Significance
As the largest class of natural products found in all living organisms, terpenoids play essential roles in plant growth, development, respiration, photosynthesis, and environmental interactions. Geranylgeranyl diphosphate (GGPP), a precursor for several terpenoid metabolic branches including chlorophyll, carotenoid, and gibberellin biosynthesis, is produced by GGPP synthase (GGPPS) in plastids. We discovered that rice GGPPS recruiting protein (GRP), which forms a heterodimer with the only plastidic GGPPS, controls GGPPS dimerization state and enhances its catalytic properties. By interacting with GGPPS, GRP determines its allocation from stroma to thylakoid membranes, where the heterodimer exists in a complex with chlorophyll biosynthetic proteins. GGPPS recruitment to thylakoids by GRP represents a mechanism directing metabolic flux toward a specific product in the terpenoid metabolic network.
Keywords: terpenoids, geranylgeranyl diphosphate synthase, chlorophyll biosynthesis, rice, protein complex formation
Abstract
In plants, geranylgeranyl diphosphate (GGPP) is produced by plastidic GGPP synthase (GGPPS) and serves as a precursor for vital metabolic branches, including chlorophyll, carotenoid, and gibberellin biosynthesis. However, molecular mechanisms regulating GGPP allocation among these biosynthetic pathways localized in the same subcellular compartment are largely unknown. We found that rice contains only one functionally active GGPPS, OsGGPPS1, in chloroplasts. A functionally active homodimeric enzyme composed of two OsGGPPS1 subunits is located in the stroma. In thylakoid membranes, however, the GGPPS activity resides in a heterodimeric enzyme composed of one OsGGPPS1 subunit and GGPPS recruiting protein (OsGRP). OsGRP is structurally most similar to members of the geranyl diphosphate synthase small subunit type II subfamily. In contrast to members of this subfamily, OsGRP enhances OsGGPPS1 catalytic efficiency and specificity of GGPP production on interaction with OsGGPPS1. Structural biology and protein interaction analyses demonstrate that affinity between OsGRP and OsGGPPS1 is stronger than between two OsGGPPS1 molecules in homodimers. OsGRP determines OsGGPPS1 suborganellar localization and directs it to a large protein complex in thylakoid membranes, consisting of geranylgeranyl reductase (OsGGR), light-harvesting-like protein 3 (OsLIL3), protochlorophyllide oxidoreductase (OsPORB), and chlorophyll synthase (OsCHLG). Taken together, genetic and biochemical analyses suggest OsGRP functions in recruiting OsGGPPS1 from the stroma toward thylakoid membranes, thus providing a mechanism to control GGPP flux toward chlorophyll biosynthesis.
Terpenoids are one of the largest and most structurally diverse classes of plant primary and secondary metabolites and are involved in numerous physiological and ecological processes ranging from growth and development to communication, environmental adaptation, and defense (1, 2). All terpenoids derive from the universal five-carbon (C5) units, isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP), which are synthesized in plants by two compartmentally separated, but metabolically cross-talking, isoprenoid pathways, the mevalonate and methylerythritol phosphate pathways (3). IPP and DMAPP are subsequently used by short-chain prenyltransferases (PTSs) to produce prenyl diphosphate precursors: geranyl diphosphate (GPP, C10), farnesyl diphosphate (FPP, C15), and geranylgeranyl diphosphate (GGPP, C20) (2, 4, 5). Short-chain PTSs determine not only the chain length of their products but also the compartmental localization of product biosynthesis. By catalyzing the branch-point reactions, they control precursor flux toward various classes of terpenoids.
Among the prenyl diphosphate intermediates, GGPP biosynthesis represents an essential metabolic node, as this precursor is shared by several vital metabolic pathways, particularly in plastids, including the biosynthesis of carotenoids and their derivatives (e.g., plant hormones abscisic acid and strigolactones), as well as gibberellins, plastoquinones, tocopherols, and the phytyl tail of chlorophylls (3). However, it remains largely unknown how allocation of GGPP among different downstream metabolic branches is regulated in plants, especially for pathways located in the same subcellular compartment. It has been proposed that different members of a GGPP synthase (GGPPS) multigene family provide GGPP for different metabolic branches (6, 7), or GGPPS protein–protein interactions with downstream GGPP-using enzymes allow GGPP channeling to specific downstream pathways (8).
To address this question, we used rice as a model system. Here, we report that the rice genome encodes one functionally active GGPPS in plastids, which exists as homo- and heterodimeric proteins. Its dimerization state, product specificity, and catalytic efficiency are controlled by GGPPS recruiting protein (GRP), which also determines GGPPS subchloroplast localization. Genetic analysis further revealed that targeting GGPPS to thylakoid membranes via protein–protein interactions plays a key role in regulating GGPP flux toward chlorophyll biosynthesis.
Results
Rice Has Only One Functional GGPPS in Plastids.
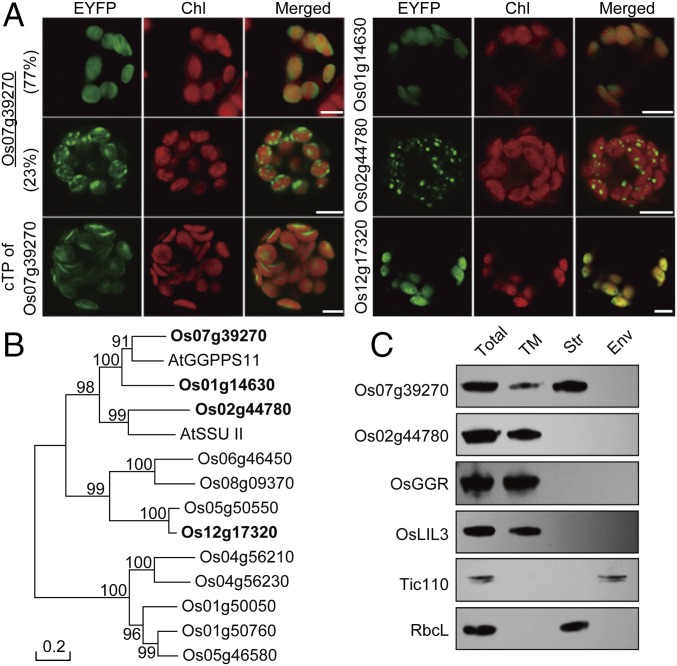
A search of the rice genome for genes with sequence similarities to PTSs from Arabidopsis thaliana revealed 12 PTS candidate genes (SI Appendix, Table S1 and Fig. S1). Analysis of their amino acid sequences using multiple subcellular localization prediction programs (PSORT, YLoc, TargetP, and Predator) predicted that three to four proteins are localized to plastids. To experimentally determine subcellular localization, ORFs of all 12 candidate genes were fused to the 5′-end of the enhanced yellow fluorescent protein (EYFP) gene and transiently expressed in rice leaf protoplasts. The EYFP signal was found in chloroplasts for four proteins encoded by Os01g14630, Os07g39270, Os02g44780, and Os12g17320 (Fig. 1A and SI Appendix, Fig. S2), thus confirming their predicted plastidic localization. Interestingly, of four proteins, Os07g39270 displayed two different fluorescent patterns: a diffused pattern in chloroplasts of most protoplasts (27 of 35 analyzed) and a speckled pattern in the remaining protoplasts (Fig. 1A). No speckled fluorescence pattern was observed when only Os07g39270 chloroplast transit peptide was fused to the N terminus of EYFP (Fig. 1A), suggesting the mature protein contributes to its thylakoid localization. In contrast, the Os02g44780-EYFP protein displayed exclusively the speckled pattern in chloroplasts (Fig. 1A). Phylogenetic analysis revealed that among all 12 homologs, only Os01g14630 and Os07g39270 cluster with functionally characterized GGPPSs from other plant species (8, 9) (Fig. 1B and clade 1 in SI Appendix, Fig. S3).
Fig. 1.
Subcellular localization and phylogenetic analysis of rice candidate GGPPSs. (A) Transient expression of four rice PTS homologs fused to EYFP in rice leaf protoplasts showing plastidic localization. Os07g39270 (OsGGPPS1) exhibits two patterns. (Upper) Representative of 77% protoplasts. (Middle) Representative of 23% protoplasts. Chl, chlorophyll autofluorescence; cTP, chloroplast transit peptide of Os07g39270 (OsGGPPS1) fused to EYFP. (Scale bars, 5 μm.) (B) Phylogenetic analysis of 12 rice PTS homologs with AtGGPPS11 and AtSSU II. Rice plastidic PTSs are shown in bold. (C) Immunoblot analysis of Os07g39270 (OsGGPPS1) and Os02g44780 (OsGRP) subchloroplast localization. Env, envelope membranes; Str, stroma; TM, thylakoid membrane. Tic110 and RbcL were used as markers for the envelope and stroma fractions, respectively. OsGGR, rice geranylgeranyl reductase; OsLIL3, rice light-harvesting-like protein 3.
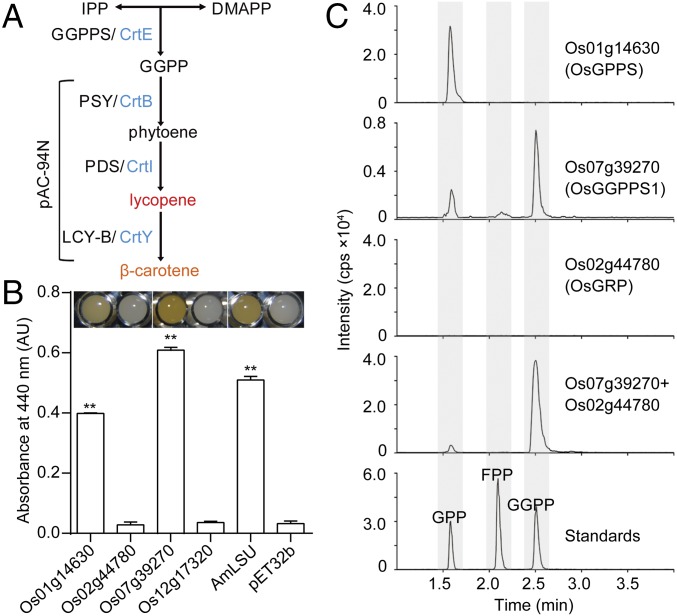
To examine the ability of these four plastidic PTS homologs to synthesize GGPP, we performed complementation assays. Each candidate gene was transformed into Escherichia coli along with the pAC-94N vector carrying a gene cluster for carotenoid biosynthesis (Fig. 2A) (10). E. coli by itself is capable of producing IPP, DMAPP, and FPP, but not GGPP; thus, coexpression of a functional GGPPS will facilitate β-carotene biosynthesis and change the white color of colonies to orange. Only colonies harboring Os01g14630 and Os07g39270 produced β-carotene, suggesting both proteins are able to synthesize GGPP in E. coli (Fig. 2B).
Fig. 2.
Functional characterization of four rice plastidic PTS homologs. (A) Diagram showing the carotenoid biosynthesis pathway in Erwinia herbicola. Plasmid pAC-94N contains the E. herbicola carotenoid biosynthetic genes, crtB, crtI, and crtY, with the exception of crtE. When coexpressed with a functional GGPPS, the pathway is complemented and β-carotene is produced. (B) Complementation assay for detecting GGPPS activity of the rice PTS homologs that have chloroplast localization. Functionally characterized GGPPS from Antirrhinum majus (AmLSU) and pET32b empty vector were used as positive and negative controls, respectively. Data are means ± SEM (n = 3 biological replicates). **P < 0.01; Student’s t test. (C) LC-MS/MS chromatograms showing the products generated by Os01g14630 (OsGPPS), Os07g39270 (OsGGPPS1), Os02g44780 (OsGRP), and a mixture of Os07g39270 and Os02g44780 (1:1) from IPP and DMAPP in vitro, as well as GPP, FPP, and GGPP standards (1 μM each).
To further functionally characterize Os01g14630 and Os07g39270, affinity-purified His-tagged recombinant proteins (SI Appendix, Fig. S4) were incubated with IPP as a substrate and DMAPP, GPP, or FPP as a cosubstrate, and products were analyzed by LC-MS/MS (Fig. 2C and SI Appendix, Fig. S5 A–C). Os07g39270 protein exhibited GGPPS activity, producing GGPP as the main product from IPP and DMAPP (67.3%) with small amounts of GPP (27.2%) and FPP (5.5%) (Fig. 2C). It could also use GPP and FPP as cosubstrates with IPP, producing a mixture of FPP (64.6%) and GGPP (35.4%), or sole GGPP, respectively (SI Appendix, Fig. S5 A and B). In contrast, Os01g14630 produced GPP as the only product from IPP and DMAPP (Fig. 2C). It was also able to produce GGPP from IPP and FPP (SI Appendix, Fig. S5B), thus explaining GGPP formation in the E. coli complementation assays (Fig. 2B). Taken together, our results indicate that Os01g14630 is a GPP synthase (GPPS) and Os07g39270 is a GGPPS (designated as OsGGPPS1).
Os02g44780 with Homology to Members of GPPS Small Subunit Type II Subfamily Enhances OsGGPPS1 Catalytic Efficiency and Specificity.
One of the four identified plastidic PTS homologs, Os02g44780, appeared to belong to a GPPS small subunit type II (SSU II) subfamily (Fig. 1B and clade 2 in SI Appendix, Fig. S3). Members of this subfamily are catalytically inactive but are able to bind GGPPS and modulate its product specificity from GGPP to GPP (11). Recombinant Os02g44780 protein (SI Appendix, Fig. S4) displayed no catalytic activity when assayed with IPP and any of the three cosubstrates (Fig. 2C and SI Appendix, Fig. S5 A and B), which was consistent with the E. coli complementation assays (Fig. 2B). Moreover, crystallographic analysis revealed that Os02g44780 contains a loop in its catalytic center that prevents substrate binding (SI Appendix, Fig. S6).
To determine whether Os02g44780 is able to interact with GGPPS and change its product specificity from GGPP to GPP, PTS activity was determined using a mixture of equal amounts of recombinant OsGGPPS1 and Os02g44780 proteins. Unlike OsGGPPS1 alone, a mixture of both proteins produced predominantly GGPP from IPP with any of the three co-substrates (DMAPP, GPP, or FPP) (Fig. 2C and SI Appendix, Fig. S5 A and B), thus displaying improved, but not modified, OsGGPPS1 product specificity. Primarily GGPP formation from GPP and IPP was a result of an increased catalytic efficiency of OsGGPPS1/Os02g44780 toward FPP as a substrate. An approximately sevenfold increase in Vmax with FPP in the mixture of OsGGPPS1 and Os02g44780 (Table 1) facilitates conversion of FPP, derived from a promiscuous OsGGPPS1 side reaction, to GGPP (SI Appendix, Fig. S5B). Moreover, in the presence of the catalytically inactive Os02g44780, OsGGPPS1 exhibits ∼14.5-fold higher affinity for IPP than OsGGPPS1 alone (Table 1). The addition of Os02g44780 to OsGGPPS1 at a 2:1 ratio did not alter kinetic parameters of OsGGPPS1/Os02g44780 (SI Appendix, Table S2). In addition, E. coli harboring both OsGGPPS1 and Os02g44780 produced more β-carotene in complementation assays than those expressing only OsGGPPS1 (SI Appendix, Fig. S5D). Taken together, these results suggest that the interaction with Os02g44780 not only increases product specificity of OsGGPPS1 but also accelerates its activity.
Table 1.
Kinetic parameters for homodimeric and heterodimeric OsGGPPS1
| Substrate | OsGGPPS1 | OsGGPPS1/Os02g44780 (OsGRP) | ||
| Km, μM | Vmax, nmol⋅min−1⋅mg−1 | Km, μM | Vmax, nmol⋅min−1⋅mg−1 | |
| DMAPP (at 60 μM IPP) | 10.87 ± 1.35 | 2.85 ± 0.09 | 14.30 ± 2.53 | 11.84 ± 0.49 |
| GPP (at 60 μM IPP) | 3.75 ± 0.93 | 10.82 ± 0.46 | 6.70 ± 1.88 | 24.89 ± 1.73 |
| FPP (at 60 μM IPP) | 12.34 ± 2.38 | 3.81 ± 0.22 | 10.16 ± 2.14 | 26.87 ± 2.03 |
| IPP (at 100 μM DMAPP) | 22.49 ± 3.40 | 4.20 ± 0.21 | 1.55 ± 0.22 | 4.76 ± 0.08 |
Data are means ± SD (n = 3 independent experiments).
OsGGPPS1 Interacts with Os02g44780 in Chloroplast Thylakoids.
To determine the subchloroplast compartments corresponding to the diffused (OsGGPPS1-EYFP only) and speckled (OsGGPPS1- and Os02g44780-EYFP) signals (Fig. 1A), isolated rice chloroplasts were fractionated into stroma, thylakoid, and envelope membrane fractions and subjected to Western blot analysis. OsGGPPS1 was found in both stroma and thylakoid membrane fractions, whereas Os02g44780 was detected solely in thylakoid membranes (Fig. 1C).
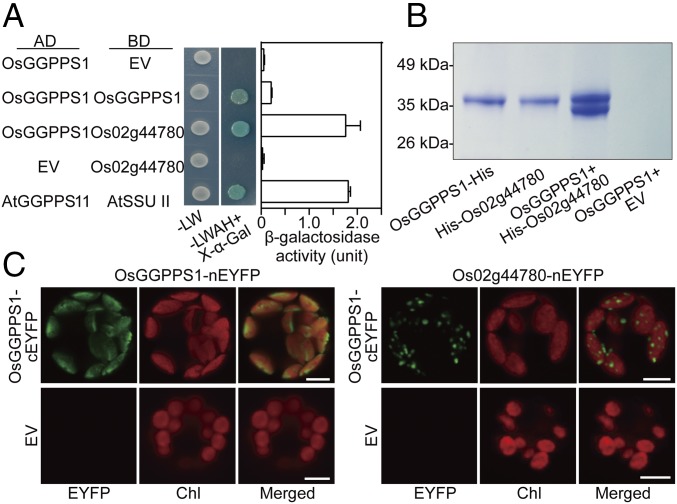
Given the thylakoid localization of Os02g44780 and OsGGPPS1, using yeast two-hybrid (Y2H), pull-down, and bimolecular fluorescence complementation (BiFC) assays (12), we analyzed whether these proteins interact with each other. Indeed, Y2H assays revealed that OsGGPPS1 could interact with itself and with Os02g44780 (Fig. 3A). Quantitative assays of the β-galactosidase activity showed that interaction between OsGGPPS1 and Os02g44780 was eightfold stronger than that between two OsGGPPS1 molecules (Fig. 3A). For pull-down assays, untagged OsGGPPS1 was coexpressed with His-tagged Os02g44780 in E. coli. OsGGPPS1 was copurified with His-Os02g44780 on Ni-nitrilotriacetic acid (NTA) agarose with a ratio of ∼1:1, demonstrating the interaction between these two proteins (Fig. 3B). Moreover, BiFC assays revealed that two molecules of OsGGPPS1 fused to the N- and C-terminal halves of EYFP (nEYFP and cEYFP, respectively) interact with each other in the stroma displaying a diffused fluorescence (Fig. 3C). In contrast, BiFC assays with OsGGPPS1 and Os02g44780 exhibited a speckled fluorescence pattern exclusively in thylakoid membranes (Fig. 3C). On the basis of these results and further genetic evidence (see following), we designated Os02g44780 as GGPPS recruiting protein (OsGRP).
Fig. 3.
Analysis of OsGGPPS1 and Os02g44780 interactions. (A) Y2H detection of protein–protein interactions. Yeast cells harboring both constructs (Left) were spotted on nonselective (−LW) and selective medium supplied with X-α-Gal (−LWAH+X-α-Gal). Interaction strength was measured by β-galactosidase activity. AD, activation domain of pGAD-T7; and BD, DNA binding domain of pGBK-T7, to which OsGGPPS1 and Os02g44780 were fused; EV, empty vector. Known interaction between Arabidopsis AtGGPPS11 and AtSSU II was used as a positive control. Data are means ± SEM (n = 3). (B) Pull-down analysis of OsGGPPS1 binding to His-tagged Os02g44780. (C) BiFC detection of protein–protein interactions in rice leaf protoplasts. Coexpressed fusion constructs are indicated. Fluorescence of reconstructed EYFP is shown in the “EYFP” panel. Chl, chlorophyll autofluorescence. (Scale bars, 5 μm.)
High Affinity of OsGRP Toward OsGGPPS1 Favors Heterodimer Formation.
To investigate the molecular mechanism of the protein interactions, we carried out structural analysis. Crystal structures of both the OsGGPPS1/OsGGPPS1 and OsGGPPS1/OsGRP dimers were solved in apo forms at 2.0- and 3.6-Å resolutions, respectively (SI Appendix, Fig. S7A). Statistics of data collection and model refinement are summarized in SI Appendix, Table S3. The homodimeric OsGGPPS1 structure was similar to other established GGPPS structures (SI Appendix, Fig. S7A) (13–15). The heterodimeric rice GGPPS was formed by an OsGGPPS1 and an OsGRP monomer (SI Appendix, Fig. S7A). The superimpositions of the structures of OsGRP and OsGGPPS1 yielded the root-mean-square deviations of 1.52 Å, indicating similar overall conformations. In addition, superimposition of the structures of OsGGPPS1 in the homodimer and heterodimer yielded root-mean-square deviations of only 0.64 Å, indicating the binding with OsGRP did not alter the structure of OsGGPPS1 (SI Appendix, Fig. S7B).
Because no significant differences between the homo- (OsGGPPS1/OsGGPPS1) and heterodimers (OsGGPPS1/OsGRP) at the conformation level were observed, we studied the interfaces of both dimers (SI Appendix, Fig. S8 A and B). We found that two hydrophilic residues (His145 and Asp177 in OsGGPPS1, His126* and Asp158* in OsGRP, * denotes residues from OsGRP) are highly conserved in GGPPSs and OsGRP, as well as in SSUs from different plant species (SI Appendix, Fig. S8C and S1). In the homodimer, His145 from each OsGGPPS1 molecule faces Asp177 from the other molecule, and in the heterodimer, His126* and Asp158* from OsGRP face the corresponding OsGGPPS1 residues as well (SI Appendix, Fig. S8C), forming hydrogen bonding interactions at the dimer interfaces.
However, homo- and heterodimers contain several different amino acid residues at their interfaces. Residue Glu231 of OsGGPPS1 is close to Ala93 in the homodimer, but close to Arg68* of OsGRP in the heterodimer (SI Appendix, Fig. S8D). The latter could facilitate an additional hydrogen bond. In addition, three hydrophobic Phe residues (Phe132*, Phe161*, Phe204*) from OsGRP are substituted by other hydrophobic residues (Met151, Leu180, Val227) in the OsGGPPS1 homodimer, which may decrease the hydrophobic interactions (SI Appendix, Fig. S8E).
The contribution of these OsGRP residues to the heterodimer formation was investigated by site-directed mutagenesis. Untagged OsGGPPS1 was coexpressed in E. coli with His-tagged wild-type OsGRP or its different mutants containing one or more amino acid substitutions (SI Appendix, Fig. S8F). Ni-NTA agarose was used to purify His-OsGRP, and the amount of untagged OsGGPPS1 copurified with His-OsGRP was quantified to evaluate affinity of OsGRP and its mutants to OsGGPPS1. These analyses revealed that OsGGPPS1 was captured by wild-type OsGRP at the highest ratio (0.968, reflecting ∼1:1 ratio of OsGGPPS1 to OsGRP), demonstrating their strong interaction (SI Appendix, Fig. S8G, lane 1). Substitution of conserved His126* or Asp158* of OsGRP with Ala drastically reduced the binding ability of OsGRPH126A* to OsGGPPS1 and almost completely eliminated binding ability of OsGRPD158A* (SI Appendix, Fig. S8G, lanes 2 and 3), indicating that the evolutionarily conserved His126* and Asp158* are crucial for the protein–protein interaction of OsGRP with OsGGPPS1.
When the remaining four residues of OsGRP were individually substituted with their corresponding OsGGPPS1 counterparts (OsGRPR68A*, OsGRPF132M*, OsGRPF161L*, and OsGRPF204V*), OsGRP ability to interact with OsGGPPS1 was reduced to different levels (SI Appendix, Fig. S8G, lanes 4–7). When three or all four residues were replaced simultaneously, OsGRP could hardly interact with OsGGPPS1 (SI Appendix, Fig. S8G, lanes 8 and 9). Thus, structural analyses with site-directed mutagenesis and quantitative Y2H studies (Fig. 3A) suggest OsGRP has a structural advantage over OsGGPPS1 in the interaction with another OsGGPPS1 molecule, making the heterodimer formation more favorable than the homodimer formation.
OsGGPPS1/OsGRP Heterodimer Exists in a Multiprotein Complex Containing OsGGR, OsLIL3, OsPORB, and OsCHLG.
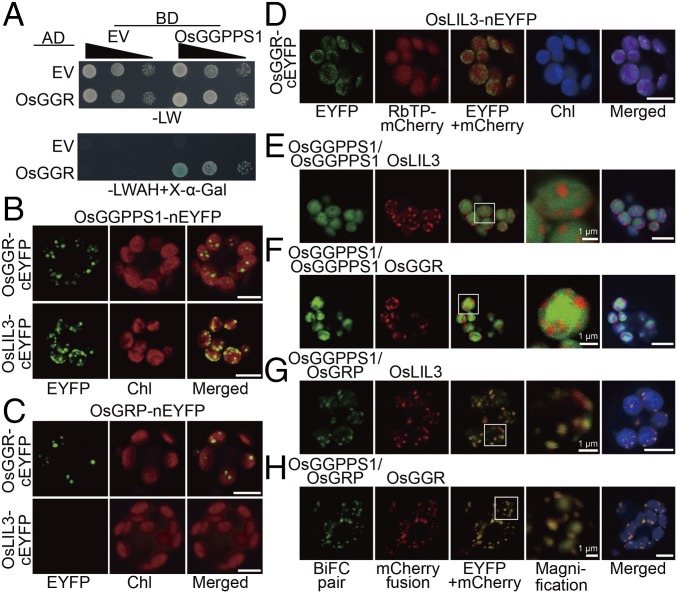
Previous studies showed that GGPPS can interact with several enzymes involved in isoprenoid biosynthesis in plastids downstream of the GGPP node, thereby directing metabolic flux toward specific downstream branches (8). To determine whether the OsGGPPS1/OsGRP dimer is also part of a multiprotein complex, we performed Y2H analyses. As potential interacting partners, we used phytoene synthase (PSY), copalyl diphosphate synthase (CPS), homogentisate geranylgeranyl transferase (HGGT), geranylgeranyl reductase (GGR), and chlorophyll synthase (CHLG), which are involved in carotenoid, diterpene/gibberellin, α-tocotrienol, and chlorophyll biosynthesis, respectively (SI Appendix, Fig. S9). Of the analyzed potential partners, only OsGGR (Os02g51080), an enzyme that reduces GGPP into phytyl diphosphate for chlorophyll biosynthesis in thylakoid membranes (16, 17), interacted with OsGGPPS1 (Fig. 4A and SI Appendix, Fig. S10A). To further examine protein–protein interactions in planta, we performed pairwise BiFC by fusing OsGGPPS1 or OsGRP to nEYFP and the other potential partners (OsPSY, OsCPS, OsHGGT, OsGGR, and OsCHLG) to cEYFP. As in Y2H analysis, interactions were found only between OsGGR and OsGGPPS1 (Fig. 4B and SI Appendix, Fig. S10B). Although interactions between GGR and GGPPS were shown previously in Arabidopsis (8), we found that OsGGR also interacts with OsGRP in rice (Fig. 4C and SI Appendix, Fig. S10C).
Fig. 4.
Interactions among OsGGPPS1/OsGRP heterodimer, OsGGR, and OsLIL3. (A) Y2H assay of OsGGPPS1 and OsGGR interaction. OsGGPPS1 and OsGGR were fused with the DNA binding domain (BD) of pGBK-T7 and the activation domain (AD) of pGAD-T7, respectively. Tenfold serial dilutions of yeast cells expressing both fusion proteins were spotted on nonselective (−LW) medium and selective medium supplied with X-α-Gal (−LWAH+X-α-Gal). (B and C) BiFC detection of the interactions of OsGGPPS1 (B) or OsGRP (C) with OsGGR and OsLIL3. (D) BiFC detection of the interaction between OsLIL3 and OsGGR. (E and F) BiFC assays of the colocalization of OsGGPPS1 homodimer with OsLIL3 (E) and OsGGR (F). (G and H) BiFC assays of the colocalization of OsGGPPS1/OsGRP heterodimer with OsLIL3 (G) and OsGGR (H). (Scale bars, 5 μm.)
GGR is anchored to thylakoid membranes by light-harvesting-like protein 3 (LIL3) in Arabidopsis (18–20). We found only one rice LIL3 ortholog (OsLIL3, Os02g03330). BiFC assays showed that, similar to Arabidopsis, there are interactions between OsLIL3 and OsGGR proteins in rice (Fig. 4D). However, these assays also revealed interactions between OsLIL3 and OsGGPPS1 (Fig. 4B), but not between OsLIL3 and OsGRP (Fig. 4C). To determine whether OsGGPPS1 dimerization state affects these protein–protein interactions, OsGGPPS1-nEYFP and OsGGPPS1-cEYFP or OsGGPPS1-nEYFP and OsGRP-cEYFP were coexpressed with OsLIL3-mCherry or OsGGR-mCherry. Although EYFP fluorescence from the OsGGPPS1 homodimer in the stroma did not overlap with that of the mCherry signals from the thylakoid localized OsLIL3 or OsGGR proteins (Fig. 4 E and F), the EYFP signal from the OsGGPPS1/OsGRP heterodimer perfectly overlapped with mCherry signals of OsLIL3 or OsGGR (Fig. 4 G and H). Taken together, our analysis revealed that OsGGPPS1 as a heterodimer with OsGRP interacts with both OsGGR and OsLIL3, OsGRP interacts only with OsGGR, and OsGGR also interacts with OsLIL3.
As Western blot analysis showed that OsGGPPS1, OsGRP, OsGGR, and OsLIL3 are all localized in thylakoid membranes (Fig. 1C), we analyzed whether these four proteins coexist in a multiprotein complex. Blue native-PAGE of isolated rice thylakoid membranes with subsequent immunoblot analysis revealed that all four proteins were detected at the same position by their corresponding antibodies (SI Appendix, Fig. S11A). On performing SDS/PAGE, as a second dimension, followed by immunoblotting, these four proteins were also found on the same vertical line (SI Appendix, Fig. S11B), confirming they are constituents of the same multiprotein complex. In addition, blue native-PAGE analysis showed that protochlorophyllide oxidoreductase (OsPORB) and OsCHLG [although the latter was not found to directly interact with OsGGPPS1 or OsGRP (SI Appendix, Fig. S10)] are components of the same complex containing the OsGGPPS1/OsGRP dimer, OsGGR, and OsLIL3 (SI Appendix, Fig. S11).
OsGRP Modulates GGPP Supply for Chlorophyll Biosynthesis.
On the basis of subchloroplast localization (Figs. 1C and 4 E–H), it can be assumed that OsGGPPS1 homodimers contribute GGPP to terpenoid metabolic branches located in the stroma, whereas OsGGPPS1/OsGRP heterodimers support branches localized in thylakoid membranes. The strong affinity between OsGGPPS1 and OsGRP implies that OsGGPPS1 allocation between stroma and thylakoid membranes could be determined by the OsGRP abundance, thereby regulating metabolic flux from GGPP toward different downstream metabolic branches. To test this hypothesis, we analyzed transgenic rice lines with down- and up-regulation of OsGGPPS1 and OsGRP expression.
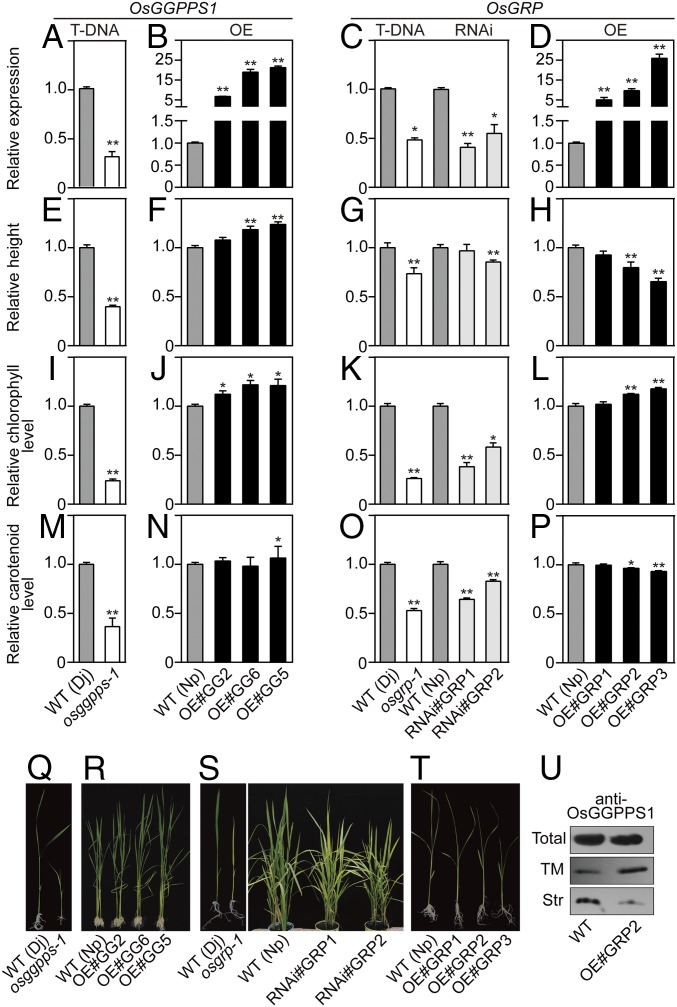
Only one T-DNA insertion osggpps-1 line was identified, which retained 30% of wild-type OsGGPPS1 mRNA level (Fig. 5A). osggpps-1 plants exhibited dwarf and leaf chlorosis phenotypes (Fig. 5 E and Q). The chlorophyll and carotenoid levels were reduced by 77% and 64% relative to wild-type plants, respectively (Fig. 5 I and M), which was not different from empty vector control (SI Appendix, Fig. S12 A, C, and D). The rice ortholog of GA4 (OsGA3OX1, Os05g08540), encoding GA 3β-hydroxylase, was used as marker for GA levels, as it has been shown that GA 3β-hydroxylase expression is negatively regulated by GA in a dose-dependent manner (21). Significantly higher OsGA3OX1 transcript levels in osggpps-1 suggest GA levels were decreased in mutant seedlings (SI Appendix, Fig. S13A). As no other OsGGPPS1 insertion lines could be identified, several attempts have been made to down-regulate OsGGPPS1 expression by RNAi. However, the resulting transgenic plants were unable to survive beyond the seedling stage. When OsGGPPS1 was overexpressed with mRNA levels more than fivefold higher than in the wild-type, rice plants showed promoted growth and enhanced chlorophyll contents (Fig. 5 B, F, J, and R). However, carotenoid levels were not affected by OsGGPPS1 overexpression, except for line OE#GG5, which showed a slight but significant increase (Fig. 5N).
Fig. 5.
Effect of down- and up-regulation of OsGGPPS1 and OsGRP expression on GGPP-derived terpenoids in rice. (A–D) Relative OsGGPPS1 (A and B) and OsGRP (C and D) transcript levels. (E–H) Relative plant height. (I–L) Relative chlorophyll levels. (M–P) Relative carotenoid levels. Analyzed lines are shown at the bottom. WT (Dj), osggpps-1 and osgrp-1: wild-type, OsGGPPS1 and OsGRP T-DNA insertion lines in Oryza sativa L. cv. Donjing (Dj) background. All overexpression and RNAi lines are in Oryza sativa L. cv. Nipponbare (Np) background. OE#GG2, 5, and 6, transgenic lines overexpressing OsGGPPS1; OE#GRP1-3, transgenic lines overexpressing OsGRP; RNAi#GRP1-2, transgenic OsGRP-RNAi lines. (Q–T) Representative plants of each of the lines. In each analyzed parameter, corresponding WT level was set as 1. Data are means ± SEM (n = 3). *P < 0.05; **P < 0.01; Student’s t test. (U) OsGGPPS1 protein levels in total chloroplast preparation and thylakoid membrane (TM) and stroma (Str) fractions of the WT and OsGRP overexpressing plants.
In the OsGRP T-DNA insertion and generated RNAi lines, OsGRP mRNA levels ranged from 40% to 55% of that in control plants (Fig. 5C). Transgenic plants exhibited significant reduction in chlorophyll levels, by 42–74% relative to controls (Fig. 5 K and S and SI Appendix, Figs. S12 B–D and 14A), as well as in the expression of chlorophyll biosynthetic genes (SI Appendix, Fig. S15A). In contrast, carotenoid levels were only slightly reduced, by 18–47% relative to control (Fig. 5O), with antheraxanthin and zeaxanthin levels even being increased (SI Appendix, Fig. S14A). In addition, expression of violaxanthin de-epoxidase gene (OsVDE) was increased (SI Appendix, Fig. S16A), which might be a response to oxidative stress caused by chlorophyll deficiency. Minor effects were observed upon plant height on OsGRP down-regulation (Fig. 5 G and S). When OsGRP mRNA levels were increased 9.6- to 25.9-fold, rice plants accumulated more chlorophyll but also showed reduced plant height (Fig. 5 D, H, L, and T). Quantification of OsGA3OX1 expression supported GA deficiency in these overexpression lines (SI Appendix, Fig. S13B). In contrast to chlorophyll, carotenoid levels were decreased in OsGRP-overexpressing lines (Fig. 5P and SI Appendix, Fig. S14B). Similar OsGGPPS1 mRNA abundances in control and OsGRP-overexpressing plants suggest that OsGRP does not affect OsGGPPS1 at the transcriptional level (SI Appendix, Fig. S15B). To further determine whether OsGRP alters OsGGPPS1 abundance and allocation, Western blot analysis was performed on thylakoid fractions isolated from OsGRP-overexpressing lines. Although no changes were detected in total OsGGPPS1 levels in OsGRP-overexpressing and control plants, higher OsGGPPS1 amounts were detected in thylakoids of OsGRP-overexpressing lines relative to control (Fig. 5U). Taken together, our results suggest OsGRP controls OsGGPPS1 dimerization state and suborganellar localization, thus increasing flux toward chlorophyll biosynthesis and reducing flux toward competing terpenoid pathways.
Discussion
Our results show that OsGRP is a protein that shares homology with members of the GPPS SSU II subfamily, but exhibits an undescribed function. Although the SSU II subfamily is present in both angiosperms and gymnosperms, to date, only one member of this subfamily was functionally characterized (11). It has been shown that Arabidopsis SSU II, although forming heterodimers with GGPPS, modifies the product profile and promotes the GPPS activity of GGPPS (11). In contrast, OsGRP, a type II SSU homolog, does not modify, but improves product specificity of OsGGPPS1 and enhances its catalytic efficiency (Table 1). OsGRP also changes OsGGPPS1 subchloroplast localization from stroma to thylakoid membranes (Fig. 3C), suggesting a diversity of functions for the type II SSU subfamily. Structural elucidation and site-directed mutagenesis studies (SI Appendix, Fig. S8) revealed that OsGRP has higher affinity toward OsGGPPS1 than one OsGGPPS1 molecule toward the other OsGGPPS1 in homodimers.
GGPP is a precursor of a variety of essential plant metabolites including carotenoids, tocopherols, phylloquinones, plastoquinones, gibberellins, and chlorophylls. However, the mechanism allocating GGPP among competing metabolic pathways leading to formation of these compounds is still unclear. Here, we demonstrated that rice contains only one functionally active GGPPS in plastids (Figs. 1 and 2 and SI Appendix, Fig. S5 A and B). OsGGPPS1 exists as homodimeric and heterodimeric protein (the latter in complex with OsGRP), which have different subchloroplast localization (Fig. 3C). Although OsGGPPS1 homodimers are localized in the stroma, OsGGPPS1/OsGRP heterodimers were found exclusively in thylakoid membranes in which both OsGGPPS1 and OsGRP interact with OsGGR, and all are constituents of a multiprotein complex (Fig. 4 B and C and SI Appendix, Fig. S11). The interaction of OsGGPPS1/OsGRP with OsGGR and OsLIL3 (Fig. 4 G and H), and indirectly with OsPORB and OsCHLG (SI Appendix, Fig. S11), suggests the GGPP produced by the OsGGPPS1/OsGRP heterodimer might be directly converted to phytyl-PP, the immediate precursor for the phytyl side chain of chlorophylls. Indeed, down- or up-regulation of either OsGGPPS1 or OsGRP mRNAs leads to decreased or increased chlorophyll levels, respectively (Fig. 5 I–L), suggesting both proteins are required for chlorophyll formation. Moreover, OsGRP overexpression led to decreases in carotenoid and GA formation, which compete with chlorophyll biosynthesis for GGPP supply (Fig. 5P and SI Appendix, Fig. S13B). Reduced carotenoid and GA levels are likely the result of an overwhelming recruitment of OsGGPPS1 to thylakoid membranes by OsGRP (Fig. 5U), and thus insufficient supply of GGPP produced by the remaining homodimeric OsGGPPS1 in the stroma for their biosynthesis. Thus, by controlling the different dimerization states of OsGGPPS1, OsGRP modulates the ratio between OsGGPPS1 homo- and heterodimer distribution in stroma and thylakoid membranes, respectively, and regulates GGPP flux toward downstream terpenoid metabolites.
The situation observed in rice is different from what has been described earlier in Arabidopsis (8), where several plastidic GGPPS paralogs with GGPPS activity in vitro were identified. Unfortunately, the functions of most of these GGPPS isoforms in planta remain unknown. However, similar to rice, one isozyme, AtGGPPS11, is a hub protein that interacts with PSY and GGR, thus channeling GGPP toward carotenoids and chlorophylls, respectively (8). Although interactions between OsGGPPS1 and OsGGR were found in rice (Fig. 4 A and B), binding of OsPSY to OsGGPPS1 was not detected (SI Appendix, Fig. S10).
Overall, we demonstrate that the thylakoid membrane-localized OsGRP possesses a strong affinity toward OsGGPPS1, and that its abundance determines the OsGGPPS1 dimerization state and localization. By interacting with OsGGPPS1, OsGRP directs it to a multiprotein complex containing chlorophyll biosynthetic proteins, thus providing a mechanism controlling GGPP allocation between different terpenoid pathways localized in the same subcellular compartment.
Materials and Methods
Details about plant material and growth conditions are described in SI Materials and Methods. Generation of constructs, gene expression, protein crystallization, plant transformation, subcellular localization analysis, Y2H, BiFC, pull-down assays, site-directed mutagenesis, enzymatic assays, blue native-PAGE and Western blot analysis, and targeted metabolic profiling were carried out according to protocols described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Alain Tissier (Leibniz Institute for Plant Biochemistry) for the help with LC-MS/MS analysis. We also thank Prof. Jun Chen, President of Nanjing University, for his help. This study was supported by the State Key Basic Research Project of China (#2013CB127004 to S.L.). M.G. is supported by the Ray Marsh and Arthur Pingree Dye Professorship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structural factors and coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5XN6 (OsGGPPS1-OsGRP), 5XN5 (OsGGPPS1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705689114/-/DCSupplemental.
References
- 1.Hemmerlin A, Harwood JL, Bach TJ. A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog Lipid Res. 2012;51:95–148. doi: 10.1016/j.plipres.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Tholl D. Biosynthesis and biological functions of terpenoids in plants. In: Schrader J, Bohlmann J, editors. Advances in Biochemical Engineering/Biotechnology. Vol 148. Springer; New York: 2015. pp. 63–106. [DOI] [PubMed] [Google Scholar]
- 3.Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- 4.Ashour M, Wink M, Gershenzon J. Biochemistry of terpenoids: Monoterpenes, sesquiterpenes and diterpenes. In: Wink M, editor. Annual Plant Reviews. 2nd Ed. Vol 40. Blackwell; Singapore: 2010. pp. 258–303. [Google Scholar]
- 5.Wang KC, Ohnuma S. Isoprenyl diphosphate synthases. Biochim Biophys Acta. 2000;1529:33–48. doi: 10.1016/s1388-1981(00)00136-0. [DOI] [PubMed] [Google Scholar]
- 6.Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y. Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 2000;122:1045–1056. doi: 10.1104/pp.122.4.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Schie CCN, Haring MA, Schuurink RC. Prenyldiphosphate synthases and gibberellin biosynthesis. In: Bach JT, Rohmer M, editors. Isoprenoid Synthesis in Plants and Microorganisms: New Concepts and Experimental Approaches. Springer; New York: 2013. pp. 213–232. [Google Scholar]
- 8.Ruiz-Sola MÁ, et al. Arabidopsis geranylgeranyl diphosphate synthase 11 is a hub isozyme required for the production of most photosynthesis-related isoprenoids. New Phytol. 2016;209:252–264. doi: 10.1111/nph.13580. [DOI] [PubMed] [Google Scholar]
- 9.Tholl D, et al. Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell. 2004;16:977–992. doi: 10.1105/tpc.020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham FX, Jr, Gantt E. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth Res. 2007;92:245–259. doi: 10.1007/s11120-007-9210-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang G, Dixon RA. Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc Natl Acad Sci USA. 2009;106:9914–9919. doi: 10.1073/pnas.0904069106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citovsky V, Gafni Y, Tzfira T. Localizing protein-protein interactions by bimolecular fluorescence complementation in planta. Methods. 2008;45:196–206. doi: 10.1016/j.ymeth.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Chang TH, Guo RT, Ko TP, Wang AH, Liang PH. Crystal structure of type-III geranylgeranyl pyrophosphate synthase from Saccharomyces cerevisiae and the mechanism of product chain length determination. J Biol Chem. 2006;281:14991–15000. doi: 10.1074/jbc.M512886200. [DOI] [PubMed] [Google Scholar]
- 14.Kloer DP, Welsch R, Beyer P, Schulz GE. Structure and reaction geometry of geranylgeranyl diphosphate synthase from Sinapis alba. Biochemistry. 2006;45:15197–15204. doi: 10.1021/bi061572k. [DOI] [PubMed] [Google Scholar]
- 15.Ohnuma S, et al. Conversion of product specificity of archaebacterial geranylgeranyl-diphosphate synthase. identification of essential amino acid residues for chain length determination of prenyltransferase reaction. J Biol Chem. 1996;271:18831–18837. doi: 10.1074/jbc.271.31.18831. [DOI] [PubMed] [Google Scholar]
- 16.Wang P, et al. Identification of a geranylgeranyl reductase gene for chlorophyll synthesis in rice. SpringerPlus. 2014;3:201. doi: 10.1186/2193-1801-3-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. Mutation of the light-induced yellow leaf 1 gene, which encodes a geranylgeranyl reductase, affects chlorophyll biosynthesis and light sensitivity in rice. PLoS One. 2013;8:e75299. doi: 10.1371/journal.pone.0075299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohscheider JN, et al. Altered levels of LIL3 isoforms in Arabidopsis lead to disturbed pigment-protein assembly and chlorophyll synthesis, chlorotic phenotype and impaired photosynthetic performance. Plant Cell Environ. 2015;38:2115–2127. doi: 10.1111/pce.12540. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi K, Takabayashi A, Tanaka A, Tanaka R. Functional analysis of light-harvesting-like protein 3 (LIL3) and its light-harvesting chlorophyll-binding motif in Arabidopsis. J Biol Chem. 2014;289:987–999. doi: 10.1074/jbc.M113.525428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka R, et al. LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci USA. 2010;107:16721–16725. doi: 10.1073/pnas.1004699107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowling RJ, Kamiya Y, Seto H, Harberd NP. Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 1998;117:1195–1203. doi: 10.1104/pp.117.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.