Significance
Protein purification is a primary step and the basis for numerous biochemical and biomedical studies. It is particularly crucial for high-resolution structural analysis and industrial protein production, where it has to meet the high-yield, high-purity, and high-activity (HHH) requirement. However, the HHH purification of many proteins or protein complexes remains a difficult, target-dependent, and multistep process. The ultra-high-affinity (CL7/Im7) purification system described in this work allows for one-step HHH purification of a wide range of traditionally challenging biological molecules, including eukaryotic, membrane, toxic, and DNA/RNA-binding proteins and complexes. It might emerge as an efficient, universal tool for high-throughput isolation of many significant biological systems to advance modern biological studies as well as manufacturing of therapeutic proteins.
Keywords: Colicin E7/Im7 affinity chromatography, one-step protein purification, RNA polymerases, membrane proteins, condensin
Abstract
Protein purification is an essential primary step in numerous biological studies. It is particularly significant for the rapidly emerging high-throughput fields, such as proteomics, interactomics, and drug discovery. Moreover, purifications for structural and industrial applications should meet the requirement of high yield, high purity, and high activity (HHH). It is, therefore, highly desirable to have an efficient purification system with a potential to meet the HHH benchmark in a single step. Here, we report a chromatographic technology based on the ultra-high-affinity (Kd ∼ 10−14–10−17 M) complex between the Colicin E7 DNase (CE7) and its inhibitor, Immunity protein 7 (Im7). For this application, we mutated CE7 to create a CL7 tag, which retained the full binding affinity to Im7 but was inactivated as a DNase. To achieve high capacity, we developed a protocol for a large-scale production and highly specific immobilization of Im7 to a solid support. We demonstrated its utility with one-step HHH purification of a wide range of traditionally challenging biological molecules, including eukaryotic, membrane, toxic, and multisubunit DNA/RNA-binding proteins. The system is simple, reusable, and also applicable to pulldown and kinetic activity/binding assays.
Obtaining protein samples with high yield, high purity, and high activity (HHH purification) is the foundation for most modern biological studies, such as proteomics, interactomics, and in vitro drug screening. In most cases, it also raises such studies to new heights. Moreover, HHH-grade samples are indispensable for industrial production of therapeutic proteins and for determining high-resolution 3D protein structures that are crucial for deep understanding of protein function. In recent years, investigations of large multisubunit complexes (transcription and translation machineries, for example) and challenging eukaryotic and membrane proteins have emerged as a major focus of proteomics and structural analyses. An efficient, ideally one-step protocol for HHH purification is highly desirable to facilitate all these crucial studies.
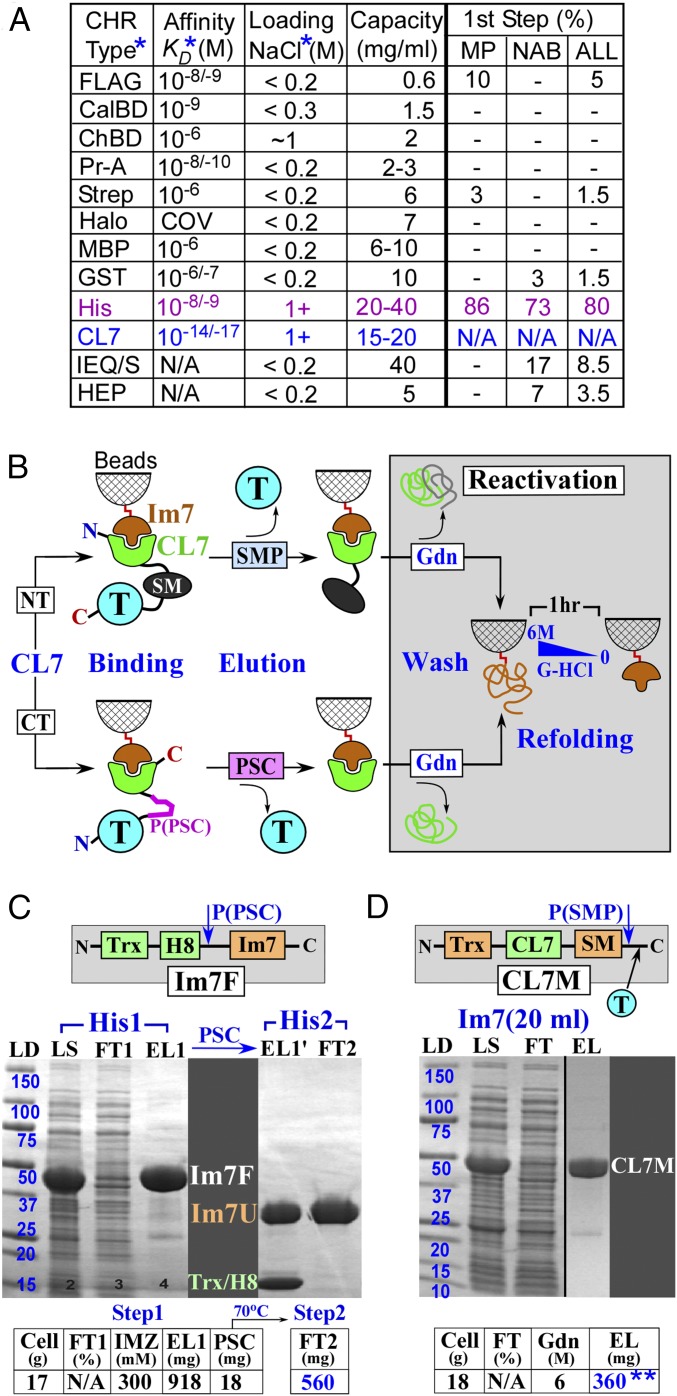
Commercial purification systems have complementary advantages that allow the purification of target proteins (1–3). However, each has certain critical disadvantages that affect their efficiencies (1–3). Some columns have low capacity and often command a high price (SI Appendix, Fig. S1A) such that they barely meet the requirement of high yield necessary for structural studies or for commercial protein production. Salt sensitivity is another notable limitation of many (protein/ligand) affinity systems; it often results in significant flow through (90–95%) of the tagged proteins even when medium salt (0.2–0.5 M) is used during lysate loading on the columns (Fig. 1A). On the other hand, if the sample is loaded in a low-salt buffer, impurities often could not be removed afterward even when very high-salt buffer is applied. This salt dilemma is well illustrated by our purification trials with the two DNA-binding proteins fused to the maltose-binding protein (MBP-Tag; SI Appendix, Fig. S2 A and B). Importantly, these results demonstrate that even loading buffer with 0.5 M salt is not sufficient to obtain high purity for some DNA-binding proteins (SI Appendix, Fig. S2B). Consistently, we obtained similar results with other low-salt systems, including GST (GST-Tag; SI Appendix, Fig. S2C) and streptavidin-binding peptide (Strep-Tag; noted in SI Appendix, Fig. S2) columns. Thus, a one-step HHH purification using these “low-salt” columns is limited to only a subset of biological targets that do not have nonspecific interactions with untagged molecules (proteins, DNA/RNA) under the low-salt conditions (see examples in SI Appendix, Fig. S3 B and C).
Fig. 1.
The CL7/Im7 purification system. (A) Comparison of the chromatographic systems. (B) Protocols of the CL7/Im7 purification. (C and D) Purification of the Im7 unit (Im7) (C) and the model CL7M protein (D). ALL, combined statistics for both (MP+NAB) sets; CalBD, calmodulin-binding domain; ChBD, chitin-binding domain; CL7, engineered CE7 tag; first step, a frequency (%), with which a technique was used on the most crucial, first (lysate) purification step in the analyzed sets of the MPs and NABs; FLAG, antibody-binding peptide; Im7F, expression vector for Im7 fusion; MBP, maltose-binding protein; MP/NAB, the analyzed sets of the membrane/NA-binding proteins (SI Appendix, Figs. S5 and S6); Pr-A, protein A tag; Strep, streptavidin-binding peptide. The cleaved IM7 recovered from FT 2 was used for all of the trials in this study. EL, eluate; FT, flow through; Gdn, guanidine hydrochloride; H8, 8 histidine tag; LD, ladder (kDa); LYS, lysate; P(PSC)/P(SMP), PSC/SMP cleavage sites; PSC, PreScission protease; SM, SMP, SUMO domain and protease; T, target protein; Trx, thioredoxin. The same abbreviations are also used in other figures. *The details of the protocols and Kd values are provided in the legend of SI Appendix, Fig. S1. **At saturation conditions (visible FT), ∼400 mg of CL7M is eluted from the 20 mL Im7-column—that is, ∼20 mg/1 mL Im7 beads.
Only the chitin-binding (ChBD) and His-Trap affinity systems tolerate high salt-containing buffers (∼1 M NaCl) at the crucial step of crude lysate loading onto the columns (Fig. 1A). However, the chitin columns have a low capacity, and in our experience, some contaminants always remained in the purified samples (SI Appendix, Fig. S4). Accordingly, this approach is rarely used for crystallographic studies (4) (SI Appendix, Figs. S5 and S6). To date, His-Trap (Ni2+-based) is the most commonly used approach. It demonstrates a high-yield capacity and also allows for high-salt loading of the lysates without losing binding affinities (Fig. 1A). Additional advantages include the low costs of the Ni2+-charged beads (SI Appendix, Fig. S1A) that can be put through multiple regeneration cycles, the small size of the His-tag, and efficient target elution with the inexpensive imidazole. However, our results and those of other investigators show that this approach possesses several drawbacks. It has relatively high, nonspecific, but cooperative affinity to DNA and to DNA-binding proteins (5) as well as to some cellular proteins (4), including many membrane (6, 7) and eukaryotic proteins (8). These contaminants occur largely in a salt-independent manner, sometimes resulting in substantial impurities in the recovered target protein. For instance, according to the combined statistics of the Structural Genomics Projects, all of which use the His-Trap technique as the first purification step, additional purification steps were always required to obtain high-purity samples (4), significantly compromising their yields and increasing purification time. In addition, according to the commercial protocols, the His-Trap Ni2+ base cannot resist metal chelating and reducing agents at concentrations that are required to maintain physiological properties of the certain categories of target proteins.
To gain a more detailed insight into the present state of purification of complex proteins, we have analyzed the published protocols for a number of biologically significant membrane and DNA-binding proteins, for which high-resolution crystal structures have been determined (SI Appendix, Figs. S5 and S6). The purification of these proteins is usually difficult, especially for crystallographic purposes, and may serve as a benchmark that a newly devised HHH purification strives to exceed. The analysis revealed several general observations. First, in agreement with the major advantages of the His-Trap approach, it was used as a first-step purification in the overwhelming majority (∼80%) of the studies (Fig. 1A). Other affinity approaches were used as a first step only in a few (∼8%) cases overall and only in 3% of cases of DNA-binding proteins, a reflection of their relatively poor capacities or “low-salt loading” limitations (Fig. 1A). Second, none of purifications could be completed in one step, consistent with the above noted disadvantages of the His-Trap and other affinity techniques. In fact, an average purification protocol includes ∼3 chromatographic steps and takes over 3 d at best. Third, most protocols include one or more nonspecific columns [gel filtration (GF), ion exchange, nucleic acid (NA) mimicking resins] that are usually substantially less predictable than those based on specific affinity. As a result, additional efforts are required to customize the conditions for each particular protein. Finally, even the simplest, two-step (His-Trap → GF) protocol (∼10% of analyzed studies) takes at least 2 d and typically have protein loss (∼50%) during the GF step (SI Appendix, Figs. S5 and S6).
In conclusion, the HHH-grade purification of challenging protein or protein complexes remains a target-dependent and multistep process. The biochemical and biomedical field would greatly benefit from a new approach, which combines the major advantages of the His-tag technique (high capacity, high-salt tolerance, and low cost) with a stringent protein/ligand-based affinity system. Here we report the development of an original, ultra-high-affinity purification system based on the small protein/protein complex, Colicin E7 DNase (∼16 kDa) and Immunity Protein 7 (∼10 kDa) (CE7/Im7) (9). With a Kd ∼ 10−14–10−17 M, the affinity of this complex is approaching that of a covalent bond and is 4–7 orders of magnitude higher than those of any other available analogs (1, 10–16) (Fig. 1A). Because the DNase activity of CE7 is lethal to the cells, we designed a catalytically inactive variant, termed CL7, based on molecular modeling. We also engineered an Im7 immobilization unit to allow efficient coupling to agarose beads without compromising its association with the CL7 tag. We demonstrate that, unlike other approaches, this CL7/Im7 system allows for a one-step HHH purification (∼97–100% purity) of the most challenging biological targets, including large multisubunit complexes, DNA/RNA-binding proteins, membrane proteins, as well as the proteins poorly expressed in the native host cells. Further, the Im7 affinity column can be easily and repeatedly regenerated without losing its affinity to the CL7 affinity tag.
Results
The Design of the CL7/Im7 Chromatographic System.
The family of colicin DNases (CE2, CE7, CE8, CE9) belongs to the category of highly toxic proteins because of their DNase domains (∼16 kDa) (9, 17, 18). Their toxic activity in the host cells is suppressed by the respective natural cognate inhibitors, the Ims (∼10 kDa). The stringent control depends on an ultra-high affinity typical for natural inhibitors of all toxic enzymes. This ultra-high affinity in theory can serve as a solid basis for a chromatographic approach. However, the native enzymes can be expressed only in the presence of their inhibitors. Even then, their expression levels have to be kept low due to their high toxicity. These restrictions complicate their large-scale production essential for preparation of high-capacity chromatographic columns. For example, a wild-type (CE7/Im7) complex has been successfully used for antibody capture in surface plasmon resonance (SPR) biosensors, an assay that requires only very small amounts of CE7 (19) for chemical coupling to the sensor chips. However, no successful attempts to use the wild-type complex for high-yield purifications have been reported.
An apparent option to bypass a toxicity obstacle is to construct the inactive variants of these enzymes. However, inhibitors usually target the active sites. Thereby, genetic inactivation of enzymes often results in substantial loss of affinity to their inhibitors, thereby affecting efficiencies of the respective chromatographic techniques. The colicin DNases are unique in this regard. Their cognate Ims bind to sites that are remotely located from the catalytic centers of the enzymes. The bound Ims sterically block DNA binding rather than inhibit the activity of the enzymes (SI Appendix, Fig. S7 A and B) (9, 20). Using this unique feature of the CE7/Im7 affinity pair and structural modeling, we have constructed a CE7 variant, CL7, which is completely devoid of the DNA binding and DNase activities (SI Appendix, Fig. S7C) while retaining the full affinity for its Im7 inhibitor.
In our design, the CL7 domain serves as a tag on a target protein, whereas the Im7 inhibitor is covalently coupled to a solid, agarose support (Fig. 1B). The Im7 was engineered to contain an “immobilization” unit (∼26 kDa; detailed in SI Appendix, Fig. S8) for fast, stable, and efficient coupling to iodo-acetyl agarose beads. We also designed an expression vector and protocol for its large-scale expression/purification (Fig. 1C). This approach yielded 300 mg of purified Im7, which was covalently cross-linked to ∼20 mL of the beads (∼15 mg/mL or ∼0.6 mM). We then designed and tested several CL7-tagged vectors to express a specific class of proteins of interest, as described in detail in our trials (Figs. 3, 6, and 7B). The target protein was inserted either C-terminal or N-terminal to the CL7-tag (Fig. 1B). The CL7-tag was fused to a SUMO domain (∼10 kDa) for the SUMO protease (SMP) cleavage or contains the PreScission protease (PSC) cleavage site (8 aa residues) to enable proteolytic elution of target proteins (1, 2) (Fig. 1B). Large-scale expression/purification protocols for both PSC and SMP were established (SI Appendix, Fig. S3). Alternatively, the target proteins with an uncleaved CL7-tag could be eluted with a low pH buffer (pH 3) at ∼85–95% recovery. Under these conditions, some proteins (for example, antibodies) are not entirely denatured and are known to restore their functional properties upon transfer into physiological buffers. After cleavage and elution of a target protein, the CL7-tag, which remained bound to the Im7 agarose beads, was removed under denaturing conditions (6 M Guanidine hydrochloride, Gdn). The column was then easily regenerated via in-column Im7 refolding (described in Fig. 1B).
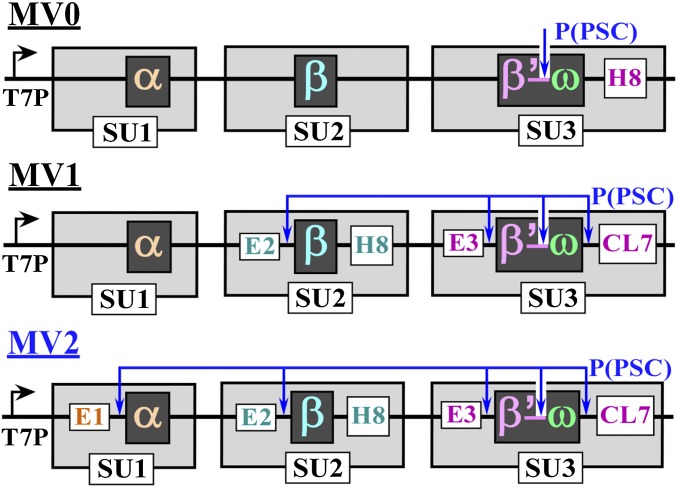
Fig. 3.
Multisubunit expression vectors. E1/E2/E3, the short N-terminal “expression” peptides; P(PSC), PSC cleavage sites; SU, subunit, each has its own ribosome binding site and stop codon; T7P, T7 promoter (SI Appendix, Fig. S9B).
Fig. 6.
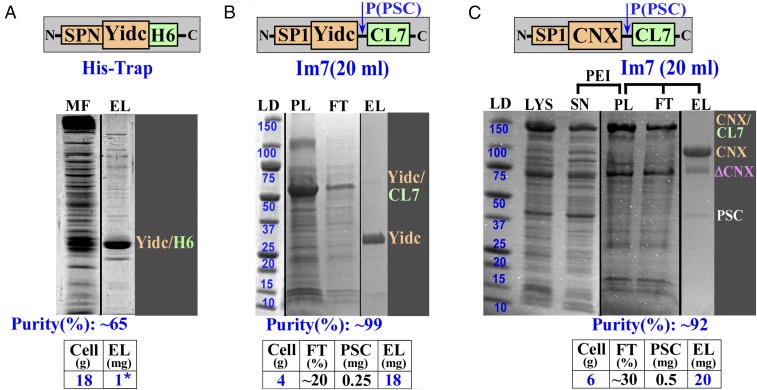
Expression and purification of membrane proteins. (A) Expression and a His-Trap purification of B. halodurans membrane integrase, YidC, using a native gene sequence, including the signal peptide (SPN) (6). (B) A one-step YidC Im7 purification following the PEI precipitation. The gene sequence was adjusted to E. coli codon use, and the signal peptide (SP1) has a Cys to Ser mutation. (C) Expression and a one-step Im7 purification of human chaperon CNX. ΔCNX, truncation of the CNX protein; EL, eluate; FT, flow through; LD, molecular mass standards in kilodaltons; LYS, lysate; MF, membrane fraction (pellet fraction after ultracentrifugation); PL, supernatant after PEI pellet was washed with the buffer containing 0.6 M NaCl and 1.5% DDM; SN, supernatant (soluble fraction after PEI). *1 mg of YidC protein in A was obtained after the last (fifth) purification step (see Fig. 2).
Fig. 7.
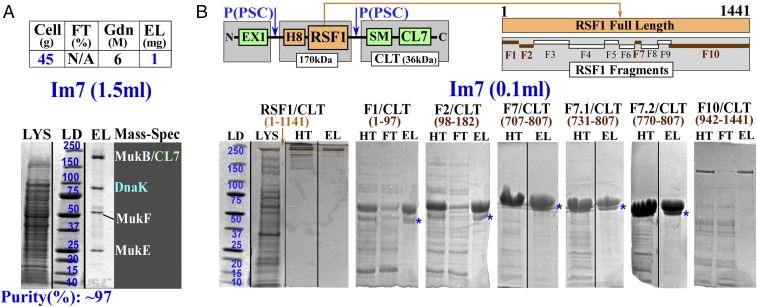
Purification of condensin complex and RSF1 protein for pull-down assays. (A) A one-step Im7 purification of bacterial CL7-tagged MukBEF condensin complex expressed from the host chromosome in S. typhimurium. (B) Expression and analytical purification of dual (H8/CL7)-tagged human RSF1 and its fragments (FN/CLT), where N is the number of the serial fragments used in pull-down assays. CLT, C-terminal (SM-CL7) tag; SM, SUMO domain. Purification is shown for the full-length protein (RSF1; orange) and some fragments (FN/CLT; brown). *A second (lower) band observed in the Im7-purified samples of all small RSF1 fragments are likely due to proteolytic (by some cellular proteases) excision of the N-terminal, flexible EX1 expression tag, as processing of these fragments with PSC converted the upper band to the lower band. EL, eluate; EX1, expression tag 1 (SI Appendix, Fig. S9B); FT, flow through; Gdn, guanidine hydrochloride; HT, the samples purified using the His-Trap columns were loaded on the Im7 beads and eluted with SDS; LD, molecular mass standards in kilodaltons; LYS, lysate.
Verification of the CL7/Im7 Affinity System with a Model Protein.
As proof of principle, we expressed in Escherichia coli a model protein (CL7M, ∼40 kDa) consisting of the thioredoxin–CL7–SUMO domain. The entirety of CL7M may also serve as an N-terminal tag for a target protein inserted after the SMP cleavable SUMO domain (indicated by a blue T in Fig. 1D) at the customized HindIII/XhoI restriction sites. The CL7M lysate was loaded onto the Im7 column. After washing with alternate cycles of high/low salt-containing buffers, the model protein was eluted from the column with 6 M Gdn. The eluted protein had no contaminants detectible by staining in SDS/PAGE (Fig. 1D). We achieved purifications of 300–400 mg of the model protein under a variety of conditions (see details in SI Appendix, SI Materials and Methods), corresponding to a capacity of ∼15–20 mg/mL for our 20-mL Im7 column (Fig. 1D). Thus, CL7M binds with nearly a 1:1 molar ratio to the immobilized Im7 unit. This result demonstrates that the immobilized Im7 unit retains the full binding activity upon chemical coupling to the beads. The column was then reactivated via a 1-h in-column Im7 protein-refolding step (Fig. 1B). This protocol stably restored ∼100% binding capacity as revealed by a nearly identical purification yield for this model protein after the column had been reused over 100 times. With its high capacity and high recycle numbers, the price/performance factor of the Im7 column is comparable or even superior to that of the most inexpensive commercial His-Trap products (SI Appendix, Fig. S1).
To challenge this new system, we applied it to a number of complex targets from the major categories (multisubunit DNA-binding, membrane, toxic, and poorly characterized proteins) that are most refractory to HHH purification.
Expression and Purification of DNA/RNA-Binding, Multisubunit RNA Polymerases.
Multisubunit RNA polymerase (RNAP) is the heart of the transcription machinery with high biological and medical significance. RNAPs and various transcription complexes are the subjects of functional and structural studies of many groups worldwide. All these studies require purified samples for the wild-type and numerous variants. Being both a large multisubunit complex and a DNA/RNA-binding enzyme, RNAP purification is exceedingly challenging. Although RNAPs from different organisms have been studied for decades, no unified protocol exists and none allows for a one-step purification (5, 21, 22) for a number of reasons (Fig. 2). First, RNAPs are large (molecular mass, ∼400 kDa) protein complexes of five subunits (α2ββ’ω); stoichiometric overexpression from vectors is virtually impossible to achieve. Unbalanced expression creates the first line of complications for purification. Second, the largest β/β’ subunits usually undergo transcription/translation-coupled truncations during overexpression. Incomplete RNAP molecules with truncated subunit(s) could affect the enzyme activity and lead to a protein purification problem. Third, RNAPs contain at least four spatially distinct DNA-binding sites. Nonspecific but cooperatively strong binding to cellular NAs upon cell lysis results in major contamination. The NA-related impurities cannot be eliminated by any known single-step chromatography. Significance and challenges of RNAP purification are emphasized by the fact that the RNAP-specific antibodies were developed to allow for isolation of untagged RNAPs from different species (23). The respective chromatography showed good performance for several bacterial RNAPs and was used in structural studies of yeast Pol II polymerase (24). However, in the latter case, the crystallization quality samples could be obtained only through the complex, six-step protocol (24) (see example 9 in SI Appendix, Fig. S6). Thus, HHH purification of RNAPs has remained a multistep process, which has to be customized and adjusted for each enzyme from each particular organism (Fig. 2).
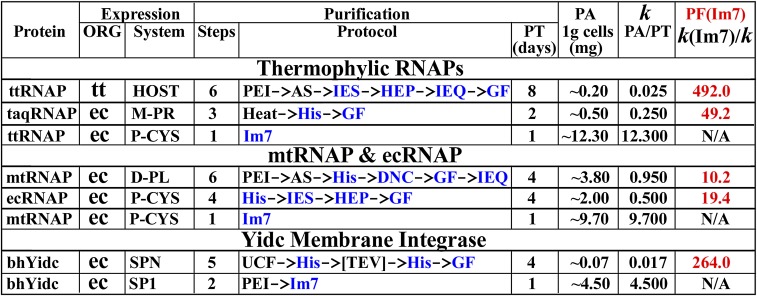
Fig. 2.
Comparative performance of the CL7/Im7 system versus conventional protocols. bh, B. halodurans (6, 7); DNC, DNA cellulose; D-PL, dual plasmid expression approach; ec, E. coli (21); GF, gel filtration; Heat, heating at 60 °C; HEP, heparin; His, His-Trap (Ni2+); HOST, expression from the host chromosome; IEQ/IES, anion/cation exchange; k, productivity factor [PF = protein amount (PA)/purification time (PT)]; M-PR, multipromoter expression vector; mt, M. tuberculosis (5); ORG, host organism; P-CYS, single promoter/polycystronic expression vector; PEI/AS, polyethyleneimine/ammonium sulfate precipitations; PF(Im7), PF of the Im7 approach with respect to the other protocols; SPN/SP1, E. coli overexpression using the Native/Engineered (with C/S mutation) YidC signal peptides; taq, T. aquaticus (22); [TEV], TEV protease cleavage; tt, T. thermophilus (28); UCF, ultracentrifugation. PT and PA for the published protocols were estimated based on our own experience with the same or similar protocols.
We selected two bacterial RNAP core enzymes (25) from Thermus thermophilus (ttRNAP) (26, 27) and the pathogenic Mycobacteria tuberculosis (mtRNAP) (5). These RNAPs are evolutionarily distinct, sharing only ∼42% identity with substantial differences in configuration and overall surface properties where homology drops off significantly. For example, the charge on mtRNAP is almost twice that on ttRNAP (–135 vs. –70). Thus, these two RNAPs represent essentially distinct targets for both expression and purification. Indeed, the conventional, multistep purification protocols for these two RNAPs differ substantially (Fig. 2). Our laboratory has extensively studied these RNAPs in the past decade using various techniques, including high-resolution crystallographic analysis, for which HHH purification is of central importance (26–29). Our objective, therefore, was not solely to test the purification system but also importantly to establish a simple and straightforward approach for production and HHH purification of RNAPs from different organisms suitable for mutagenesis and successful high-throughput crystallization.
To develop an efficient protocol of the large-scale production and HHH isolation of RNAPs, we have designed several multisubunit, polycystronic expression vectors. Each used a T7 promoter inducible by IPTG, and the ORF of each subunit was preceded by a ribosome-binding site. There are two major criteria for their designs. The vector should be easily used to clone RNAPs from various species, and it should be applicable to other multisubunit proteins as well. It should also possess enhanced expression levels of the key (or each) individual subunits.
We began the vector design using ttRNAP, as it is the most difficult target for overexpression in E. coli. There are no efficient expression/purification protocols for recombinant ttRNAP because the T. thermophilus genes have exceedingly high G/C (∼70%) content and contain a high frequency of E. coli rare codons. These features result in overall poor expression levels along with many translational truncations. To minimize these obstacles, we synthesized the genes of the RNAP subunits; the rare codons were eliminated, whereas the GC content was decreased to a reasonable level of ∼59% for expression in E. coli.
To provide a reference point and to compare the new CL7/Im7 purification system with the most popular His-Trap approach, we created three expression vectors (Fig. 3). In each, we fused the C terminus of the largest β’ subunit to the N terminus of the smallest ω subunit through a flexible (removable by PSC) linker (SI Appendix, Fig. S9A). This approach has two notable advantages. It reduces the number of coexpressed subunits while maintaining a perfect stoichiometry of the β’ and ω subunits during purification. The second is especially important. Our experience with RNAP purifications indicates that the high-salt (∼1 M NaCl) loading conditions dissociate these two subunits. Thus, their expression as a fusion protein would alleviate the loss of the ω subunit in this initial step.
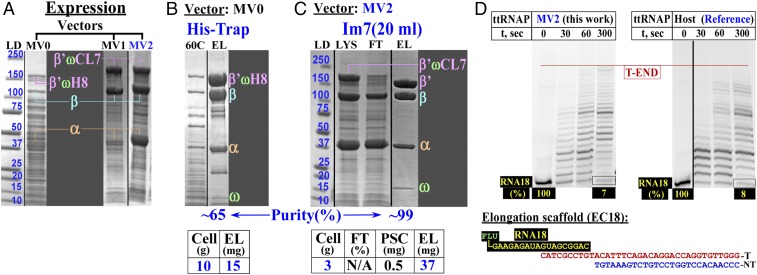
In the MV0 construct (Fig. 3, top line), we have introduced only a His-tag at the C terminus of the largest (β’ω) subunit to test the expression and purification performance. The vector demonstrated a modest expression level of the intact RNAP subunit molecules (Fig. 4A, left lane). However, multiple attempts under different loading and washing conditions failed to provide a reasonably pure sample even though a heated lysate was used to eliminate most of the E. coli proteins (Fig. 4B).
Fig. 4.
Expression, purification, and activity of ttRNAP. (A) Expression levels of ttRNAP using expression vectors developed in this study (Fig. 3). (B and C) A one-step purification of ttRNAP using the His-tag (B) or CL7/Im7 (C) approaches. (D) Transcription elongation assays for the purified, overexpressed ttRNAP enzyme (Left) and the reference WT enzyme isolated from the host without overexpression (Right). 60C, lysate (LYS) heated at 60 °C for ∼45 min; β’ωH8/β’ωCL7, β’ω fusion with the His8 or CL7 tags; EL, eluate; FT, flow through; LD, molecular mass standards in kilodaltons; RNA18, synthetic 18-mer RNA with fluorescein (FLU) at the 5′-end; T/NT, DNA template/nontemplate strands.
The vector was then modified to relocate the His-tag to the second largest β subunit and to add the CL7-tag (cleavable by PSC) at the C terminus of the β’ω subunit. In addition, we have introduced N-terminal short PSC cleavable “expression” tags (SI Appendix, Fig. S9B) designed to increase the expression levels of the two largest β and β’ω-CL7 subunits (Fig. 3, MV1, middle line) or all three (α, β, β’ω-CL7) coexpressed subunits (Fig. 3, MV2, bottom line).
As expected, the expression of two (from MV1; Fig. 4A, middle lane) or all (from MV2; Fig. 4A, right lane) RNAP subunits was substantially improved. The production levels are (α > β > β’ω-CL7) from the superior MV2 expression system (Fig. 4C). We were able to obtain a large amount (∼37 mg) of a pure and intact ttRNAP sample in a single Im7 purification step from lysates of 3 g of cells (Fig. 4C). Notably, the best purity of ttRNAP using either the His-Trap or the CL7/Im7 approaches was achieved if the lysates were first treated with DNase before loading onto the columns in high (1–1.2 M) NaCl-containing buffers. Loading the lysates at a lower (0.5–0.8 M) salt concentrations or without prior DNase treatment always produced somewhat contaminated and only partially active products, even when the column was washed with extra-high-salt (2 M) buffers after loading. The activity tests using the synthetic elongation scaffold showed that the recombinant ttRNAP was as active as enzyme purified from the host thermophilic cells, which were previously used in successful high-resolution structural studies (26–28) (Fig. 4D).
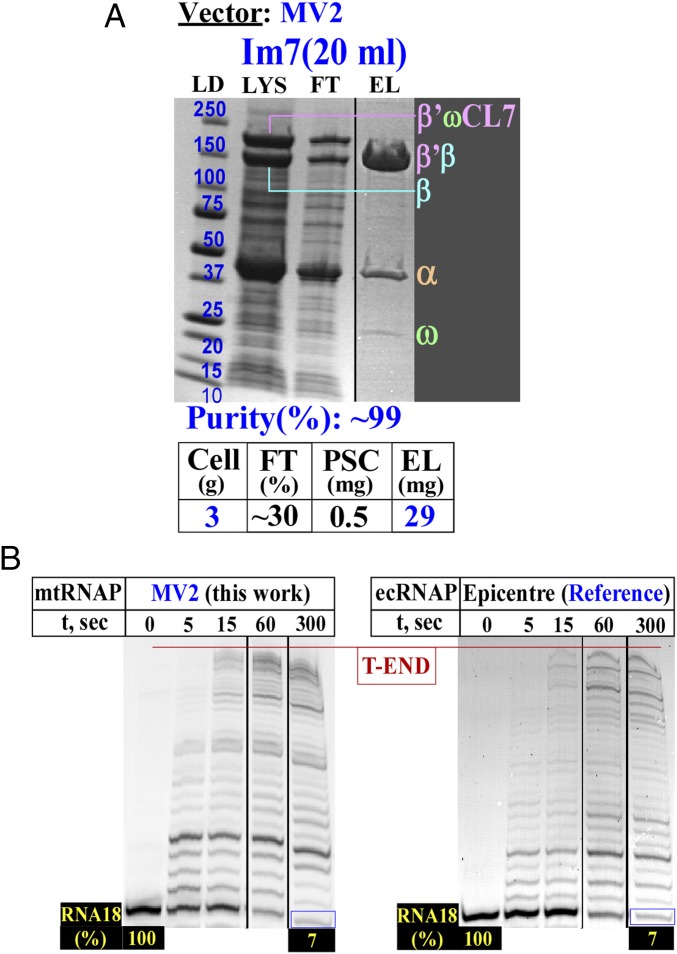
Given the successful one-step HHH purification protocol for ttRNAP, we adjusted the sequence of mtRNAP to the E. coli codons and used the identical expression (Fig. 3, MV2) and purification approaches. We obtained essentially the same results (Fig. 5A). The purified enzyme possessed high activity in the elongation assay similar to that of commercial E. coli RNAP (Fig. 5B).
Fig. 5.
Expression, purification, and activity of mtRNAP. (A) A one-step purification of mtRNAP using the CL7/Im7 approach. (B) Transcription elongation assays for the overexpressed mtRNAP (Left) and commercial E. coli RNAP (ecRNAP; Right). β'ωCL7, β'ω fusion with the His8 or CL7 tags; EL, eluate; FT, flow through; LD, molecular mass standards in kilodaltons; LYS, lysate; RNA18, synthetic 18-mer RNA with fluorescein at the 5'-end.
In summary, our results demonstrate that HHH samples of the large, multisubunit DNA/RNA-binding proteins (RNAPs) can be obtained in one step within only 5–7 h. Our CL7/Im7 expression/purification system represents a dramatic (10- to 500-fold) improvement over the previously used approaches (Fig. 2).
Expression and Purification of Membrane Proteins.
Transmembrane proteins constitute up to 40% of the total protein pool in living cells. Most of them are of high functional and clinical significance, yet only a few have been studied at a high-resolution structural level. This deficit is largely attributable to challenges in expression or purification (30) (SI Appendix, Fig. S5). For our trials, we selected two membrane proteins, one from prokaryotic and one from eukaryotic organisms. These two proteins differ drastically in size, function, and configuration in the membrane.
Bacillus halodurans YidC membrane integrase (molecular mass, ∼32 kDa) is an “all”-membrane protein, without bulky extramembrane domains. The structure of YidC has already been determined (6, 7). It was selected as a reference in our purification. The lengthy original purification protocol (five steps; ∼4 d) (Fig. 2) is one of the most complex used to purify a selected subset of the significant membrane proteins, the crystal structures of which have been determined (SI Appendix, Fig. S5). In particular, the first His-Trap step resulted in only ∼65% purity protein, because other untargeted membrane proteins have substantial binding affinities to the Ni2+-activated column (Fig. 6A). The protocol was also characterized by a rather poor yield (∼1 mg protein from ∼20 g cells). This low yield is likely caused by a low overexpression level typical for membrane proteins and is further compounded by a loss of material during multiple purification steps (Fig. 2).
To test our new system, we first adjusted the YidC gene to the E. coli codons and constructed a vector with the PSC-cleavable CL7-tag fused to the C terminus (SI Appendix, Fig. S10A, schematic drawing). Upon IPTG induction, the YidC expression level was greatly improved (SI Appendix, Fig. S10A, LYS, Left) compared with published studies (6) (Fig. 6A, MF lane). However, almost all YidC remained in the supernatant (SI Appendix, Fig. S10A, SN, Left) upon ultracentrifugation, a traditional first step in membrane protein purification away from cytosolic proteins and NAs (6). One explanation was that, upon lysis, abundant YidC led to very small pieces of membrane fragments, preventing them from sedimenting to the bottom of the tube. Indeed, in the absence of IPTG induction, the YidC expression level was reduced, but the membrane fraction increased upon ultracentrifugation (MF; SI Appendix, Fig. S10B). The MF was then loaded onto the Im7 column in a high concentration of NaCl (0.9 M) and detergent (1.5% dodecyl-maltopyranoside; DDM). A high-purity YidC preparation (∼14 mg protein from ∼20 g cells) was obtained in one-step elution with PSC, much higher than the yield in the published work (6). A small (∼5–7%) but visible impurity was observed in the purified sample (EL; SI Appendix, Fig. S10B). This impurity might be attributable to the covalent linkage of the protein to the membrane components through the Cys residue, which becomes the N-terminal residue upon cleavage by a signal peptidase (31). It was removed by introducing a TEV protease cleavage site immediately after this Cys residue (6). In our case, we mutated this Cys residue to Ser, which often occurs in this position in signal peptides and, thus, is unlikely to affect the susceptibility to the signal peptidase. Indeed, this was the case (Fig. 6B).
With a goal to eliminate the ultracentrifugation step and to simplify the protocol, we found, in an additional trial, that YdiC had significant affinity for NAs and was almost entirely precipitated with DNA by polyethyleneimine (PEI) in ∼0.3–0.35 M salt (SI Appendix, Fig. S10A, Right). A wash of the PEI pellet of an IPTG-induced lysate with the higher salt (0.6 M) buffer containing 1.5% DDM fully released the YidC protein into the buffer solution, largely eliminating the NA component (SI Appendix, Fig. S10A, Right). Before loading on the Im7 column, it was necessary to dilute the supernatant 10-fold with high- (0.9 M) salt buffer, reducing the detergent concentration to 0.15%. The dilution reduced protein micelles that might have formed large dynamic aggregates in solution and on the column, sterically blocking the Im7-active groups from binding the CL7-tag. The incorporation of this dilution step achieved a ∼fourfold improvement in yield (∼18 mg from 4 g of cells) (Fig. 6B). The purified preparation was of the same size (on a gel) as the wild-type protein cleaved off its signal peptide and was free of any impurity (Fig. 6B). Thus, the Cys to Ser mutation did not affect the signal peptide cleavage. The amount of eluted protein, albeit excellent, was lower than the capacity of the column. About 20% of protein remained in the flow-through fraction. A very similar result was obtained for another membrane protein under the same purification conditions (Fig. 6C), suggesting that a fraction of the overexpressed protein might be improperly folded. Overall, we conclude that these results provide a major (∼250-fold) advance in “productivity factor” (mg protein/d purification) over the previous, conventional protocol (6) (Fig. 2).
The human calnexin (CNX) protein is a transmembrane chaperon with a molecular mass of ∼65 kDa. CNX contains a ∼35 residue-long transmembrane segment and two soluble domains on either side of the membrane (32–34). We recently showed that, in vivo, CNX likely forms a physiological complex with the HIV Nef protein. Therefore, it might be a promising candidate for anti-AIDS drug design (34). Detailed functional and structural studies of these interactions as well as inhibitor screening can be performed in vitro if HHH purification of CNX is achieved. To this end, only the N-terminal luminal domain of CNX was previously expressed in the insect cells and purified through a complex, multistep protocol (32). This domain contains two disulfide bonds (33) and undergoes significant conformational changes (oligomerization) dependent on Ca2+, ATP, or both (32). All these properties significantly complicate the expression and purification of the full-length protein. Accordingly, it has never been overexpressed in E. coli and is commercially available only in very small quantities (SI Appendix, Fig. S10C).
To design the CNX overexpression vector and a purification protocol, we used an identical approach to that for YidC, except for one modification. We replaced the natural CNX signal peptide with the mutated YidC signal peptide, which contributed to improved expression of YidC. Consistently, the final yield was ∼20 mg of high-purity CNX protein from 6 g of cells (Fig. 6C). The only minor contamination (∼7%) observed in the purified CNX sample is not related to purification itself but rather represents a truncated fragment of CNX. The recombinant full-length CNX protein likely forms a physiological complex with HIV Nef in vitro as confirmed by kinetic binding assays (34).
Purification of the Multisubunit Condensin Protein Complex Expressed from the Chromosome in the Native Host.
Many functionally significant proteins are not suitable for overexpression approach in alien hosts for a number of reasons. For example, some proteins may be toxic when expressed at high levels. Alternatively, the protein may be poorly soluble or possess defects in folding due to the absence of natural accessory chaperons or important yet unknown natural binding partners. All these difficulties could be alleviated if the affinity tag is directly introduced into the target gene in the native host chromosome and the fusion protein is expressed in the natural context.
However, purification of the regulatory proteins expressed in the host organisms without overexpression is challenging because of their low expression levels (0.001–0.0001% or lower at protein mass/cell mass). This major disadvantage makes it practically impossible to achieve the HHH purification either due to insufficient affinity of the existing chromatographic techniques to the target or to the nonspecific association of affinity columns to untagged cellular proteins. In most cases, such trials are limited to analytical-scale purification or produce substantially contaminated samples.
To test whether the sensitivity of the CL7/Im7 affinity system is sufficient to achieve efficient HHH purification of complex proteins expressed from the native host cells, we chose a condensin complex as a target. Condensins are intricate protein machines that fold and compact the cellular DNA in bacterial and eukaryotic cells into nucleoids or chromosomes, respectively (35–37). Their expression levels are very low (Fig. 7A). The bacterial MukBEF condensin complex consists of three protein subunits (MukB, MukE, and MukF) and has a molecular mass of ∼540 kDa in a proposed B2(E2F)2 configuration (37). An overexpression protocol established for a full E. coli MukBEF complex or its reconstitution from the individually expressed subunits has allowed for protein purification only on a small analytical scale (37). The complex requires metal ions for stabilization and has a tendency to dissociate even upon small variations of salt concentrations or under certain chromatographic conditions (37). Accordingly, the crystal structure of only a partial MukBEF complex has been determined by using relatively short, truncated subunits (35).
We have introduced the PSC-cleavable CL7-tag coding sequence directly to the 3′ end of the Salmonella typhimurium chromosomal mukB gene, which encodes the largest subunit. The derived S. typhimurium was cultivated using a fermenter facility. One milligram of the condensin complex was purified from 45 g of cells in essentially the same manner to that of RNAPs, except that PEI precipitation was applied to remove DNA from the lysate in place of the DNase digestion and that a lower (0.5 M NaCl) salt concentration was used during loading. We used a small 1.5-mL Im7 gravity column because of the anticipated low amount of the target complex.
The chief purpose of this purification trial was to evaluate the sensitivity of the Im7 column, the target protein purity, the physiological configuration of the condensin complex, and the stoichiometry of its components. Thus, after washing, the bound MukBEF complex was stripped from the column using 6 M Gdn in place of proteolytic elution by PSC (Fig. 7A). This simple procedure yielded 1 mg of high-purity (∼97%) protein free of contaminating DNA (OD 260/280 ratio, ∼0.55–0.65). Stoichiometry (BE2F) of the complex subunits in the purified sample was as expected (Fig. 7A). Importantly, the identities of the bands were confirmed by mass spectrometry. Unexpectedly, mass spectrometry also identified a prominent band of a molecular chaperone DnaK, which was present at an equimolar abundance as MukB. DnaK is likely an integral part of the functional condensin complex, as it resisted dissociation by 0.8 M NaCl applied to the column during purification.
There are three major implications from these results. First, the CL7/Im7 approach can efficiently achieve >1,000-fold purification of the large, multisubunit DNA-binding protein complex of very low abundance from the native host in a one-step/1-d protocol. Second, in a striking contrast to the overexpressed/reconstituted E. coli MukBEF (37), the Salmonella complex was extremely stable, resistant to both high- and low-salt conditions. The difference in stability might reflect intrinsic properties of the condensin complex of different organisms. Alternatively, it could suggest that the overexpressed/reconstituted samples from E. coli may have some folding/assembly defects due to unnatural expression conditions. One distinct possibility is that the overexpression/reconstitution systems lack some essential cellular component(s) required for their proper folding, for complex integrity, or both. The molecular chaperone DnaK is known to assist protein folding and complex assembly. The stoichiometric presence of DnaK in the purified Salmonella MukBEF complex is consistent with its functional role. Finally, only a small amount of the Im7 beads (1.5 mL) were used to obtain 1 mg of purified complex. We therefore do not anticipate significant difficulties to obtain crystallization quantities (10+ mg) of the protein complex via the same protocol through scaling up the amount of beads and cell lysates.
Expression and Purification of Human RSF1.
The human remodeling and spacing factor (RSF) complex consists of two subunits (hSNF2H and RSF1) and was previously implicated in mediating nucleosome assembly (38). RSF1 is 1,441 amino acids in length with a molecular mass of ∼164 kDa. Expression and purification of RSF1 was previously conducted in the insect sf9 cells using the baculovirus system (38). The yield was rather small, limiting in vitro functional studies. Due to the large size and unknown characteristics, there has not been an attempt to express and purify RSF1 in prokaryotic cells.
To elucidate the function of RSF1, to identify its functional domain, and to test whether the His-Trap or the CL7/Im7 approach is suitable for purification and functional (pull-down) assays for this eukaryotic protein, we established an expression and purification protocol for RSF1 for E. coli cells. The NA sequence of the RSF1 gene was adjusted to the E. coli codons. We then constructed a dual-tagged (His8/CL7) vector for expressing the full-length RSF1 and 10 serial fragments (Fig. 7B). In addition, a PSC-cleavable expression tag was introduced to the N terminus. The serial fragments differ substantially in sequence or size from one another. They therefore possess distinct chemical and structural properties and essentially represent unrelated purification targets.
RSF1 is likely a multidomain protein, and the exact domain boundaries are difficult to identify based on only the sequence or homology information. Therefore, there is the possibility that some of the arbitrary fragments might harbor structurally incomplete and unfolded domains, affecting their solubility. To accommodate for this possibility, we introduced to the C terminus the SUMO domain, which is known to increase solubility of the protein targets (Fig. 7B). Consistently, no solubility problems were detected for any of the constructs in either the expression or purification trials.
In this first experimental design, we used a two-step purification protocol with His-Trap in the first step to evaluate the amount of proteins recovered (Fig. 7B). A second Im7 step was immediately used in pull-down assays to identify the functionally active fragments capable of interacting with proteins in mammalian cells. The minimal functional domain was then further delineated upon additional dissection (Fig. 7B, fragments F7.1 and F7.2).
Our results can be summarized as follows. First, the full-length RSF1 and several fragments purified through the His-Trap column demonstrated significant contaminants (Fig. 7B). Different fragments demonstrate distinct patterns of impurities, consistent with our prediction that they represent essentially unrelated purification targets (Fig. 7B). Second, the His-Trap could not be used for pull-down assays, as the Ni2+ beads exhibit intrinsic affinities to heavily charged protein, such as histones. Furthermore, a one-step purification via Im7 consistently provided similar quality samples to that of the two-step protocol. In the one-step purification, lysates containing the full-length RSF1 or its largest fragment (F10; Fig. 7B) were directly loaded on the Im7 column in high salt. Thus, a single Im7-based purification protocol could be scaled up for high-yield protein production with no major obstacles. Finally, the CL7/Im7 approach proved to be efficient in pull-down assays, allowing for detailed functional characterization of RSF1 and its 12 distinct fragments at the molecular level. We suggest that it could also be effectively used in functional studies of the other biological systems.
Discussion
We have developed a CL7/Im7 ultra-high-affinity system, which is capable of a one-step/1-d HHH purification of proteins from the most challenging, distinct “purification” classes (Figs. 4–6). No conventional technique was able to achieve a one-step HHH purification of proteins studied in this work, nor of proteins used for high-throughput crystallographic analysis (4) or other high-impact structural studies (SI Appendix, Figs. S5 and S6). Overall, our HHH purification via the CL7/Im7 system results in a 10–500-fold improvement in productivity over the previously published, multistep protocols (Fig. 2). A notable illustration of the superior performance of the CL7/Im7 approach is that the CNX protein sample, which we purified in a few hours and from only a few grams of E. coli cells, would have a market value of ∼$400,000, according to the current commercial prices (SI Appendix, Fig. S11C).
Importantly, due to its ultrahigh sensitivity, the CL7/Im7 technique is able to achieve a one-step HHH purification for low-abundance proteins expressed from the context of the chromosomes of the native hosts. This capability is crucial for a big pool of functionally important proteins for which overexpression is not possible. The modern recombineering technologies (39, 40) are capable of genetically introducing CL7 tags into any proteins in bacterial or eukaryotic chromosomes, as we have demonstrated with the MukB gene used to purify the MukBEF–DnaK complex (Fig. 7A).
The CL7/Im7 schema can also be applied to the study of essential genes with unknown structures and function. This is exemplified by the application to the study of human RSF1. In this study, the CL7/Im7 approach is readily used to establish analytical purifications and functional characterization using pull-down assays (Fig. 7B), an essential research tool in most biochemical studies and related disciplines.
Our results with the three distinct multisubunit, RNA/DNA-binding proteins (two evolutionary distant RNAPs and MukBEF) and two different membrane proteins show that once a purification protocol is established for one protein, it can be successfully applied to other proteins from the same purification category with no major modifications. Based on this experience, it is reasonable to predict that the CL7/Im7 approach would work equally well for other targets from multiple purification categories, including proteins of little known structure and property such as the relatively large human RSF1 expressed in E. coli (Fig. 7B).
The CL7/Im7 approach combines the distinctive advantages of His-Trap and other affinity techniques. Similar to His-Trap, but unlike other affinity methods, the Im7 column has a high capacity and binds stably to the CL7-tag during the first and critical step of the crude lysate loading in high-salt buffers. The high-salt resistance is imperative for achieving HHH purification of proteins with intrinsic NA-binding affinity, such as RNAPs (Figs. 4C and 5A) and MukBEF (Fig. 7A). However, unlike His-Trap, but similar to other (protein/ligand) affinity systems, the Im7 unit possesses no affinity for untagged cellular components (Fig. 1D) and no sensitivity to some reagents widely used for protein purification, such as PEI, metal chelating agents, and reducing compounds.
Our purification trials showed that the Im7-based chromatography operates successfully under a wide range of conditions (see SI Appendix, SI Materials and Methods). This adaptability allows a user to identify the optimal setup common to a large group of proteins as well as to adjust them individually for each particular protein, if necessary. Nevertheless, additional original purification trials should be performed to verify these predictions. For example, we cannot rule out the possibility that poor folding, poor solubility, or both of some targets upon their overexpression will make their purification inefficient, if not impossible. In those cases, the ultrahigh affinity and sensitivity of the CL7/Im7 system offer a rational alternative plan by tagging a target protein in the context of a host chromosome (Fig. 7A). However, it is still possible that there will be new expression- or target-specific challenges to overcome and that additional improvements of the CL7/Im7 system would be required.
The success in the development and application of the CL7/Im7 purification system has additional important implications. First, the family of colicins includes three additional enzyme/inhibitor pairs (CE2/Im2, CE8/Im8, and CE9/Im9). Members of this family share high sequence/structural similarities and ultrahigh affinity yet lack cross-affinity (17, 18). Thus, by using an analogous approach to that developed for the CL7/Im7 affinity column, this protein family could offer three additional, mutually orthogonal, affinity-chromatography systems. The availability of several such systems for multitag purification is gaining attention and importance, as a rapidly increasing number of complex multisubunit biological systems are under study by many research groups worldwide. In general, the necessity for purification through multitag protocols increases with the number of subunits.
Second, SPR is a critical technique to characterize binding partners in the physiological complexes and to validate results of inhibitory drug screening (41). Technically, SPR is quite similar to chromatography. One of the binding partners should be covalently attached to a solid support (sensor), whereas the other in solution flows over the sensor chip. Given the earlier successful application of the WT CE7/Im7 complex in the SPR experiments (19), the advantages of the engineered CL7/Im7 system over its WT precursor would additionally facilitate SPR studies. We suggest that the CL7/Im7 affinity approach would be applicable to constructing a new generation of efficient, reusable Im7-activated SPR biosensors to which various CL7-tagged targets may be immobilized under physiological conditions.
In conclusion, the CL/Im purification approach overcomes significant weaknesses of current commercially available chromatography systems and provides several crucial advantages. It may emerge as the most efficient and potentially universal tool for high-throughput studies of many significant biological systems. The CL/Im chromatography would also facilitate large-scale production of biologically active proteins by the pharmaceutical industry and form a basis for high-throughput protocols to characterize intermolecular interactions. In addition to crystallography, the CL/Im approach may also facilitate (single molecule) cryo-EM studies (42), another high-resolution structural technique for which homogeneous, high-purity protein samples are of central importance.
Methods
Full details of the methods used are presented in SI Appendix, SI Materials and Methods.
Cloning, Protein Expression, and Purification.
Cloning and protein expression used standard methods. Purification of the proteins included in this work was carried out by using the conventional protocols, the newly developed CL7/Im7 technique, or both.
Fluorescent Transcription Assays.
The components of the transcription elongation scaffold, the 18-mer RNA (RNA18) labeled with fluorescein (FLU) at the 5′-end, and template (T) and nontemplate (NT) oligonucleotides were ordered from IDT and then assembled as described previously (43). The elongation complexes (ECs) formed by RNAPs at this scaffold were trapped in a stable posttranslocated state as revealed by no signs of the exo-/endonucleolytic RNA degradation upon their incubation with an NTP-free reaction buffer and excess (10 mM) of Mg2+ ions for ∼5 h. The transcription elongation reactions were carried out under the following conditions: 0.1 M NaCl, 50 mM Tris, pH 8.0, 5 mM MgCl2, 100 μM NTPs, ∼2.83 μM RNAP, and ∼2.55 μM RNA18 (in the scaffold) at 20 °C. The reactions were stopped by adding 8 M urea and run on denaturing 20% polyacrylamide gels also containing urea. The gels were then processed by the fluorescent imager, Typhoon Trio, and quantified with the respective ImageQuant program (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Drs. David Bedwell and David Schneider for the helpful comments and discussions during manuscript preparation. The D.G.V. laboratory is supported by setup/discretionary funds from UAB; M.B.R. is supported by NIH Grant GM98539; H. Wang is supported by grants from the Leukemia and Lymphoma Society and NIH Grant GM081489; and L.T.C. is supported by funds from the Anderson Family Endowed Chair.
Footnotes
Conflict of interest statement: The CL7/Im7 purification technology is protected by a PCT patent (PCT/US2016/065843) filed by the UAB Research Foundation.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704872114/-/DCSupplemental.
References
- 1.Kimple ME, Brill AL, Pasker RL. Overview of affinity tags for protein purification. Curr Protoc Protein Sci. 2013;73:9. doi: 10.1002/0471140864.ps0909s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fong BA, Wu WY, Wood DW. The potential role of self-cleaving purification tags in commercial-scale processes. Trends Biotechnol. 2010;28:272–279. doi: 10.1016/j.tibtech.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Lichty JJ, Malecki JL, Agnew HD, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein purification. Protein Expr Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 4.Gräslund S, et al. Structural Genomics Consortium; China Structural Genomics Consortium; Northeast Structural Genomics Consortium Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee R, Rudra P, Prajapati RK, Sengupta S, Mukhopadhyay J. Optimization of recombinant Mycobacterium tuberculosis RNA polymerase expression and purification. Tuberculosis (Edinb) 2014;94:397–404. doi: 10.1016/j.tube.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Kumazaki K, et al. Crystallization and preliminary X-ray diffraction analysis of YidC, a membrane-protein chaperone and insertase from Bacillus halodurans. Acta Crystallogr F Struct Biol Commun. 2014;70:1056–1060. doi: 10.1107/S2053230X14012540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumazaki K, et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 8.Ohana RF, et al. HaloTag-based purification of functional human kinases from mammalian cells. Protein Expr Purif. 2011;76:154–164. doi: 10.1016/j.pep.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7:91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 10.Wegner GJ, Lee HJ, Corn RM. Characterization and optimization of peptide arrays for the study of epitope-antibody interactions using surface plasmon resonance imaging. Anal Chem. 2002;74:5161–5168. doi: 10.1021/ac025922u. [DOI] [PubMed] [Google Scholar]
- 11.Manjeet K, Purushotham P, Neeraja C, Podile AR. Bacterial chitin binding proteins show differential substrate binding and synergy with chitinases. Microbiol Res. 2013;168:461–468. doi: 10.1016/j.micres.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Saha K, Bender F, Gizeli E. Comparative study of IgG binding to proteins G and A: Nonequilibrium kinetic and binding constant determination with the acoustic waveguide device. Anal Chem. 2003;75:835–842. doi: 10.1021/ac0204911. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt TG, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 14.Miller DM, 3rd, Olson JS, Pflugrath JW, Quiocho FA. Rates of ligand binding to periplasmic proteins involved in bacterial transport and chemotaxis. J Biol Chem. 1983;258:13665–13672. [PubMed] [Google Scholar]
- 15.Tessema M, et al. Glutathione-S-transferase-green fluorescent protein fusion protein reveals slow dissociation from high site density beads and measures free GSH. Cytometry A. 2006;69:326–334. doi: 10.1002/cyto.a.20259. [DOI] [PubMed] [Google Scholar]
- 16.Knecht S, Ricklin D, Eberle AN, Ernst B. Oligohis-tags: Mechanisms of binding to Ni2+-NTA surfaces. J Mol Recognit. 2009;22:270–279. doi: 10.1002/jmr.941. [DOI] [PubMed] [Google Scholar]
- 17.Wallis R, Moore GR, James R, Kleanthous C. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion-controlled association and femtomolar binding for the cognate complex. Biochemistry. 1995;34:13743–13750. doi: 10.1021/bi00042a004. [DOI] [PubMed] [Google Scholar]
- 18.Wallis R, et al. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry. 1995;34:13751–13759. doi: 10.1021/bi00042a005. [DOI] [PubMed] [Google Scholar]
- 19.Hosse RJ, et al. Kinetic screening of antibody-Im7 conjugates by capture on a colicin E7 DNase domain using optical biosensors. Anal Biochem. 2009;385:346–357. doi: 10.1016/j.ab.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 20.Wang YT, Yang WJ, Li CL, Doudeva LG, Yuan HS. Structural basis for sequence-dependent DNA cleavage by nonspecific endonucleases. Nucleic Acids Res. 2007;35:584–594. doi: 10.1093/nar/gkl621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svetlov V, Artsimovitch I. Purification of bacterial RNA polymerase: Tools and protocols. Methods Mol Biol. 2015;1276:13–29. doi: 10.1007/978-1-4939-2392-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuznedelov K, et al. Recombinant Thermus aquaticus RNA polymerase for structural studies. J Mol Biol. 2006;359:110–121. doi: 10.1016/j.jmb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Thompson NE, Foley KM, Stalder ES, Burgess RR. Identification, production, and use of polyol-responsive monoclonal antibodies for immunoaffinity chromatography. Methods Enzymol. 2009;463:475–494. doi: 10.1016/S0076-6879(09)63028-7. [DOI] [PubMed] [Google Scholar]
- 24.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 25.Vassylyev DG. Elongation by RNA polymerase: A race through roadblocks. Curr Opin Struct Biol. 2009;19:691–700. doi: 10.1016/j.sbi.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassylyev DG, et al. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 27.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 28.Vassylyeva MN, et al. Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus. Acta Crystallogr D Biol Crystallogr. 2002;58:1497–1500. doi: 10.1107/S0907444902011770. [DOI] [PubMed] [Google Scholar]
- 29.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 30.Pandey A, Shin K, Patterson RE, Liu XQ, Rainey JK. Current strategies for protein production and purification enabling membrane protein structural biology. Biochem Cell Biol. 2016;94:507–527. doi: 10.1139/bcb-2015-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resh MD. Covalent lipid modifications of proteins. Curr Biol. 2013;23:R431–R435. doi: 10.1016/j.cub.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou WJ, Bergeron JJ, Li Y, Kang CY, Thomas DY. Conformational changes induced in the endoplasmic reticulum luminal domain of calnexin by Mg-ATP and Ca2+ J Biol Chem. 1995;270:18051–18059. doi: 10.1074/jbc.270.30.18051. [DOI] [PubMed] [Google Scholar]
- 33.Schrag JD, et al. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 34.Hunegnaw R, et al. Interaction between HIV-1 Nef and calnexin: From modeling to small molecule inhibitors reversing HIV-induced lipid accumulation. Arterioscler Thromb Vasc Biol. 2016;36:1758–1771. doi: 10.1161/ATVBAHA.116.307997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo JS, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 36.She W, Wang Q, Mordukhova EA, Rybenkov VV. MukEF is required for stable association of MukB with the chromosome. J Bacteriol. 2007;189:7062–7068. doi: 10.1128/JB.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrushenko ZM, Lai CH, Rybenkov VV. Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J Biol Chem. 2006;281:34208–34217. doi: 10.1074/jbc.M606723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loyola A, et al. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23:6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Singh V, Braddick D, Dhar PK. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene. 2017;599:1–18. doi: 10.1016/j.gene.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Piliarik M, Vaisocherová H, Homola J. Surface plasmon resonance biosensing. Methods Mol Biol. 2009;503:65–88. doi: 10.1007/978-1-60327-567-5_5. [DOI] [PubMed] [Google Scholar]
- 42.Neyer S, et al. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016 doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- 43.Kashkina E, et al. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 2006;34:4036–4045. doi: 10.1093/nar/gkl559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.