of residual viral DNA. On days 1, 3, 6, and 9 after infection, the cells were washed with PBS and resuspended in 100 µL of lysis buffer (10 mM Tris⋅HCl pH 8.0, 1 mM EDTA, 0.2 mM CaCl2, 0.001% Triton X-100, 0.001% SDS, 1 mg/mL proteinase K). The resuspended cells were incubated at 58 °C for 1 h and then the protease was heat inactivated at 95 °C for 10 min. Aliquots (4 μL) were analyzed by real-time quantitative PCR based on the MESA GREEN qPCR MasterMix Plus for SYBR Assay Kit (Eurogentec) and a CFX96 system for qPCR detection (Bio-Rad). Quantifications of viral transcripts were performed with primers 5′-GCCAGGGAACCTTTGGATTA) and 5′-CCCTTAAAATCACCTTCGGAAA in env gene or 5′-ACGTAAACGGCCACAAGTTC and 5′- TGCAGATGAACTTCAGGGTCAG in gfp. Results were normalized to the amount of mGAPDH genomic locus, which was measured with primers 5′-AACTTTGGCATTGTGGAAGG and 5′-ATCCACAGTCTTCTGGGTGG. The volume of the reaction mixture was 20 μL with 400 nM final concentration of each primer. Cycling conditions were 95 °C for 5 min, 40 cycles of 95 °C for 15 s, 61 °C for 20 s, and 72 °C for 20 s. Calibration curves were prepared by amplification of diluted plasmid samples ranging from 102 to 107 copies per reaction.
Determination of Viral Titers.
PR-RSV-C and H20-RSV viruses were titrated by infectious center assay described previously (6). RCAS-EnvC-GFP, RCAS-EnvH20-GFP, and all other env-mutant viruses derived from these two vectors were titrated using flow cytometry. DF-1 and HEK293 cells were seeded at a density of 5 × 104 per well in a 24-well plate; NIL, NIL-Tvc, and RPE1-hTERT were seeded at a density of 2.5 × 104 per well in a 24-well plate. Five hours after seeding, the cells were pretreated for 30 min with 200 µL of medium with polybrene (20 µg/mL) and infected with 200 µL of virus-containing medium. Next day, the medium was replaced and in the case of DF-1 cells, AZT (final concentration 5 µM) was added into new media 24 h postinfection. The percentage of GFP+ cells was quantified by fluorescence-activated cells sorting (FACS) using an LSRII analyzer (Becton, Dickinson) 2 d (in the case of DF-1 and HEK293 cells) or 3 d (in the case of NIL, NIL-Tvc, and RPE1 cells) postinfection. The cells were trypsinized and washed with PBS before the analysis. The viral titer was determined from the following formula: TU/mL = (1/dilution) × (1/volume used to infect) × (cells per well at time of infection) × [−ln (1 − positive fraction)] (14).
Virus Purification.
Viruses used for the pH inactivation assay, PMB inhibition, and TM oligomerization assays were concentrated from culture supernatants by ultracentrifugation through a 25% sucrose cushion in HN buffer [10 mM Hepes, 130 mM NaCl (pH 8.0)], and then resuspended in a small volume of HN buffer overnight at 4 °C.
Temperature-Sensitivity Assay of Virions.
Virus stocks in culture medium were diluted to a similar titer and aliquoted into five samples, which were incubated for 0, 2, 4, 6, or 8 h at 44 °C and transferred to ice. DF-1 cells and NIL-Tvc cells were infected as described above. The next day, DF-1 cells were harvested for quantification of newly made viral DNA. NIL-Tvc cells were analyzed for GFP expression by flow cytometry 3 d after infection.
pH Inactivation Assay.
Virus inactivation by low pH was performed as described previously (20). To modify the pH, virions in HN buffer were diluted 100-fold in medium that was kept at pH 7.4 or adjusted to pH 5.0 with HCl. The samples were then incubated either on ice or at 37 °C for 30 min before neutralization with an equal volume of medium buffered with 25 mM Hepes (pH 7.4). The virus was then added to DF-1 cells in 24-well plates, and the next day the medium was replaced and AZT (final concentration 5 µM) was added. Two days after infection, GFP expression was analyzed by flow cytometry.
PMB Inhibition.
PMB inhibition of virus infection was performed as described previously (23). PMB (Thermo Fisher Scientific) was dissolved in HN buffer and added into the virus preparation in HN buffer to the indicated final concentrations. Samples were incubated at 37 °C for 30 min, cooled on ice, diluted in growth medium supplemented with 40 mM Hepes, and added on ice to 24-well plates of DF-1 cells that were washed with chilled PBS. Plates were centrifuged for 2.5 h at 1,520 × g, 4 °C. Medium was changed on ice and plates were then transferred to a 37 °C incubator. The next day, the medium was changed and AZT (final concentration 5 µM) was added. Two days after infection, GFP expression was analyzed by flow cytometry.
Production of Liposomes.
Liposomes were produced by a modification of the protocol described previously (67). Liposomes were always freshly prepared before each experiment as a 2:1 molar ratio mixture of egg phospholipids and cholesterol (Sigma) in chloroform. Hen egg phospholipids were isolated according to method 2 by Gladkowski et al. (68). The concentration of phospholipids was determined by molybdenum blue reaction (69). The lipid mixture was dried down to a thin film in a round-bottom glass tube under a constant stream of nitrogen at room temperature. After resuspension in PBS by extensive vortexing, liposomes were sonicated twice for 60 s at 43 °C in a water bath sonicator and then extruded 25 times through a 0.1-μm pore size polycarbonate membrane (Avestin) in a LiposoFast Basic apparatus (Avestin).
Product-Enhanced Reverse Transcriptase Assay.
The product-enhanced reverse transcriptase (PERT) assay was performed as described previously (70). In each run, one positive control (ASLV of defined titer) and one negative control (culture medium of uninfected cells) were included.
Acknowledgments
We thank J. Hejnar, T. Hron, and especially D. Elleder for critical reading of the manuscript, corrections, and suggestions; W. Mothes for providing us with antibody against the ASLV C-terminal part of Env; M. Federspiel for antibodies against ASLV N-and C-terminal parts of Env; M. Dvořák and M. Dvořáková for their help with phospholipids preparation; and L. Mikušová for technical assistance. This project was supported by Grant 15-22207S of the Czech Science Foundation and also institutionally by RVO: 68378050 and NPU I: LO1419.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704750114/-/DCSupplemental.
References
- 1.Parrish CR, et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin H. The early history of tumor virology: Rous, RIF, and RAV. Proc Natl Acad Sci USA. 2011;108:14389–14396. doi: 10.1073/pnas.1108655108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svoboda J, Chyle P, Simkovic D, Hilgert I. Demonstration of the absence of infectious Rous virus in rat tumour XC, whose structurally intact cells produce Rous sarcoma when transferred to chicks. Folia Biol (Praha) 1963;9:77–81. [PubMed] [Google Scholar]
- 4.Temin HM. The DNA provirus hypothesis. Science. 1976;192:1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- 5.Lounková A, et al. Molecular events accompanying rous sarcoma virus rescue from rodent cells and the role of viral gene complementation. J Virol. 2014;88:3505–3515. doi: 10.1128/JVI.02761-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svoboda J, Dourmashkin R. Rescue of Rous sarcoma virus from virogenic mammalian cells associated with chicken cells and treated with Sendai virus. J Gen Virol. 1969;4:523–529. doi: 10.1099/0022-1317-4-4-523. [DOI] [PubMed] [Google Scholar]
- 7.Weiss RA. In: Cellular Receptors and Viral Glycoproteins Involved in Retrovirus Entry. The Retroviridae. Levy JA, editor. Plenum Press; New York: 1993. pp. 1–108. [Google Scholar]
- 8.Young JA, Bates P, Varmus HE. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J Virol. 1993;67:1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elleder D, et al. The receptor for the subgroup C avian sarcoma and leukosis viruses, Tvc, is related to mammalian butyrophilins, members of the immunoglobulin superfamily. J Virol. 2005;79:10408–10419. doi: 10.1128/JVI.79.16.10408-10419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai N, Bates P. Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc Natl Acad Sci USA. 2006;103:5531–5536. doi: 10.1073/pnas.0509785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adkins HB, Brojatsch J, Young JA. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol. 2000;74:3572–3578. doi: 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brojatsch J, Naughton J, Rolls MM, Zingler K, Young JA. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 13.Hunter E. Viral entry and receptors. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1997. pp. 71–120. [PubMed] [Google Scholar]
- 14.Rainey GJ, Natonson A, Maxfield LF, Coffin JM. Mechanisms of avian retroviral host range extension. J Virol. 2003;77:6709–6719. doi: 10.1128/JVI.77.12.6709-6719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taplitz RA, Coffin JM. Selection of an avian retrovirus mutant with extended receptor usage. J Virol. 1997;71:7814–7819. doi: 10.1128/jvi.71.10.7814-7819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amberg SM, Netter RC, Simmons G, Bates P. Expanded tropism and altered activation of a retroviral glycoprotein resistant to an entry inhibitor peptide. J Virol. 2006;80:353–359. doi: 10.1128/JVI.80.1.353-359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz DE, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 18.Aydin H, Smrke BM, Lee JE. Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2-step entry mechanism. FASEB J. 2013;27:5059–5071. doi: 10.1096/fj.13-232371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bova CA, Olsen JC, Swanstrom R. The avian retrovirus env gene family: Molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitsialis SA, Katz RA, Svoboda J, Guntaka RV. Studies on the structure and organization of avian sarcoma proviruses in the rat XC cell line. J Gen Virol. 1983;64:1885–1893. doi: 10.1099/0022-1317-64-9-1885. [DOI] [PubMed] [Google Scholar]
- 21.Hughes SH. The RCAS vector system. Folia Biol (Praha) 2004;50:107–119. [PubMed] [Google Scholar]
- 22.Bova-Hill C, Olsen JC, Swanstrom R. Genetic analysis of the Rous sarcoma virus subgroup D env gene: Mammal tropism correlates with temperature sensitivity of gp85. J Virol. 1991;65:2073–2080. doi: 10.1128/jvi.65.4.2073-2080.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 24.Smith JG, Mothes W, Blacklow SC, Cunningham JM. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J Virol. 2004;78:1403–1410. doi: 10.1128/JVI.78.3.1403-1410.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez LD, et al. Activation of a retroviral membrane fusion protein: Soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J Cell Biol. 1997;139:1455–1464. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JG, Cunningham JM. Receptor-induced thiolate couples Env activation to retrovirus fusion and infection. PLoS Pathog. 2007;3:e198. doi: 10.1371/journal.ppat.0030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainey GJ, Coffin JM. Evolution of broad host range in retroviruses leads to cell death mediated by highly cytopathic variants. J Virol. 2006;80:562–570. doi: 10.1128/JVI.80.2.562-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsichlis PN, Conklin KF, Coffin JM. Mutant and recombinant avian retroviruses with extended host range. Proc Natl Acad Sci USA. 1980;77:536–540. doi: 10.1073/pnas.77.1.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffin JM. Virions at the gates: Receptors and the host-virus arms race. PLoS Biol. 2013;11:e1001574. doi: 10.1371/journal.pbio.1001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorner AJ, Coffin JM. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986;45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 31.Postler TS, Desrosiers RC. The tale of the long tail: The cytoplasmic domain of HIV-1 gp41. J Virol. 2013;87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimitrov DS. Virus entry: Molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barros M, et al. Membrane binding of HIV-1 matrix protein: Dependence on bilayer composition and protein lipidation. J Virol. 2016;90:4544–4555. doi: 10.1128/JVI.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Damico RL, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damico R, Rong L, Bates P. Substitutions in the receptor-binding domain of the avian sarcoma and leukosis virus envelope uncouple receptor-triggered structural rearrangements in the surface and transmembrane subunits. J Virol. 1999;73:3087–3094. doi: 10.1128/jvi.73.4.3087-3094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert JM, Hernandez LD, Balliet JW, Bates P, White JM. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnard RJ, Elleder D, Young JA. Avian sarcoma and leukosis virus-receptor interactions: From classical genetics to novel insights into virus-cell membrane fusion. Virology. 2006;344:25–29. doi: 10.1016/j.virol.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Barnard RJ, Narayan S, Dornadula G, Miller MD, Young JA. Low pH is required for avian sarcoma and leukosis virus Env-dependent viral penetration into the cytosol and not for viral uncoating. J Virol. 2004;78:10433–10441. doi: 10.1128/JVI.78.19.10433-10441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melikyan GB, Barnard RJ, Markosyan RM, Young JA, Cohen FS. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J Virol. 2004;78:3753–3762. doi: 10.1128/JVI.78.7.3753-3762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narayan S, Barnard RJ, Young JA. Two retroviral entry pathways distinguished by lipid raft association of the viral receptor and differences in viral infectivity. J Virol. 2003;77:1977–1983. doi: 10.1128/JVI.77.3.1977-1983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netter RC, et al. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup a infection and identify a fusion intermediate. J Virol. 2004;78:13430–13439. doi: 10.1128/JVI.78.24.13430-13439.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Damico R, Bates P. Soluble receptor-induced retroviral infection of receptor-deficient cells. J Virol. 2000;74:6469–6475. doi: 10.1128/jvi.74.14.6469-6475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myszka DG, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Babel AR, Bruce J, Young JA. The hr1 and fusion peptide regions of the subgroup B avian sarcoma and leukosis virus envelope glycoprotein influence low pH-dependent membrane fusion. PLoS One. 2007;2:e171. doi: 10.1371/journal.pone.0000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melder DC, Yin X, Delos SE, Federspiel MJ. A charged second-site mutation in the fusion peptide rescues replication of a mutant avian sarcoma and leukosis virus lacking critical cysteine residues flanking the internal fusion domain. J Virol. 2009;83:8575–8586. doi: 10.1128/JVI.00526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorner AJ, Stoye JP, Coffin JM. Molecular basis of host range variation in avian retroviruses. J Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konstantoulas CJ, Lamp B, Rumenapf TH, Indik S. Single amino acid substitution (G42E) in the receptor binding domain of mouse mammary tumour virus envelope protein facilitates infection of non-murine cells in a transferrin receptor 1-independent manner. Retrovirology. 2015;12:43. doi: 10.1186/s12977-015-0168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrarone J, Knoper RC, Li R, Kozak CA. Second site mutation in the virus envelope expands the host range of a cytopathic variant of Moloney murine leukemia virus. Virology. 2012;433:7–11. doi: 10.1016/j.virol.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves JD, Schulz TF. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubay JW, Roberts SJ, Brody B, Hunter E. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J Virol. 1992;66:4748–4756. doi: 10.1128/jvi.66.8.4748-4756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haim H, et al. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 2011;7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: Strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McRoy WC, Baric RS. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J Virol. 2008;82:1414–1424. doi: 10.1128/JVI.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forni D, et al. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci Rep. 2015;5:14480. doi: 10.1038/srep14480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duran-Reynals F. The reciprocal infection of ducks and chickens with tumor inducing viruses. Cancer Res. 1942;2:343–369. [Google Scholar]
- 56.Geryk J, Sainerová H, Sovová V, Svoboda J. Characterization of cryptovirogenic, virus-productive and helper-dependent virogenic hamster tumour cell lines. Folia Biol (Praha) 1984;30:152–164. [PubMed] [Google Scholar]
- 57.Svoboda J, Chyle P. Malignization of rat embryonic cells by Rous sarcoma virus in vitro. Folia Biol (Praha) 1963;9:329–342. [PubMed] [Google Scholar]
- 58.Roberts CT, Jr, Kurre P. Vesicle trafficking and RNA transfer add complexity and connectivity to cell-cell communication. Cancer Res. 2013;73:3200–3205. doi: 10.1158/0008-5472.CAN-13-0265. [DOI] [PubMed] [Google Scholar]
- 59.Sattentau QJ. Cell-to-cell spread of retroviruses. Viruses. 2010;2:1306–1321. doi: 10.3390/v2061306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wurdinger T, et al. Extracellular vesicles and their convergence with viral pathways. Adv Virol. 2012;2012:767694. doi: 10.1155/2012/767694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Svoboda J, et al. Characterization of exogenous proviral sequences in hamster tumor cell lines transformed by Rous sarcoma virus rescued from XC cells. Virology. 1983;128:195–209. doi: 10.1016/0042-6822(83)90330-6. [DOI] [PubMed] [Google Scholar]
- 62.Svoboda J. Presence of chicken tumour virus in the sarcoma of the adult rat inoculated after birth with Rous sarcoma tissue. Nature. 1960;186:980–981. doi: 10.1038/186980b0. [DOI] [PubMed] [Google Scholar]
- 63.Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 64.Diamond L. Two spontaneously transformed cell lines derived from the same hamster embryo culture. Int J Cancer. 1967;2:143–152. doi: 10.1002/ijc.2910020209. [DOI] [PubMed] [Google Scholar]
- 65.Ogert RA, Lee LH, Beemon KL. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Federspiel MJ, Hughes SH. Retroviral gene delivery. Methods Cell Biol. 1997;52:179–214. [PubMed] [Google Scholar]
- 67.Earp LJ, Delos SE, Netter RC, Bates P, White JM. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J Virol. 2003;77:3058–3066. doi: 10.1128/JVI.77.5.3058-3066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gladkowski W, Chojnacka A, Kielbowicz G, Trziszka T, Wawrzenczyk C. Isolation of pure phospholipid fraction from egg yolk. J Am Oil Chem Soc. 2012;89:179–182. [Google Scholar]
- 69.Rouser G, Fkeischer S, Yamamoto A. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- 70.Fábryová H, Hron T, Kabíčková H, Poss M, Elleder D. Induction and characterization of a replication competent cervid endogenous gammaretrovirus (CrERV) from mule deer cells. Virology. 2015;485:96–103. doi: 10.1016/j.virol.2015.07.003. [DOI] [PubMed] [Google Scholar]