Significance
Benzo[a]pyrene (BaP) is a widespread potent carcinogen found in food, coal tar, cigarette smoke, and industrial smoke. Cigarette smoking is the leading cause of lung cancer, and the mutagenesis in smoking-associated lung cancer is determined by multiple factors, including nucleotide excision repair. We have developed a general method for genome-wide mapping of nucleotide excision repair at single-nucleotide resolution and applied it to generate repair maps of UV- and BaP-induced DNA damage in human. Results show a novel sequence specificity of BaP diol epoxide-deoxyguanosine repair. This general method can be used to study repair of all types of DNA damages that undergo nucleotide excision repair.
Keywords: tXR-seq, nucleotide excision repair, UV, benzo[a]pyrene diol epoxide, lung cancer
Abstract
Benzo[a]pyrene (BaP), a polycyclic aromatic hydrocarbon, is the major cause of lung cancer. BaP forms covalent DNA adducts after metabolic activation and induces mutations. We have developed a method for capturing oligonucleotides carrying bulky base adducts, including UV-induced cyclobutane pyrimidine dimers (CPDs) and BaP diol epoxide-deoxyguanosine (BPDE-dG), which are removed from the genome by nucleotide excision repair. The isolated oligonucleotides are ligated to adaptors, and after damage-specific immunoprecipitation, the adaptor-ligated oligonucleotides are converted to dsDNA with an appropriate translesion DNA synthesis (TLS) polymerase, followed by PCR amplification and next-generation sequencing (NGS) to generate genome-wide repair maps. We have termed this method translesion excision repair-sequencing (tXR-seq). In contrast to our previously described XR-seq method, tXR-seq does not depend on repair/removal of the damage in the excised oligonucleotides, and thus it is applicable to essentially all DNA damages processed by nucleotide excision repair. Here we present the excision repair maps for CPDs and BPDE-dG adducts generated by tXR-Seq for the human genome. In addition, we report the sequence specificity of BPDE-dG excision repair using tXR-seq.
Nucleotide excision repair is a versatile repair pathway that removes a variety of DNA damages, including UV- and benzo[a]pyrene (BaP)-induced DNA damages. BaP, a widespread carcinogen, is the major cause of lung cancer (1). It is produced by incomplete combustion of organic materials and converted to the ultimate mutagen, BaP diol epoxide (BPDE), through enzymatic metabolism (2). BPDE preferentially forms bulky covalent DNA adducts at N2 position of guanines and causes mutations if these BPDE-deoxyguanosines (BPDE-dGs) are not efficiently eliminated by nucleotide excision repair (3). Various methods of varying resolutions have been developed for mapping DNA damage and repair genome-wide (4–9). We previously reported a method, termed excision repair-sequencing (XR-seq), for mapping nucleotide excision repair (6). This method has been used to generate excision repair maps for UV-induced cyclobutane pyrimidine dimers (CPDs) and (6-4)pyrimidine-pyrimidone photoproducts [(6-4)PPs], as well as cisplatin and oxaliplatin-induced Pt-d(GpG) diadducts for the human genome and CPDs for the Escherichia coli genome (10–12).
Although the XR-seq method has been quite useful in determining the effects of various factors, such as genetic background, transcription, posttranscriptional histone modification, and chromatin states, on the timing and efficiency of repair (13), the method in its original form requires the reversal of damage in the excised oligonucleotide (26–27 mers in humans and 12–13 mers in E. coli) by enzymatic or chemical means before it can be processed for next-generation sequencing (NGS) and generation of repair maps. As such, this method has limitations, because it is not possible to reverse, either enzymatically or chemically, most DNA lesions removed from the duplex by nucleotide excision repair, which is a necessary step for PCR amplification and NGS. We have overcome this obstacle by using appropriate translesion DNA synthesis (TLS) polymerases to convert the excised ssDNA oligomer to the dsDNA form and then amplification by PCR, followed by NGS and alignment of the sequences to the genome. Here we present the application of this approach to the mapping of CPD repair and BPDE-dG repair in the human genome.
Results
tXR-seq of CPD and BPDE-dG.
To perform tXR-seq, the human lymphocyte cell line GM12878, which has been extensively characterized by the ENCODE project (14), was either exposed to UV (20 J/m2 at 254 nm) or treated with (±)-anti-BPDE (2 μM). After allowing 4 h or 1 h for repair, respectively, cells were lysed gently, and low molecular weight DNA, consisting mostly of excised 26–27 mers, was separated from chromosomal high molecular weight DNA by centrifugation.
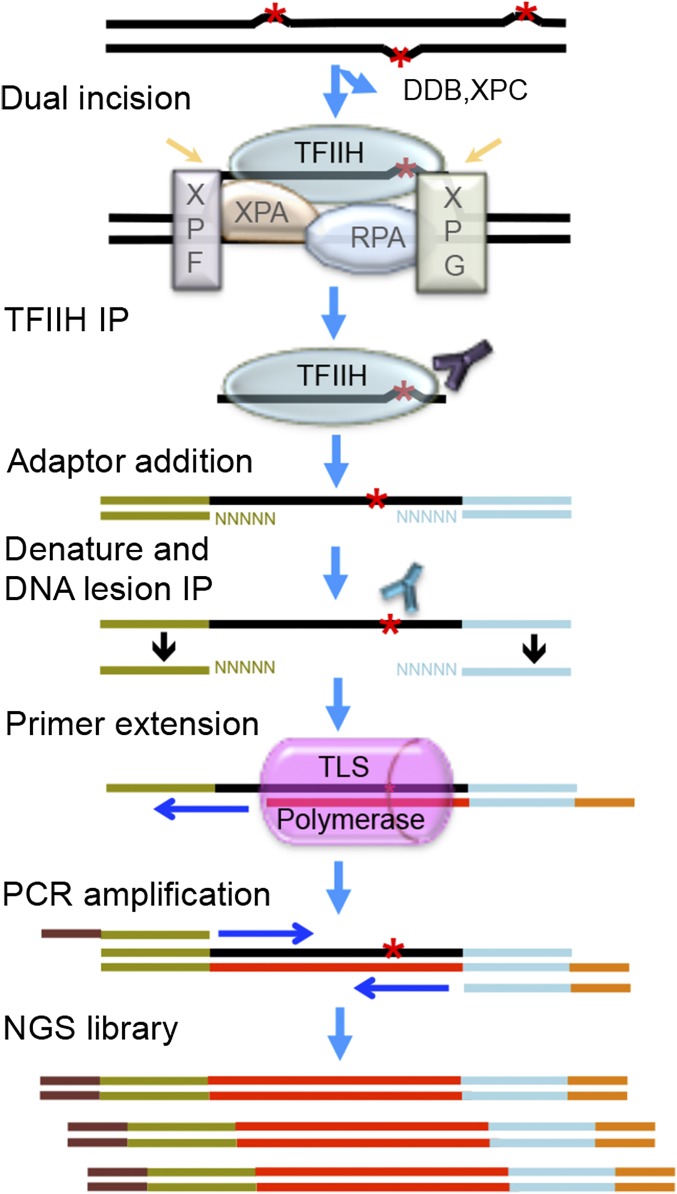
The basic features of sequencing library generation for tXR-seq are shown in Fig. 1. In brief, the excised oligomers are immunoprecipitated with anti-TFIIH (anti-XPB and anti-p62) antibodies, because the excision product is released in a relatively stable complex with TFIIH, which also protects the excision product from degradation by nonspecific nucleases. This is followed by extraction of the excised oligomers and ligation to adaptors and a second immunoprecipitation with either anti-CPD antibody or anti-BPDE antibody. Human DNA polymerases η and κ are used for primer extension in the presence of purified excised oligonucleotides containing either CPD or BPDE-dG, because polymerases η and κ can bypass CPD and BPDE-dG damage, respectively, in an error-free manner (15–17). This is followed by PCR amplification with index primers and NGS on the Illumina HiSeq 2500 platform.
Fig. 1.
Schematic of sequencing library construction strategy for tXR-seq. The red asterisk represents a DNA lesion processed by nucleotide excision repair. Excised oligomers are precipitated with TFIIH antibodies, extracted, and ligated to adaptors. Then, the adaptor-containing oligomers are precipitated with anti-lesion antibody, the primer with index is annealed and extended by an appropriate TLS DNA polymerase (pink), and the extension product is amplified by PCR to obtain the dsDNA library for NGS.
Length Distribution and Nucleotide Frequency Analysis of Excision Products.
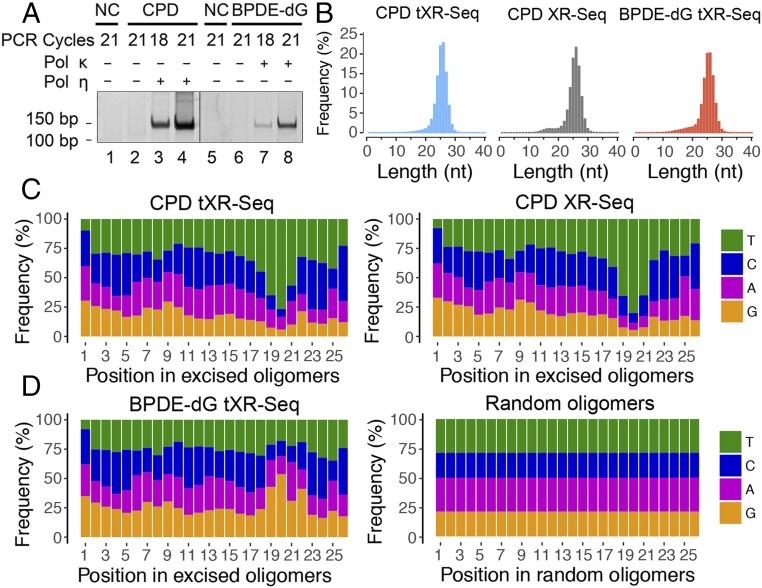
The PCR products, approximately 145 bp long, can be seen only in the presence of polymerases η or κ, indicating that the resulting dsDNA libraries exclusively contain the sequence information from the previously excised oligonucleotides attributed to efficient lesion bypass (15–17) (Fig. 2A). Fig. 2B shows the length distributions of the oligomers obtained by tXR-seq and conventional CPD XR-seq in which the CPD is repaired by CPD photolyase. In both CPD tXR-seq and BPDE-dG tXR-seq cases, oligomers in the size range of 24–28 nt predominate, with a median size of 26 nt, in agreement with the well-established dual incision mode of the human nucleotide excision repair system (18–20). Importantly, the length distribution of the oligomers obtained by tXR-seq for CPD is in remarkable agreement with that obtained by conventional CPD XR-seq, affirming the validity of tXR-seq.
Fig. 2.
Analyses of length distribution and single-nucleotide frequencies for the excised oligomers obtained from tXR-seq and conventional XR-seq. (A) dsDNA library for tXR-seq analyzed by 10% native polyacrylamide gel electrophoresis. Primer extension products (1%) were amplified by PCR using the indicated cycles. NC, nontemplate control. (B) Length distribution of excised oligomers from CPD and BPDE-dG tXR-seq (∼20 million each) and CPD XR-seq 6 (∼10 million) after removal of adaptors and duplicate reads. (C) Single-nucleotide frequencies for 26-mers obtained by CPD tXR-seq (∼13 million) and conventional CPD XR-seq 6 (∼10 million). (D) Single-nucleotide frequencies for the 26-mers from BPDE-dG tXR-seq (∼12 million) along with single-nucleotide distribution of randomly selected 26-mers from the human genome (10 million).
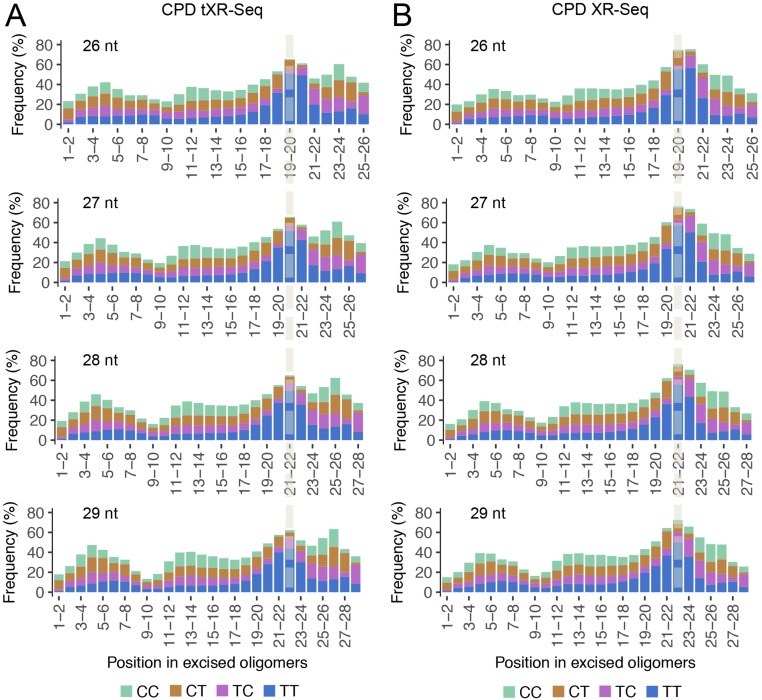
We further analyzed the base sequence distributions along the excised CPD oligomers obtained by tXR-seq and conventional XR-seq (Fig. 2C). As expected, for CPD oligomers isolated by both conventional XR-seq and tXR-seq methods, positions 19–21 from the 5′ end were enriched for thymines (Ts), in agreement with the dual incision pattern of the human excision nuclease system (21, 22). In contrast, for the BPDE-dG oligomers, guanines (Gs) are enriched at positions 19–22, consistent with BPDE-dG monoadducts at one of these positions (Fig. 2D). As a control, when randomly chosen oligomers (10 million) of 26 nt from the human genome were analyzed for sequence composition, all four nucleotides were uniformly distributed throughout the length of the oligomers, with ∼60% AT and 40% GC at all positions, consistent with Chargaff's rules (23) (Fig. 2D). We also analyzed the frequencies of di-pyrimidines (TT, TC, CT, and CC) at each position along the excised oligomers of 26–29 nt from both tXR-seq and conventional XR-seq for CPD (Fig. S1 A and B). In both experiments, there was strong TT enrichment at the fixed position of 6 nt from the 3′ end, further confirming the validity of tXR-seq.
Fig. S1.
Dipyrimidine frequencies along the reads of 26–29 nt from CPD tXR-seq in GM12878 cells and conventional CPD XR-seq in NHF1 cells. The enrichment of TT frequency is at a fixed position of 6 nt from 3′ end in both CPD tXR-seq (A) and conventional CPD XR-seq (B). The fixed position in all reads of different lengths is highlighted in dashed line.
Sequence Specificity of BPDE-dG Repair.
Several studies have been conducted on the effects of nearest-neighbor and distant-neighbor sequences on the repair rate of BPDE-dG. The data have been interpreted within the context of structural deformity and duplex backbone flexibility caused by the adduct in various sequence contexts, and some general rules have been derived (24–27). Although these pioneering studies have been quite useful, some of the rules were derived from the excision rates of the BPDE-dG adduct from in vitro experiments with human cell-free extracts and oligonucleotide duplexes 135 bp in length (28).
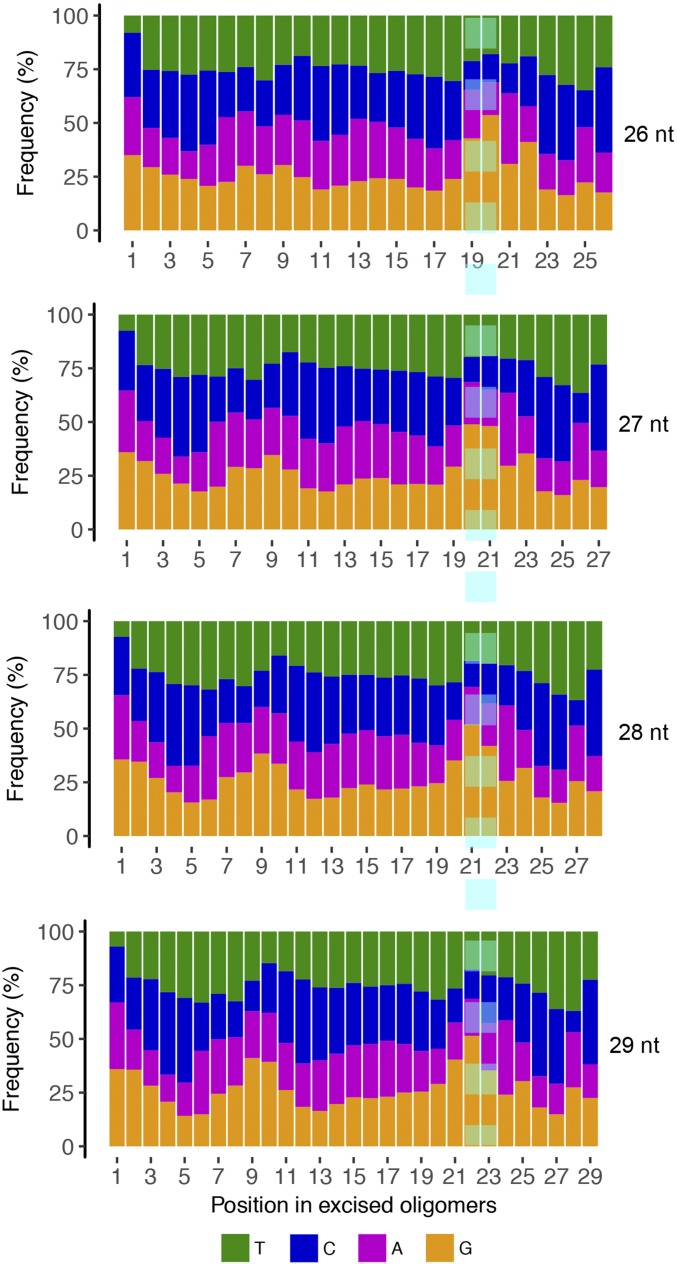
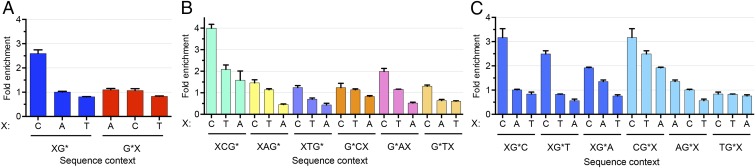
The tXR-seq method, which produces at least ∼20 million reads in each biological replicate, provides a model-independent means for analyzing the effects of nearest and distant (one base flanking the nearest bases) sequence neighbors on BPDE-dG excision efficiencies. To secure the vast majority of the excised oligonucleotides used in this analysis containing BPDE-dG at the same position, we classified the sequencing reads from tXR-seq based on read lengths. An analysis of single-nucleotide frequencies along reads 26–29 nt in length showed Gs enriched at two fixed positions (Fig. S2). For 26, 27, and 28 oligomers, we selected those with nucleotide is G at position 20, 21, and 22, respectively. We further filtered out those reads containing G within three bases upstream and four bases downstream of the specific G position to eliminate other possible BPDE-dG sites that could possibly interfere with our analysis. We applied the same criteria to the randomly selected 26, 27, and 28 oligomers from human genomic sequences and then calculated the frequencies of dinucleotides and trinucleotides for both excised oligomers and randomly selected oligomers. Those frequencies for excised oligomers were normalized to their respective frequencies for randomly selected oligomers. We plotted the average fold enrichment of nearest- and distant-neighbor base sequence context frequencies for the damaged G base that had been excised in 26–28 oligomers (Fig. 3).
Fig. S2.
Single-nucleotide frequencies for 26–29 oligomers obtained by BPDE-dG tXR-seq. The enrichment of Gs at the two positions of 6 nt from 3′ end is highlighted by the turquoise dashed line. The figures represent data from merged two biological replicates.
Fig. 3.
Sequence specificity of BPDE-dG repair. (A) Average fold enrichment of dinucleotide excision frequencies in 26, 27, and 28 oligomers derived from BPDE-dG tXR-seq. To calculate the fold enrichment, the dinucleotide excision frequencies from 26, 27, and 28 excised oligomers were divided by the dinucleotide frequencies from randomly selected 26, 27, and 28 oligomers in the human genome. “X” stands for the possible A, C and T bases. “G*” represents the BPDE-dG lesion. Error bars indicate SDs. (B and C) Same as in A, except that the trinucleotide frequencies were used for excised oligomers and random oligomers. All dinucleotide and trinucleotide frequencies were calculated only in those oligomers that met our selection criteria. The figures represent data from two merged biological replicates.
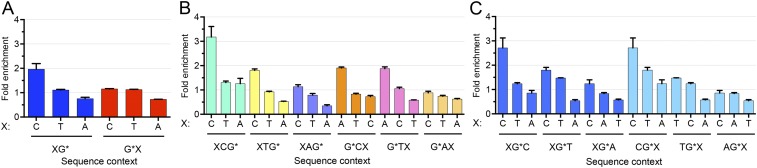
Based on the results, we can draw the following conclusions: First, the fold enrichment of CG context was ∼2.5, whereas there was no apparent enrichment of the other dinucleotide contexts (Fig. 3A). Second, there was a significant effect of the nearest and distant neighbors on the frequency of BPDE-dG excision. In trinucleotide frequency analysis, the excision frequency generally followed the “C>T>A” rule at both the 5′ and 3′ distant-neighbor positions (Fig. 3 B and C and Table 1). The 3′ nearest-neighbor base also followed the same rule, except for AG*, which had an A>C>T order. For G*A and G*T, the 5′ nearest-neighbor bases with the second-highest excision frequency were A and T, respectively. We also selected the specific G at positions 19, 20, and 21 for 26, 27, and 28 oligomers, respectively, and analyzed the sequence context effect in the same way (Fig. S3 and Table S1). The results suggest that the excision frequency was similar and also generally followed the C>T>A rule. Finally, these conclusions must be qualified, because they are based on the assumption of no sequence effect on adduct formation. In fact, BPDE preferentially binds at methylated CpG sites over any other NpG sites (29). In view of these reported sequence preferences for damage formation, genome-wide BPDE-dG damage distribution must be experimentally demonstrated by Damage-seq (10) for a definitive generalization of the sequence effect on repair.
Table 1.
Sequence specificity of BPDE-dG repair
| 5′ distant | XG* | 3′ nearest | 5′ nearest | G*X | 3′ distant |
| C>T>A | CG* | C>T>A | C>A>T | G*C | C>T>A |
| C>T>A | AG* | A>C>T | C>A>T | G*A | C>T>A |
| C>T>A | TG* | C>T>A | C>T>A | G*T | C>A>T |
G* represents the BPDE-dG lesion, and X represents the possible A, C, and T bases.
Fig. S3.
Sequence specificity of BPDE-dG repair. (A) Same as in Fig. 3A, except specific Gs were chosen at positions 19, 20, and 21 for 26, 27, and 28 oligomers, respectively. (B and C) Same as in A, except that the trinucleotide frequencies were used for excised oligomers and random oligomers. The figures represent data from merged two biological replicates.
Table S1.
Sequence specificity of BPDE-dG repair
| 5′ distant | XG* | 3′ nearest | 5′ nearest | G*X | 3′ distant |
| C>T>A | CG* | C>T>A | C>T>A | G*C | A>T>C |
| C>T>A | TG* | T>C>A | C>T>A | G*T | A>C>T |
| C>T>A | AG* | C>A>T | C>A>T | G*A | C>A>T |
G* represents the BPDE-dG lesion, and X represents the possible A, C, and T bases.
Effect of Transcription on BPDE-dG Repair.
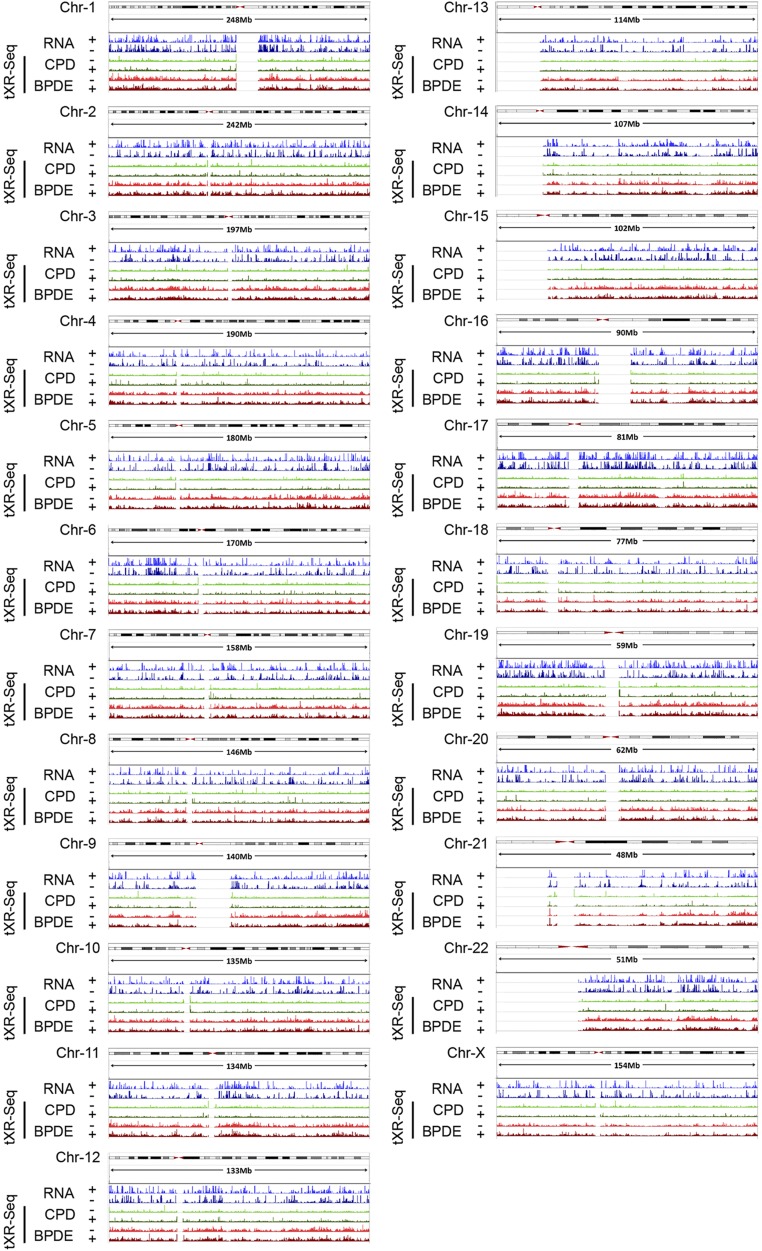
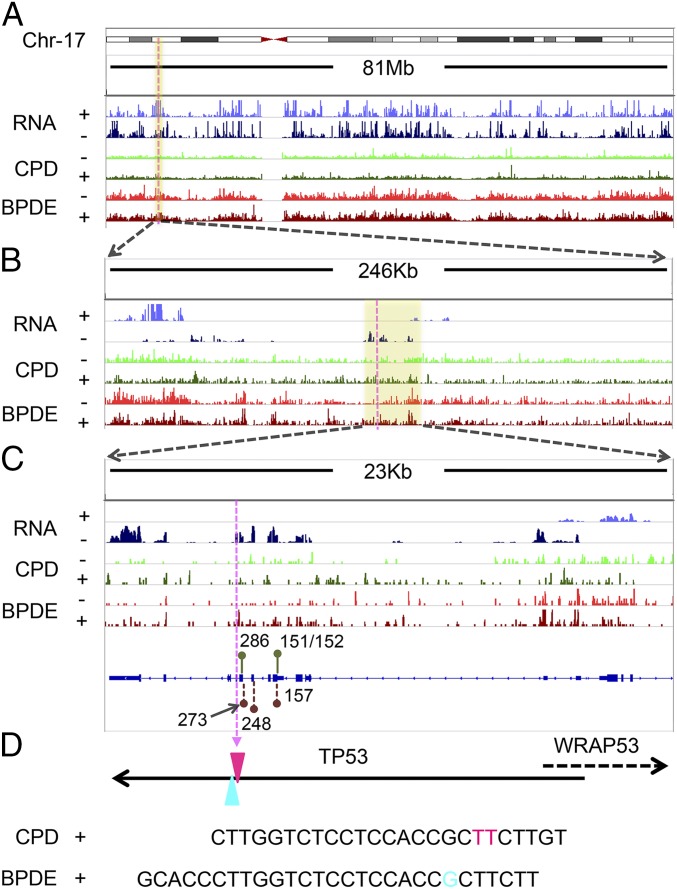
Transcription stimulates repair on the transcribed strand (TS) (30). As a general rule, this effect is more apparent in highly transcribed genes and for lesions that are poorly recognized by the core nucleotide excision repair machinery. To explore the effect of transcription on BPDE-dG repair, we first generated both CPD and BPDE-dG repair maps and integrated the total RNA-seq signal across the entire human genome using tXR-seq reads and ENCODE RNA-seq data from GM12878 cells (Fig. S4). We chose the TP53 gene to analyze the effect of transcription on BPDE-dG repair for several reasons. First, it is mutated as a driver both in UV-induced and cigarette smoke-induced cancers. Second, it is transcribed at moderately high levels in many cell lines, including the GM12878 cell line used in this study. Finally, previous high-resolution repair studies by ligation-mediated PCR (LM-PCR) on short fragments of TP53 may be used as references for tXR-seq data (31). Fig. 4 shows the transcription and repair maps of human chromosome 17, which carries the TP53 gene, at resolutions ranging from megabase to single base or dinucleotide in TP53. This figure also shows tXR-seq data for both CPD and BPDE-dG repair in GM12878 cells, as well as two excision products that can be unambiguously assigned to specific TT and G damages, respectively (Fig. 4). As is apparent from the repair data at all resolutions, the TS for both TP53 and the adjacent WRAP53 gene transcribed in opposite direction were repaired at higher efficiency than the nontranscribed strand (NTS). In addition, for both damage types, there appeared to be hotspots for repair, although deeper sequencing is needed to be able to comment on the cause and significance of these repair hotspots.
Fig. S4.
Whole genome map of CPD and BPDE-dG repair generated using the tXR-seq method in GM12878 cells. ENCODE nonpolyA RNA-seq signals in blue, separated by strands, are plotted on top of the tXR-seq tracks.
Fig. 4.
Screenshots of CPD and BPDE-dG tXR-seq signals for chromosome 17 along with the RNA-seq profile in GM12878 cells. (A–C) Representative CPD and BPDE-dG repair map at increasing resolution, zooming in on the TP53 and WRAP53 genes. The yellow shaded boxes indicate the zoomed-in regions. For the RNA signal, the “+” indicates genes transcribed from left to right, and the “−” indicates genes transcribed from right to left. For the tXR-seq signal, “+” indicates plus-strand DNA (5′ to 3′ direction) and “−” represents minus-strand DNA (3′ to 5′ direction). Green and dark-red lollipops indicate the CPD hotspot codons (286 and 151/152) (41) and BPDE-dG hotspot codons (273, 248, and 157) (31), respectively. (D) Two representative oligomers in CPD tXR-seq and BPDE-dG tXR-seq excised from the TP53. The purple dashed line indicates the excision positions in TP53 at different scales. Pink TT (chr17: 7,576,999–7,577,000) and blue G (chr17: 7,576,997) indicate the positions of the respective damages in the TP53.
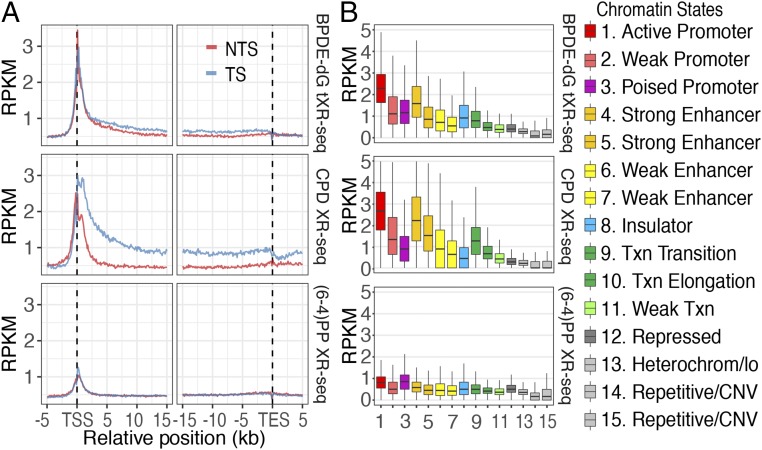
In addition, we analyzed CPD and BPDE-dG repair around the transcription start sites (TSS) and transcription end sites (TES) in NHF1 and GM12878 cell lines, respectively (Fig. 5). The trend was in general agreement with the data obtained from the NHF1 cell line for the repair of CPD and (6-4)PP as measured by the conventional XR-seq method. As expected, all of the repair profiles had repair peaks around TSS, in agreement with the documented high RNA polymerase II density and nascent RNA levels in this region (13, 32–34). In addition, as we reported previously (13), a low level of enriched excision repair near TSS was also seen in CSB cell line, which can be linked to the open chromatin structure close to TSS. In fact, the repair ratio at 1 h for BPDE-dG fell somewhere between that for CPD and (6-4)PP, indicating that BPDE-dG is recognized by the core excision repair complex at an affinity between that of (6-4)PP and CPD (Fig. 5A).
Fig. 5.
Effect of transcription and chromatin states on CPD, BPDE-dG, and (6-4)PP repair. (A) BPDE-dG repair profile around the transcription start and end sites of 5,025 selected genes at 1 h in GM12878 cells, and CPD and (6-4)PP repair profiles at 1 h in NHF1 cells. The TS/NTS repair ratio for CPD is higher than that for BPDE-dG. NTS, nontranscribed strand; TS, transcribed strand. (B) Effect of chromatin states on BPDE-dG repair at 1 h in GM12878, and on CPD and (6-4)PP repair at 1 h in NHF1 cells. The subclassified strong enhancer and weak enhancer differ in their distance from the TSS. States 4 and 6 are closer to the TSS, but states 5 and 7 are farther from the TSS. The figures represent data from merged two biological replicates.
Effect of Chromatin States on Repair.
Based on DNA sequence elements and histone posttranslational modifications, 15 chromatin states have been defined (35). In previous studies, we found that although these states did not affect cisplatin-induced damage formation, they did affect repair efficiencies, particularly in active promoters, enhancers, and transcribed gene bodies (10, 13). We analyzed all 15 chromatin states for CPD, BPDE-dG, and (6-4)PP repair (Fig. 5B). Again, the general pattern observed in tXR-seq was similar to the patterns seen in CPD and (6-4)PP repair obtained by conventional XR-seq. Interestingly, for BPDE-dG and CPD repair at 1 h after UV treatment, both exhibited high repair levels over active chromatin states but had distinct repair levels over TS and NTS around TSS and TES (Fig. 5 A and B). For BPDE-dG repair, there was only a minor increase on TS compared with NTS, whereas there was much higher repair on TS than on NTS in CPD repair. This finding further confirms the unique BPDE-dG repair characteristics.
The foregoing findings not only show that BPDE-dG as a bulky adduct is processed by the cell in a manner similar to other bulky DNA lesions, but also show that tXR-seq, as a simpler and more versatile method, can be used for damages not suitable for analysis by the XR-seq method.
Discussion
The recently developed XR-seq method has been quite useful for mapping nucleotide excision repair of UV and cisplatin and oxaliplatin damage. However, the method in its original form is applicable only to damages that can be either enzymatically or chemically reversed. Here we have used TLS polymerases to circumvent this obstacle and made the method more widely applicable. In doing so, we have also generated a human genome repair map for one of the major carcinogens, BaP. Interestingly, we found that transcription-coupled repair has only a moderate effect on BPDE-dG adduct, because this lesion, like the UV-induced (6-4)PP, is relatively efficiently recognized by the core human nucleotide excision repair machinery. We also note that LM-PCR analysis of TP53 hotspots for cigarette smoke-related mutagenesis has led to the conclusion that BPDE adducts form preferentially at G residues in codons 157, 248, and 273 of TP53, and that these cancer-causing mutagenic adducts are in the nontranscribed strand and are repaired at slower rates than other BPDE-dG adducts in the same strand in TP53 gene, in the context of G*TC (157), CG*G (248), and CG*T (273) (31). The sequencing depth in this tXR-seq was insufficient to detect repair at these sites and thus make a meaningful comparison with repair at other sites to confirm these findings. With new improvements in NGS technology, we expect that deeper sequencing will be affordable and will allow us to address the issue of whether carcinogenic mutations at these sites are isolated with high damage frequency combined with poor repair efficiency to give rise to lung cancers.
We note that although the DNA polymerases η and κ that we used in this study are not applicable to all lesions processed by nucleotide excision repair. However, many TLS polymerases have been identified and characterized in recent years (36, 37); thus, for virtually any bulky DNA lesion, it is possible to use an appropriate TLS polymerase or a combination of two TLS polymerases to accomplish translesion synthesis and construct genomic DNA repair maps. In conclusion, the tXR-seq method opens the door to genome-wide mapping repair of all types of DNA damages processed by nucleotide excision repair at single-nucleotide resolution.
Nonetheless, we note that although tXR-seq is a significant improvement over XR-seq, further technical and computational optimizations are needed for rigorous and quantitative analyses of all damages processed by nucleotide excision repair. The translesion synthesis efficiency and accuracy of the TLS polymerases used need to be taken into consideration, as does the neighboring sequence effect on TLS. In the analysis of BPDE-dG adducts, the elimination of sequences of reads containing Gs three bases upstream and four bases downstream of the adducted G residue precludes the analysis of neighboring Gs on excision efficiency of the adducted G. Although these limitations are important, we believe that we will be able to address them in the near future. The translesion synthesis efficiency and neighboring sequence effects for most TLS polymerases are known, and we expect to determine these factors for the TLS polymerases that have not yet been analyzed. Regarding the neighboring G effects on BPDE-dG adducts, we are developing computational tools to predict with precision the adducted G even when there are neighboring Gs, and thus we will be able to analyze the effect of all sequence contexts on the repair of BPDE-dG damage and the damage induced by other agents, such as N-acetoxy-2-acetylaminofluorene, that specifically attack G residues.
Materials and Methods
Antibodies and TLS DNA Polymerases.
The following antibodies were used in this study: anti-mouse IgG (sc-2025), anti-rabbit IgG (sc-2027), anti-XPB (sc-293), and anti-p62 (sc-292) from Santa Cruz Biotechnology; rabbit anti-mouse IgG (ab46540) from Abcam; anti-CPD from Kamiya Biomedical; and anti-BPDE (clone 8E11) from Trevigen. TLS DNA polymerases η and κ were obtained from Enzymax.
Cell Line and Culture Conditions.
Human GM12878 cells were cultured in RPMI medium 1640 (no phenol red) with 15% FBS and 2 mM glutamine at 37 °C in a 5% CO2 humidified chamber.
UV Irradiation and BPDE Treatment.
GM12878 cells were grown to ∼80% confluence before UV irradiation or (±)-anti-BPDE (MRIGlobal) treatment. The UV (20 J/m2 at 254 nm) irradiation was performed as described previously (6), except that 20 mL of GM12878 cells were irradiated in RPMI medium 1640 (no phenol red) in a 150-mm Petri dish. BPDE stock solution (4 mM), which was freshly made by dissolving BPDE in DMSO, was added to the GM12878 cell suspension at a final concentration of 2 μM, and cells were incubated for 1 h at 37 °C in a 5% CO2 humidified chamber.
tXR-seq Library Preparation and Sequencing.
After UV or BPDE treatment, GM12878 cells were incubated for 4 h and 1 h respectively. At appropriate time points, cells were collected by centrifugation and lysed in ice-cold Buffer A (25 mM Hepes pH 7.9, 100 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 12.5% glycerol, 0.5% Nonidet P-40) for 10 min, followed by 40 strokes in a glass hand homogenizer. The chromatin fraction was pelleted by centrifugation at 16,800 × g for 30 min at 4 °C, and the supernatant was subjected to TFIIH immunoprecipitation by adding 2 µg of anti-XPB, 1 µg of anti-p62, and 200 µg of RNase A (Sigma-Aldrich; R4642) per 20 mL of original cell culture. After 3–5 h rotation at 4 °C, 15 µL of recombinant protein A/G Plus-agarose (Santa Cruz Biotechnology; sc2003) per 20 mL of original cell culture was added, and the mixture was gently rotated overnight at 4 °C. The excised oligonucleotides were eluted from the recombinant protein A/G Plus-agarose beads by 100 µL of Buffer C (10 mM Tris⋅Cl pH 7.5, 1 mM EDTA, and 1% SDS) for 15 min at 65 °C after two washes with Buffer A and Buffer B (25 mM Hepes pH 7.9, 100 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 12.5% glycerol, and 1% Nonidet P-40), respectively. The eluted excised oligonucleotides were extracted by phenol-chloroform, and the ethanol-precipitated excised oliogos were incubated with 5 µL of RNase A/T1 mixture (Thermo Fisher Scientific; EN0551) for 1 h at 37 °C. Before adaptor ligation, another round of phenol-chloroform extraction was applied, and a G50 filtration column (GE Healthcare) was used to further purify the excised oligonucleotides.
For one adaptor ligation reaction, 1 µL of 5′ adaptor (20 pmol), 2 µL of 3′ adaptor (40 pmol), and 1.2 µL of 10 × hybridization buffer (20 mM Tris⋅HCl, pH 7.5, 200 mM NaCl, and 0.2 mM EDTA) were added into the 5.8 µL of purified excised oligonucleotides, and the reaction mixture was incubated at 60 °C for 10 min, followed by a 5-min incubation at 16 °C. Then 4 µL of 5 × ligase buffer, 1 µL of T4 DNA ligase HC (Thermo Fisher Scientific; 15224-041), 1 µL of 50% PEG8000 (New England BioLabs), and 4 µL of ddH2O were added into the mixture, and the ligation reaction mixture was incubated overnight at 16 °C. The 5′ adaptor and 3′ adaptor were the same as described previously (6).
After phenol-chloroform extraction and ethanol precipitation, the excised oligonucleotides with adaptors were boiled for 5 min and then immediately placed in ice water. Either 2 µL of anti-CPD or 4 µL of anti-BPDE was preincubated with 5 µL of Protein G Dynabeads (Thermo Fisher Scientific; 1004D), 5 µL of anti-rabbit Dynabeads (Thermo Fisher Scientific; 11203D) and 4 µL of 50% rabbit anti-mouse IgG (Abcam; ab46540) for 2–3 h, followed by mixing with 100 µL of Reaction Buffer (20 mm Tris⋅Cl at pH 8.0, 2 mm EDTA, 150 mm NaCl, 1% Triton X-100, and 0.5% sodium deoxycholate) containing the denatured excised oligonucleotides with adaptors. The mixtures were rotated at 4 °C overnight. The subsequent washing, elution, phenol-chloroform extraction, and ethanol precipitation steps were performed as described previously (4).
In the primer extension step, human DNA polymerases η and κ were used to bypass CPD and BPDE-dG damage, respectively. The 10 µL of excised oligonucleotides with adaptors was mixed with 15 µL of 2× TLS polymerase buffer (50 mM potassium phosphate pH 7.0, 10 mM MgCl2, 5 mM DTT, 200 µg/mL BSA, 20% glycerol, and 200 µM dNTP) and 3 µL of 10 µM RPIn primer (where n represents different index sequences). The single-cycle primer extension procedure is as follows: initial denaturing at 98 °C for 3 min; ramping to 65 °C at 0.1 °C/s and holding for 10 min; ramping to 37 °C at 0.1 °C/s; addition of either 2 µL of polymerase η or 1 µL of polymerase κ into the reaction with mixing; and incubation at 37 °C for 30 min. Then the primer extension product was purified by phenol-chloroform extraction and ethanol precipitation.
The dsDNA library was constructed by PCR amplification using Kapa HotStart ReadyMix with RP1 and RPIn primers, and purified from 10% native polyacrylamide gel as described previously (6). In CPD tXR-seq and BPDE-dG tXR-seq, the primer extension products were amplified for 11 and 14 PCR cycles, respectively. All tXR-seq sequencing libraries were sequenced on the Illumina HiSeq 2500 platform.
Read Processing and Genome Alignment.
At least 22 million reads were obtained in tXR-seq. All adaptors in reads were trimmed using Trimmomatic, and duplicate reads were removed by the FASTX-Toolkit (38). Reads >50 mer are filtered before further analysis. The processed reads were aligned to the hg19 human female genome using bowtie with the following command options: -x -q–nomaqround–phred33-quals -m 4 -n 2 -e 70 -l 20–best -p 4–seed = 123 –S (39).
Data Visualization.
All sequencing data were obtained from two biological replicates of tXR-seq dsDNA libraries. To compare the repair signal, we normalized all count data by the sequencing depth and visualized them in the Integrative Genomics Viewer (40). Sequencing data for both CPD XR-seq and (6-4)PP XR-seq at 1 h in NHF1 cells were obtained from a previously published dataset (6) [Gene Expression Omnibus (GEO) accession no. GSE67941]. The raw data and bigwig tracks for tXR-seq in this study were deposited in the GEO database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE97675).
Encode Data.
GM12878 long nonpolyA RNA-seq [University of California Santa Cruz (UCSC) genome browser accession no. wgEncodeEH000148], chromatin state datasets for GM12878 (accession no. wgEncodeEH000784), and NHLF (accession no. wgEncodeEH000792) were downloaded from the ENCODE portal (genome.ucsc.edu/ENCODE/).
Length Distribution and Nucleotide Frequencies.
We obtained the length distribution and nucleotide frequencies using custom scripts after adaptor trimming and removal of duplicate reads. To normalize nucleotide frequencies in excised oligonucleotides to human whole genome nucleotide frequencies, we used our published undamaged human genomic sequencing dataset (10) (GEO accession no. GSE82213) and selected size-specific genomic sequences at random for further analysis.
Repair Profiles of Strands and Chromatin State Analysis.
The transcript coordinates from hg19 reference genome were downloaded from the UCSC genome browser. The transcripts with an expression score of ≥300 were taken into account. Among those transcripts, those with another transcript in the 6 kb up-downstream vicinity and those <15 kb were filtered out. Regions 5 kb upstream and 15 kb downstream from the transcription start and end sites were binned into 100-bp windows.
The dataset of chromatin states for GM12878 mapped on the hg19 reference genome was downloaded from ENCODE. Reads per kilobase per mapped million reads values were computed and plotted. Two biological replicate sets were combined to generate the boxplots. Bedtools software was used to count the reads for each analysis. All quantitative data were analyzed and plotted using R or GraphPad Prism 6 software.
Acknowledgments
This work was supported by NIH Grants GM118102 and ES027255.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE97675).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706021114/-/DCSupplemental.
References
- 1.Alexandrov LB, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354:618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 3.Seeberg E, Steinum AL, Nordenskjöld M, Söderhäll S, Jernström B. Strand-break formation in DNA modified by benzo[alpha]pyrene diolepoxide: Quantitative cleavage by Escherichia coli uvrABC endonuclease. Mutat Res. 1983;112:139–145. doi: 10.1016/0167-8817(83)90036-6. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, et al. Nucleotide excision repair in human cells: Fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi JH, et al. Highly specific and sensitive method for measuring nucleotide excision repair kinetics of ultraviolet photoproducts in human cells. Nucleic Acids Res. 2014;42:e29. doi: 10.1093/nar/gkt1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29:948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi JH, Kim SY, Kim SK, Kemp MG, Sancar A. An integrated approach for analysis of the DNA damage response in mammalian cells: Nucleotide excision repair, DNA damage checkpoint, and apoptosis. J Biol Chem. 2015;290:28812–28821. doi: 10.1074/jbc.M115.690354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zavala AG, Morris RT, Wyrick JJ, Smerdon MJ. High-resolution characterization of CPD hotspot formation in human fibroblasts. Nucleic Acids Res. 2014;42:893–905. doi: 10.1093/nar/gkt912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao P, Smerdon MJ, Roberts SA, Wyrick JJ. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc Natl Acad Sci USA. 2016;113:9057–9062. doi: 10.1073/pnas.1606667113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J, Lieb JD, Sancar A, Adar S. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc Natl Acad Sci USA. 2016;113:11507–11512. doi: 10.1073/pnas.1614430113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canturk F, et al. Nucleotide excision repair by dual incisions in plants. Proc Natl Acad Sci USA. 2016;113:4706–4710. doi: 10.1073/pnas.1604097113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adebali O, Chiou YY, Hu J, Sancar A, Selby CP. Genome-wide transcription-coupled repair in Escherichia coli is mediated by the Mfd translocase. Proc Natl Acad Sci USA. 2017;114:E2116–E2125. doi: 10.1073/pnas.1700230114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adar S, Hu J, Lieb JD, Sancar A. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc Natl Acad Sci USA. 2016;113:E2124–E2133. doi: 10.1073/pnas.1603388113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 16.Masutani C, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Wu X, Guo D, Rechkoblit O, Wang Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: Error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair (Amst) 2002;1:559–569. doi: 10.1016/s1568-7864(02)00055-1. [DOI] [PubMed] [Google Scholar]
- 18.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 19.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 20.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 22.Sancar A. Mechanisms of DNA repair by photolyase and excision nuclease (Nobel Lecture) Angew Chem Int Ed Engl. 2016;55:8502–8527. doi: 10.1002/anie.201601524. [DOI] [PubMed] [Google Scholar]
- 23.Chargaff E, Zamenhof S, Green C. Composition of human desoxypentose nucleic acid. Nature. 1950;165:756–757. doi: 10.1038/165756b0. [DOI] [PubMed] [Google Scholar]
- 24.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: Enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J Mol Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: Hierarchies of recognition signals. J Mol Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Y, et al. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J Mol Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menzies GE, Reed SH, Brancale A, Lewis PD. Base damage, local sequence context and TP53 mutation hotspots: A molecular dynamics study of benzo[a]pyrene-induced DNA distortion and mutability. Nucleic Acids Res. 2015;43:9133–9146. doi: 10.1093/nar/gkv910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kropachev K, et al. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: Insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen JX, Zheng Y, West M, Tang MS. Carcinogens preferentially bind at methylated CpG in the p53 mutational hot spots. Cancer Res. 1998;58:2070–2075. [PubMed] [Google Scholar]
- 30.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 31.Denissenko MF, Pao A, Tang M, Pfeifer GP. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 32.Gyenis A, et al. UVB induces a genome-wide acting negative regulatory mechanism that operates at the level of transcription initiation in human cells. PLoS Genet. 2014;10:e1004483. doi: 10.1371/journal.pgen.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nojima T, et al. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 37.Goodman MF, Woodgate R. Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tornaletti S, Rozek D, Pfeifer GP. Mapping of UV photoproducts along the human P53 gene. Ann N Y Acad Sci. 1994;726:324–326. doi: 10.1111/j.1749-6632.1994.tb52840.x. [DOI] [PubMed] [Google Scholar]