Abstract
Extraintestinal pathogenic Escherichia coli (ExPEC) strains are important pathogens that cause diverse diseases in humans and poultry. Some E. coli isolates from chicken feces contain ExPEC-associated virulence genes, so appear potentially pathogenic; they conceivably could be transmitted to humans through handling and/or consumption of contaminated meat. However, the actual extraintestinal virulence potential of chicken-source fecal E. coli is poorly understood. Here, we assessed whether fecal E. coli isolates from healthy production chickens could cause diseases in a chicken model of avian colibacillosis and three rodent models of ExPEC-associated human infections. From 304 E. coli isolates from chicken fecal samples, 175 E. coli isolates were screened by PCR for virulence genes associated with human-source ExPEC or avian pathogenic E. coli (APEC), an ExPEC subset that causes extraintestinal infections in poultry. Selected isolates genetically identified as ExPEC and non-ExPEC isolates were assessed in vitro for virulence-associated phenotypes, and in vivo for disease-causing ability in animal models of colibacillosis, sepsis, meningitis, and urinary tract infection. Among the study isolates, 13% (40/304) were identified as ExPEC; the majority of these were classified as APEC and uropathogenic E. coli, but none as neonatal meningitis E. coli. Multiple chicken-source fecal ExPEC isolates resembled avian and human clinical ExPEC isolates in causing one or more ExPEC-associated illnesses in experimental animal infection models. Additionally, some isolates that were classified as non-ExPEC were able to cause ExPEC-associated illnesses in animal models, and thus future studies are needed to elucidate their mechanisms of virulence. These findings show that E. coli isolates from chicken feces contain ExPEC-associated genes, exhibit ExPEC-associated in vitro phenotypes, and can cause ExPEC-associated infections in animal models, and thus may pose a health threat to poultry and consumers.
Introduction
The primary and secondary habitats of Escherichia coli are the intestinal tract of warm-blooded animals and the environment, respectively. In poultry, as in humans, E. coli resides in the lower digestive tract, which it colonizes in the first 24 h after hatching [1] or birth [2]. Although many E. coli strains are harmless commensals, a subset have acquired the ability to cause intestinal or extraintestinal diseases. Extraintestinal pathogenic E. coli (ExPEC) strains cause diverse infections outside of the intestinal tract in humans and animals [3–5]. Based on the host and the site of infection, different ExPEC strains are subclassified as neonatal meningitis E. coli (NMEC), sepsis-associated E. coli (SEPEC), uropathogenic E. coli (UPEC), which cause newborn meningitis, sepsis, and urinary tract infections (UTI), respectively; and avian pathogenic E. coli (APEC), which mainly causes respiratory and systemic disease in poultry.
ExPEC infections are important to human health and are a major cause of economic loss to the poultry industry. In the United States, the costs associated with ExPEC infections in humans and poultry exceeds $4 billion per year [3, 6]. ExPEC strains can colonize the intestine, similar to non-pathogenic commensal E. coli [3, 6], but are equipped with virulence factors that allow them to cause disease in extraintestinal sites. In addition to the intestine, poultry houses serve as a reservoir for APEC [7], and this environment allows strains to persist for many months over successive flocks [8].
Epidemiological studies have documented the presence of ExPEC, as defined by molecular criteria, in the intestine of healthy poultry and in poultry meat, with some strains being genetically similar to those responsible for human infections [7, 9, 10]. Based on epidemiological analysis and molecular typing, it is suspected that food-producing animals are a source of bacteria capable of causing human ExPEC infections [11]. However, the frequency with which humans acquire ExPEC through consumption or handling of ExPEC-contaminated foods, become colonized intestinally, and subsequently develop infection at extraintestinal sites, is undefined [12].
Chicken-to-chicken ExPEC transmission, through pecking or inhalation of contaminated fecal dust could result in carcass condemnation and severe disease or death of poultry [3, 8]. In addition, ExPEC transmission among chickens may increase the presence of ExPEC colonized chickens, and thus increase the frequency of ExPEC transmission onto poultry products. Fecal contamination of poultry carcasses at slaughter, including from rupture of the digestive system during processing, is likely a major source of meat contamination with ExPEC [7, 10, 13]. Such organisms could be transmitted to humans through consumption of contaminated meat, cross-contamination of non-meat items during food preparation, hand-mouth contamination by the food preparer, or direct human-animal contact [11].
Improved understandings of the risk of chicken-source fecal E. coli are needed to guide the development of preventative measures to reduce infection in poultry and subsequent food contamination. Accordingly, this study's objectives were (i) to characterize E. coli isolates from chicken fecal samples both genetically and phenotypically for virulence-associated traits and (ii) to determine the virulence of selected isolates in animal models of chicken colibacillosis and human ExPEC diseases (sepsis, meningitis, and UTI).
Materials and methods
Human and animal ethics statement
With approval from the Arizona State University (ASU) Institutional Review Board (#1012005820) and the subjects' written informed consent, voided urine was collected from 2 male and 2 female healthy adult human volunteers. Animal infection experiments were performed in dedicated animal facilities in accordance with protocols approved by the ASU or Iowa State University (ISU) Institutional Animal Care and Use Committee (ASU Protocol number 1168R and ISU Protocol number 1-16-8159G). Appropriate procedures were used to reduce potential pain, distress, and discomfort. Animals were acclimated for 7 days before each experiment and received enrichment devices. Animals were housed in groups in order to promote social behavior. Humane endpoint criteria were set for all animals such that any moribund animal, animals exhibiting immobility (unable to feed or drink) or failure to groom (rodents only) were euthanized immediately according to the recommendations of the American Veterinary Medical Association 2013 Guidelines, and all remaining animals were euthanized at specific time points post-inoculation as described below. Animals exhibiting signs of illness but not meeting endpoint were not treated to maintain critical experimental data (e.g., bacterial loads), instead specific early endpoints were used as described below to minimize suffering.
Bacterial strains and growth media
Bacterial strains were routinely grown at 37°C in Luria Bertani (LB) broth, on LB agar, or on MacConkey agar unless stated otherwise. Freezer stocks were maintained at -80°C in peptone-glycerol medium. Positive control E. coli strains for the following disease models included urosepsis isolate CFT073 [14] for sepsis and UTI, cystitis isolate UTI89 [15] for UTI, neonatal meningitis isolate RS218 [16] for meningitis, and avian-source χ7122 [17] and APEC-O2 [18] for avian colibacillosis. Negative control strains included E. coli K-12 MG1655 for sepsis, UTI, and colibacillosis, and laboratory E. coli strain DH5α for meningitis. For colicin production, E. coli K-12 χ6092 was used as a sensitive indicator [19].
Three-hundred and four fecal E. coli isolates were obtained from conventionally-raised commercial chickens. For this, fresh fecal samples from the pen floor were collected from 7 different broiler chicken farms in Quebec, Canada. Each farm housed 20,000 to 30,000 chickens aged from 35 to 50 days at the time of sampling From each farm, 5–15 pooled fecal samples were suspended 1/10 (weight/volume) in buffered peptone water and enriched overnight at 37°C. Boiled DNA extracts from these cultures were tested by PCR [20] for the presence of the virulence genes tsh, papC, iucD, and cnf, which are associated with E. coli causing extraintestinal infections in one or more production animal species or humans [21–25], thereby permitting a rapid and inexpensive initial screen for a wide spectrum of possible ExPEC strains.
For screen-positive samples, enriched broths were streaked onto MacConkey agar. Three to 10 lactose-positive (i.e., presumptive E. coli) colonies were picked randomly per MacConkey agar plate and tested individually by PCR for tsh, papC, and iucD (no broth samples were positive for cnf). All isolates positive for any of these virulence genes were confirmed as E. coli by PCR detection of the E. coli-specific housekeeping gene uidA and underwent a more extensive virulence gene screen, as described below.
Genotypic and phylogenetic screening
The 175 E. coli isolates identified by this initial screen as containing ≥ 1 of tsh, papC, and iucD were further screened by multiplex PCR for ExPEC status, based on detection of ≥ 2 of the following 5 ExPEC-defining traits: papA and/or papC (P fimbriae: counted as 1), sfa/foc (S and F1C fimbriae), afa/dra (Dr-binding adhesins), kpsM II (group 2 capsule), and iutA (aerobactin system) [26]. All isolates qualifying as ExPEC (n = 40), and a similar number of randomly selected non-ExPEC isolates (n = 37) from the remaining 135 E. coli isolates that did not qualify as ExPEC, underwent further analysis for major E. coli phylogenetic groups (A, B1, B2, and D) by triplex PCR [27] and were screened by multiplex PCR for 50 ExPEC-associated virulence genes (Table 1) [21, 28, 29].
Table 1. Prevalence of extraintestinal pathogenic Escherichia coli (ExPEC)-associated genes among chicken fecal E. coli isolates.
| Functional category | Gene | No. of isolates positivea (%) | P-valueb | |
|---|---|---|---|---|
| ExPEC (n = 40) | Non-ExPEC (n = 37) | |||
| Adhesin | F10 | 1 (3) | 0 (0) | 1.00 |
| F14 | 7 (18) | 0 (0) | 0.01 | |
| fimH | 36 (90) | 36 (97) | 0.36 | |
| hra | 15 (38) | 9 (24) | 0.23 | |
| iha | 1 (3) | 1 (3) | 1.00 | |
| papA | 11 (28) | 0 (0) | < 0.001 | |
| papC | 14 (35) | 0 (0) | < 0.001 | |
| papEF | 14 (35) | 1 (3) | < 0.001 | |
| papG2 | 38 (95) | 1 (3) | < 0.001 | |
| papG3 | 39 (98) | 0 (0) | < 0.001 | |
| sfa | 0 (0) | 1 (3) | 0.48 | |
| Protectin | cvaC | 17 (43) | 24 (65) | 0.07 |
| iss | 30 (75) | 29 (78) | 0.79 | |
| kpsMT3 | 4 (10) | 5 (14) | 0.73 | |
| kpsMT K1 | 2 (5) | 0 (0) | 0.49 | |
| kpsM II | 37 (93) | 1 (3) | < 0.001 | |
| rfc | 0 (0) | 1 (3) | 0.48 | |
| traT | 0 (0) | 1 (3) | 0.48 | |
| Siderophore | fyuA | 7 (18) | 7 (19) | 1.00 |
| ireA | 8 (20) | 11 (30) | 0.43 | |
| iroN | 19 (48) | 27 (73) | 0.04 | |
| iutA | 40 (100) | 35 (95) | 0.23 | |
| Toxin | astA | 13 (33) | 11 (29) | 0.81 |
| hlyF | 36 (90) | 37 (100) | 0.12 | |
| pic | 2 (5) | 1 (3) | 1.00 | |
| tsh | 21 (53) | 36 (97) | < 0.001 | |
| Miscellaneous | ibeA | 1 (3) | 0 (0) | 1.00 |
| malX | 3 (8) | 0 (0) | 0.24 | |
| ompT | 13 (33) | 14 (38) | 0.64 | |
| usp | 17 (43) | 25 (68) | 0.04 | |
aAll isolates were negative for the following adhesins (afa/draBC, afaE, bmaE, clpG, focG, F11, F12, F16, F17, gafD, papG1, sfaS), protectins (kpsMT K2, kpsMT K15, kfiC K5), toxins (cdt, cnf1, hlyA, saT), and fliC H7 gene.
bP-values determined by Fisher’s exact test, two-tailed.
In vitro phenotypic screening
Study isolates underwent phenotypic screening for siderophore and colicin production, biofilm formation, complement resistance, growth in human urine, swimming motility, and cell association ability. Siderophore production was analyzed using Chrome azurol S agar as described previously [30]. A positive result consisted of bacterial colonies displaying orange haloes on blue agar after overnight incubation at 37°C; halo diameters were recorded. Total colicin production was tested using the double-agar diffusion method [31] on trypticase soy agar. E. coli K-12 χ6092 was used as a sensitive indicator for colicin production.
Biofilms were quantified in 96-well microtiter plates (Microtest™ U-Bottom, Becton Dickenson, Franklin Lakes, NJ) as described previously [32]. Bacterial strains were grown overnight to an optical density (OD) at 600 nm of 1.0, diluted 1:100 in PBS, and 200 μl of the culture was added to 96-well plates in quadruplicate. After overnight incubation at 37°C, plates were stained with crystal violet. Individual experiments were performed at least three times. A crystal violet-stained biofilm with an OD600 at least 3-fold greater than the negative control well containing only growth medium was considered a positive result.
Resistance to guinea pig serum complement was determined using a standard quantitative microtiter plate method [33]. Briefly, 104 CFU of bacteria in 100 μl of PBS was mixed with an equal volume of 50% serum. After 4 h at 37°C, the OD492 was determined spectrophotometrically. Isolates were considered complement-resistant if the OD492 in serum-containing wells equaled or exceeded that of the no-serum control well. Heat-inactivated sera was used as a control.
Growth in human urine was assessed as described previously [32]. Urine was filter sterilized, pooled, and frozen in aliquots. Diluted bacterial suspensions in urine were prepared by adding to urine a 1:100 volume of an overnight LB culture after it had been adjusted to an OD600 of 1.0. The turbidity of the bacterial suspensions was measured using a wideband filter (420–580 nm) every 15 min for 8 h at 37°C. E. coli K-12 strain χ6092 was used as a negative control and UPEC strain CFT073 as a positive control.
For swimming motility assays, a toothpick was used to stab-inoculate overnight LB cultures adjusted to an OD600 of 1.0 onto 0.25% agar plates containing 0.7% sodium chloride and 1.3% tryptone. Plates were incubated for 8 h at 37°C.
T24 human bladder carcinoma (ATCC HTB-4) and A498 human renal carcinoma (ATCC HTB-44) cell lines were obtained from American Type Culture Collection (ATCC) and maintained in growth media as specified by ATCC. For inoculation onto cell monolayers, bacterial cultures were prepared from an LB overnight culture, diluted 1:100 in freshly pooled (4 individual samples) filter-sterilized human urine, and then incubated statically for 24 h at 37°C. Approximately 105 CFU of bacteria were inoculated onto cells at a multiplicity of infection of 10. For bacterial association assays, the inoculated cells were incubated at 37°C in 5% CO2 for 1 h, then rinsed three times with PBS. Cells were lysed with 0.1% deoxycholic acid sodium salt for enumeration of viable colonies by serial dilution plating on MacConkey agar. For persistence assays, after the cells had been incubated with bacteria for 1 h and rinsed with PBS, medium containing 100 μg/ml gentamicin (Sigma-Aldrich) was added and cells were incubated at 37°C for an additional 1 or 3 h. Cells were then washed three times with PBS and lysed for serial dilution plating. Association was calculated as the ratio of the number of cell-associated bacteria at 1 h to the initial inoculum size, and persistence as the ratio of the number of intracellular bacteria at 3 h vs. 1 h.
Virulence in chickens
Female white leghorn chickens (VALO BioMedia, Adel, IA) were raised on the floor in pens containing deep wood shavings to mimic cage-free conditions, and separate rooms were used for each bacterial challenge strain. Animals were maintained on a Purina® non-medicated feed containing prebiotics and probiotics throughout the study. During the acclimation period prior to infection, 2 animals found with pecking wounds had died. At 5 weeks of age, chickens were inoculated with 107 CFU via the air sac from an overnight LB culture suspended in PBS [17]. All experimental and control groups contained at least 7 animals. Chickens were monitored twice daily for 2 days and euthanized at 48 h post-infection by carbon dioxide inhalation. No chickens died following infection prior to the experimental endpoint. At 2 days post-challenge blood, heart, liver, lung, spleen, and an air sac swab were collected for detection and quantification of E. coli using MacConkey agar. Gross colibacillosis lesions in the air sac, heart, and liver were scored using an established scoring scheme [17].
Virulence in mammals
Rodent models of human ExPEC infections, including sepsis, meningitis, and UTI, were used to evaluate the isolates' virulence potential for humans. Seven-week-old female BALB/c mice (Charles River Laboratories, Wilmington, MA) were injected intraperitoneally with approximately 108 CFU of a log-phase LB culture suspended in PBS. All experimental and control groups contained 5 mice. Mice were observed daily over 7 days and scored for illness severity using an established scoring scheme [34] as follows: 1, healthy; 2, minimally ill; 3, moderately ill; 4, severely ill; 5, dead. All animals meeting endpoint criteria were euthanized by carbon dioxide inhalation, death was not considered an endpoint criterion. However, some animals died following infection prior to the experimental endpoint due to sepsis. On day 7, surviving mice were euthanized by carbon dioxide inhalation.
The ability of chicken fecal isolates to enter the central nervous system was tested in an established rat model of E. coli meningitis [35]. Briefly, outbred pregnant Sprague-Dawley rats (Charles River Laboratories) with timed conception were used to give birth to neonatal rats. Five-day-old Sprague-Dawley rats were divided randomly into groups of 10 to 12 rats and received approximately 102 CFU intraperitoneally. No infected rats died prior to the experimental endpoint. At 18 h post-inoculation, rats were euthanized by carbon dioxide inhalation followed by cervical dislocation, and blood and cerebrospinal fluid specimens were collected, serially diluted, and plated on MacConkey agar.
The ability of bacteria to cause UTI was tested in mice, as described previously [36]. Seven to eight-week-old female CBA/J mice (Jackson Laboratories, Bar Harbor, ME) were inoculated via a urethral catheter with approximately 108 CFU of bacteria. Mice were catheterized following anesthesia with an intraperitoneal injection of a ketamine—xylazine—acepromazine cocktail. Three isolates that grew in human urine (MM242, MM243, and MM244) and two that failed to grow (MM248 and MM259) were selected as experimental isolates. All experimental and control groups contained at least 9 mice. Mice were monitored twice daily for 2 days. No animals died following infection but prior to the experimental endpoint. Mice were euthanized 48 h post-infection by carbon dioxide inhalation and CFU/g of bladder, kidney, liver, and spleen were determined by serial dilution plating of organ homogenates on MacConkey agar.
Statistical analysis
Fisher's exact test (two-tailed) was used to compare ExPEC and non-ExPEC isolates for the prevalence of ExPEC virulence genes and virulence-associated phenotypes, and experimental and control strains for the proportion of tissues positive for E. coli in the chicken colibacillosis model. A t-test was used to compare ExPEC and non-ExPEC isolates for colicin and siderophore production. An ANOVA followed by Dunnett’s method for multiple means comparison was used to compare experimental and control strains in cell association and persistence assays, and in the colibacillosis, meningitis, and UTI models. The Log-rank (Mantel-Cox) test was used to compare survival curves from the sepsis model. Analyses were performed using Graphpad Prism 6.0. P values < 0.05 were considered significant.
Results
Prevalence of ExPEC virulence genes
Fecal E. coli isolates (n = 304) from healthy chickens were prescreened for 4 genes (tsh, papC, iucD, and cnf) and 175 tested positive for one or more of these genes. Among the 175 isolates, 40 qualified as ExPEC using a PCR-based ExPEC screening method [37]. Extended virulence genotyping of the 40 ExPEC and 37 randomly selected non-ExPEC isolates identified 26 of the 50 genes investigated in at least one isolate each (Table 1 and S1 Table).
Phylogenetic groups and subpathotypes
Prevalence of phylogroups differed for isolates classified as ExPEC vs. non-ExPEC for group A (63% vs. 8%, respectively: P < 0.001), B1 (0% vs. 78%: P < 0.001), and D (33% vs. 11%: P = 0.03), but not group B2 (5% vs. 3%: P = 1.0).
ExPEC isolates were classified into subpathotypes based on previously described criteria [32] (Table 2). Of the 40 ExPEC isolates, 32 (80%) qualified for one or more of the defined subpathotypes, including 24 (60%) as APEC (18% APEC only) and 15 (38%) as UPEC (18% UPEC only). In contrast, none qualified as NMEC, and 8 (20%) fit none of the defined pathotypes. Of the 32 APEC and UPEC isolates, 15 (47%) qualified additionally as SEPEC.
Table 2. Criteria and prevalence of extraintestinal pathogenic Escherichia coli (ExPEC) subpathotypes.
| Subpathotypea | Selection-based criteria | No. (%)c | |
|---|---|---|---|
| Phenotype | Genotypeb | ||
| APEC | None | ExPEC and ≥ 4 of 5 selected APEC genes | 7 (18) |
| NMEC | None | ExPEC plus kpsMT K1 and ibeA | 0 (0) |
| UPEC | Growth in urine | ExPEC | 7 (18) |
| Undefined | None | ExPEC | 4 (10) |
| APEC/SEPEC | Complement resistant | ExPEC and ≥ 4 of 5 selected APEC genes | 10 (25) |
| APEC/UPEC | Growth in urine | ExPEC and ≥ 4 of 5 selected APEC genes | 3 (8) |
| APEC/UPEC/SEPEC | Growth in urine and complement resistant | ExPEC and ≥ 4 of 5 selected APEC genes | 4 (10) |
| UPEC/SEPEC | Growth in urine and complement resistant | ExPEC | 1 (3) |
| Undefined/SEPEC | Complement resistant | ExPEC | 4 (10) |
aAPEC, avian pathogenic E. coli; NMEC, neonatal meningitis E. coli; SEPEC, sepsis-associated E. coli; Undefined, classified as ExPEC but does not correspond with any of the three major subpathotypes (APEC, NMEC, or UPEC); UPEC, uropathogenic E. coli.
bExPEC defined by ≥ 2 of the following genes: papA and/or papC (counted as 1), sfa/foc, afa/dra, kpsM II, and iutA. For APEC, genes included: (1) kpsM II; (2) iss; (3) tsh; (4) one of the 5 genes: sfa, foc, papA, papC, and papEF; and (5) one of the 2 genes iutA and fyuA.
cThe number of isolates positive for a given subpathotype only.
In vitro virulence-associated phenotypes
PCR-confirmed ExPEC isolates and randomly selected non-ExPEC isolates were compared for virulence-associated phenotypes. For this, siderophore and colicin production, biofilm formation, complement resistance, and growth in human urine were assessed by standard assays (S2 Table). The results are summarized in Table 3.
Table 3. Prevalence of virulence-associated in vitro phenotypes among chicken fecal Escherichia coli isolates.
| ExPECa | No. of isolates | Siderophore production | Mean CASb zone diameter (mm) | Colicin production | Mean colicin zone diameter (mm) | Biofilm production | Complement resistance | Growth in urine |
|---|---|---|---|---|---|---|---|---|
| Yes | 40 | 100% | 18.4c | 93% | 18.5 | 100%c | 48% | 38%c |
| No | 37 | 100% | 14.4 | 92% | 15.6 | 81% | 24% | 0% |
aExtraintestinal pathogenic E. coli.
bChrome azurol S (zone diameter indicates extent of siderophore production).
cStatistically significant difference, ExPEC vs. non-ExPEC (P < 0.05) determined by a t-test for mean zone diameter of colicin and siderophore production, and Fisher’s exact test (two tailed) for siderophore, colicin, and biofilm production, complement resistance, and growth in urine.
Selected isolates—chosen based on differing ExPEC status, genotype, phylogroup, and in vitro phenotypes—were additionally characterized for swimming motility, ability to associate with and persist within human A498 and T24 cells, and virulence in animal models of ExPEC-associated infections. These isolates were selected based on applicability to the animal challenge models. Thus, all APEC and two APEC/UPEC isolates were tested in chickens, two of three complement-resistant isolates were selected for the sepsis model, and APEC/UPEC isolates were selected for the UTI model. Since, no fecal E. coli isolates were classified as NMEC, isolates containing virulence factors (K1 capsule or ibeA) associated with NMEC were selected. In addition, non-ExPEC isolates were selected to determine if isolates not classified as ExPEC based on molecular typing could still cause ExPEC-associated diseases. Table 4 summarizes the isolates' relevant in vitro phenotypes. Since bladder and kidney cell lines were used for cell association and persistence assays, only isolates tested in the UTI mouse model were characterized in these assays.
Table 4. Characteristics of selected Escherichia coli isolates from chicken fecal samples used for in vivo experiments.
| Isolate | ExPEC subpathotype or non-ExPEC | Phylo-group | Virulence genotype | CR | Urine growth | Siderophore | Colicin | Biofilm | Swim | A498 cells | T24 cells | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | P | A | P | ||||||||||
| MM149 | APEC | B2 | astA, cvaC, fimH, fyuA, hlyF, ibeA, iroN, iss, iutA, kpsM II, malX, ompT, tsh, usp | + | - | + | + | + | NS | NT | NT | NT | NT |
| MM218 | APEC | A | astA, fyuA, hlyF, hra, ire, iroN, iss, iutA, K1, kpsM II, tsh | - | - | + | + | + | Sig- | NT | NT | NT | NT |
| MM225 | Non-ExPEC | A | hlyF, iss, ompT, tsh | - | - | + | + | + | Sig- | NT | NT | NT | NT |
| MM242 | APEC/UPEC | A | cvaC, fimH, hlyF, iss, iutA, kpsM II, tsh, usp | + | + | + | + | + | Sig- | Sig+ | NS | Sig+ | NS |
| MM243 | APEC/UPEC | A | cvaC, fimH, hlyF, iss, iutA, kpsM II, tsh, usp | - | + | + | + | + | Sig- | NS | NS | NS | NS |
| MM244 | APEC/UPEC | A | cvaC, fimH, hlyF, iss, iutA, kpsM II, tsh, usp | + | + | + | + | + | NS | NS | NS | NS | NS |
| MM248 | Non-ExPEC | B1 | cvaC, fimH, hlyF, iroN, iss, iutA, tsh, usp | - | - | + | + | - | Sig+ | Sig+ | NS | Sig+ | Sig+ |
| MM259 | Non-ExPEC | B2 | fimH, hlyF, tsh | - | - | + | + | + | Sig+ | NS | NS | NS | Sig+ |
| MM299 | APEC | D | astA, fimH, hlyF, iroN, iss, iutA, kpsM II, tsh | + | - | + | + | + | Sig+ | NT | NT | NT | NT |
A, cell association assay; APEC, avian pathogenic E. coli; CR, complement resistance; ExPEC, extraintestinal pathogenic E. coli; P, cell persistence assay; Phylo, phylogenetic group; NS, not significantly (P < 0.05) different compared to negative control MG1655; NT, not tested; Sig+, significantly (P < 0.05) greater than negative control MG1655; Sig-, Significantly (P < 0.05) less than negative control MG1655; Swim, swimming motility; UPEC, uropathogenic E. coli.
Ability to cause chicken airsacculitis
Using a chicken airsacculitis model, six chicken fecal E. coli isolates and positive controls APEC-O2 and χ7122 were compared with negative control MG1655 for invasion of the internal organs of chickens after inoculation via the air sac (Table 5). Isolates classified as APEC (MM149, MM218, and MM299), two of three APEC/UPEC (MM242 and MM243), and one non-ExPEC (MM259) were selected. Some test isolates and both positive controls, but not the negative control, yielded positive cultures for multiple internal organs. Bacterial counts for chicken fecal isolates exceeded those for negative control strain MG1655 in the spleen for isolates MM149 and MM299, and in the air sac and heart for isolate MM218.
Table 5. Ability of Escherichia coli isolates to cause systemic infection in chickens.
| Strain | Mean lesion score | Air sac | Blood | Heart | Liver | Lung | Spleen | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Air sac | Heart and liver | Pro-portion positive | Pro-portion positive | Mean log10 CFU/ml | Pro-portion positive | Mean log10 CFU/g | Pro-portion positive | Mean log10 CFU/g | Pro-portion positive | Mean log10 CFU/g | Pro-portion positive | Mean log10 CFU/g | |
| Controls | |||||||||||||
| χ7122 | 2.1a | 2.4a | 8/10a | 6/10 | 1.1 ± 1.2 | 9/10a | 3.4 ± 1.6a | 8/10a | 1.9 ± 1.1a | 7/10a | 1.8 ± 1.3 | 9/10a | 3.2 ± 1.3a |
| APEC-O2 | 1.0 | 1.6 | 3/8 | 1/8 | 0.6 ± 1.6 | 3/8 | 2.2 ± 3.2a | 4/8 | 1.2 ± 1.4 | 4/10 | 1.8 ± 2.5 | 5/8a | 2.1 ± 1.9a |
| MG1655 | 0.6 | 0.5 | 0/8 | 1/8 | 0.3 ± 0.7 | 0/8 | 0.0 ± 0.0 | 0/8 | 0.0 ± 0.0 | 0/8 | 0.0 ± 0.0 | 0/8 | 0.0 ± 0.0 |
| Fecal isolates | |||||||||||||
| MM149 | 0.6 | 1.4 | 3/10 | 2/10 | 0.8 ± 1.6 | 2/10 | 0.5 ± 1.1 | 2/10 | 0.8 ± 1.8 | 4/10 | 1.2 ± 1.6 | 5/10a | 1.5 ± 1.8 |
| MM218 | 1.0 | 1.9 | 4/7a | 1/7 | 0.5 ± 1.2 | 4/7a | 2.2 ± 2.3a | 3/7 | 1.3 ± 1.8 | 3/7 | 1.7 ± 2.1 | 3/7 | 1.4 ± 1.9 |
| MM242 | 0.0 | 0.4 | 3/8 | 0/8 | 0.0 ± 0.0 | 1/8 | 0.2 ± 0.5 | 1/8 | 0.2 ± 0.7 | 1/8 | 0.4 ± 1.2 | 1/8 | 0.4 ± 1.2 |
| MM243 | 0.1 | 0.6 | 0/7 | 1/7 | 0.4 ± 1.0 | 1/7 | 0.3 ± 0.7 | 1/7 | 0.2 ± 0.6 | 1/7 | 0.4 ± 0.9 | 1/7 | 0.4 ± 1.1 |
| MM259 | 0.3 | 0.1 | 1/8 | 1/8 | 0.6 ± 1.7 | 1/8 | 0.3 ± 0.7 | 1/8 | 0.4 ± 1.1 | 1/8 | 0.5 ± 1.4 | 2/8 | 0.7 ± 1.5 |
| MM299 | 0.3 | 0.9 | 3/10 | 5/10 | 1.0 ± 1.3 | 1/10 | 0.3 ± 0.8 | 3/10 | 0.8 ± 1.3 | 4/10 | 1.4 ± 2.0 | 6/10a | 1.8 ± 1.7 |
Concentration data is represented by mean values ± standard deviation. Counts were determined at 48 h post-inoculation
aSignificant difference (P < 0.05) compared with MG1655 (negative control) determined by a Fisher’s exact test (two tailed) for the proportion positive, or by an ANOVA followed by Dunnett’s method for mean bacterial loads.
Ability to cause sepsis
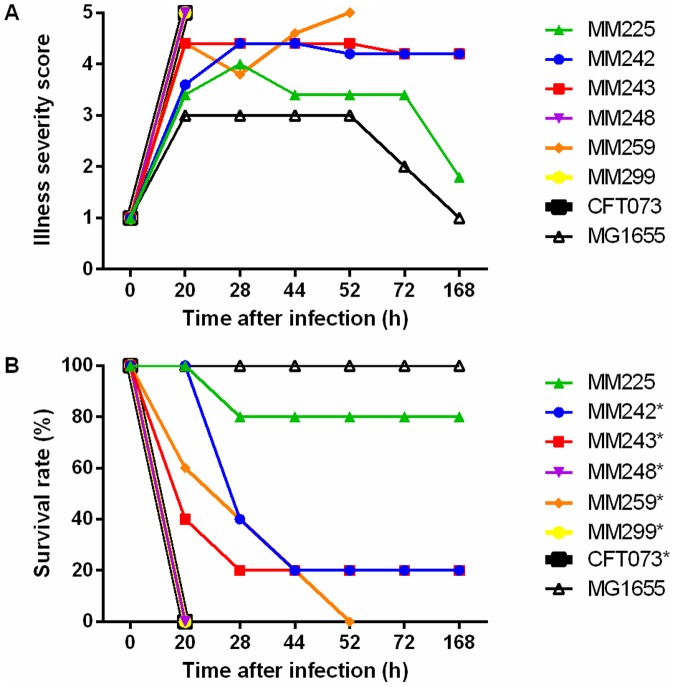
Using a mouse sepsis model, three chicken fecal isolates classified as ExPEC (MM242, MM243, and MM299) and three as non-ExPEC (MM225, MM248, and MM259) were randomly selected, and along with positive control CFT073 were compared with negative control MG1655 for illness severity score and survival (Fig 1). During the experiment, 29 of 40 mice died from sepsis infection or were euthanized due to meeting endpoint criteria. Survival curves (Fig 1) were significantly different from negative control strain MG1655 for positive control strain CFT073 (P = 0.003) and fecal ExPEC isolates MM242 (P = 0.01), MM243 (P = 0.01), and MM299 (P = 0.003), and non-ExPEC isolates MM248 (P = 0.003) and MM259 (P = 0.002). In contrast, the non-ExPEC isolate MM225 was lethal in only one of five mice and the survival curve was not significantly different (P = 0.3) from the negative control.
Fig 1. Ability of fecal Escherichia coli isolates to cause lethal sepsis in mice.
A BALB/c mouse sepsis model was used to evaluate the ability of E. coli isolates to cause lethal sepsis within 7 days of intraperitoneal challenge with 108 CFU. Five mice were used per strain. (A) Severity scores, as recorded over the week using a 5-point scoring scheme (1, healthy; 2, minimally ill; 3, moderately ill; 4, severely ill; 5, dead). (B) Survival rate over 7 d. Human ExPEC isolate CFT073 was used as a positive control and E. coli K-12 MG1655 as a negative control. An asterisk (*) represents a significantly (P < 0.05) different survival curve determined by The Log-rank (Mantel-Cox) test for experimental isolates or positive control strain CFT073 compared with the negative control MG1655.
Ability to cause meningitis
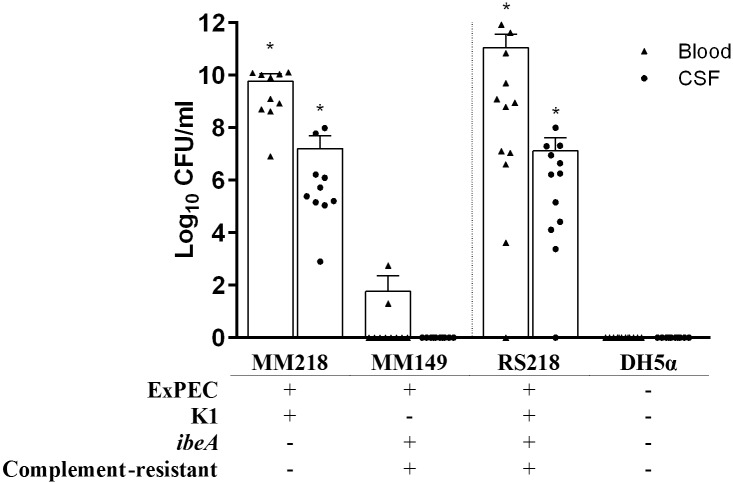
Since no chicken fecal E. coli isolates were classified as NMEC based on molecular identification of both K1 and ibeA, isolates that were positive for K1 or ibeA were selected to be tested in the rat meningitis model. Chicken fecal isolates MM149 and MM218, which qualify as ExPEC but differ for complement resistance, K1 capsule, and the NMEC-associated invasin gene ibeA (Fig 2), were tested for their ability to cause meningitis in a neonatal rat model in comparison with human NMEC isolate RS218 and negative control DH5α. MM218 was recovered from the blood and cerebral spinal fluid at a similar level to NMEC isolate RS218, and at a significantly higher level compared with negative control DH5α. In contrast, MM149 was recovered inconsistently from blood (< 102 CFU/ml) and not at all from CSF, and for neither endpoint differed significantly from negative control DH5α (Fig 2).
Fig 2. Ability of fecal Escherichia coli isolates to cause meningitis in rats.
E. coli MM149 and MM218 isolated from chicken feces, positive control neonatal meningitis strain RS218, and negative control strain DH5α were assessed for their abilities to induce septicemia and meningitis in 5 day-old Sprague-Dawley rats. Isolates were characterized for extraintestinal pathogenic E. coli (ExPEC) status, K1 capsule, meningitis-associated gene ibeA, and complement resistance. Rats were challenged intraperitoneally with 102 CFU and assessed 18 h later for bacterial concentration in blood (triangles) and cerebral spinal fluid (CSF) (circles). Each experimental group contained at least 10 rats. Each symbol represents an individual animal and the vertical dashed line separates chicken fecal E. coli isolates from control strains. An asterisk (*) represents significantly (P < 0.05) higher mean values determined by an ANOVA followed by Dunnett’s method for experimental isolates or positive control strain RS218 compared with the negative control DH5α.
Ability to cause urinary tract infection
In the mouse model of ascending UTI, bacterial loads were quantified in the bladder, kidney, liver, and spleen of mice 48 h after inoculation of 108 CFU of the challenge strain into the bladder (Fig 3). Of the fecal E. coli isolates, three ExPEC isolates (MM242, MM243, and MM244) that could grow in urine and two non-ExPEC (MM248 and MM259) that failed to grow were selected. Some chicken fecal E. coli isolates equalled or exceeded one or both positive controls for bacterial counts in the internal organs. Significantly greater bacterial loads than observed with negative control MG1655 were observed in the bladder for both positive control strains and for the non-ExPEC fecal isolate MM248; in the kidney for positive control strain CFT073 and for APEC/UPEC fecal isolates MM242 and MM243; in the liver for fecal isolates MM242, MM243, and MM248; and in the spleen no significant differences were observed.
Fig 3. Ability of fecal Escherichia coli isolates to urinary tract infection.
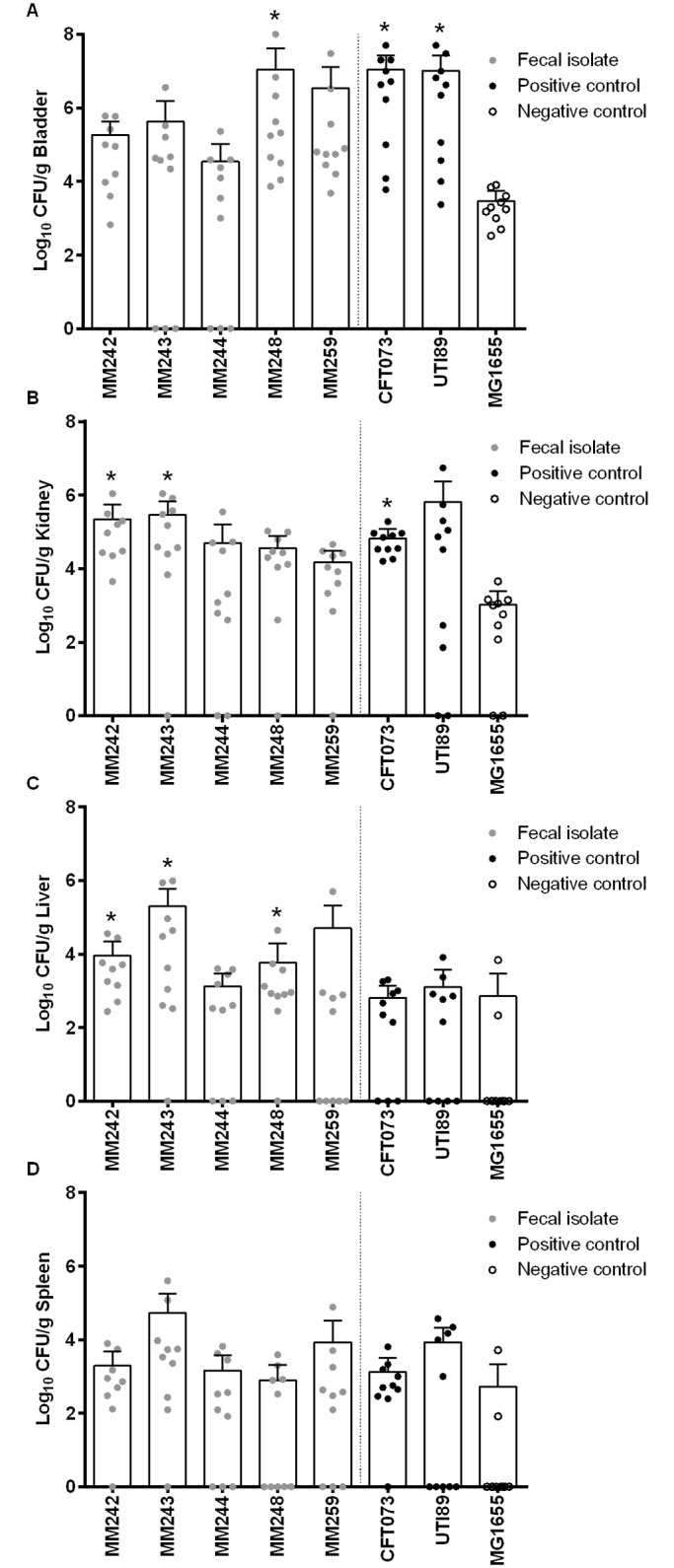
E. coli isolates MM242, MM243, MM244, MM248, and MM259, positive controls CFT073 and UTI89, and negative control MG1655 were assessed for their ability to colonize the (A) bladder and (B) kidney, and to invade in the (C) liver and (D) spleen of CBA/J mice. Mice were challenged with 108 CFU via a urethral catheter and monitored for 2 days. Each experimental group contained at least 9 mice. Each dot represents an individual animal; the vertical dashed line separates chicken fecal E. coli isolates from control strains. An asterisk (*) represents significantly higher mean values determined by an ANOVA followed by Dunnett’s method for fecal E. coli isolates and positive control strains compared with the negative control.
Discussion
The presence and characteristics of pathogenic E. coli colonizing healthy production chickens could be important to both animal and human health. Here, we characterized E. coli isolates from the feces of healthy production chickens both genotypically and phenotypically, including for their ability to cause disease in animal models of chicken and human infections. Based on the molecular criteria of Johnson et al. [37], 13% (40/304) of the present chicken fecal E. coli isolates qualified as ExPEC. Varying isolation methods, classification methods, geographic locations, and management practices likely contribute to differences in frequency of ExPEC isolation between studies. In a previous study using methods different than that of the current study, 10% of E. coli isolates from feces of commercial egg layer and meat chickens qualified molecularly as ExPEC [38]. These findings indicate that commercial chickens can harbor E. coli isolates with virulence characteristics of ExPEC that could be transmitted to other chickens in the production house or contaminate carcasses during processing. Notably, one study recovered E. coli from 87% (691/798) of post-chill chicken carcasses at large commercial harvest facilities [39]. Although concentrations decreased with subsequent processing steps, low counts persisted, suggesting the possibility of contaminated retail poultry products, as documented in multiple retail market surveys [32, 40].
We analyzed for major E. coli phylogenetic groups to further characterize the virulence potential of the present study isolates. As found previously for isolates from chicken meat and eggs [32], phylogroup distribution varied in relation to ExPEC status, with most ExPEC isolates representing phylogroups A and D, and non-ExPEC isolates phylogroup B1. This is consistent with the fact that APEC strains belong predominantly to phylogroups A and D, whereas human-source ExPEC strains belong mainly to phylogroups B2 and D [13, 41, 42].
Genotypic tests that distinguish ExPEC from non-ExPEC isolates have been proposed [21, 37, 43–45], and have been used in previous studies to predict the zoonotic potential of animal-source ExPEC isolates [10, 13, 21]. However, certain in vitro phenotypes (e.g., biofilm formation, colicin production, complement resistance) for which straightforward genetic screens are unavailable also contribute to, or correspond with, the ability of E. coli to cause extraintestinal infections. We showed previously a correlation between complement resistance and the ability of APEC to invade the internal organs of experimentally challenged chickens [46]. Here, we found that virulence-associated in vitro phenotypes were more prevalent among ExPEC isolates than non-ExPEC isolates, and significantly so for biofilm formation and growth in urine. Biofilm formation, which has been identified as important for UPEC colonization [47], provides bacteria with protection from detergents, antibiotics, and host defense elements [48]. Here, we detected biofilm formation for all ExPEC isolates. Analogous to the contribution of biofilm to bacterial persistence in the genitourinary tract [49], biofilm formation may also allow bacteria to persist on surfaces of poultry products such as raw meat and eggs, a suitable topic for future study.
To survive in extraintestinal sites, bacteria must not only overcome harsh conditions but also acquire nutrients, including micronutrients such as iron. Iron acquisition is critical in the iron-limited environment of the urinary tract. Most E. coli produce the catecholate siderophore enterobactin [50], possibly explaining why most of the present study isolates exhibited siderophore production. However, the ExPEC isolates produced larger haloes in that assay than did non-ExPEC isolates, suggesting that ExPEC produce more enterobactin and/or additional siderophores, e.g., aerobactin, salmochelin, and yersiniabactin [51, 52]. Whether siderophore production also contributes to bacterial persistence and survival on raw poultry meat and eggs warrants study.
Animal models of poultry and human infections were used to assess the ability of the study isolates to cause ExPEC-associated infections. To our knowledge, this is the first study to test E. coli from the feces of healthy production chickens in four ExPEC disease models, including avian colibacillosis, sepsis, meningitis, and UTI. In the avian colibacillosis model, chicken-source E. coli isolates were recovered from multiple internal organs, supporting that initially uncolonized chickens in production houses could acquire potentially invasive E. coli that are shed by colonized birds. Our findings confirm those of a previous study in which E. coli isolates from chicken feces and the poultry house environment invaded the internal organs of challenged chickens [7].
In previous studies of poultry-source E. coli in rodent models of ExPEC-associated human infections, Johnson et al. found that an APEC turkey lung isolate lacked full virulence in a mouse sepsis model and failed to cause bacteremia or meningitis in a rat meningitis model [53], whereas Tivendale et al. found that some avian colibacillosis isolates caused bacteremia and meningitis in the rat meningitis model [54]. Production chickens with colibacillosis may die because of infection or have their carcasses condemned when the lesions are identified during processing [3], which could reduce the risk of transfer of APEC strains from infected chickens to meat products during processing. However, if ExPEC are harbored in chicken feces, they may pose a less apparent but nonetheless real risk to food safety. To our knowledge, the present study is the first to show that a fecal E. coli isolate (MM218) from a healthy production chicken can cause bacteremia and meningitis in a rat meningitis model. The same isolate invaded internal organs of chickens demonstrating the potential of E. coli from chickens to cause disease in both poultry and humans. In addition, some of the studied fecal isolates caused lethal sepsis in ≤ 20 h, similar to human ExPEC strain CFT073. Fecal isolate MM299 caused lethal sepsis ≤ 20 h and invaded in the spleen of challenged chickens at significant levels, further demonstrating the potential of E. coli from chickens to cause disease in both poultry and humans. These findings have potentially important implications for food safety, since they suggest that chickens without colibacillosis could, via fecal contamination at harvest, transfer to poultry meat E. coli isolates with the ability to cause human meningitis and sepsis.
In the United States, UTI-related healthcare costs exceed $1 billion per year [6], and food-producing animals have been identified as a potential source of human ExPEC infection [11]. We found that 19% (15/77) of the tested chicken fecal E. coli isolates had the potential to cause UTI, based on their in vitro growth in urine, and that these isolates were mostly ExPEC per molecular criteria. These findings support a previous study that identified 23% of ExPEC isolates from raw chicken meat as UPEC [32]. We found that some chicken-source isolates could cause UTI in a mouse model, and with similar intensity as observed for positive control strains from humans with cystitis and pyelonephritis. This confirms in a very different geographical region the findings of previous studies that E. coli isolates from the feces of healthy Danish broiler chickens were virulent in the UTI mouse model [41, 55]. To further implicate bacterial isolates from chickens as a cause of disease in humans, another study [56] demonstrated nearly identical pulsed-field gel electrophoresis profiles between isolates from chickens and humans. However, additional studies are needed to establish that ExPEC are transferred from animals to humans via contaminated meat and to define the frequency of such transfer.
Some fecal isolates that caused diseases in animal models tested were classified as non-ExPEC by the ExPEC-genotypic and phenotypic associated criteria. Future characterization such as genomic and high-throughput sequencing of these isolates could elucidate their mechanisms of virulence and improve ExPEC detection criteria.
Conclusions
Our study provides an in-depth assessment of virulence-related genotypes and phenotypes, including in vivo virulence, of fecal ExPEC isolates from healthy production chickens. Multiple methods were used to identify isolates with presumptive zoonotic potential. Some isolates were able to cause one or several diseases in animal models of septicemia, meningitis, UTI, and avian colibacillosis. Thus, this study provides the strongest evidence to date that chicken feces could be a source of virulent ExPEC that are able to infect humans and poultry. Interventions that reduce these pathogens in the chicken intestine and on carcasses and meat products could help to reduce transmission via poultry products and thus prevent clinical ExPEC infections and humans.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors thank Mr. Brian Johnston (Veterans Affairs Medical Center and University of Minnesota), Ms. Natalie Mitchell (Biodesign Institute, Arizona State University), and Mr. Jocelyn Bernier-Lachance and Ms. Ghyslaine Vanier (Faculty of Veterinary Medicine, Université de Montréal) for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by a grant from the U.S. Department of Agriculture National Research Initiative USDA-NIFA-AFRI grant 2011-67005-30182 (M.M. and R.C.), and by Office of Research and Development, Medical Research Service, Department of Veterans Affairs grant #1 I01 CX000192 01 (J.R.J.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ballou AL, Ali RA, Mendoza MA, Ellis JC, Hassan HM, Croom WJ, et al. Development of the chick microbiome: how early exposure influences future microbial diversity. Front Vet Sci. 2016;3:2 doi: 10.3389/fvets.2016.00002 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettelheim KA, Lennox-King SM. The acquisition of Escherichia coli by new-born babies. Infection. 1976;4(3):174–9. doi: 10.1007/BF01638945 . [DOI] [PubMed] [Google Scholar]
- 3.Mellata M. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog Dis. 2013;10(11):916–32. Epub 2013/08/22. doi: 10.1089/fpd.2013.1533 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo TA, Johnson JR. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis. 2000;181(5):1753–4. Epub 2000/05/24. doi: 10.1086/315418 . [DOI] [PubMed] [Google Scholar]
- 5.Wu G, Ehricht R, Mafura M, Stokes M, Smith N, Pritchard GC, et al. Escherichia coli isolates from extraintestinal organs of livestock animals harbour diverse virulence genes and belong to multiple genetic lineages. Vet Microbiol. 2012;160(1–2):197–206. Epub 2012/05/30. doi: 10.1016/j.vetmic.2012.05.029 . [DOI] [PubMed] [Google Scholar]
- 6.Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 2003;5(5):449–56. . [DOI] [PubMed] [Google Scholar]
- 7.Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl Environ Microbiol. 2009;75(1):184–92. Epub 2008/11/11. doi: 10.1128/AEM.01324-08 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer RS, Jeffrey JS, Carpenter TE, Cooke CL, Atwill ER, Johnson WO, et al. Persistence of cellulitis-associated Escherichia coli DNA fingerprints in successive broiler chicken flocks. Vet Microbiol. 2000;75(1):59–71. . [DOI] [PubMed] [Google Scholar]
- 9.Manges AR, Johnson JR. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis. 2012;55(5):712–9. Epub 2012/05/23. doi: 10.1093/cid/cis502 . [DOI] [PubMed] [Google Scholar]
- 10.Delicato ER, de Brito BG, Gaziri LC, Vidotto MC. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet Microbiol. 2003;94(2):97–103. . [DOI] [PubMed] [Google Scholar]
- 11.Bélanger L, Garenaux A, Harel J, Boulianne M, Nadeau E, Dozois CM. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol Med Microbiol. 2011;62(1):1–10. Epub 2011/03/24. doi: 10.1111/j.1574-695X.2011.00797.x . [DOI] [PubMed] [Google Scholar]
- 12.Singer RS. Urinary tract infections attributed to diverse ExPEC strains in food animals: evidence and data gaps. Front Microbiol. 2015;6:28 Epub 2015/02/04. doi: 10.3389/fmicb.2015.00028 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson TJ, Logue CM, Wannemuehler Y, Kariyawasam S, Doetkott C, DebRoy C, et al. Examination of the source and extended virulence genotypes of Escherichia coli contaminating retail poultry meat. Foodborne Pathog Dis. 2009;6(6):657–67. Epub 2009/07/08. doi: 10.1089/fpd.2009.0266 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao JS, Stucker DM, Warren JW, Mobley HL. Pathogenicity island sequences of pyelonephritogenic Escherichia coli CFT073 are associated with virulent uropathogenic strains. Infect Immun. 1997;65(7):2812–20. Epub 1997/07/01. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103(15):5977–82. Epub 2006/04/03. doi: 10.1073/pnas.0600938103 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y, Xie Y, Kim KS. Genomic comparison of Escherichia coli K1 strains isolated from the cerebrospinal fluid of patients with meningitis. Infect Immun. 2006;74(4):2196–206. doi: 10.1128/IAI.74.4.2196-2206.2006 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, Brown PK, Arné P, et al. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect Immun. 2003;71(1):536–40. doi: 10.1128/IAI.71.1.536-540.2003 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. J Bacteriol. 2006;188(2):745–58. doi: 10.1128/JB.188.2.745-758.2006 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellata M, Touchman JW, Curtiss R. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli chi7122 (O78:K80:H9). PLoS One. 2009;4(1):e4232 doi: 10.1371/journal.pone.0004232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maluta RP, Fairbrother JM, Stella AE, Rigobelo EC, Martinez R, de Avila FA. Potentially pathogenic Escherichia coli in healthy, pasture-raised sheep on farms and at the abattoir in Brazil. Vet Microbiol. 2014;169(1–2):89–95. doi: 10.1016/j.vetmic.2013.12.013 [DOI] [PubMed] [Google Scholar]
- 21.Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol. 2008;46(12):3987–96. doi: 10.1128/JCM.00816-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao E-M, et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int J Med Microbiol. 2007;297(3):163–76. doi: 10.1016/j.ijmm.2007.01.003 [DOI] [PubMed] [Google Scholar]
- 23.Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, et al. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol. 2008;74(22):7043–50. doi: 10.1128/AEM.01395-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dezfulian H, Batisson I, Fairbrother JM, Lau PCK, Nassar A, Szatmari G, et al. Presence and characterization of extraintestinal pathogenic Escherichia coli virulence genes in F165-positive E. coli strains isolated from diseased calves and pigs. J Clin Microbiol. 2003;41(4):1375–85. doi: 10.1128/JCM.41.4.1375-1385.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngeleka M, Brereton L, Brown G, Fairbrother JM. Pathotypes of avian Escherichia coli as related to tsh-, pap-, pil-, and iuc-DNA sequences, and antibiotic sensitivity of isolates from internal tissues and the cloacae of broilers. Avian Dis. 2002;46(1):143–52. doi: 10.1637/0005-2086(2002)046[0143:POAECA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 26.Johnson JR, Kuskowski MA, Owens K, Gajewski A, Winokur PL. Phylogenetic origin and virulence genotype in relation to resistance to fluoroquinolones and/or extended-spectrum cephalosporins and cephamycins among Escherichia coli isolates from animals and humans. J Infect Dis. 2003;188(5):759–68. Epub 2003/08/23. doi: 10.1086/377455 . [DOI] [PubMed] [Google Scholar]
- 27.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–8. Epub 2000/09/30. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JR, Delavari P, O'Bryan TT, Smith KE, Tatini S. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2005;2(1):38–49. doi: 10.1089/fpd.2005.2.38 . [DOI] [PubMed] [Google Scholar]
- 29.Johnson JR, Kuskowski MA, Smith K, O'Bryan TT, Tatini S. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis. 2005;191(7):1040–9. doi: 10.1086/428451 . [DOI] [PubMed] [Google Scholar]
- 30.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. Epub 1987/01/01. . [DOI] [PubMed] [Google Scholar]
- 31.Davies JK, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol. 1975;123(1):102–17. Epub 1975/07/01. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell NM, Johnson JR, Johnston B, Curtiss R, Mellata M. Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl Environ Microbiol. 2015;81(3):1177–87. doi: 10.1128/AEM.03524-14 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MD, Wooley RE, Brown J, Spears KR, Nolan LK, Shotts EB Jr. Comparison of a quantitative microtiter method, a quantitative automated method, and the plate-count method for determining microbial complement resistance. Avian Dis. 1991;35(4):892–6. Epub 1991/10/11. . [PubMed] [Google Scholar]
- 34.Johnson JR, Porter SB, Zhanel G, Kuskowski MA, Denamur E. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun. 2012;80(4):1554–62. Epub 2012/02/06. doi: 10.1128/IAI.06388-11 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang SH, Wass C, Fu Q, Prasadarao NV, Stins M, Kim KS. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63(11):4470–5. ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thai KH, Thathireddy A, Hsieh MH. Transurethral induction of mouse urinary tract infection. J Vis Exp. 2010;(42). Epub 2010/08/05. doi: 10.3791/2070 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JR, Murray AC, Gajewski A, Sullivan M, Snippes P, Kuskowski MA, et al. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob Agents Chemother. 2003;47(7):2161–8. Epub 2003/06/25. doi: 10.1128/AAC.47.7.2161-2168.2003 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obeng AS, Rickard H, Ndi O, Sexton M, Barton M. Antibiotic resistance, phylogenetic grouping and virulence potential of Escherichia coli isolated from the faeces of intensively farmed and free range poultry. Vet Microbiol. 2012;154(3–4):305–15. Epub 2011/07/21. doi: 10.1016/j.vetmic.2011.07.010 . [DOI] [PubMed] [Google Scholar]
- 39.Altekruse SF, Berrang ME, Marks H, Patel B, Shaw WK, Saini P, et al. Enumeration of Escherichia coli cells on chicken carcasses as a potential measure of microbial process control in a random selection of slaughter establishments in the United States. Appl Environ Microbiol. 2009;75(11):3522–7. Epub 2009/04/10. doi: 10.1128/AEM.02685-08 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, et al. Prevalence of Camplyobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, D.C., area. Appl Environ Microbiol. 2001;67(12):5431–6. doi: 10.1128/AEM.67.12.5431-5436.2001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jakobsen L, Spangholm DJ, Pedersen K, Jensen LB, Emborg HD, Agerso Y, et al. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int J Food Microbiol. 2010;142(1–2):264–72. Epub 2010/07/27. doi: 10.1016/j.ijfoodmicro.2010.06.025 . [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Fakhr MK, Nolan LK. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151(Pt 6):2097–110. Epub 2005/06/09. doi: 10.1099/mic.0.27499-0 . [DOI] [PubMed] [Google Scholar]
- 43.Moulin-Schouleur M, Répérant M, Laurent S, Brée A, Mignon-Gasteau S, Germon P, Rasschaert D, Schouler C. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol. 2007;45(10):3366–76. Epub 2007/07/15. doi: 10.1128/JCM.00037-07 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ewers C, Janssen T, Kiessling S, Philipp HC, Wieler LH. Rapid detection of virulence-associated genes in avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian Dis. 2005;49(2):269–73. Epub 2005/08/13. doi: 10.1637/7293-102604R . [DOI] [PubMed] [Google Scholar]
- 45.Schouler C, Schaeffer B, Bree A, Mora A, Dahbi G, Biet F, et al. Diagnostic strategy for identifying avian pathogenic Escherichia coli based on four patterns of virulence genes. J Clin Microbiol. 2012;50(5):1673–8. Epub 2012/03/02. doi: 10.1128/JCM.05057-11 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R 3rd, Lehoux B, Fairbrother JM. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect Immun. 2003;71(1):494–503. Epub 2002/12/24. doi: 10.1128/IAI.71.1.494-503.2003 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenke P, Koves B, Nagy K, Hultgren SJ, Mendling W, Wullt B, et al. Update on biofilm infections in the urinary tract. World J Urol. 2012;30(1):51–7. Epub 2011/05/19. doi: 10.1007/s00345-011-0689-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall MR, McGillicuddy E, Kaplan LJ. Biofilm: basic principles, pathophysiology, and implications for clinicians. Surg Infect. 2014;15(1):1–7. Epub 2014/01/31. doi: 10.1089/sur.2012.129 . [DOI] [PubMed] [Google Scholar]
- 49.Berry RE, Klumpp DJ, Schaeffer AJ. Urothelial cultures support intracellular bacterial community formation by uropathogenic Escherichia coli. Infect Immun. 2009;77(7):2762–72. Epub 2009/05/20. doi: 10.1128/IAI.00323-09 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. Epub 2000/10/06. doi: 10.1146/annurev.micro.54.1.881 . [DOI] [PubMed] [Google Scholar]
- 51.Garenaux A, Caza M, Dozois CM. The Ins and Outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Veterinary microbiology. 2011;153(1–2):89–98. Epub 2011/06/18. doi: 10.1016/j.vetmic.2011.05.023 . [DOI] [PubMed] [Google Scholar]
- 52.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–51. Epub 2007/09/07. doi: 10.1128/MMBR.00012-07 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson TJ, Wannemuehler Y, Kariyawasam S, Johnson JR, Logue CM, Nolan LK. Prevalence of avian-pathogenic Escherichia coli strain O1 genomic islands among extraintestinal and commensal E. coli isolates. J Bacteriol. 2012;194(11):2846–53. Epub 2012/03/30. doi: 10.1128/JB.06375-11 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, et al. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun. 2010;78(8):3412–9. Epub 2010/06/03. doi: 10.1128/IAI.00347-10 ; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakobsen L, Garneau P, Bruant G, Harel J, Olsen SS, Porsbo LJ, et al. Is Escherichia coli urinary tract infection a zoonosis? Proof of direct link with production animals and meat. Eur J Clin Microbiol Infect Dis. 2012;31(6):1121–9. Epub 2011/10/28. doi: 10.1007/s10096-011-1417-5 . [DOI] [PubMed] [Google Scholar]
- 56.Johnson JR, Kuskowski MA, Menard M, Gajewski A, Xercavins M, Garau J. Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J Infect Dis. 2006;194(1):71–8. Epub 2006/05/31. doi: 10.1086/504921 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.