Abstract
Clinical evidence suggests that mood and behavioral symptoms in Premenstrual Dysphoric Disorder (PMDD), a common, recently recognized, psychiatric condition among women, reflect abnormal responsivity to ovarian steroids. This differential sensitivity could be due to an unrecognized aspect of hormonal signaling or a difference in cellular response. In this study, lymphoblastoid cell line cultures (LCLs) from women with PMDD and asymptomatic Controls were compared via whole transcriptome sequencing (RNA-seq) during untreated (ovarian steroid-free) conditions and following hormone treatment. The women with PMDD manifested ovarian steroid-triggered behavioral sensitivity during a hormone suppression and add-back clinical trial, and Controls did not, leading us to hypothesize that women with PMDD might differ in their cellular response to ovarian steroids. In untreated LCLs, our results overall suggest a divergence between mRNA (e.g., gene transcription) and protein (e.g., RNA translation in proteins) for the same genes. Pathway analysis of the LCL transcriptome revealed, among others, over-expression of ESC/E(Z) complex genes (an ovarian steroid-regulated gene silencing complex) in untreated LCLs from women with PMDD, with more than half of these genes over-expressed as compared to Controls, and with significant effects for MTF2, PHF19, and SIRT1 (p<0.05). RNA and protein expression of the 13 ESC/E(Z) complex genes were individually quantitated. This pattern of increased ESC/E(Z) mRNA expression was confirmed in a larger cohort by qRT-PCR. In contrast, protein expression of ESC/E(Z) genes was decreased in untreated PMDD LCLs with MTF2, PHF19, and SIRT1 all significantly decreased (p<0.05). Finally, mRNA expression of several ESC/E(Z) complex genes were increased by progesterone in Controls only, and decreased by estradiol in PMDD LCLs. These findings demonstrate that LCLs from women with PMDD manifest a cellular difference in ESC/E(Z) complex function both in the untreated condition and in response to ovarian hormones. Dysregulation of ESC/E(Z) complex function could contribute to PMDD.
Introduction
Premenstrual dysphoric disorder (PMDD) is a clinically distinct affective disorder that was recently included in DSM 5 [1]. Women with PMDD have recurrent and distressing mood and behavioral symptoms during the luteal phase of the menstrual cycle that remit within a few days of menses. The prevalence of PMDD is 2–5% of women of reproductive age [2–5].
PMDD symptoms are triggered by ovarian steroids, estradiol and progesterone (hereafter referred to as “E/P”) but peripheral E/P levels and hypothalamic-pituitary-ovarian axis function are normal [6]. Moreover, symptoms in PMDD are reported to occur mainly in ovulatory menstrual cycles [7]. When E/P production is suppressed by gonadotropin releasing hormone (GnRH) receptor agonists, approximately 55–70% of women with PMDD experience remission of symptoms [8–13], which recur when physiologic doses of E/P are added back [8]. In contrast, asymptomatic Controls undergoing identical hormone manipulations experience no changes in mood [8]. These findings indicate that PMDD may be a disorder of cellular response to E/P. Neuroimaging studies have identified behaviorally-relevant brain regions that are modulated by E/P, and some are differentially responsive in women with PMDD, including the amygdala, striatum, medial orbitofrontal cortex, anterior cingulate cortex, and prefrontal cortex [14–23] (and for review see [24]. This suggests that the differential behavioral response to E/P in PMDD is accompanied by ovarian steroid-related alterations in the activity of neuronal circuits underlying reward, social cognition, and affective states [25].
Despite these observations, a cellular basis for PMDD has not been identified. In postpartum depression (PPD), another form of reproductive endocrine-related depression in which normal physiologic levels of E/P have been reported to precipitate depressive symptoms [26], abnormalities in estrogen receptor (ER) signaling [27] and estrogen-mediated DNA methylation patterns in genes regulating hippocampal synaptic plasticity have been observed [28].
Cellular models involving neuronal cells and lymphoblastoid cell lines (LCLs) have many advantages, but also distinct limitations for the study of brain diseases [29–31]. Studies employing LCLs have investigated several forms of both inherited diseases [32,33] and neuropsychiatric conditions [29,34–36]. Cultured LCLs offer the opportunity to identify trait-like biochemical characteristics potentially related to disease phenotypes, and enable comparisons between women with and without PMDD previously studied in our clinical trials, which would not have been possible if we employed cultured neurons. We hypothesized that women with PMDD are genetically predisposed to altered cellular response to physiologic E/P changes. To test this, we used LCLs from women with PMDD, in whom symptom remission and recurrence (i.e., E/P sensitivity) was confirmed by E/P manipulation, and asymptomatic Controls [8]. In this study, we addressed the following questions: 1) Are ovarian steroid hormone receptor mRNA and protein expressed in LCLs? LCLs express 35–80% of genes expressed in the CNS [37], thus we first attempted to demonstrate that LCLs from women with PMDD and controls express ovarian steroid receptors such that they would potentially be capable of responding to E and P. 2) Does the transcriptome of untreated (i.e., no exposure to E/P) LCLs different in those from women with PMDD compared to control women? and 3) Does ovarian steroid treatment of LCLs differentially alter Extra Sex Combs/Enhancer of Zeste (ESC/E(Z)) complex gene expression in cells from PMDD women compared with control women?
Materials and Methods
Participants and Cell Lines
Women between the ages of 18 and 48 years who were medication-free (at time of recruitment), with regular menstrual cycles (range, 21 to 35 days), not medically ill and not pregnant were included in the clinical study[8].
Women with PMDD were self-referred in response to newspaper advertisements or were referred by their physician. The diagnosis of PMDD was confirmed prospectively prior to entry into this study by self-administered symptom ratings (a 100mm visual analog scale) completed daily over three consecutive menstrual cycles. In addition to meeting DSM IV criteria for PMDD [38], each woman had an increase of at least 30 percent (relative to the range of the scale she used) in average self-ratings of negative moods (irritability, depression, anxiety, and mood stability) in the seven days before menses compared with the ratings for the seven days after the end of menses, in at least two of the three baseline cycles, and as described previously [8,39]. Functional impairment was assessed through self-reports of distress and functional disability on the daily rating form (DRF) [40]. The DRF criteria for functional impairment were as follows: a DRF score of 2 (minimal) or higher on one of 4 questions related to functional impairment (i.e., stayed at home or avoided social activities, had conflicts or problems with people, symptoms interfered with relationships at work or home, or symptoms interfered with work productivity) in at least 3 days out of 7 days pre-menses. Finally, DRF ratings and the results of both a semi-structured interview (Menstrual Screening Form, unpublished) and a self-report questionnaire (Menstrual Assessment Form, unpublished) were employed to confirm that all women met the required number of symptoms specified in the DSM criteria for PMDD. Additionally, women with significant negative mood symptoms (on the DRF) occurring during the follicular phase of the menstrual cycle were excluded. These are more stringent criteria than those of DSM-IV[38].
Women without PMDD (henceforth referred to as Control women) were recruited, and studied, in parallel. Absence of premenstrual mood symptoms was confirmed using the same daily rating scales during a two-month baseline period.
Women with PMDD had no other current Axis I psychiatric diagnosis within the past two years per Structured Clinical Interview for DSM-IV (SCID) [41], while Control women had neither current nor past Axis I diagnosis as confirmed by SCID.
All women received remuneration according to guidelines from the NIH Healthy Volunteer Office. The study protocol was reviewed and approved by the National Institute of Mental Health Institutional Review Board, and all women gave written informed consent.
Selection of participants for generation of lymphoblastoid cell lines (LCLs), and Hormone Manipulation Protocol
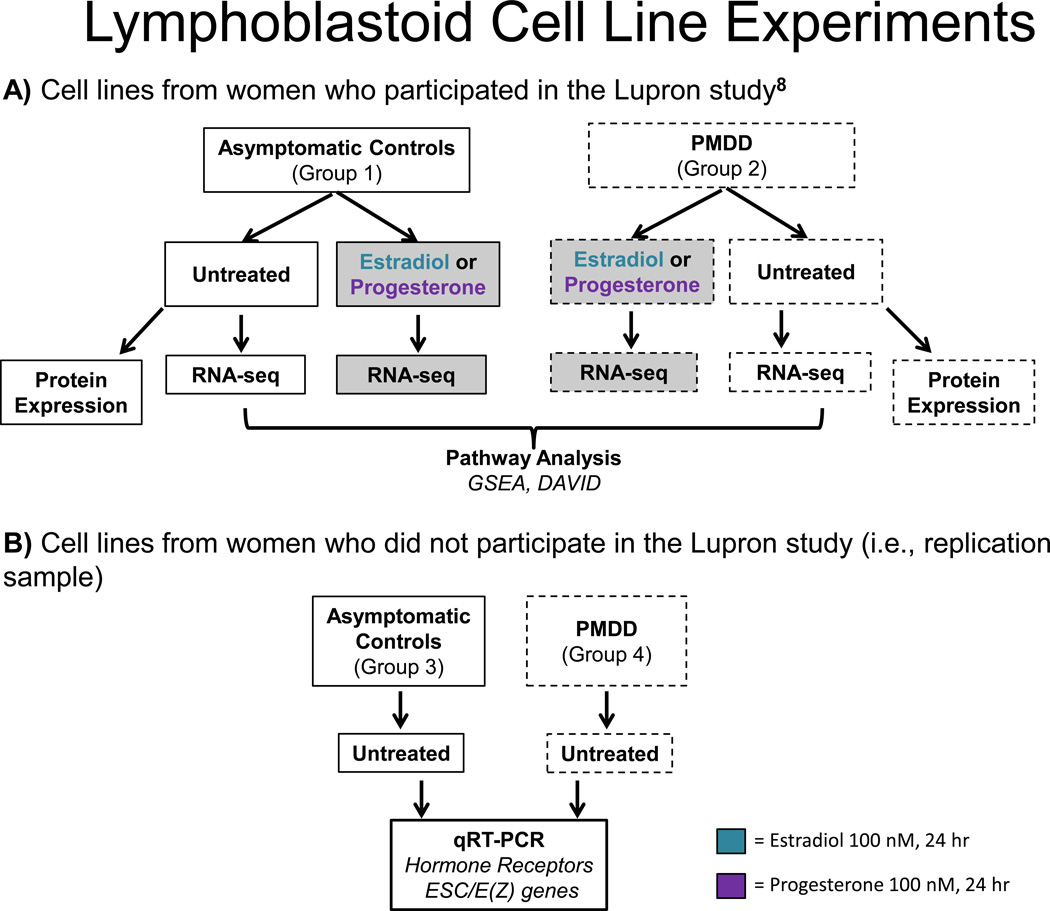
Two main sets of LCLs were created for this study: a focus sample (Group 1- women with PMDD and who underwent the GnRH-agonist hormone intervention, Group 2 – Controls, asymptomatic women who also underwent the GnRH-agonist hormone intervention), and a larger replication sample (Group 3 – women with PMDD, Group 4 – Controls; neither of whom were selected on the basis of their symptom response to the GnRH-agonist hormone intervention but who did meet the same inclusion criteria [and diagnostic evaluation] as Groups 1 and 2). All women in the focus sample had completed a GnRH agonist-induced hormone manipulation protocol (see Figure S1). Within the focus sample, Group 1 (n=10) consisted of PMDD women whose symptoms were suppressed by leuprolide (Lupron) and recurred during either of the E/P add-back conditions [8]. Thus all women with PMDD in Group 1 met criteria for response to Lupron and experienced a recurrence of PMDD symptoms during addback. The criteria employed to characterize women with PMDD as responders to Lupron and experiencing a recurrence of symptoms on either estradiol or progesterone addback were as follows (and for additional details of outcome measures and response patterns please see [8]): responders were defined by self-reported improvement during the last month of Lupron confirmed by the absence of weekly [7–day] average DRF scores of >3 for irritability, sadness or anxiety (indicating less than moderate severity of the particular symptom)[42,43]. Significant recurrence of PMDD symptoms was defined by weekly average DRF scores of greater than three for either irritability, anxiety, or sadness [5]. Group 2 (n=9) consisted of asymptomatic Controls with no PMDD and who were asymptomatic throughout the entire protocol. Controls were defined by absence of affective symptoms throughout the 6 month hormone manipulation protocol (i.e., no weekly average DRF score ≥2). Groups 1 and 2 were matched for age and ethnicity (see Table 1).
Table 1. Demographics for each study component.
Age at time of study, BMI, past history of Major Depressive Episode (MDE), PMDD treatment history, and parity (which refers to the number of times a woman has carried a pregnancy to a viable gestational age) of the subjects chosen for cell line generation. qRT-PCR for Hormone Receptors: Demographics of the 15 Control women and 15 PMDD women chosen (subset from Groups 3 and 4) to complete qRT-PCR analysis of the four hormone receptors. No significant differences in age (t28=0.7, p=0.5) or BMI (t28=0.0, p>0.99) between groups. RNAseq for untreated conditions and hormone exposure (and protein): Demographics of the 9 Control women and 10 PMDD women chosen (from Groups 1 and 2) to complete RNA-sequencing studies of untreated or hormone-treated cell lines as well as all protein analyses. No significant differences in age (t17=1.3, p=0.2) or BMI (t17=0.2, p=0.8) between groups. qRT-PCR replication: Demographics of the 24 control women and 24 PMDD women chosen (Groups 3 and 4) to complete the replication/validation study of mRNA expression in untreated cell lines for the 13 ESC/E(Z) genes. No significant differences in BMI (t35=1.39, p=0.173) between groups, though Age was significantly different (t46=4.4, p<0.0001).
| From Groups 3 and 4 – used in qRTPCR for Hormone Receptors | |||||||||
| Diagnosis | N | Ethnicity |
Agea (years) |
BMIa (kg/m2) |
Past History of MDEb |
Past PMDD Treatment Historyb |
Parityc | ||
| Control | 15 | Caucasian (10) African American (3) Asian (2) |
39.7(6.6) | 25.1(6.7) 1 missing |
0 (0%) | NA | 10 (1–3) | ||
| PMDD | 15 | Caucasian (10) African American (5) |
38.0(6.7) | 25.1(4.9) 2 missing |
6 (40%) | Anti-depressants 3 (20%) |
8 (1–5) | ||
| Groups 1 and 2 – used in RNAseq for untreated and hormone exposures (and protein) | |||||||||
| Diagnosis | N | Ethnicity |
Agea (years) |
BMIa (kg/m2) |
Past History of MDEb |
Past PMDD Treatment Historyb |
Parityc |
Age of onset of PMDDa (years) |
Duration of PMDDa (years) |
| Control | 9 | Caucasian (4) African American (5) |
36.9(6.5) | 27.8(7.4) | 0 (0%) | NA | 4 (2–3) | NA | NA |
| PMDD | 10 | Caucasian (6) African American (4) |
40.6(5.7) | 27.2(4.6) | 4 (40%) | Anti-depressants 3 (30%) |
8 (1–5) | 27.9(10.5) | 12.7(8.4) |
| Groups 3 and 4 – used in qRTPCR replication | |||||||||
| Diagnosis | N | Ethnicity |
Agea (years)* |
BMIa (kg/m2) |
Past History of MDEb |
Past PMDD Treatment Historyb |
Parityc | ||
| Control | 24 | Caucasian (15) African American (6) Asian (2) Hispanic (1) |
29.9(7.2) | 23.9(5.1) 4 missing |
0 (0%) | NAd | 6 (1–3) | ||
| PMDD | 24 | Caucasian (17) African American (7) |
37.9(5.3) | 26.2(5.1) 7 missing |
6 (25%) | Anti-depressants 3 (12.5%) Supplements 1 (4.1%) |
12 (1–3) | ||
numbers listed as mean(standard deviation)
numbers listed as number(percentage) – All women in this study were medication free. Medication use in the past (i.e., prior to enrollment in the clinical studies) is listed in the Past PMDD Treatment History column for each group. Anti-depressants include Prozac, SSRI, and Zoloft; Supplement is EPO
numbers listed as number of parous women (range of number of live births)
note one Control subject has a history of Effexor treatment
significant difference between Control and PMDD (students t-test, p<0.05)
The replication sample consisted of a totally independent sample of women relative to those women included in Groups 1 and 2: Group 3 (n=24) - women with prospectively confirmed PMDD and Group 4 (n=24) - Controls confirmed by absence of menstrual cycle-related mood symptoms by two cycles of daily ratings. Although Groups 3 and 4 represent less restricted PMDD/Control phenotypes (i.e., PMDD not required to demonstrate symptom remission and recurrence under ovarian suppression and hormone add-back, respectively), studies indicate that approximately 70% of women with PMDD experience symptom remission with GnRH agonist treatment [8,44] and that 90% of controls do not develop clinically significant mood symptoms on leuprolide [45]. Samples from women with PMDD and controls were collected from 1997–2010 (mean 2003) and year of collection and subsequent storage did not significantly differ between the two groups. Nor did year of collection predict levels of gene expression in the ESC/E(Z) complex genes.
Experimental and Technical Methods
Culturing and hormone treatments of LCLs
Epstein Barr virus (EBV)-transformed LCLs were made as described [46], cultured under identical conditions, and studied during untreated conditions or following treatment with estradiol or progesterone for 24 hours, as described in supplemental methods. Henceforth, the term untreated LCLs will refer to those cells that were not exposed to ovarian steroids and treated cells to those that were exposed to ovarian steroids. For generation and maintenance of LCLs (immortalized lymphoblastoid B cell lines), venous blood samples from PMDD and Control women were obtained at the same times, and cultured in the same way for similar intervals as described in more detail in Supplementary Material.
mRNA quantification using whole transcriptome RNA sequencing and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
First, mRNA was quantitated by qRT-PCR (see below) to demonstrate that LCLs express sex steroid receptor genes (and therefore express the machinery to respond to E and P. Fifteen cell lines each from Groups 3 and 4 were examined, thus, addressing a main mechanism by which ovarian steroids could alter cellular function. Results are shown in Figure 2.
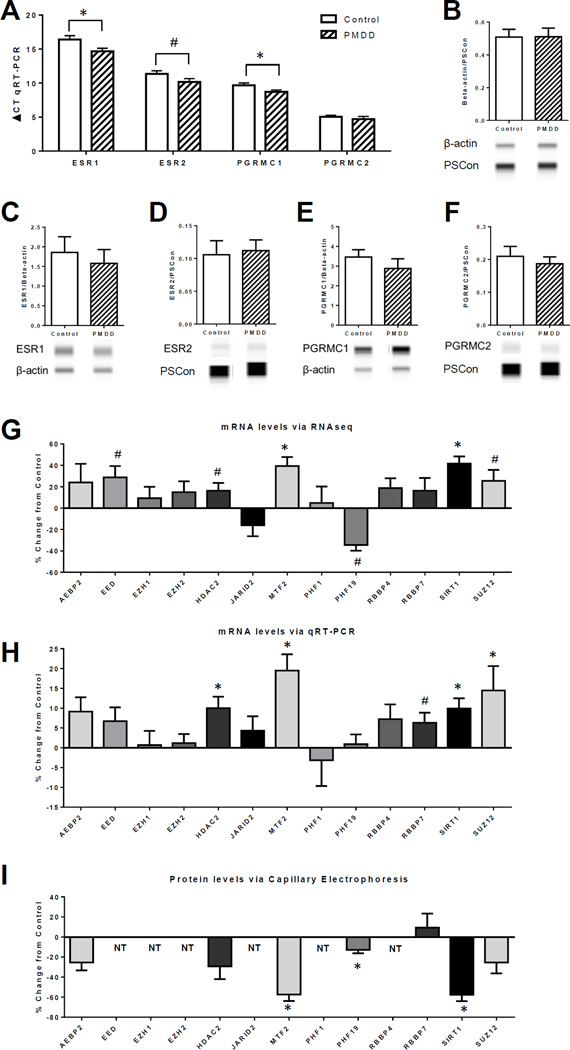
Figure 2. mRNA and protein expression for hormone receptors and ESC/E(Z) Complex genes comparing untreated Control and PMDD lymphoblastoid cell lines.
All graphs are presented as group mean ± SEM. (A–F) Student’s t-test was performed for each comparison. (A) qRT-PCR analysis of mRNA expression for hormone receptors ESR1, ESR2, PGRMC1, and PGRMC2 in lymphoblastoid cell lines (LCLs), n=15. Relative gene expression levels were calculated for ∆CT value with β-actin as the reference control for normalization; note a lower ∆CT value indicates higher expression. Both ESR1 and PGRMC1 were significantly higher in the PMDD group than the Control group (*p<0.05), and ESR2 was higher at a trend level of significance in the PMDD group than the Control group (#p<0.1). (B) Verification of β-actin as a normalizing protein for ProteinSimple© western analysis of protein expression in LCLs. The chemiluminescense ratio of β-actin over the ProteinSimple© Loading Control (PSCon) was calculated for each group and no significant difference was found (n=7, t12=0.02, p=0.9). (C–F) ProteinSimple© western analysis of protein expression for hormone receptors ESR1, ESR2, PGRMC1, and PGRMC2 (n=7). Target protein chemiluminescense was normalized to β-actin or PSCon if there was band interference with β-actin, as was the case for ESR2 and PGRMC2. There were no significant differences for any of the proteins. For (B–F), representative images of protein “bands” from the Compass software are shown below each group. (G–H) All graphs are calculated as percent change of values of the PMDD group from the Control group mean. Statistical analysis comes from Student’s t-test of the original group means (see Supplemental Figure 2). (G) Percent change from control of mRNA expression via RNAseq for the 13 genes of the ESC/E(Z) complex (n=9–10). Both MTF2 and SIRT1 were significantly higher in the PMDD group than the Control group (*p<0.05), and EED, HDAC2, and SUZ12 were higher at a trend level of significance in the PMDD group than the Control group (#p<0.1), while PHF19 was lower at a trend level of significance in the PMDD group than the control group (#p<0.1). (H) Percent change from control of mRNA expression via qRT-PCR for the 13 genes of the ESC/E(Z) complex (n=29–30). HDAC2 (t43=2.6), MTF2 (t37=3.7), SIRT1 (t46=3.2), and SUZ12 (t42=2.1) were significantly higher in the PMDD group than the Control group (*p<0.05), and RBBP7 (t33=1.8) was trending higher in the PMDD group than the Control group (#p<0.1). (I) Percent change from control of protein expression via ProteinSimple© western analysis for select genes of the ESC/E(Z) complex (n=7). NT indicates that protein was not tested. MTF2 (t11=3.4), PHF19 (t12=2.5), and SIRT1 (t11=3.2) were significantly lower in the PMDD group than the Control group (*p<0.05).
Second, mRNA sequencing was performed on untreated (i.e., steroid free) PMDD and control LCLs from the focus set (Groups 1 and 2) [see Figure 1]. Results are in Figures 2, 3, S2–4, and Tables 2, S2–7, OL1, and OL2. Pathway analyses of the PMDD and control LCL transcriptomes were performed with Gene Set Enrichment Analysis (GSEA) and Database for Annotation, Visualization, and Integrated Discovery (DAVID). We used the following criteria to prioritize significant gene sets for further confirmation and more detailed analysis of physiological relevance: priority to gene sets with smaller numbers of genes, modulation by ovarian hormones, and published involvement in other mood disorders.
Figure 1. LCL treatments and analyses.
Flow chart of LCL experiments. Blood is collected from women with PMDD or asymptomatic Controls and transformed into LCLs with Epstein Barr virus. Cells are then left untreated or treated with hormone (estradiol or progesterone, 100 nM each) and undergo RNA-sequencing. Pathway analyses were applied to results, and selected genes from the indicated LCL groups are followed up with qRT-PCR and protein analysis via ProteinSimple©.
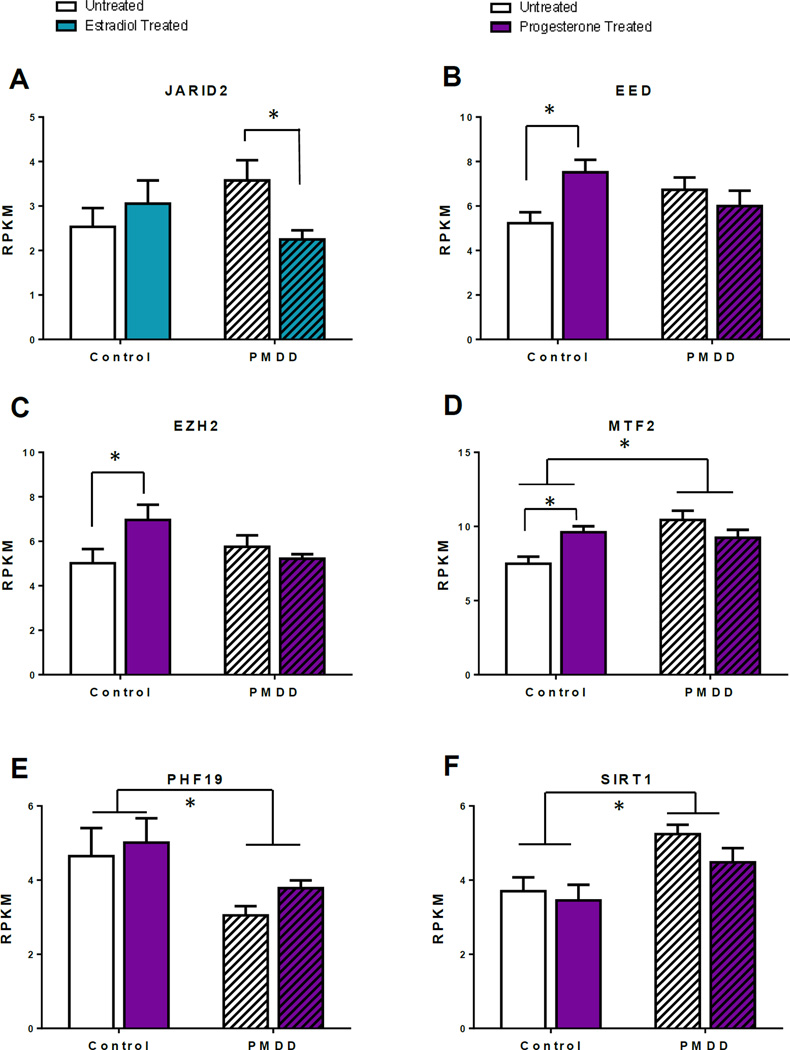
Figure 3. Selected ESC/E(Z) gene expressions from RNAseq: ANOVAs comparing untreated and estradiol- or progesterone-treated Control and PMDD lymphoblastoid cell lines.
All graphs are presented as group mean ± SEM using RPKM values from RNA-sequencing. Two-way ANOVAs were performed for each gene of interest, followed by Sidak’s multiple comparisons post-hoc test in cases of a significant interaction. Untreated Control and PMDD lymphoblastoid cell lines compared with estradiol treatment (A) or progesterone treatment (B-F). (A) For JARID2 there was a significant interaction effect between diagnosis and estradiol treatment (F1,24=5.5, *p<0.05). Expression was significantly decreased after estradiol only in the PMDD group (t24=2.5, adjusted p-value<0.05). (B) For EED there was a significant interaction effect between diagnosis and progesterone treatment (F1,29=6.8, *p<0.05). Expression was significantly increased after progesterone only in the Control group (t29=2.8, adjusted p-value<0.05). (C) For EZH2 there was a significant interaction effect between diagnosis and progesterone treatment (F1,29=4.8, *p<0.05). Expression was significantly increased after progesterone only in the Control group (t29=2.4, adjusted p-value<0.05). (D) For MTF2 there was a significant interaction effect between diagnosis and progesterone treatment (F1,29=9.2, *p<0.05). Expression was significantly increased after progesterone only in the Control group (t29=2.7, adjusted p-value<0.05). Additionally, there was also a significant diagnosis effect (F1,29=5.5, *p<0.05) where MTF2 is more highly expressed in the PMDD group than the Control group (post-hoc untreated Control vs untreated PMDD t29=4.1, p<0.001). (E) For PHF19 there was a significant diagnosis effect (F1,29=16.1, *p<0.05) where expression is lower in the PMDD group compared with the Control group, but there was no interaction effect between diagnosis and progesterone treatment (F1,29=0.1, p=0.7). (E) For SIRT1 there was a significant diagnosis effect (F1,29=13.1, *p<0.05) where expression was higher in the PMDD group compared to the Control group, but there was no interaction effect between diagnosis and progesterone treatment (F1,29=0.5, p=0.5).
Table 2. Top 20 GSEA results from RNAseq comparing untreated Control and PMDD lymphoblastoid cell lines.
Results are from the Gene Set Enrichment Analysis (GSEA) by CLC Genomics Workbench (version 6.5) on transformed RNA expression values from RNA sequencing of untreated Control (n=9) and PMDD (n=10) lymphoblastoid cell lines using gene ontologies (GO) for cellular component with 10,000 permutations. Lower tail values reflect a permutation-based p-value, indicating significant difference of the gene set expression between groups. The top 20 ranked GO categories showing enrichment in PMDD cell lines over Control cell lines are presented. The ESC/E(Z) complex (GO category 35098) has a significant permutation-based p-value (lower tail) of 0.007 and has been highlighted here in yellow. Full GSEA results are listed in Table S4.
| Category | Description | # of Genes |
Test statistic |
Lower tail |
|---|---|---|---|---|
| 502 | proteasome complex (GO_REF:0000037 [IEA] UniProtKB-KW:KW-0647) |
51 | −9.245 | 0 |
| 5730 | nucleolus (GO_REF:0000052 [IDA]) | 1049 | −19.582 | 0 |
| 5654 | nucleoplasm (PMID:16687569 [IDA]) | 709 | −16.791 | 2.00E-05 |
| 15030 | Cajal body (PMID:16687569 [IDA]) | 40 | −7.516 | 5.00E-05 |
| 22624 | proteasome accessory complex (GO_REF:0000024 [ISS] UniProtKB:P62334) |
15 | −6.323 | 8.00E-05 |
| 5737 | cytoplasm (GO_REF:0000052 [IDA]) | 2521 | −25.887 | 4.20E-04 |
| 5634 | nucleus (GO_REF:0000052 [IDA]) | 3131 | −28.385 | 5.80E-04 |
| 5839 | proteasome core complex (GO_REF:0000019 [IEA] Ensembl:ENSMUSP00000129767) |
17 | −5.906 | 6.10E-04 |
| 5681 | spliceosomal complex (PMID:7935475 [TAS]) | 65 | −7.412 | 9.80E-04 |
| 781 | chromosome, telomeric region (PMID:12768206 [IDA]) | 30 | −5.967 | 1.89E-03 |
| 5669 | transcription factor TFIID complex (PMID:15899866 [IDA]) |
17 | −5.294 | 2.30E-03 |
| 5813 | centrosome (PMID:21399614 [IDA]) | 261 | −10.45 | 3.61E-03 |
| 19005 | SCF ubiquitin ligase complex (PMID:20347421 [IDA]) | 21 | −5.146 | 5.43E-03 |
| 151 | ubiquitin ligase complex (GO_REF:0000002 [IEA] InterPro:IPR003613) |
52 | −6.246 | 6.72E-03 |
| 35098 | ESC/E(Z) complex (PMID:20075857 [IDA]) | 13 | −4.622 | 6.92E-03 |
| 5720 | nuclear heterochromatin (PMID:10570185 [TAS]) | 14 | −4.604 | 7.62E-03 |
| 71013 | catalytic step 2 spliceosome (PMID:11991638 [IDA]) | 56 | −6.204 | 9.23E-03 |
| 31461 | cullin-RING ubiquitin ligase complex (GO_REF:0000002 [IEA] InterPro:IPR001373) |
10 | −4.275 | 9.69E-03 |
| 242 | pericentriolar material (PMID:21211617 [IDA]) | 15 | −4.576 | 9.87E-03 |
| 33116 | ER-Golgi intermediate compartment membrane (GO_REF:0000039 [IEA] UniProtKB-SubCell:SL-0099) |
24 | −4.945 | 0.012 |
Third, LCLs from Groups 1 and 2 were exposed 24 hours in vitro to either estradiol or progesterone (100 nmol each). RNA-seq analyses were performed in hormone-treated and untreated LCLs from women with PMDD and Controls. Experimental results are in Figures 3, S5 and S7. Thus whole transcriptome mRNA-sequencing was performed in the focus sample under untreated conditions and after E/P exposures.
Finally, qRT-PCR assays were used in the replication samples (Groups 3, 4) for validation and replication of differences in mRNA expression of the 13 ESC/E(Z) complex genes identified in the initial pathway analyses of mRNA expression in untreated LCLs. qRT-PCR was performed in these groups during untreated conditions only.. In the replication study, which targeted 13 genes implicated in gene set analysis, we elected to measure these transcripts in a larger, independent sample, and via an independent method. The estimated sample size for the replication sample was based on the effect sizes observed in the differences in mRNA expression between PMDD and controls in untreated cells (see Statistics section below).
Total RNA of high quality (A260/A280 ratio in the range of 1.8–2.0) and high integrity (RIN>9.0), as measured by an Agilent® 2100 Bioanalyzer, was isolated from LCLs as described in supplemental methods. Transcriptome analysis was performed on cDNA libraries from ribosomal RNA-depleted total RNA, using an Illumina Genome Analyzer (GAIIx). An average of 17.7 million 36 bp RNA-seq reads for each sample were retrieved and parsed. Sequence read mapping and quantification was performed with the Illumina pipeline CASAVA_v1.8.1 and CLC Genomics Workbench (version 6.5) as described in supplemental methods. TaqMan qRT-PCR assays of 4 hormone receptor genes and 13 ESC/E(Z) genes were performed on a 7900HT Real-Time PCR System as described in supplemental methods.
Pathway analysis of transcriptome data
GSEA was performed on transformed mRNA-expression (reads per kilobase of transcript per million mapped reads [RPKM]) values using gene ontologies (GO) database in the CLC Genomics Workbench (version 6.5) program; results were sorted by enrichment score. Additionally, the genes identified as differentially expressed by RNA sequencing were independently analyzed using DAVID, version 6.7. For gene set analyses, reflecting gene pathways or networks, and as described in more detail in Supplementary Methods, we used two different tools, GSEA and DAVID, that access pre-annotated gene ontology terms and calculate significance based on over-representation of transcripts within pre-defined gene sets, and is corrected for multiple testing using the False Discovery Rate (FDR, Benjamini Hochberg) method. [47].
Interaction network of ESC/E(Z) complex genes and hormone receptors
A gene network was generated using GeneMANIA (genemania.org) by inputting genes of the ESC/E(Z) complex and 4 ovarian steroid receptor genes identified as expressed in LCLs. Weights of interactions between input and edge genes were determined by linear regression. The gene network analyses were not based on the present data, but on existing data bases, using the current data only for selecting the genes included in these network analyses. The search was performed in September, 2015, and reflects all published works in the GeneMANIA database at that time.
Protein quantification using ProteinSimple© western analysis
Protein levels were quantitated both to confirm the presence of sex steroid receptor proteins in LCLs from women with PMDD and controls, and to measure differences in protein products between untreated cells from women with PMDD and those from controls (Groups 1 and 2) in the ESC/E(Z) complex genes identified by pathway analysis (see Figure 1). Protein was extracted from LCLs with radio-immunoprecipitation buffer, and total protein from each sample was measured by Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., CA, cat # 500-0006). Target protein levels were quantified using an automated capillary-based size sorting system “WES” from ProteinSimple© [48]. All procedures were performed with manufacturer’s reagents according to the user manual (primary antibody information is in Table S1); any changes from standard protocol also are noted in Table S1.
Statistical Analyses
The sample size estimates for the replication sample were based on the effect sizes (Cohen’s d = 1.6 – 1.7) for MTF2 and SIRT1 in the case/control RNA-seq comparisons.
For RNA-seq, multiple comparisons were corrected using the False Discovery Rate (FDR, Benjamini Hochberg) method. RNA-seq was analyzed using normalized RPKM values. CLC Genomics Workbench (version 6.5) was used for statistical analyses including pathway analyses described earlier.
After identifying significant case/control differences in expression of ESC/E(Z) complex genes through pathway analyses, differences between RNA expression (i.e., RNA-seq and qRT-PCR) in untreated LCLs from both the focus sample (Groups 1 and 2) and the replication sample (Groups 3 and 4), were compared by Student’s t-test (two-tailed) with a nominal uncorrected p value of < 0.05. Similarly, diagnosis-related differences in both RNA expression (qRT-PCR) and protein levels of sex steroid receptor genes were compared by Student’s t-test (two-tailed) with a nominal uncorrected p value of < 0.05. Protein levels of ESC/E(Z) complex genes in untreated LCLs were compared between PMDD and controls in an identical manner in the focus sample.
Finally, comparisons of RNAseq results between untreated and hormone-treated cell cultures for the 13 genes of the ESC/E(Z) complex, two-way ANOVAs were performed for each gene of interest, followed by Sidak’s multiple comparisons post-hoc test in cases of a significant interaction, with hormone condition (i.e., untreated versus hormone exposure) as the within-groups factor and diagnostic group (i.e., PMDD versus Control) as the between-groups factor. Analyses were performed using Prism6 software (GraphPad, Inc.).
RESULTS
Cultured LCLs express ovarian steroid receptors
Ovarian steroid receptors, which represent important components of E/P cellular response, were present in LCLs. Using qRT-PCR we found mRNA expression for ER alpha (ESR1) and ER beta (ESR2), and the progesterone membrane component receptors, PGRMC1 and PGRMC2 (Figure 2A). We did not detect mRNA for the progesterone nuclear receptor (PR). We confirmed presence of proteins for these four genes using ProteinSimple© western analysis (Figure 2C–F).
Global differences in mRNA expression between untreated (no hormone treatment) PMDD and Control LCLs
Whole transcriptome (RNA-seq) analysis of untreated Control and PMDD LCLs was used to identify genes whose mRNA expression differed between the two groups. Pooling sequencing reads from all samples, mRNA expression was detected for 15,055 RefSeq genes. The total number of mapped reads in PMDD and Control LCLs was 13,552 and 13,488, respectively (Table S2). The scatter and volcano plots of levels of all gene transcripts in untreated PMDD versus Control LCLs are shown in Figure S2.
Exactly 1000 gene transcripts were differentially expressed in untreated LCLs from PMDD versus Control at an uncorrected p-value of <0.05 (Table OL1). Among these transcripts, the 117 that were >2 fold different in expression are listed in Table S3. Approximately equal numbers were up- and down-regulated.
Pathway analysis using GSEA of the untreated LCL transcriptome of PMDD (Group 1 vs Group 2) identified the ESC/E(Z) complex (Extra Sex Combs/Enhancer of Zeste complex) as one of the most significant pathways (lower tail permutation-based p=0.007) from among the top GSEA pathway hits listed in Table 2, and the full list of pathway hits in Table S4. We focused on genes in the ESC/E(Z) pathway, in accordance with pre-defined criteria (see Methods), and due to the previously reported roles for these genes in ovarian steroid response [49–57]Transcripts representing the ESC/E(Z) complex were upregulated in the LCLs from the PMDD group compared with those from the control group.
We also subjected the PMDD transcriptome data to pathway analysis using DAVID. DAVID returned 645 pathways significantly different between untreated PMDD and Control LCLs (Table OL2) and again identified the ESC/E(Z) complex as one of the more significant pathways (p=0.003). Additionally, two of the ESC/E(Z) genes, MTF2 (nominal p=0.001, fold change −1.14) and SIRT1 (nominal p=0.0039, fold change −1.2) were also present in the list of 1000 genes differentially expressed in untreated LCLs (Table OL1, highlighted in yellow).
At the individual gene level in the untreated transcriptome data for the ESC/E(Z) complex there was an overall pattern of up-regulation in PMDD LCLs: both MTF2 and SIRT1 were significantly higher in PMDD (uncorrected p=0.001 and p=0.004, respectively), and EED, HDAC2, and SUZ12 were higher (trend) in PMDD (uncorrected p=0.06, p=0.09, and p=0.05, respectively). Although not significant, AEBP2, EZH1, EZH2, RBBP4, and RBBP7 also showed a pattern of increased mRNA expression in PMDD LCLs. Of the 13 ESC/E(Z) transcripts, only PHF19 and JARID2 were lower in PMDD (uncorrected p=0.09, p=0.62, respectively). No directional difference was observed in PHF1. Original group data are shown in Figure S3A and presented as percent difference between the PMDD and Control group means in Figure 2G.
Quantitative real-time PCR (qRT-PCR) in the larger replication set of untreated LCLs (Groups 3 and 4) validated many of the RNA-seq findings for the ESC/E(Z) complex. Overall, the pattern of up-regulation of mRNA for the ESC/E(Z) complex genes in PMDD LCLs was again observed. HDAC2 (p=0.012), MTF2 (p=0.001), SIRT1 (p=0.003), and SUZ12 (p=0.045) were significantly higher in PMDD, and RBBP7 (p=0.076) was higher at a trend level of significance in PMDD. Although not significant, AEBP2, EED, and RBBP4 also showed increased mRNA expression in PMDD. In contrast to the RNA-seq results, we did not observe significant down-regulation of either JARID2 or PHF19 in PMDD LCLs by qRT-PCR. Original group data are shown in Figure S3B and presented as percent difference between the PMDD and Control group means in Figure 2H.
Finally, the protein levels of 7 of the 13 ESC/E(Z) complex genes in untreated LCLs (Group 1 versus 2), were down-regulated in PMDD compared with Control LCLs (Figure 2I). MTF2 (p=0.01), PHF19 (p=0.02), and SIRT1 (p=0.01) had significantly lower expression in PMDD. No significant differences were observed in AEBP2 (p=0.1), HDAC2 (p=0.1), RBBP7 (p=0.6), and SUZ12 (p=0.3), although expression of AEBP2, HDAC2, and SUZ12 were lower in PMDD LCLs.
Global changes in transcript expression in PMDD and Control LCLs in response to sex-steroid (E/P) exposure
Scatter and volcano plots for RNA-seq comparisons of PMDD and Control LCLs (Group 1 and Group 2) untreated or treated with either E or P after 3–5 days in hormone-free media are shown in Figures S4 and S6. The number of transcripts assayed and the number differentially expressed (at a nominal p value < 0.05) are reported in Table S2.
For the 13 genes of the ESC/E(Z) complex, RNA-seq differences and results from ANOVAs and post hoc comparisons for the E/P treatments are shown in Figures S5 and S7, respectively, with statistical results in Table S7. Significant diagnosis by treatment interactions emerged for 4 of the 13 ESC/E(Z) complex genes: JARID2 (Figure 3A, p<0.05) with decreased expression after E2-treatment in PMDD but not Control LCLs, and EED, EZH2, and MTF2 (Figure 3B–D, p<0.05), with increased expression after P4-treatment in Control but not PMDD LCLs.
Gene network analyses
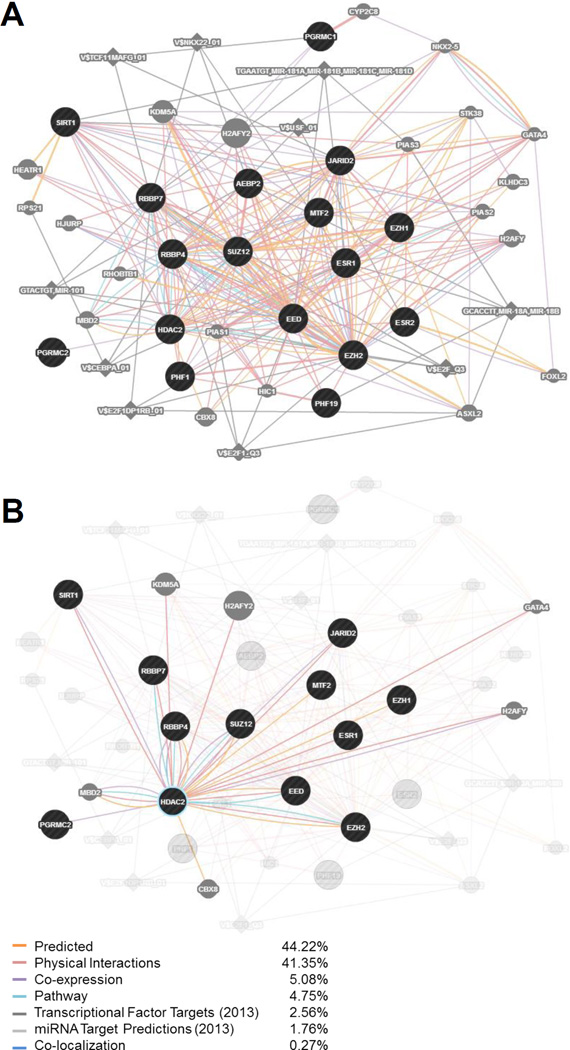
We preliminarily explored connections between ER/PGRMC1/2 and the ESC/E(Z) complex to elucidate potential mechanisms of the differential response to hormone we had observed. Via GeneMANIA (genemania.org) a gene interaction network was generated from inputting the 13 ESC/E(Z) genes and 4 ovarian steroid receptor genes expressed in our LCLs. This analysis computes connection density and strength of the input genes and identifies “edge” genes that are also strongly connected to the network (Figure 4A) and that are potential candidates for future studies of mechanisms of differential E/P response in PMDD. This analysis revealed that HDAC2 strongly interacts with 9 of the 13 ESC/E(Z) complex genes, and is the only gene of the ESC/E(Z) complex whose protein also interacts with a receptor for both ESR1 and PGRMC2 (Figure 4B). As indicated in Figure 4B, these HDAC2 interactions are predominantly protein/protein in nature.
Figure 4. GeneMANIA interaction analysis of ESC/E(Z) and steroid hormone receptor genes.
GeneMANIA (genemania.org) was used to generate a gene network analysis inputting the 13 genes of the ESC/E(Z) complex (AEBP2, EED, EZH1, EZH2, HDAC2, JARID2, MTF2, PHF1, PHF19, RBBP4, RBBP7, SIRT1, SUZ12) and the 4 hormone receptor genes (ESR1, ESR2, PGRMC1, PGRMC2), whose expression was identified in LCLs. Genes and targets are included in the network for the input genes based on weights of interactions with the input genes, determined automatically by linear regression. Black circles indicate the input genes. Gray circles indicate additional genes identified in the network – a larger circle reflects a higher rank of connection with the input genes. Diamonds indicate miRNA or transcription factor targets. A thicker connecting bar between nodes reflects a stronger relationship between those two nodes. (A) The expanded network based on a search of these 17 input genes (September, 2015). The strongest relationships driving this network are Predicted Interactions (44.22% weight) and Physical Interactions (41.35% weight). (B) Selection of HDAC2 and highlighting its interactions within the predicted network. HDAC2 shows interactions with 9 of the 13 genes of the ESC/E(Z) complex (7 of them physical), as well as several other genes in the network. Importantly, it is the only component of the ESC/E(Z) complex that interacts with a receptor for both estradiol (ESR1) and progesterone (PGRMC2).
DISCUSSION
We examined the hypotheses that alterations in in vitro cellular function could serve as a substrate for the observed ovarian steroid-related behavioral sensitivity in PMDD, and whether cellular responses to E/P exposure distinguish women with PMDD from asymptomatic matched Controls. The women with PMDD not only met clinical criteria for the presence of PMDD, but additionally, all were behaviorally sensitive to E/P based on absence of behavioral symptoms (i.e., PMDD symptoms) after ovarian suppression and emergence of negative affective symptoms after E/P addback [8]. Controls lacked any significant menstrual cycle-related behavioral symptoms. Using an in vitro experimental strategy we attempted to detect differences in gene expression in untreated LCLs (i.e., absence of ovarian steroid exposure) and differences in steroid sensitivity at the cellular level in these highly selected clinical phenotypes. Our findings suggest that LCLs from women with PMDD are differentially sensitive to ovarian steroids (i.e., respond differently to ovarian steroids) compared with LCLs from Controls, and differ in both untreated and ovarian steroid-stimulated mRNA and protein expression of ESC/E(Z) complex genes that could mediate this difference.
Analysis of the 13 gene members of the ESC/E(Z) complex across diagnosis and hormone exposure revealed a paradoxical pattern. Twelve of thirteen ESC/E(Z) gene transcripts were increased in expression in PMDD, several significantly. However, protein expression of these genes tended to be decreased in PMDD LCLs, several significantly so. Significant diagnosis-by-hormone effects were observed for four of the members of this gene family.
The ESC/E(Z) complex is a multimeric protein complex that can methylate lysine-27 and lysine-9 residues of histone H3, interacts with several noncoding RNAs [58], and is an essential gene silencing complex regulating transcription [59,60]. The ESC/E(Z) complex consists of 4 core proteins (EZH2, SUZ12, EED, and RBBP4), as well as 9 additional proteins (AEBP2, EZH1, HDAC2, JARID2, MTF2, PHF1, PHF19, RBBP7 and SIRT1) that can be included or interchanged to create the entire functioning complex. Depending on the target tissue or the presence of other activating genes, EZH1 can be substituted for EZH2, and RBBP7 can be substituted for RBBP4. The core protein assembly then interacts with a polycomb-like protein (PCL) which is either PHF1, PHF19, or MTF2. The four core proteins plus the PCL then interact with AEBP2 and/or JARID2, which both bind DNA directly to sit down on the target gene, and then is capable of recruiting HDAC2 and/or SIRT1 to aid in the ESC/E(Z) complex function of modifying histones through either deacetylation or methylation at H3K9 or H3K27. Interestingly, HDAC2 mRNA expression was significantly (qRT-PCR) or trending (RNAseq) higher in untreated LCLs from women with PMDD compared to Controls, and our GeneMANIA network search identified HDAC2 as a potential hub connecting sex steroid receptors and members of the ESC/E(Z) complex. It is important to note that not all proteins interact all the time, but in fact variation and substitution of these components can allow for different tissue-specific functionalities [59,61], which could contribute to a differential cellular processing of steroid signaling within the CNS and be potentially relevant to the pathophysiology of PMDD. Thus, it will be important for future work to study any differences in protein interaction between women with PMDD and Controls, and in several different cell types. In vitro studies show that ESC/E(Z) complex function can be both regulated by E/P [49–54] and also regulate E/P signaling in a range of tissues [55–57]. Therefore, the observed alterations in ESC/E(Z) complex gene expression potentially could translate into altered steroid signaling and cellular function within the CNS and contribute to differential steroid sensitivity in women with PMDD.
PMDD remains a potentially stress-related condition [62], albeit modified by ovarian steroids. Pathway analyses may yield some results that are of less obvious relevance than others. For example, spliceosomal complexes, ER Golgi compartment membrane, and ubiquitin ligase were among the “top hits” in our pathway analyses. Although these may be pursued in other “omics” studies, we chose to pursue the ESC/E(Z) complex because of the already converging evidence to support the role of this pathway in steroid signal regulation and in stress-and affective-related behaviors. A role for the ESC/E(Z) complex in differential behavioral responsivity to E/P is thus far indirect, being based on measures of transcript and protein expression in an in vitro LCL model, but is further supported by several other findings. First, the ESC/E(Z) complex is present in neuronal tissue, where PHF1 modifies GABA system function [63]. Modulation of this system by neurosteroid metabolites of progesterone has been suggested to be important in the pathophysiology of PMDD [64–66]. Second, the function of the ESC/E(Z) complex is differentially regulated in stress-related conditions, in both human and animals [67–74]. Third, ESC/E(Z) complex genes (and their target H3K27 methylation) are implicated in depression-like behaviors, circadian function [75], antidepressant responsivity [68,76], neuroplasticity [77,78], and neuroprotection [79]. In mice, alterations in the expression of SIRT1, a member of the ESC/E(Z) complex, leads to changes in anxiety-related behaviors and 5HT system function [80]. In humans, genetic variation in SIRT1 has been associated with dimensionally measured anxiety and major depression in certain populations [71,80,81]. H3K27 methylation of the trkB receptor is increased (postmortem) within the medial orbital frontal cortex of suicide completers [70]. Thus, members of the ESC/E(Z) complex have been implicated, more directly in model organisms than in human studies, in modulation of many of the systems mediating or underlying the regulation of affective adaptation. Our findings in women with PMDD of altered ESC/E(Z) complex gene mRNA and protein relative to Controls suggest a biological substrate for the risk of recurrent affective destabilization in these women. Alternatively, it is possible that the changes in ESC/E(Z) complex we observed could reflect the long-term sequelae of a recurrent mood disorder experienced by these women. Finally, there is a growing literature documenting the relevance of inflammation in the development, course, and possibly treatment response characteristics of mood disorders [82–84]. Several epidemiologic studies have reported an association between alterations in specific inflammatory factors and the severity of PMS (but not PMDD) symptoms [85,86]. Nonetheless, the relevance of these reports to PMDD remains to be determined. LCLs are derived from cells of the immune system, and so may over-represent gene pathways related to immune response. In our dataset, however, there were no differences in immune pathways between control and PMDD LCLs (in neither GSEA nor DAVID analyses). Further, an analysis of our RNA-seq data in InnateDB.com, a pathway analysis tool that looks for enrichment of innate immunity interactions and pathways, no significant differences between groups were found. Nonetheless, our findings of altered gene expression within immune-based LCLs could reflect an abnormal immune cell physiology that contributes to the pathophysiology of PMDD.
Our findings of a diagnosis-by-hormone interaction of several genes suggest that ESC/E(Z) complex function may be differentially expressed in the absence of ovarian steroids and differentially regulated in PMDD compared with Controls after short term exposures to E/P. These findings showing differences in cellular function, therefore, are analogous to both behavioral [8] and neuroimaging [16,17] observations in PMDD in that the same ovarian steroid hormonal stimulus elicits a different response in women with PMDD compared with controls: affective and behavioral symptoms in women with PMDD (but not controls), as well as differential activation of brain regions involved with a range of social and affective behaviors. However, the cellular findings represent an in vitro model in which effects of many extrinsic factors, such as recent nutrition, stress, or physical activity, have been eliminated.
In this study, we also observed differences in steroid receptor gene expression between LCLs from women with PMDD and control women. Specifically, cells from women with PMDD had significantly increased gene expression (but not protein expression) in ESR1 and PGRMC1 (Figure 2A). The difference in the ESC/E(Z) gene complex in PMDD women may be attributable to inherited differences in genes that encode ovarian steroid receptors (ESR1, ESR2, PGRMC1 and PGRMC2), all of which we observed are expressed in LCLs. For example ESR1, variants were associated to PMDD by Huo et al, [87]. Other genes that can modulate ovarian steroid signaling, including genes encoding components of the ESC/E(Z) complex may also have variants altering ESC/E(Z) function.
We employed an unbiased genome-wide approach, rather than examining changes in expression levels of presumed relevant genes. Nonetheless, several limitations of this study deserve mention. First, although gene expression of the ESC/E(Z) complex was identified to differ in cells from women with PMDD compared to controls in our pathway analyses, comparisons of expression levels from individual ESC/E(Z) complex genes between cases and controls resulted in only a few genes (MTF2 and SIRT1) in which diagnostic differences in expression would remain significant after correction for the multiple comparisons. This pattern was observed in both the focus and the replication samples; nonetheless, potential diagnostic differences in gene expression (and protein) of individual ESC/E(Z) complex genes should be considered preliminary. Finally, the biological relevance of these findings in PMDD should be interpreted cautiously until future studies have more thoroughly identified the role of ESC/E(Z) complex genes in this condition. Second, we used an LCL-based in vitro cellular model that would not be fully informative for functional consequences in neurons. We think that the cellular difference we found captures an important component of the vulnerability to PMDD. Having said that there are many important elements (both upstream and downstream) within the CNS that are not present in LCLs. Furthermore, validating these findings in a more authentic model will be a focus of future studies. Cellular models, including neuronal cell lines and LCLs, have many advantages, but also distinct limitations for the study of brain diseases [29–31]. Compared to fresh samples, for example peripheral blood, cultured cells offer the opportunity to identify long-lasting, trait-like, or “intrinsic” characteristics. This has been a cornerstone of the use of fibroblast and lymphoblastoid cell lines in biochemical genetics. Here, the use of LCLs enabled us to compare some 34 women with PMDD and 33 Control women, as would not have been possible if we had studied cultured commercially available human neuronal cell lines. LCLs have been employed to investigate several forms of neuropsychiatric illness including epilepsy, major depression, Alzheimer’s disease, and stress-related conditions [29,34–36]. However, important limitations exist in extending our findings both to neuronal cells, to the brain, and to our understanding of behavior (as described above). Although the percentage of the genes expressed in common between LCLs and neuronal cells is important, the validity of the model depends on the system and its functionality. Third, we have not investigated the paradoxical relationship between ESC/E(Z) transcript and protein levels that we observed, which could reflect post-transcriptional regulation of ESC/E(Z) complex genes (e.g., SIRT1 [88,89] and MTF2 [90]), or degradation of mRNA or protein. Fourth, we did not study the dynamics or downstream molecular effects of the ESC/E(Z) complex itself. Fifth, the twenty-four hour exposure to E/P may have been too brief to adequately activate the systems involved in the differential expression of ESC/E(Z) complex genes. The choice of a 24-hour exposure time is consistent with the time course for cellular steroid signaling and previous cellular studies of steroid nuclear signaling [91–93]. LCLs are not intended to be models of behavior and there are downstream effects on behavior that could take days to weeks to emerge. Sixth, it is possible that women who do not meet the DSM criteria for PMDD could show a similar pattern of symptom remission during ovarian suppression and symptom recurrence after ovarian steroid addback. Certainly, all women in this study would meet criteria for PMS. Nonetheless, our findings cannot be generalized to the larger group of women with PMS who do not meet criteria for PMDD, and it would be only a matter of speculation to suggest that these data could apply to women with PMS. Finally, we were unable to identify the nuclear PR in LCLs which also could reflect the many downstream elements important in brain but not present in LCLs. Nonetheless, we did identify expression of both PGRMC1 and PGRMC2, both of which have been identified to serve as important transcriptional modifiers within the central nervous system (e.g., hippocampus) to mediate neurosteroid signaling, the regulation of synaptic remodeling, neurogenesis, neuroprotection and neuroendocrine functions [94–100].
Several issues require future exploration. First, the roles of ER/PR in the mediation of the effects of E or P on ESC/E(Z) complex genes remain to be clarified. Our GeneMANIA network search identified HDAC2 as the only ESC/E(Z) complex gene that has connections to both receptors for ESR1 and PGRMC2 as well as many ESC/E(Z) genes, suggesting HDAC2 could serve as the connection between hormone signaling and ESC/E(Z) complex function. This is further supported by our significant finding of higher HDAC2 mRNA in PMDD LCLs over Control LCLs. Thus the function of HDAC2 in PMDD is potentially relevant since HDAC2 plays a pivotal role in transcriptional regulation and is one of the primary features of the ESC/E(Z) complex’s operations [101,102]. It is possible that HDAC2 acts as a hub between hormone signaling and ESC/E(Z) complex stability and/or function, which is disrupted in women with PMDD. Second, the apparent discrepancy between higher mRNA but lower protein in PMDD compared with Control LCLs may be due to differences in miRNA regulation of gene expression and translation, but could also be due to differences in assembly, trafficking, or binding of the complex to any of its interacting proteins and DNA. Third, the specific set of genes differentially silenced by the ESC/E(Z) complex in PMDD and control women were not characterized here. However, several candidate systems relevant to PMDD are suggested by recent studies of SIRT1 [80,81] and of enhanced estrogen transcriptional regulation in postpartum depression [27]. Finally, the future use of iPSC (induced pluripotent stem cells)-derived neurons or glia should be more informative about the CNS than LCLs.
It is unclear what allows ovarian steroids to trigger PMDD and why they do so in only a susceptible subgroup of women. Certainly, the role of a potential genetic abnormality in PMDD was suggested by observations that the heritability of premenstrual syndrome (a less severe form of PMDD, cohorts of which would contain women with PMDD) was approximately 56% [103]. The findings in this study offer possible explanations for a cellular substrate that could mediate PMDD including the following: 1) altered baseline expression in PMDD of factors within a major gene silencing pathway suggests a means by which steroid signals may be translated - as an enduring trait - into abnormal transcriptional events, which may underlie the genesis of pathological behavioral states; 2) although disturbed steroid signaling has been presumed to underlie the differential behavioral response to changes in ovarian steroids in PMDD, the findings from this study present for the first time cellular evidence for abnormal steroid signaling in cells from women with PMDD; and 3) as estradiol regulates transcription and translation at multiple levels, previously identified structural variants in ESR1 in PMDD may combine with or contribute to the disturbances in the family of transcriptional regulators identified in the LCLs from women with PMDD. We have identified an intrinsic cellular difference and pathway that could be responsible for the vulnerability to ovarian steroid-triggered affective destabilization in women with PMDD. Our findings provide the first evidence of a plausible biological substrate for the differential behavioral response to E/P in women with PMDD. Further clarification of the role of the ESC/E(Z) complex in PMDD and other reproductive endocrine-related mood disorders could identify substrates of risk, and targets for intervention, in these prevalent conditions.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIMH and NIAAA NIH; NIMH Protocols NCT00001259 and NCT00001322; NIMH Project # MH002865; NIAAA Project # AA000301.
We would like to thank Cheryl Marietta, Longina Akhtar, Bani Mukhopadhyay, and Elisa Moore of NIAAA for their technical assistance and expertise in conducting this study. We also would like to thank Karla Thompson and Linda Schenkel of NIMH for their clinical support and data management, and Catherine Roca who initiated the PMDD genetics project and was responsible for collecting many of the samples employed in the replication sample for this study. Finally, we wish to thank Dr. Shailaja Mani of Baylor College of Medicine for her guidance and consultation on this project.
Footnotes
The Authors have no potential conflicts of interest or financial support regarding this manuscript.
This work was written as part of Peter J. Schmidt’s official duties as a Government employee.
Reference List
- 1.American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition. Fifth. Arlington, VA: 2013. [Google Scholar]
- 2.Yonkers K, O'Brien PMS, Eriksson E. Premenstrual syndrome. Lancet. 2008;371:1200–1210. doi: 10.1016/S0140-6736(08)60527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epperson CN, Steiner M, Hartlage SA, Eriksson E, Schmidt PJ, Jones I, et al. Premenstrual dysphoric disorder: evidence for a new category for DSM-5. Am J Psychiatry. 2012;169:465–475. doi: 10.1176/appi.ajp.2012.11081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearlstein T, Steiner M. Premenstrual dysphoric disorder: burden of illness and treatment update. J Psychiatry Neurosci. 2008;33:291–301. [PMC free article] [PubMed] [Google Scholar]
- 5.Halbreich U, Borenstein J, Pearlstein T, Kahn LS. The prevalence, impairment, impact, and burden of premenstrual dysphoric disorder (PMS/PMDD) Psychoneuroendocrinology. 2003;28:1–23. doi: 10.1016/s0306-4530(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 6.Schiller CE, Johnson AL, Abate AC, Rubinow DR, Schmidt PJ. Reproductive steroid regulation of mood and behavior. Comprehensive Physiology. 2016 doi: 10.1002/cphy.c150014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammarback S, Ekholm UB, Backstrom T. Spontaneous anovulation causing disappearance of cyclical symptoms in women with the premenstrual syndrome. Acta Endocrinol (Copenh) 1991;125:132–137. doi: 10.1530/acta.0.1250132. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt PJ, Nieman LK, Danaceau MA, Adams LF, Rubinow DR. Differential behavioral effects of gonadal steroids in women with and in those without premenstrual syndrome. N Engl J Med. 1998;338:209–216. doi: 10.1056/NEJM199801223380401. [DOI] [PubMed] [Google Scholar]
- 9.Helvacioglu A, Yeoman RR, Hazelton JM, Aksel S. Premenstrual syndrome and related hormonal changes: long-acting gonadotropin releasing hormone agonist treatment. J Reprod Med. 1993;38:864–870. [PubMed] [Google Scholar]
- 10.Brown CS, Ling FW, Andersen RN, Farmer RG, Arheart KL. Efficacy of depot leuprolide in premenstrual syndrome: effect of symptom severity and type in a controlled trial. Obstet Gynecol. 1994;84:779–786. [PubMed] [Google Scholar]
- 11.Freeman EW, Sondheimer SJ, Rickels K, Albert J. Gonadotropin-releasing hormone agonist in treatment of premenstrual symptoms: with and without comorbidity of depression: a pilot study. J Clin Psychiatry. 1993;54:192–195. [PubMed] [Google Scholar]
- 12.West CP, Hillier H. Ovarian suppression with the gonadotrophin-releasing hormone agonist goserelin (Zoladex) in management of the premenstrual tension syndrome. Hum Reprod. 1994;9:1058–1063. doi: 10.1093/oxfordjournals.humrep.a138633. [DOI] [PubMed] [Google Scholar]
- 13.Hussain SY, Massil JH, Matta WH, Shaw RW, O'Brien PMS. Buserelin in premenstrual syndrome. Gynecol Endocrinol. 1992;6:57–64. doi: 10.3109/09513599209081007. [DOI] [PubMed] [Google Scholar]
- 14.Protopopescu X, Pan H, Altemus M, Tuescher O, Polanecsky M, McEwen B, et al. Orbitofrontal cortex activity related to emotional processing changes across the menstrual cycle. Proc Natl Acad Sci U S A. 2005;102:16060–16065. doi: 10.1073/pnas.0502818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Protopopescu X, Butler T, Pan H, Root J, Altemus M, Polanecsky M, et al. Hippocampal structural changes across the menstrual cycle. Hippocampus. 2008;18:985–988. doi: 10.1002/hipo.20468. [DOI] [PubMed] [Google Scholar]
- 16.Protopopescu X, Tuescher O, Pan H, Epstein J, Root J, Chang L, et al. Toward a functional neuroanatomy of premenstrual dysphoric disorder. J Affective Disord. 2007;108:87–94. doi: 10.1016/j.jad.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Baller EB, Wei SM, Kohn P, Rubinow DR, Alarcon G, Schmidt PJ, et al. Abnormalities of dorsolateral prefrontal function in women with premenstrual dysphoric disorder: a multimodal neuroimaging study. Am J Psychiatry. 2013;170:305–314. doi: 10.1176/appi.ajp.2012.12030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreano JM, Cahill L. Menstrual cycle modulation of medial temporal activity evoked by negative emotion. Neuroimage. 2010;53:1286–1293. doi: 10.1016/j.neuroimage.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein JM, Jerram M, Abbs B, Whitfield-Gabrieli S, Makris N. Sex differences in stress response circuitry activation dependent on female hormonal cycle. J Neurosci. 2010;30:431–438. doi: 10.1523/JNEUROSCI.3021-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gingnell M, Bannbers E, Wikstrom J, Fredrikson M, Sundstrom-Poromaa I. Premenstrual dysphoric disorder and prefrontal reactivity during anticipation of emotional stimuli. Eur Neuropsychopharmacol. 2013;23:1474–1483. doi: 10.1016/j.euroneuro.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Sundstrom PI, Gingnell M. Menstrual cycle influence on cognitive function and emotion processing-from a reproductive perspective. Front Neurosci. 2014;8:380. doi: 10.3389/fnins.2014.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreher J, Schmidt PJ, Kohn P, Furman D, Rubinow D, Berman KF. Menstrual cycle phase modulates reward-related neural function in women. Proc Natl Acad Sci U S A. 2007;104:2465–2470. doi: 10.1073/pnas.0605569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman KF, Schmidt PJ, Rubinow DR, Danaceau MA, Van Horn JD, Esposito G, et al. Modulation of cognition-specific cortical activity by gonadal steroids: a positron-emission tomography study in women. Proc Natl Acad Sci U S A. 1997;94:8836–8841. doi: 10.1073/pnas.94.16.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toffoletto S, Lanzenberger R, Gingnell M, Sundstrom-Poromaa I, Comasco E. Emotional and cognitive functional imaging of estrogen and progesterone effects in the female human brain: a systematic review. Psychoneuroendocrinology. 2014;50:28–52. doi: 10.1016/j.psyneuen.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt PJ, Ben Dor R, Martinez PE, Guerrieri GM, Harsh VL, Thompson K, et al. Effects of Estradiol Withdrawal on Mood in Women With Past Perimenopausal Depression: A Randomized Clinical Trial. JAMA Psychiatry. 2015;72:714–726. doi: 10.1001/jamapsychiatry.2015.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bloch M, Schmidt PJ, Danaceau M, Murphy J, Nieman L, Rubinow DR. Effects of gonadal steroids in women with a history of postpartum depression. Am J Psychiatry. 2000;157:924–930. doi: 10.1176/appi.ajp.157.6.924. [DOI] [PubMed] [Google Scholar]
- 27.Mehta D, Newport DJ, Frishman G, Kraus L, Rex-Haffner M, Ritchie JC, et al. Early predictive biomarkers for postpartum depression point to a role for estrogen receptor signaling. Psychol Med. 2014;44:2309–2322. doi: 10.1017/S0033291713003231. [DOI] [PubMed] [Google Scholar]
- 28.Guintivano J, Arad M, Gould TD, Payne JL, Kaminsky ZA. Antenatal prediction of postpartum depression with blood DNA methylation biomarkers. Mol Psychiatry. 2014;19:560–567. doi: 10.1038/mp.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:559–576. doi: 10.1016/j.pnpbp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson AC, Unger ER, Mangalathu R, Ojaniemi H, Vernon SD. Exploration of neuroendocrine and immune gene expression in peripheral blood mononuclear cells. Brain Res Mol Brain Res. 2004;129:193–197. doi: 10.1016/j.molbrainres.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci Res. 2009;87:1953–1959. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- 33.de Mello AS, Provin F, Michelin-Tireli K, Camelier MV, Coelho JC. Feasibility of using cryopreserved lymphoblastoid cells to diagnose some lysosomal storage diseases. Cell Prolif. 2010;43:164–169. doi: 10.1111/j.1365-2184.2010.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahn S, Chan MK. What can we learn about depression from gene expression in peripheral tissues? Biol Psychiatry. 2015;77:207–209. doi: 10.1016/j.biopsych.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 35.Iga J, Ueno S, Ohmori T. Molecular assessment of depression from mRNAs in the peripheral leukocytes. Ann Med. 2008;40:336–342. doi: 10.1080/07853890802082088. [DOI] [PubMed] [Google Scholar]
- 36.Karsten SL, Kudo LC, Bragin AJ. Use of peripheral blood transcriptome biomarkers for epilepsy prediction. Neurosci Lett. 2011;497:213–217. doi: 10.1016/j.neulet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain "-omes". Am J Med Genet B Neuropsychiatr Genet. 2013;162B:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 38.American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders Fourth Edition. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 39.Rubinow DR, Roy-Byrne PP, Hoban MC, Gold PW, Post RM. Prospective assessment of menstrually related mood disorders. Am J Psychiatry. 1984;141:684–686. doi: 10.1176/ajp.141.5.684. [DOI] [PubMed] [Google Scholar]
- 40.Endicott J, Nee J, Cohen J, Halbreich U. Premenstrual changes: patterns and correlates of daily ratings. J Affective Disord. 1986;10:127–135. doi: 10.1016/0165-0327(86)90035-2. [DOI] [PubMed] [Google Scholar]
- 41.First MB, Spitzer RL, Gibbon M, Williams JBW. Biometrics Research. New York: New York State Psychiatric Institute; 2002. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition (SCID-IP) [Google Scholar]
- 42.Halbreich U, Backstrom T, Eriksson E, O'Brien S, Calil H, Ceskova E, et al. Clinical diagnosis criteria for premenstrual syndrome and guidelines for their quantification for research studies. Gynecol Endocrinol. 2007;23:123–130. doi: 10.1080/09513590601167969. [DOI] [PubMed] [Google Scholar]
- 43.Halbreich U. The diagnosis of premenstrual syndromes and premenstrual dysphoric disorder - clinical procedures and research perspectives. Gynecol Endocrinol. 2004;19:320–334. doi: 10.1080/0951590400018215. [DOI] [PubMed] [Google Scholar]
- 44.Freeman EW, Sondheimer SJ, Rickels K. Gonadotropin-releasing hormone agonist in the treatment of premenstrual symptoms with and without ongoing dysphoria: a controlled study. Psychopharmacol Bull. 1997;33:303–309. [PubMed] [Google Scholar]
- 45.Ben Dor R, Harsh VL, Fortinsky P, Koziol DE, Rubinow DR, Schmidt PJ. Effects of pharmacologically-induced hypogonadism on mood and behavior in healthy young women. Am J Psychiatry. 2013;170:426–433. doi: 10.1176/appi.ajp.2012.12010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh HM, Oh JM, Choi SC, Kim SW, Han WC, Kim TH, et al. An efficient method for the rapid establishment of Epstein-Barr virus immortalization of human B lymphocytes. Cell Prolif. 2003;36:191–197. doi: 10.1046/j.1365-2184.2003.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmans P. Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Adv Genet. 2010;72:141–179. doi: 10.1016/B978-0-12-380862-2.00007-2. [DOI] [PubMed] [Google Scholar]
- 48.O'Neill RA, Bhamidipati A, Bi X, Deb-Basu D, Cahill L, Ferrante J, et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc Natl Acad Sci U S A. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Histone methyltransferase EZH2 is transcriptionally induced by estradiol as well as estrogenic endocrine disruptors bisphenol-A and diethylstilbestrol. J Mol Biol. 2014;426:3426–3441. doi: 10.1016/j.jmb.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 50.Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, et al. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep. 2013;3:411–426. doi: 10.1016/j.celrep.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 51.Bredfeldt TG, Greathouse KL, Safe SH, Hung MC, Bedford MT, Walker CL. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol Endocrinol. 2010;24:993–1006. doi: 10.1210/me.2009-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Izzo F, Mercogliano F, Venturutti L, Tkach M, Inurrigarro G, Schillaci R, et al. Progesterone receptor activation downregulates GATA3 by transcriptional repression and increased protein turnover promoting breast tumor growth. Breast Cancer Res. 2014;16:491. doi: 10.1186/s13058-014-0491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamm-Rosenstein K, Simm J, Suhorutshenko M, Salumets A, Metsis M. Changes in the transcriptome of the human endometrial Ishikawa cancer cell line induced by estrogen, progesterone, tamoxifen, and mifepristone (RU486) as detected by RNA-sequencing. PLoS One. 2013;8:e68907. doi: 10.1371/journal.pone.0068907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han L, Wang P, Zhao G, Wang H, Wang M, Chen J, et al. Upregulation of SIRT1 by 17beta-estradiol depends on ubiquitin-proteasome degradation of PPAR-gamma mediated by NEDD4-1. Protein Cell. 2013;4:310–321. doi: 10.1007/s13238-013-2124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore RL, Dai Y, Faller DV. Sirtuin 1 (SIRT1) and steroid hormone receptor activity in cancer. J Endocrinol. 2012;213:37–48. doi: 10.1530/JOE-11-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang S, Jia Y, Liu X, Winters C, Wang X, Zhang Y, et al. Systematic dissection of the mechanisms underlying progesterone receptor downregulation in endometrial cancer. Oncotarget. 2014;5:9783–9797. doi: 10.18632/oncotarget.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang C, Giri VN, Wilkinson JC, Wright CW, Wilkinson AS, Cooney KA, et al. EZH2 regulates the transcription of estrogen-responsive genes through association with REA, an estrogen receptor corepressor. Breast Caner Res Treat. 2008;107:235–242. doi: 10.1007/s10549-007-9542-7. [DOI] [PubMed] [Google Scholar]
- 58.Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 59.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di CL, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 61.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 62.Crowley SK, Girdler SS. Neurosteroid, GABAergic and hypothalamic pituitary adrenal (HPA) axis regulation: what is the current state of knowledge in humans? Psychopharmacology (Berl) 2014;231:3619–3634. doi: 10.1007/s00213-014-3572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saha S, Hu Y, Martin SC, Bandyopadhyay S, Russek SJ, Farb DH. Polycomblike protein PHF1b: a transcriptional sensor for GABA receptor activity. BMC Pharmacol Toxicol. 2013;14:37. doi: 10.1186/2050-6511-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Epperson CN, Haga K, Mason GF, Sellers E, Gueorguieva R, Zhang W, et al. Cortical γ-aminobutyric acid levels across the menstrual cycle in healthy women and those with premenstrual dysphoric disorder: a proton magnetic resonance spectroscopy study. Arch Gen Psychiatry. 2002;59:851–858. doi: 10.1001/archpsyc.59.9.851. [DOI] [PubMed] [Google Scholar]
- 65.Smith MJ, Adams LF, Schmidt PJ, Rubinow DR, Wassermann EM. Abnormal luteal phase excitability of the motor cortex in women with premenstrual syndrome. Biol Psychiatry. 2003;54:757–762. doi: 10.1016/s0006-3223(02)01924-8. [DOI] [PubMed] [Google Scholar]
- 66.Schiller CE, Schmidt PJ, Rubinow DR. Allopregnanolone as a mediator of affective switching in reproductive mood disorders. Psychopharmacology (Berl) 2014;231:3557–3567. doi: 10.1007/s00213-014-3599-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol Med. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Machado-Vieira R, Ibrahim L, Zarate CA., Jr Histone deacetylases and mood disorders: epigenetic programming in gene-environment interactions. CNS Neurosci Ther. 2011;17:699–704. doi: 10.1111/j.1755-5949.2010.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ernst C, Chen ES, Turecki G. Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol Psychiatry. 2009;14:830–832. doi: 10.1038/mp.2009.35. [DOI] [PubMed] [Google Scholar]
- 71.Kishi T, Yoshimura R, Kitajima T, Okochi T, Okumura T, Tsunoka T, et al. SIRT1 gene is associated with major depressive disorder in the Japanese population. J Affect Disord. 2010;126:167–173. doi: 10.1016/j.jad.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 72.Kenworthy CA, Sengupta A, Luz SM, Ver Hoeve ES, Meda K, Bhatnagar S, et al. Social defeat induces changes in histone acetylation and expression of histone modifying enzymes in the ventral hippocampus, prefrontal cortex, and dorsal raphe nucleus. Neuroscience. 2014;264:88–98. doi: 10.1016/j.neuroscience.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 73.Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, III, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 75.Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 76.Nasca C, Xenos D, Barone Y, Caruso A, Scaccianoce S, Matrisciano F, et al. L-acetylcarnitine causes rapid antidepressant effects through the epigenetic induction of mGlu2 receptors. Proc Natl Acad Sci U S A. 2013;110:4804–4809. doi: 10.1073/pnas.1216100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qi C, Liu S, Qin R, Zhang Y, Wang G, Shang Y, et al. Coordinated regulation of dendrite arborization by epigenetic factors CDYL and EZH2. J Neurosci. 2014;34:4494–4508. doi: 10.1523/JNEUROSCI.3647-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan W, Fang Z, Yang Q, Dong H, Lu Y, Lei C, et al. SirT1 mediates hyperbaric oxygen preconditioning-induced ischemic tolerance in rat brain. J Cereb Blood Flow Metab. 2013;33:396–406. doi: 10.1038/jcbfm.2012.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Libert S, Pointer K, Bell EL, Das A, Cohen DE, Asara JM, et al. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147:1459–1472. doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Licinio J, Wong ML. The role of inflammatory mediators in the biology of major depression: central nervous system cytokines modulate the biological substrate of depressive symptoms, regulate stress-responsive systems, and contribute to neurotoxicity and neuroprotection. Mol Psychiatry. 1999;4:317–327. doi: 10.1038/sj.mp.4000586. [DOI] [PubMed] [Google Scholar]
- 83.Gold PW. The organization of the stress system and its dysregulation in depressive illness. Mol Psychiatry. 2015;20:32–47. doi: 10.1038/mp.2014.163. [DOI] [PubMed] [Google Scholar]
- 84.Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol. 2016;16:22–34. doi: 10.1038/nri.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bertone-Johnson ER, Ronnenberg AG, Houghton SC, Nobles C, Zagarins SE, Takashima-Uebelhoer BB, et al. Association of inflammation markers with menstrual symptom severity and premenstrual syndrome in young women. Hum Reprod. 2014;29:1987–1994. doi: 10.1093/humrep/deu170. [DOI] [PubMed] [Google Scholar]
- 86.Gold EB, Wells C, Rasor MO. The Association of Inflammation with Premenstrual Symptoms. J Womens Health (Larchmt) 2016 doi: 10.1089/jwh.2015.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huo L, Straub RE, Schmidt PJ, Shi K, Vakkalanka R, Weinberger DR, et al. Risk for premenstrual dysphoric disorder is associated with genetic variation in ESR1, the estrogen receptor alpha gene. Biol Psychiatry. 2007;62:925–933. doi: 10.1016/j.biopsych.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tseng RC, Wang YC. SIRT1 (sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae)) Atlas of Genetics and Cytogenetics in Oncology and Haematology. 2010;14:1152–1156. [Google Scholar]
- 89.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2014;33:1609–1620. doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li X, Isono K, Yamada D, Endo TA, Endoh M, Shinga J, et al. Mammalian polycomb-like Pcl2/Mtf2 is a novel regulatory component of PRC2 that can differentially modulate polycomb activity both at the Hox gene cluster and at Cdkn2a genes. Mol Cell Biol. 2011;31:351–364. doi: 10.1128/MCB.00259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kininis M, Kraus WL. A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and DNA sequence analysis. Nucl Recept Signal. 2008;6:e005. doi: 10.1621/nrs.06005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hah N, Kraus WL. Hormone-regulated transcriptomes: lessons learned from estrogen signaling pathways in breast cancer cells. Mol Cell Endocrinol. 2014;382:652–664. doi: 10.1016/j.mce.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Villablanca AC, Lewis KA, Rutledge JC. Time- and dose-dependent differential upregulation of three genes by 17 beta-estradiol in endothelial cells. J Appl Physiol (1985) 2002;92:1064–1073. doi: 10.1152/japplphysiol.00374.2001. [DOI] [PubMed] [Google Scholar]
- 94.Brinton RD, Thompson RF, Foy MR, Baudry M, Wang J, Finch CE, et al. Progesterone receptors: form and function in brain. Front Neuroendocrinol. 2008;29:313–339. doi: 10.1016/j.yfrne.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bali N, Arimoto JM, Iwata N, Lin SW, Zhao L, Brinton RD, et al. Differential responses of progesterone receptor membrane component-1 (Pgrmc1) and the classical progesterone receptor (Pgr) to 17β-estradiol and progesterone in hippocampal subregions that support synaptic remodeling and neurogenesis. Endocrinology. 2012;153:759–769. doi: 10.1210/en.2011-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mani SK, Blaustein JD. Neural progestin receptors and female sexual behavior. Neuroendocrinology. 2012;96:152–161. doi: 10.1159/000338668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thomas P, Pang Y. Membrane progesterone receptors: evidence for neuroprotective, neurosteroid signaling and neuroendocrine functions in neuronal cells. Neuroendocrinology. 2012;96:162–171. doi: 10.1159/000339822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peluso JJ, DeCerbo J, Lodde V. Evidence for a genomic mechanism of action for progesterone receptor membrane component-1. Steroids. 2012;77:1007–1012. doi: 10.1016/j.steroids.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thomas P. Characteristics of membrane progestin receptor alpha (mPRalpha) and progesterone membrane receptor component 1 (PGMRC1) and their roles in mediating rapid progestin actions. Front Neuroendocrinol. 2008;29:292–312. doi: 10.1016/j.yfrne.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schumacher M, Mattern C, Ghoumari A, Oudinet JP, Liere P, Labombarda F, et al. Revisiting the roles of progesterone and allopregnanolone in the nervous system: resurgence of the progesterone receptors. Prog Neurobiol. 2014;113:6–39. doi: 10.1016/j.pneurobio.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 101.Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 102.Van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 103.Kendler KS, Silberg JL, Neale MC, Kessler RC, Heath AC, Eaves LJ. Genetic and environmental factors in the aetiology of menstrual, premenstrual and neurotic symptoms: a population-based twin study. Psychol Med. 1992;22:85–100. doi: 10.1017/s0033291700032761. [DOI] [PubMed] [Google Scholar]
- 104.Endicott J, Halbreich U. Retrospective report of premenstrual depressive changes: Factors affecting confirmation by daily ratings. Psychopharmacol Bull. 1982;18:109–112. [Google Scholar]
- 105.Garcia-Gonzalo FR, Izpisua Belmonte JC. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS One. 2008;3:e1384. doi: 10.1371/journal.pone.0001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.