Abstract
The human hepatitis B virus (HBV) has a compact genome encoding four major overlapping coding regions: the core, polymerase, surface and X. The polymerase initiation codon is preceded by the partially overlapping core and four or more upstream initiation codons. There is evidence that several mechanisms are used to enable the synthesis of the polymerase protein, including leaky scanning and ribosome reinitiation. We have examined the first AUG in the pregenomic RNA, it precedes that of the core. It initiates an uncharacterized short upstream open reading frame (uORF), highly conserved in all HBV subtypes, we designated the C0 ORF. This arrangement suggested that expression of the core and polymerase may be affected by this uORF. Initiation at the C0 ORF was confirmed in reporter constructs in transfected cells. The C0 ORF had an inhibitory role in downstream expression from the core initiation site in HepG2 cells and in vitro, but also stimulated reinitiation at the polymerase start when in an optimal context. Our results indicate that the C0 ORF is a determinant in balancing the synthesis of the core and polymerase proteins.
INTRODUCTION
Hepatitis B virus (HBV) remains a major human pathogen. Although new infections are preventable through vaccination, new antiviral targets are being sought for the treatment of the estimated 350 million affected individuals worldwide (1–3). The expression of several HBV proteins is modulated at the translational level and these regulatory mechanisms are potential targets. HBV replication is carried out by the polymerase (P) protein through reverse transcription of the pregenomic RNA (pgRNA) and this protein is the target of current treatments (4). The pgRNA transcript is encapsidated and then serves as the template for reverse transcription of the DNA genome. It is also the template for the synthesis of the core (C) and P proteins (4–6). In the pgRNA, the C open reading frame (ORF) precedes and overlaps the P ORF (Figure 1B). Since both coding regions are in a single mRNA, the question arises of how the P protein in the second reading frame is expressed.
Figure 1.
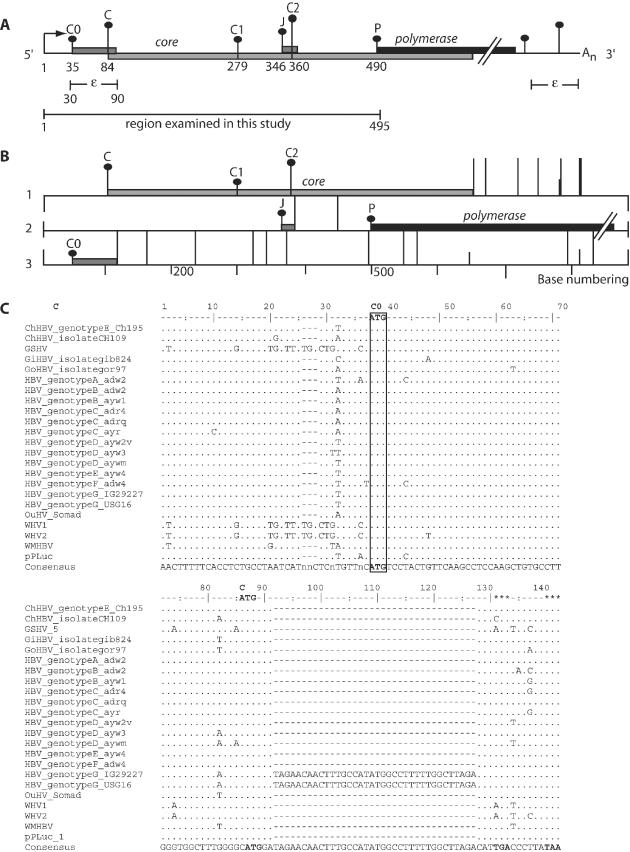
The conservation of the C0 ORF in all HBV genotypes and other members of the orthohepadnaviridae. (A) Schematic representation of the HBV pregenomic RNA transcript. The numbering in the scheme is such that nucleotide 1 is the pgRNA start site, indicated by the arrow (this number corresponds to nucleotide number 1816 in the ayw subtype). The pgRNA contains an epsilon structure, near the 5′ and 3′ end of the RNA. This transcript also contains a redundant sequence of 117 bases in its 3′ end, found in the 5′ end of the transcript. Vertical bars with dots represent the initiation codons within the pgRNA labeled C0, C, C1, J, C2 and P. The height of each vertical bar with black dots represents the match to the ‘ideal’ initiation context of Kozak's consensus. ORFs encoded by each initiation codon are represented by filled boxes while the poly(A) tail is denoted by An. (B) Schematic representation of the three different ORFs in the pgRNA in which the C0, C, J and P protein are made. The initiation codons (vertical bars with black dots) and termination codons (long vertical bars without dots) are marked in each of the three possible reading frames with the corresponding ORFs represented by filled boxes. (C) The comparison of DNA sequences in the region of the C0 ORF from representative members of the orthohepadnaviruses. The alignment of these sequences revealed a well-conserved ‘weak’ initiation codon (C0) and its 19 amino acid ORF which overlaps the C ORF by 8 bases.
Most translation in eukaryotic cells is postulated to proceed according to the ribosome scanning model (7–9). First, the 43S small ribosomal subunit complex binds to the capped 5′ end of the mRNA and scans in the 3′ direction until it encounters the first AUG in a suitable Kozak's context. The resulting complex is joined by the large subunit to form a complete ribosome, and polypeptide synthesis begins (7).
Although translation usually initiates at the first AUG codon (9), many transcripts contain at least one AUG codon before the major open reading frame (10–13). The scanning model predicts that an upstream AUG codon will interfere with translation of downstream ORF, but the scanning complex may bypass AUG codons by ‘leaky scanning’ if the surrounding nucleotide context is suboptimal or very close to the 5′ cap (9,14). Recent studies have identified an increasing number of uORFs that have key translational regulatory properties (13,15–19). Small regulatory upstream ORFs (uORFs) are found in the transcripts of viruses (20), proto-oncogenes, genes encoding growth factors and cellular receptors (9). They have a range of effects on the translation of subsequent ORFs. In most cases, the peptides encoded have not been detected directly, although function may depend on their sequences (21,22). Some function at the site of synthesis on the ribosome (23,24).
There is no specific transcript for P protein (6,25,26) and previous studies of HBV showed that P is not synthesized as a capsid–polymerase fusion protein (27) nor by internal entry of ribosomes (6,26,28). These studies supported a complex model in which the HBV P protein is translated by ribosomes which scan from the capped 5′ end of the pgRNA. Four upstream AUG codons are avoided by leaky scanning past the C and C1 AUG codons, translation of the J ORF, termination then reinitiation at the P AUG codon (6,26,28).
In this study, we examine the function of the previously overlooked first initiation codon in the pgRNA. It initiates a highly conserved uORF. Previous studies have examined four AUG codons, designated C, C1, C2 and J preceding the P gene in the pgRNA (Figure 1A) (28). Thus, we designated the novel initiation codon C0 as it precedes these. We established a sensitive cell culture based assay system to examine the role of this first AUG and uORF in the pgRNA and their impact on the core and polymerase initiation codons.
MATERIALS AND METHODS
Plasmid construction and mutagenesis
All DNA constructs containing HBV sequences were subcloned into the pGL3-control reporter vector (Promega) under the control of an SV40 promoter. The HBV test sequence from ATCC 40103 clone and its subsequent mutations were generated through PCR using Expand™ High Fidelity DNA polymerase (Roche) with mutagenic primers. Amplified fragments were subcloned into the pGL-3 control using a unique HindIII and a newly introduced AatII sites. The unique AatII restriction site introduced via site-directed-mutagenesis PCR (SDM-PCR) replaced the NcoI site within pGL3-control encoding the ATG codon directing luciferase expression. Consequently, expression of the luciferase gene would then be directed by the desired in-frame ATG codon within the pgRNA leader introduced upstream of the AatII site. All the constructs used in this study contain an SV40 and T7 promoter upstream of the HBV sequence (Table 1). In order that the DNA constructs mimic the pgRNA transcript, the 117 nt of terminally redundant sequence present in the 3′ end of the pgRNA was inserted immediately after the luciferase gene (Figure 2A). The terminally redundant sequence was amplified from ATCC 40103 HBV genomic DNA clone using primer XbaIF and XbaIR. The amplified product also included a 42 nt long poly(A) tract in the 3′ end, which conferred stability to the in vitro synthesized RNA transcripts (Figure 2A). This amplified product was subcloned into the XbaI site immediately after the luciferase gene of pGL-3 control. All plasmids were confirmed by sequencing. The AUG series constructs consisted of C0AUG, CAUG, C1AUG, JAUG, C2AUG, PAUG and CAUGKO. They were generated by inserting a constant leader sequence of 31 bases followed by sequences flanking respective initiation codon between the −6 and +6 position (Table 2). As a negative control (CAUGKO), the C AUG codon from the HBV pgRNA sequence was mutated to AAG. The pg group of constructs consisted of the C0LUC, CLUC, C1LUC, JLUC, C2LUC and PLUC. Each construct in this series contain the epsilon structure at both the 5′ and 3′ end. Plasmid C0LUC (Table 2), contains HBV sequence comprising the HBV pgRNA authentic start site and the C0 uORF, which is fused in-frame with the luciferase reporter gene. The CLUC construct consisted of the HBV pgRNA leader sequence until the C AUG codon fused in-frame with the luciferase reporter (Table 2). This construct also contains the C0 uORF which overlaps the C AUG codon. Likewise, the C1LUC, JLUC and C2LUC plasmid would have the HBV pgRNA leader sequence from the transcript start until their respective initiation codon fused in-frame to the luciferase gene. Plasmid PLUC (Table 2) mimics the pgRNA consisted of the HBV pgRNA leader sequence (495 bp) starting from the authentic pgRNA transcript start site and ending at the internal P AUG codon fused in-frame with the luciferase reporter gene. The leader sequence was amplified using the following primers: H3T7PGRNAF containing the HindIII site and the T7 promoter followed by corresponding bases from the authentic pgRNA transcript start and primer AatIILUCR with AatII restriction enzyme site. This sequence was submitted to DDBJ, accession no. AB037684. As a result of the P AUG codon fusion to the luciferase gene, the C ORF was modified at the 3′ end with an overlap of 143 nt over the luciferase ORF. The constructs used to study the role of C0 (Figure 4A) were derivatives of the pg series construct. They include C0KOCLUC, C0KOC1LUC, C0KOJLUC, C0KOC2LUC and C0KOPLUC. Changes were introduced using SDM–PCR in which the C0 initiation codon was changed to AAG by converting a T to A at position 36. All the alterations of initiation codons to AAG in this study were based on studies by Peabody (29), ensuring that non-AUG initiation codons were not introduced by this mutation. Two other constructs, C0CJLUC and C0stopKOJLUC containing modified C0 ORF were also made to study the effect of the C0 ORF on downstream J expression. C0CJLUC had the C0 ORF abolished through fusion to the C ORF via a single A base deletion at position 42. C0stopKOJLUC contained a lengthened C0 ORF via deletion of two immediate in-frame stop codons (TGA to CGA and TAA to CAA) to generate a modified C0 ORF with 38 codons instead of 20. Alternatively, constructs used to address reinitiation potential of a strong C0 AUG include C0d3CLUC, C0d3C1LUC, C0d3JLUC, C0d3C2 and C0d3PLUC. These constructs were derived from the pg series backbone with the C0 AUG codon altered to a strong context by a single base mutation of a T to A in the −3 position of the C0 AUG context. Other constructs used to address the effects of upstream mutations on P were derived from PLUC. They include C0stopKOPLUC, C0CPLUC, CKOPLUC, C1KOPLUC, C1d3PLUC, C2KOPLUC, JKOPLUC and uAUGKOPLUC. Plasmid C0stopKOPLUC had two of the subsequent stop codons (TGA to CGA and TAA to CAA) of the C0 ORF removed to extend the C0 ORF to 38 codons (20–38 codons). Plasmid C0CPLUC has an A base deletion at position 42 causing the C0 reading frame to be shifted in-frame with the C ORF. The deletion at base 42 also caused the C0 coding sequence to be different starting at codon 3. Plasmid CKOPLUC contains the disrupted C initiation codon (base change of a T to A at position 85). In C1KOPLUC, the C1 initiation codon was changed to AAG by converting a T to A base while in C1d3PLUC, the C1 initiation codon was changed to an optimal context by introducing an A in the −3 position. Plasmid C2KOPLUC consisted of the C2 AUG codon changed to AAG to abolish initiation at C2. JKOPLUC was designed to study the importance of the J uORF in P translation. In JKOPLUC, the JAUG was mutated to ggG to avoid introduction of premature stop codon in the C ORF. Plasmid uAUGKOPLUC had all upstream AUGs prior to P removed to understand what roles these upstream AUG may have towards P synthesis. Other derivatives from PLUC include negative control constructs namely PKOLUC and UKOLUC. The PKOLUC construct has the P initiation codon in-frame to the luciferase gene is altered to AAG. It is designed to rule out the possibility of initiation from internal luciferase AUG giving rise to luciferase activity. The other control construct UKOLUC consisted of all the pgRNA AUG codons mutated to account for any in-frame AUGs present in the luciferase gene, which might result in luciferase activity. The pcPLUC mimics the precore RNA transcript. It is identical to the PLUC construct but contains an additional 34 bases at its 5′ end from the precore RNA, this section includes the precore initiation codon (PC). Plasmid PCKOpcPLUC was derived from pcPLUC with the PC AUG codon removed by mutation to AAG. The Renilla luciferase reporter, phRL-SV40 was used as an internal control (Figure 2A).
Table 1.
PCR primers used to generate parent constructs
| Primers | Sequence |
|---|---|
| APH3T7F | 5′-gggcccaaAAGCTTTAATACGACTCACTATAGG-3′ |
| H3T7PCRNAF | 5′-gcgAAGCTTTAATACGACTCACTATAGGTAGGCACAAATTGGTCTGCG-3′ |
| H3T7PGRNAF | 5′-gcgatAAGCTTTAATACGACTCACTATAGGAACTTTTTCACCTCTGCCTAATC-3′ |
| AatIILUCR | 5′-gtttttggcgtcttcGACGTCgggcatTTGGTGGTCTATAGG-3′ |
| XbaIF | 5′-gcTCTAGAAACTTTTTCACCTCTGCCTAATC-3′ |
| XbaIR | 5′-gcTCTAGAATTCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGTAGCTCCAAATTCTTT-3′ |
| GLR | 5′-CCAGGGCGTATCTCTTCATAGCC-3′ |
The italicized sequence represents the T7 promoter and the underlined sequences represent the restriction enzyme site.
Figure 2.
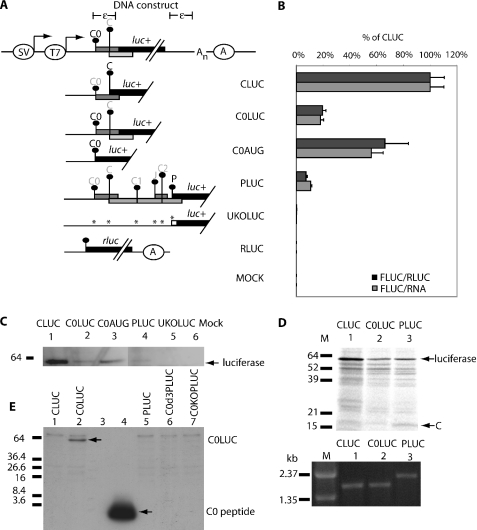
Translation initiation from the C0 initiation codon compared with that from C and P initiation codons. (A) Schematic representation of the DNA constructs used in this study (diagram not to scale). It contains the SV40 and T7 promoter at the 5′ end followed by DNA sequences corresponding to the HBV pregenomic RNA leader fused to the luciferase gene. The nucleotide sequence for the hepatitis B virus pregenomic RNA leader has been deposited in the DNA Data Bank of Japan under DDBJ accession no. AB037684. Immediately downstream of the luciferase gene is the 117 bases of repeated sequence to mimic the terminally redundant pgRNA which also contains the epsilon structure denoted by ε. At the 3′ end, An denotes the poly(A) tract while the SV40 poly(A) signal is denoted by an enclosed A. Vertical bars with dots represent the initiation codons within the pgRNA labeled C0, C in a light and dark font. The darker font represents the initiation codon that is in-frame to the luciferase reporter gene. ORFs encoded by each initiation codon are represented by filled boxes. Any mutation introduced into the leader sequence is denoted with an asterisk (*). Other labels are as in Figure 1. (B) Normalized expression level from the C0, C and P initiation codons after transfection into HepG2 cells. Expression from each initiation codon is expressed as a percentage of normalized CLUC expression. These results are averages of three replicates from two independent experiments. (C) Luciferase expression was confirmed via western blot using anti-luciferase antibody on HepG2 transfected lysates separated on 5–20% SDS–PAGE. (D) Fluorograph of the luciferase fusion proteins translated off capped transcript in rabbit reticulocyte. Proteins were separated on 5–20% SDS–PAGE. Lane 1, CLUC RNA; lane 2, C0LUC RNA; lane 3, PLUC RNA; M, protein or DNA marker. The integrity and quantitation of the capped RNA used for cell-free translation is shown on the RNA gel below. (E) Detection of the C0LUC fusion protein in HepG2 transfected lysates via immunoblot analysis using anti-C0 antibody. Lane 4 contains 50 ng of synthetic C0.
Table 2.
Plasmids used in this study
| Plasmid name | Template PCR | Mutations | Position | Effect |
|---|---|---|---|---|
| P series | Determine effect of uATGs on P expression | |||
| PLUC | ATCC40103 | NcoI to AatII | 496 | Introduction of adw HBV leader with AatII |
| C0KOPLUC | PLUC | ATG to AaG | 36 | Removal of C0 ATG codon |
| C0stopKOPLUC | PLUC | TGA to cGA, TAA to cAA | 92, 101 | Extended C0 ORF to 38 codons from 20 codons |
| C0CPLUC | PLUC | A deletion | 141 | C0 ORF removed via fusion to C ORF |
| CKOPLUC | PLUC | ATG to AaG | 85 | Removal of C ATG codon |
| C1KOPLUC | PLUC | ATG to AaG | 280 | Removal of C1 ATG codon |
| C1D3PLUC | PLUC | TTGATGA to aTcATGA | 276, 278 | Optimized C1 ATG context |
| JKOPLUC | PLUC | ATG to ggG | 347, 348 | Removal of J ATG codon |
| C2KOPLUC | PLUC | ATG to AaG | 361 | Removal of C2 ATG codon |
| PKOLUC | PLUC | ATG to AaG | 491 | Removal of P ATG codon |
| uaugkopluc | PLUC | ATGs to AaGs or ggG | 36, 85, 280, 347,348, 361 | Removal of all uATGs except P ATG codon |
| UKOLUC | PKOLUC | ATGs to AaG or ggG | 36, 85, 280, 347,348, 361, 491 | Removal of all uATGs including P ATG |
| PcPLUC | PLUC | 34 bases insertion | Upstream of pgRNA | 34 base extension at 5′ end to mimic the pcRNA leader |
| PCKOpcPLUC | pcPLUC | ATG to AaG | 32 (pcRNA numbering) | Removal of PC ATG codon |
| pg series | Initiation level from each ATG codons in respective leader sequence and uATGs | |||
| C0LUC | PLUC | C0 ORF fused to luciferase ORF | ||
| CLUC | C0LUCX | C ATG directs luciferase expression | ||
| C1LUC | PLUC | C1 ATG directs luciferase expression | ||
| JLUC | PLUC | J ATG directs luciferase expression | ||
| C2LUC | PLUC | C2 ATG directs luciferase expression | ||
| C0KO series | Derivative of pg series | Determine the effect of C0 on downstream expression | ||
| COKOCLUC | CLUC | ATG to AaG | 36 | C0 ATG codon abolished |
| COKOC1LUC | C1LUC | ATG to AaG | 36 | C0 ATG codon abolished |
| COKOJLUC | JLUC | ATG to AaG | 36 | C0 ATG codon abolished |
| COKOC2LUC | C2LUC | ATG to AaG | 36 | C0 ATG codon abolished |
| C0CJLUC | JLUC | ATG to AaG | 36 | C0 ORF abolished |
| C0stopKOJLUC | JLUC | ATG to AaG | 36 | longer C0 ORF 38 codons from 20 codons |
| C0D3 series | Derivative of pg series | Determine the effect of an optimal C0 context on downstream expression | ||
| C0D3CLUC | CLUC | GTACATGT to caACATGT | 31 and 32 | C0 ATG context optimized |
| C0D3C1LUC | C1LUC | GTACATGT to caACATGT | 31 and 32 | C0 ATG context optimized |
| C0D3JLUC | JLUC | GTACATGT to caACATGT | 31 and 32 | C0 ATG context optimized |
| C0D3C2LUC | C2LUC | GTACATGT to caACATGT | 31 and 32 | C0 ATG context optimized |
| C0D3PLUC | PLUC | GTACATGT to caACATGT | 31 and 32 | C0 ATG context optimized |
| C0D3ORF | PLUC | GTACATGT to caACATGT | 31 and 32 | C0 ATG context optimized |
| AUG series | Determine the strength of respective ATG context | |||
| COAUG | C0LUC | C0 ATG context directs luciferase expression | ||
| CAUG | COAUG | C ATG context directs luciferase expression | ||
| C1AUG | COAUG | C1 ATG context directs luciferase expression | ||
| JAUG | COAUG | J ATG context directs luciferase expression | ||
| C2AUG | COAUG | C2 ATG context directs luciferase expression | ||
| PAUG | COAUG | P ATG context directs luciferase expression | ||
| CAUGKO | CAUG | C ATG abolished | ||
Figure 4.
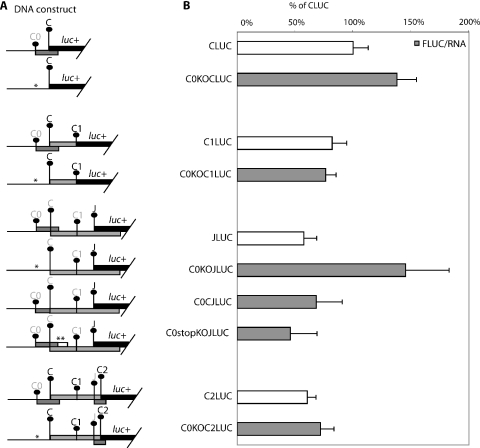
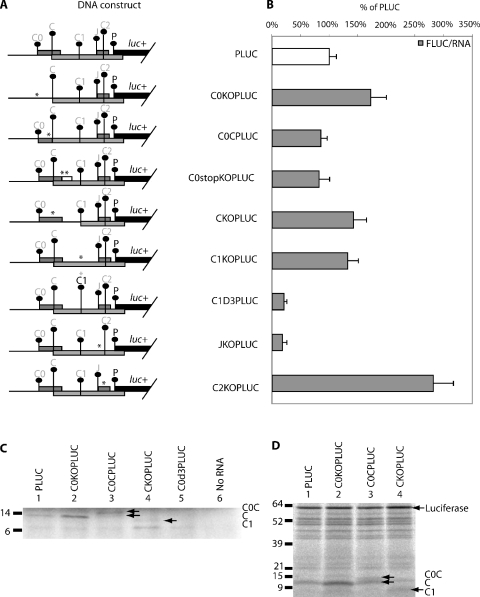
The effect of C0 initiation codon removal on expression at downstream initiation. (A) Schematic diagram of the HBV test constructs used to study the effect of C0 on downstream expression. The initiation codon with a darker font is in-frame to the luciferase reporter gene and an asterisk (*) indicates the position where a mutation is introduced. (B) Result of DNA transfection of the above constructs into HepG2 cells. The normalized luciferase activity of each construct with C0 mutation is compared with its respective parent construct without the C0 mutation. Normalized expression levels are presented as a percentage of the CLUC construct, assigned 100%. These results are averages of replicates from two independent experiments. Results from the C, C1 and C2 series represent sums of the activities of fusion proteins with similar specific activities.
In vitro transcription
The plasmids used for in vitro transcription and translation were linearized by EcoRI digestion. Linear DNA was proteinase K treated and then gel purified (Qiagen). Plasmids were transcribed in vitro by T7 RNA polymerase in the presence of the methylated cap analog m7GpppG (Roche Diagnostic) 0.6 mM. After 1 h incubation at 37°C, the in vitro synthesized capped RNA was DNase (1 U) treated for 15 min at 37°C. After purification by LiCl precipitation and a 70% ethanol wash, RNA was resuspended in 20 μl of DEPC treated water. All RNA integrity was confirmed via gel electrophoresis. RNA quantitation was done by spectrophotometry and ethidium bromide visualization. RNA transcripts were used for in vitro translation.
In vitro translation
Equal amounts of capped RNA in sub-saturating concentrations were translated in cell-free system, rabbit reticulocyte lysate (Promega). The translated proteins were separated by SDS–PAGE. The gels were treated with Amplify (Amersham), dried and exposed to imaging plate (Fuji BasIIIs) at room temperature. Quantitative phosphoimaging was performed using Fuji MacBAS version 2.0 software.
RNA electroporation into cells
HepG2 cells grown to 80% confluence in supplemented DMEM medium as described below. Cells were collected and washed twice with phosphate-buffered saline (PBS) before immediate usage. Cells (107) in 0.8 ml were mixed with 1 μg of in vitro synthesized capped RNA and electroporated in PBS (250 μF capacitance, 400 V). The quantity of RNA was determined by spectrophotometry and its integrity by gel electrophoresis. Following electroporation, the cells were incubated for 7 h in the supplemented DMEM medium. Luciferase activity increased for 7–8 h in cells before the mRNA became translationally inactive. Seven hours post-transfection, cells were harvested, washed and lysed as described and the supernatants (cytoplasmic lysates) collected and assayed for luciferase activity. Typical activities were 17 000 for C0LUC, 114 000 for CLUC in HepG2 and 47 000 and 735 000 for COS7, mock transfected cells or machine background was 5800 ± 40). All transfections were done in duplicate and the experiment repeated at least twice.
DNA transfection into hepatoma cells
All DNA transfection studies were done in HepG2 and Huh-7 cells. Hepatoma cell lines were chosen as HBV is a hepatropic virus. Prior studies have shown that most nonhepatic cultured cell lines transfected with the hepadnaviral DNA expressed incorrect genomic transcripts (4). Human hepatoma cells were grown in 24-well cell culture plates (Costar) at 37°C under 5% CO2 in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine and 0.1 mM MEM non-essential amino acids. Plasmid DNA used for transfection were quantitated twice using the NanoDrop® ND-1000 Spectrophotometer (Nanodrop Technologies, Inc.) followed by quantitation by gel electrophoresis. A total of 800 ng of each plasmid construct together with 10 ng of the internal control plasmid phRLSV40 were cotransfected into cells at 60% confluence using FuGENE 6 (Roche). Twenty-four hours post-transfection, cells were washed with PBS (10 mM phosphate buffer, pH 7.4, 140 mM NaCl) and lysed with 1× Passive Lysis Buffer (Promega). Cell debris and nuclei were removed by centrifugation and the supernatants (cytoplasmic lysates) collected and assayed for luciferase activity.
Luciferase assays
In vivo expression levels were determined by luciferase activity assays. Reactions were performed with an aliquot of the cytoplasmic lysates (10 μl) corresponding to 103 cells and 50 μl of passive lysis buffer (Promega). Photons emitted by the reaction were measured in the EG&G Berthold AutoLumat LB 953 luminometer. Luciferase activity was expressed as the number of light units detected in 10 s. Each of the constructs was assayed in at least two replicates and averages (background subtracted) reported. The luciferase activity was adjusted by Renilla luciferase activity (internal control) and expressed as normalized luciferase activity. Relative luciferase expression was calculated by dividing the average normalized ratio for each respective construct with that of a reference construct.
RNA isolation and quantitation of LUC transcripts
RNA was extracted from hepatoma cells using Qiagen RNeasy Mini Kit 24 h post-transfection. The RNA was treated with RNase-free DNase on the spin column for 10 min. Quantitation of total RNA was determined by measuring OD at 260 nm using NanoDrop® ND-1000 Spectrophotometer. RNA was then subjected to two step RT–PCR according to manufacturer's protocol (superscript II) (Applied Biosystems). Taqman assay were performed on ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The following oligonucleotides were used for the amplification of 66 bp fragments of the cDNA corresponding to the luciferase reporter. The primers and probe used are outlined in Table 3. Errors for the normalized ratios represent those of both the RNA and luciferase estimations.
Table 3.
PCR primers used for quantitative PCR
| Primers | Sequence |
|---|---|
| Taqman lucfwd | 5′-TTCTAAAACGGATTACCAGGGATT-3′ |
| Taqman lucrev | 5′-CCGGGAGGTAGATGAGATGTG-3′ |
| MGB lucprobe | 5′-CAGTCGATGTACACGTTC-3′ |
Western blot analysis
Transfected cells were harvested in 100 μl of 1× Passive Lysis Buffer (Promega) and centrifuged at 11 000 g for 1 min. The cell extracts (15 μl) were separated by SDS–PAGE on a 10% (w/v) polyacrylamide gel and electrophoretically transferred to a Hybond-P (Amersham Pharmacia) membrane in 24.8 mM Tris, 192 mM glycine, 15% methanol. Membranes were blocked for 1 h at room temperature with 5% non-fat milk powder solution in 1× TBS buffer, pH 7.6 (20 mM Tris base, 137 mM NaCl, 3.8 mM HCl) containing 0.1% Tween-20. Then blots were incubated with the anti-luciferase antibody (1/1000) in TBS for 1 h at room temperature, then washed four times in TBS containing 0.1% Tween-20, and incubated with anti-rabbit antibody (1/10 000) for 1 h at room temperature. The blots were then treated with the ECL Plus western blotting reagent (Amersham Pharmacia) and exposed to Hyperfilm™ MP X-ray film (Amersham Pharmacia).
RESULTS
The first uORF in the HBV pgRNA is highly conserved in all HBV subtypes
The pgRNA, which encodes the P gene is preceded by an overlapping C gene and the epsilon stem loop structure in its 5′ end (Figure 1A). Our detailed analysis of this region revealed a small uORF that was previously unreported, which overlaps the C ORF. The presence of this uORF within the pgRNA prompted us to examine the sequences from other HBV variants. The alignment of representative HBV sequences showed that all the HBV subtypes encoded a 19 amino acid peptide at the same position in the pgRNA (Figure 1C). We named this coding sequence the C0 ORF. The C0 initiation codon is the first in the pgRNA [base 35, sequence and numbering according to DDBJ accession no. AB037684, Figure 1A, 1852 according to the numbering in (30)]. It has a context expected to direct poor initiation as there are pyrimidines at −3 and +4 flanking this initiation codon in all subtypes (T/CT/AT/C ATG T, Figure 1C) (31,32).
This uORF is highly conserved in its position and length (Figure 1C), suggesting a functional role in HBV translation. However, this region spans the encapsidation signal (epsilon, ε, 30–90) required for packaging of the pgRNA into mature virions and this is highly conserved (30). It also encodes C in the −1 frame relative to the uORF. Although highly conserved amongst the HBV genotypes at the DNA level, several variations were observed. This included an additional ATG codon upstream and in-frame to the C0 ATG codon, also in a weak context (CTCATGT) in genotype B and C. Another variation was an insertion of 36 bases resulting in a longer C0 ORF of 32 codons, found only in the newly classified G genotype. The C0 ORF is also conserved in the genome of all orthohepadnaviridae members (Figure 1C). Despite its conserved presence in the HBV pgRNA, previous studies have either omitted the C0 ATG codon and its ORF in their test constructs, or not considered its role (5,6,26,28).
Initiation at the C0 AUG codon was detected using a reporter system in cultured human hepatoma cells
A system was constructed which models the human pgRNA, in which the P gene is replaced by the firefly luciferase (FLUC) reporter gene. The constructs contain all or part of the 495 base P leader and the 3′ UTR and polyadenylation signal of the pgRNA (Figures 1A and 2A, PLUC). Synthesis is directed in transfected human hepatoma (HepG2) cell lines by the SV40 promoter and in vitro by the T7 promoter. As controls, a renilla luciferase (RLUC) construct was cotransfected and luciferase mRNA quantitated by RT–PCR.
As an initial test of the ability of the C0 initiation codon to function, constructs containing the pgRNA leader sequence with the entire C0 ORF, which also incorporates the epsilon structure (94 nt, C0LUC, Figure 2A) and from the transcript start to the C0 AUG codon (35 nt, C0AUGLUC, Figure 2A) were made. In order to compare expression to that of C and P, constructs with their upstream leader sequences were also generated (CLUC and PLUC, Figure 2A). As a control, a derivative of PLUC was generated with all the AUG codons in the pgRNA leader altered to AAG rendering the luciferase gene without an initiation codon (UKOLUC). These knockouts will remove AUG initiation codons, but rare non-canonical initiations could still be used. Results are presented as the percentage activity of C after normalization to RLUC activity and mRNA quantity to control for transfection efficiency and effects on mRNA stability (Figure 2B). However, mRNA stability was not affected and these normalizations have little effect on the data.
The C0 initiation codon is the first AUG codon in the transcript (C0LUC) and gave 18% (normalized to RNA) or 20% (normalized to RLUC) of the initiation from the C initiation codon (CLUC, Figure 2B). When the epsilon structure is removed (C0AUGLUC) initiation rose to 60% (normalized to RNA) and 70% (normalized to RLUC). Western analysis using an antibody to luciferase showed equivalent changes in protein levels (Figure 2C, lanes 1–3). Expression from the P initiation codon was 11% of C consistent with previous findings (26). The negative control constructs (e.g. UKOLUC, RLUC, MOCK) gave negligible luciferase expression as expected (Figure 2B and C).
These constructs were also used as templates to generate capped transcripts in vitro and translated in rabbit reticulocyte lysate. The translated lysates were separated on SDS–PAGE and subjected to fluorography and luciferase assays (Figure 2D). The expression of C0LUC in vitro was similar to that in HepG2 cells. Similar results were also obtained in Hep3B cells and COS-7 cells (data not shown).
Capped RNA was prepared in vitro from the CLUC, C0LUC and pgPLUC constructs and electroporated into HepG2 cells. These mRNAs have a transcript start site identical to that of the HBV pgRNA. Activities for C0LUC were 7.5% ± 2.5% of CLUC, pgPLUC had 15% ± 0.5% (see Materials and Methods). In COS-7 cells, activities were approximately 3- to 7-fold higher, with similar ratios, C0LUC 7.5% ± 1%, and pgPLUC 12% ± 2% of CLUC. This indicates that the C0 initiation codon can be utilized near to the beginning of the mRNA (34 base leader) and in the presence of the epsilon sequence, albeit lower than the DNA constructs.
A polyclonal antibody was raised to a synthetic C0 peptide to detect the C0 peptide and C0 fusions. The luciferase fusion protein (C0LUC) could be detected in lysates (Figure 2E, lane 2). The predicted 2 kDa C0 peptide was however undetectable (e.g. lane 5, Figure 2E) in transfected cells without the stabilizing luciferase, although the synthetic peptide is readily detected in lane 4. A DNA construct with an optimal C0 initiation context (C0d3PLUC, lane 6, Figure 2E) also produced undetectable peptide, as did a cell line with higher expression, COS-7 (data not shown).
Determination of the initiation efficiency of each of the upstream AUG codons in the pgRNA leader in a controlled context
To understand how the translation of this first initiation codon can affect subsequent initiation events, we first characterized the relative efficiency at all the initiation codons. These efficiencies are an important feature of previous leaky scanning models although not measured individually in those studies (5,6,26,28). The level of initiation is likely to be inversely related to the level of leaky scanning. The relative strengths of the initiation codons were tested by inserting −6 to +6 flanking sequences in-frame with the luciferase gene. Each initiation codon was preceded by the same leader sequence of sufficient length (31 bases) to direct efficient initiation (33). In HepG2 cells, the respective initiation levels were generally consistent with the order predicted by the consensus around RefSeq initiation codons, except for J (Table 4). The most effective context was AACAUGG from C2 assigned 1.00, which has one of the closest matches to the optimal context. This is followed by C and J at 0.87 and 0.69 both consisting of purines in the −3 position, a requirement for strong initiation. J has poor matches in less significant positions, giving it an overall poor match (0.16). Next were P, C1 and the weakest, C0 (0.32). The P initiation context initiated at 0.56 relative to C2 despite lacking purine resides in the −3 and +4 positions; however, it matches well to the consensus in other position (score 0.51). The C0 initiation codon has a context associated with very weak initiation and is likely to facilitate the highest level of leaky scanning, to enable synthesis from the C and P initiation codons on the pgRNA.
Table 4.
Strength of each initiation context within the HBV pgRNA in HepG2 cells
| Sequence context surrounding ATG codon (−6 to +6) | Predicted initiation strength | Strength of each context tested in HepG2 cells | |
|---|---|---|---|
| Human RefSeq Consensus | GCCGCCATGGCC | 1.00 | |
| C0AUG | TTGTACATGTCC | 0.41 | 0.32 ± 0.05 |
| CAUG | TGGGGCATGGAC | 0.79 | 0.87 ± 0.08 |
| C1AUG | GAATTGATGACT | 0.33 | 0.44 ± 0.05 |
| JAUG | TCAATTATGTTA | 0.16 | 0.69 ± 0.06 |
| C2AUG | ACTAACATGGGT | 0.77 | 1.00 ± 0.02 |
| PAUG | CACCAAATGCCC | 0.51 | 0.56 ± 0.04 |
| CAUGKO | TGGGGCAAGGAC | 0.00 | 0.00 ± 0.00 |
The key nucleotides at positions −3 and +4 relative to the start codon (underlined) are represented in bold. Matches to an information content scoring matrix derived from 16 232 human RefSeq initiation codons are scored 0–1. When tested in HepG2 cells, the relative strength of each initiation context are represented in arbitary units relative to the C2 AUG context (designated 1.00). Note that sequences within the coding region could also affect protein stability or activity.
Determination of the initiation efficiency of each of the upstream AUG codons within the pgRNA leader
Next, we determined the ability of scanning ribosomes to initiate at each internal initiation codon with their respective upstream leader sequences (Figure 3A). According to the scanning model, the expression level from subsequent AUG codons should decrease with the increasing number of upstream AUGs. The C0 initiation context was 18% as efficient as C in this context (C0LUC, Figure 3B). The J AUG codon allowed efficient expression at 60% relative to C, a high level considering there are three preceding AUGs (see later). Initiation from the P AUG codon gave only 10% of C. Therefore, as expected, the presence of upstream AUGs caused the P AUG codon to initiate at a significantly lower level; however, J is not severely affected. When all the upstream AUG codons preceding the P initiation codon were removed (uAUGKOPLUC), the initiation level at this internal P AUG codon was similar to that of C, confirming the inhibitory roles of these upstream AUG codons (83 ± 14% in HepG2, 97 ± 9% in Huh-7 cells, normalized to RLuc). The P initiation codon was also mutated to AAG (construct PKOLUC, Figure 3B) abolishing activity indicating there is not a background due to initiation from internal AUG codons within the fluc gene.
Figure 3.
The expression levels from each respective initiation codon within the HBV pregenomic RNA. (A) A schematic diagram of the HBV test constructs used to study the levels of initiation from C0, C, J and P initiation codons. Each initiation codon is fused in-frame to the luciferase gene. The initiation codon with a darker font is in-frame to the luciferase reporter gene and an asterisk (*) indicates the position where a mutation is introduced. (B) Normalized expression level from each initiation codon as compared with that from CLUC, assigned as 100%. These results are averages from two independent experiments.
The extended mRNA pcPLUC, models the pcRNA. This does not express P as it contains the PC initiation codon, in an optimal context and first in the pcRNA, which primarily directs synthesis of the precore protein (5,26,34,35). As expected, the pcPLUC construct directed very little expression from the internal P AUG codon (3%) indicating that the PC initiation codon is not very leaky, unlike those of the pgRNA leader. Initiation at P from this DNA construct was typically <20% of pgRNA constructs. In addition, electroporation of capped pcPLUC mRNA gave only 0–2% of the activity of pgPLUC mRNA when electroporated into HepG2 or COS-7 cells. These data indicate there is not a strong internal ribosome entry site in this sequence. When the PC AUG codon is mutated, expression from P was restored and slightly increased (PCKOpcLUC, 136 ± 5%, normalized to Rluc) relative to PLUC.
An inhibitory effect of the C0 uORF on downstream translation
Since the C0 AUG has been shown to support initiation at appreciable levels, its effect on initiation at C and P was tested. Constructs were made with and without C0 AUG codons (Figure 4A). The translation of C was enhanced (140%) when the C0 AUG was mutated (C0KOCLUC, Figure 4B). This increase was also evident in the in vitro translated C protein from C0KOPLUC in the fluorographs shown in Figure 5C and D. There was little effect on C1 and C2 initiation. The removal of C0 also caused JLUC expression to increase to 250% (C0KOJLUC, Figure 4B). Fusing the C0 to C had little effect on J expression (C0CJLUC). Extending the C0 so that it terminates at a later point reduces J expression to 70% (C0stopKOJLUC).
Figure 5.
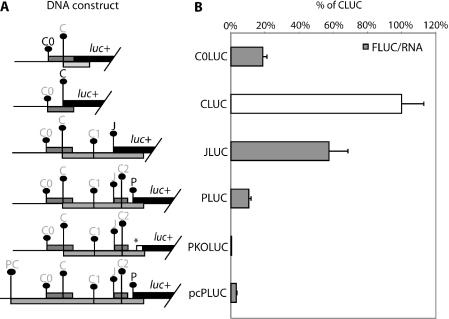
The effect of upstream AUG mutations on the expression at P initiation codon. (A) Schematic diagram of the HBV test constructs used to study the effect of upstream mutation on P expression. The initiation codon with a darker font is in-frame to the luciferase reporter gene and an asterisk (*) indicates the position where a mutation is introduced. (B) Result of DNA transfection of the above constructs into HepG2 cells. The normalized luciferase activity of each construct with upstream mutations is compared with the parent PLUC construct. Average luciferase counts for PLUC were 12 545 ± 141 RLU (relative light unit) in 103 HepG2 cells, which were normalized against respective RNA levels as determined by real-time PCR. These results are averages of two replicates from two independent experiments. (C) Fluorograph of the proteins translated in wheat germ extract in the presence of [35S]methionine from representative in vitro synthesized cap RNA. Proteins were separated on a 5–20% SDS–polyacrylamide gel. (D) Fluorograph of the proteins translated in rabbit reticulocyte lysate. Proteins were separated on a 5–20% SDS–polyacrylamide gel.
The influence of AUGs in the pgRNA on P translation
The P initiation codon is the fifth in the pgRNA (construct PLUC, Figure 5A). To characterize the usage, effect and interplay between these upstream AUGs on P, translation mutations to each of these initiation codons were made (Figure 5A).
Removal of C0 by an AUG to AAG mutation led to an increase (170%) in translation from the P initiation codon (Figure 5B) implying C0 has an inhibitory role. This increase was also observed in vitro (Figure 5D, lane 2). To further examine the mechanism of this effect, we lengthened the C0 uORF through the removal of two subsequent stop codons. This resulted in a slight decrease relative to PLUC (C0stopKOJLUC, Figure 5B). The C0 ORF was also fused in-frame to the C ORF (C0CPLUC, Figure 5A) resulting in the lengthening of this uORF, which also resulted in a small decrease in the expression from P (Figure 5B). The fusion protein (C0C) could be detected on the fluorograph of in vitro translated protein (wheat germ extract, Figure 5C and rabbit reticulocyte lysate, Figure 5D). Wheat germ extracts are very cap dependent and the activity of luciferase from PLUC, its derivatives, or pcPLUC were lower than observed in cells or rabbit reticulocyte lysate. pcPLUC was only 1 ± 0.5% that of CLUC in wheat germ extract, suggesting that the contribution of an IRES is minimal in this system.
Removal of C also resulted in increase in translation of P (142%) (CKOPLUC, Figure 5B). This was also seen in vitro (Figure 5D, lane 4). In addition, a protein corresponding in size to initiation at C1 was expressed at a higher level (Figure 5C, lane 4 and Figure 5D, lane 4). Removal of the C1 initiation codon resulted in an increase in P (132%, Figure 5B). Conversely, when C1 is strengthened to an optimal context, its inhibitory effect increased, reflected by a marked reduction of P initiation (21%). Removal of the inhibitory C2 resulted in a large increase in P (280%). Previous studies (6,28) have reported the importance of the J ORF in facilitating reinitiation at downstream P and our study revealed that the removal of J AUG codon led to a marked decrease in initiation at P (18%). Fusion of the uORF to C ORF (C0CPLUC) led to an insignificant change in downstream P initiation.
A role in reinitiation at the J and P AUGs for the C0 uORF
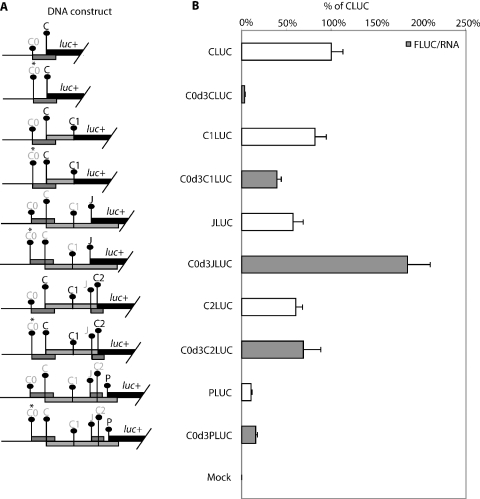
Our results have shown that the removal of the C0 uORF led to an increase in P expression suggesting a mainly inhibitory role. However, uORFs may also be involved in reinitiation following termination at their stop codons. As initiation at C0 and P is low, the effects on reinitiating ribosomes are difficult to detect and confused by simultaneous changes to leaky scanning. We therefore increased expression of C0 by optimizing the initiation context, in order to detect effects on reinitiation.
When the C0 AUG codon was put in an optimal context (Figure 6A), it markedly reduced C translation to 4% (C0d3CLUC, Figure 6B). Less inhibition was observed at C1 with a reduction of only half (82–39%, C0d3C1LUC). In contrast, an increase in initiation was observed at J (3-fold, C0d3JLUC) and P (1.5-fold, C0d3PLUC). This increase in initiation is suggestive of reinitiation occurring at J and P from ribosomes terminating the C0 uORF. The C2 initiation codon, located after J did not show an increase (C0d3C2LUC), thus the increase in P could be a consequence of the large increase in initiation/reinitiation at the J uORF.
Figure 6.
The effect of an optimal C0 initiation context on the expression at downstream initiation. (A) Schematic diagram of the HBV test constructs used to study the effect of an optimal C0 initiation context. The initiation codon with a darker font is in-frame to the luciferase reporter gene while an asterisk (*) indicates the position where a mutation is introduced. (B) Result of DNA transfection of the above constructs into HepG2 cells. The normalized luciferase activity of each construct with the improved C0 initiation context is compared with their corresponding counterpart without C0 context enhancement. Normalized expression levels are presented as a percentage of the CLUC construct, assigned 100%. These results are averages of two replicates from two independent experiments.
DISCUSSION
Our studies indicate a role for the C0 ORF in reducing initiation at the core initiation codon in the pgRNA of HBV and of facilitating polymerase initiation via a novel reinitiation step. The mechanism of P synthesis in HBV has been extensively studied. In the previously proposed model, most ribosomes would initiate at C, some would scan past the C and C1 initiation codons, initiate at J, translate the small J peptide, terminate and then reinitiate at P (6,28). However, these studies did not consider the first AUG in the mRNA, an upstream AUG (coined C0 in this study) and its associated uORF in the model. Here, we have shown that the C0 ORF within the HBV pgRNA leader is highly conserved, although this may in part be due to the highly conserved nature of this key region of the pgRNA, e.g. the epsilon structure. Another feature is the conserved critical flanking nucleotides at position −3 (T/C) and +4 (T) indicating the weak context of its initiation site is maintained.
The use of this C0 initiation site was confirmed by translation of the C0LUC fusion protein in HepG2 cells (Figure 2B and E) and in vitro (Figure 2D). Due to its weak initiation context, C0 AUG codon was used at ∼20% the efficiency of C in HepG2 cells (7–9,31). The higher initiation level at the downstream C AUG codon implies that the C0 AUG codon was bypassed via a leaky scanning mechanism (Figure 2B). Despite the observation of the C0–LUC fusion protein, the 2 kDa C0 peptide was not detected (Figure 2E). To date, very few small uORF peptides have been detected, despite many uORFs having been shown to have regulatory roles (9,17,21,36). This may be due to instability, e.g. the reported avian retroviral uORF peptide is rapidly degraded during in vitro translation with a half-life of only 2.6 min (36).
The C0 initiation codon reduced downstream initiation, particularly at the C initiation site. The removal of the C0 AUG (C0KOCLUC) caused a 1.4-fold increase in expression from the C AUG codon (Figure 4B) and strengthening C0 expression a 20-fold reduction (Figure 6B, C0d3LUC). The translation of C could be affected by C0 in several ways in vivo. First, small subunit complexes translating the C0 ORF would not scan to C. Second, the translation of C0 ORF may block initiation at C, as C0 ribosomes will be terminating in this region. In addition, ribosomes translating the uORF would also physically block scanning to later initiation codons. Taken together, these results suggest that C0 has a role in repressing C protein synthesis.
The location of this uORF in the epsilon encapsidation signal suggests a potential role in regulating RNA packaging, as translating ribosomes would melt the structure. Our data indicate competition between the structure and uORF translation as its presence reduces translation 3-fold to 20% of C (Figure 2B). In several other viruses, translation initiation at upstream AUG codons affects encapsidation of the viral genome (17). In HBV, the extended form of the pgRNA, the pcRNA transcript, is not packaged into virions, even though it includes the encapsidation signal also found in the pgRNA (37). In pcRNA translation, initiation at the PC initiation codon and the elongation for at least 13 codons prevents encapsidation (37). It was suggested that ribosomes translating through the HBV epsilon packaging signal disrupt the interaction between the signal and encapsidation proteins. Therefore, translation from the weak C0 AUG within the encapsidation signal may also regulate pgRNA packaging during HBV replication. This could present a system to prevent encapsidation until the levels of core and polymerase are high enough to outcompete translation.
The role of C0 uORF in reinitiation was established using constructs with optimized C0 initiation context. This would be expected to abolish leaky scanning and it did at C. However, there was an increase in initiation at J and P (Figure 6B) suggesting reinitiation following C0 termination. For the type member of the related avihepadnaviridae family, duck hepatitis B virus (DHBV), it had been suggested that 13 initiation codons were apparently bypassed by an IRES mechanism (25). However, a more recent study now indicates that a shunting mechanism is used (38). Ribosomes initiate at the 5′ Cap and reinitiate at the P codon [designated P1 in (38)]. This is likely to be due to a discontinuous scanning process, as structures and initiation codons in the bypassed region did not affect P initiation. It was unclear by what mechanism ribosomes shunted. However, the more distantly related caulimovirus family uses a well-characterized shunt after translation of a uORF (39).
We analysed the upstream leader of DHBV and the avihepadnaviridae for uORFs that may facilitate such shunting. There are two unreported conserved uORFs, in different frames to C, in all avihepadnaviridae. One of these has the context GGT ATG T (start 2616), and the other TTT ATG A (start 2630). It would be surprising if these were not utilized as initiation codons. The first would terminate just before the C (C1) ATG and the second soon after. These uORFs may play a similar role in DHBV virus to C0, providing a pool of ribosomes for reinitiation at P.
HBV C0 has two roles in regulating expression. First, being upstream and overlapping the C ORF, its translation slightly represses C expression, hence also affecting P. Second, it could facilitate P expression through reinitiation from J. The J uORF in the HBV pgRNA has previously been shown to be important for reinitiation at the P gene (6,28). P gene translation was mostly carried out by ribosomes that reinitiated after termination at J ORF (74%) (6). Our data are consistent with this model, as it showed that most of P initiation was from translation of the J ORF (Figure 5B, JKOPLUC).
Our results suggest a revision to the model by which the P protein is synthesized from the pgRNA (6,28). Subunits would assemble at the cap, then scan. About one-fifth of scanning subunits initiate at the C0 AUG, most would scan past and synthesize the core protein from C, with some scanning past the C and C1 initiation codons to initiate at J, translate it, then reinitiate at P to synthesize the polymerase. Reinitiating ribosomes from the C0 uORF would also contribute to the pool of ribosomes initiating at J. Thus, C0 has a pivotal role in reducing the synthesis at C to achieve a 1:10 ratio of P to C. This model is similar but more elaborate than that recently proposed for DHBV. However, we have not determined whether the bypass is by continuous or discontinuous scanning in HBV, whereas for DHBV the role of the uORFs has not been tested.
This examination of the role of the C0 initiation codon indicates the complexity of a system required to bypass multiple initiation codons. It also suggests C0 may influence the function of the epsilon element in the HBV genome.
Acknowledgments
We thank T. Hohn (Basel) for the discussion of preliminary data from this study. Funding was provided by a NZ Health Research Council grant to Warren Tate and C.M.B., NZ Lotteries Health grants to C.M.B., and a UO Postgraduate Scholarship to A.C. Funding to pay the Open Access publication charges for this article was provided by the NZ Health Research Council.
REFERENCES
- 1.Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin R., Locarnini S. Treatment of chronic hepatitis B: current challenges and future directions. Rev. Med. Virol. 2003;13:255–272. doi: 10.1002/rmv.393. [DOI] [PubMed] [Google Scholar]
- 3.Feld J., Lee J.Y., Locarnini S. New targets and possible new therapeutic approaches in the chemotherapy of chronic hepatitis B. Hepatology. 2003;38:545–553. doi: 10.1053/jhep.2003.50389. [DOI] [PubMed] [Google Scholar]
- 4.Ganem D., Varmus H.E. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 5.Ou J.H., Bao H., Shih C., Tahara S.M. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J. Virol. 1990;64:4578–4581. doi: 10.1128/jvi.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang W.L., Su T.S. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol. 1998;79:2181–2189. doi: 10.1099/0022-1317-79-9-2181. [DOI] [PubMed] [Google Scholar]
- 7.Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 9.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peri S., Pandey A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 11.Rogozin I.B., Kochetov A.V., Kondrashov F.A., Koonin E.V., Milanesi L. Presence of ATG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with ‘weak’ context of the start codon. Bioinformatics. 2001;17:890–900. doi: 10.1093/bioinformatics/17.10.890. [DOI] [PubMed] [Google Scholar]
- 12.Pesole G., Gissi C., Grillo G., Licciulli F., Liuni S., Saccone C. Analysis of oligonucleotide AUG start codon context in eukariotic mRNAs. Gene. 2000;261:85–91. doi: 10.1016/s0378-1119(00)00471-6. [DOI] [PubMed] [Google Scholar]
- 13.Wang X.Q., Rothnagel J.A. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meijer H.A., Thomas A.A. Ribosomes stalling on uORF1 in the Xenopus Cx41 5′ UTR inhibit downstream translation initiation. Nucleic Acids Res. 2003;31:3174–3184. doi: 10.1093/nar/gkg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak M. Emerging links between initiation of translation and human diseases. Mamm. Genome. 2002;13:401–410. doi: 10.1007/s00335-002-4002-5. [DOI] [PubMed] [Google Scholar]
- 16.Vilela C., McCarthy J.E. Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol. Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 17.Geballe A.P., Sachs M.S. In: Translational Control of Gene Expression. 2nd edn. Sonenberg N., Hershey J.W.B., Mathews M., editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 595–614. [Google Scholar]
- 18.Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poyry T.A., Kaminski A., Jackson R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 1996;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale M., Jr, Tan S.L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas J., Park E.C., Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 23.Moffat J.G., Tate W.P., Lovett P.S. The leader peptides of attenuation-regulated chloramphenicol resistance genes inhibit translational termination. J. Bacteriol. 1994;176:7115–7117. doi: 10.1128/jb.176.22.7115-7117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong F., Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 25.Chang L.J., Ganem D., Varmus H.E. Mechanism of translation of the hepadnaviral polymerase (P) gene. Proc. Natl Acad. Sci. USA. 1990;87:5158–5162. doi: 10.1073/pnas.87.13.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C.G., Lo S.J. Evidence for involvement of a ribosomal leaky scanning mechanism in the translation of the hepatitis B virus pol gene from the viral pregenome RNA. Virology. 1992;188:342–352. doi: 10.1016/0042-6822(92)90763-f. [DOI] [PubMed] [Google Scholar]
- 27.Schlicht H.J., Radziwill G., Schaller H. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell. 1989;56:85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- 28.Fouillot N., Tlouzeau S., Rossignol J.M., Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peabody D.S. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 30.Flodell S., Cromsigt J., Schleucher J., Kidd-Ljunggren K., Wijmenga S. Structure elucidation of the hepatitis B virus encapsidation signal by NMR on selectively labeled RNAs. J. Biomol. Struct. Dyn. 2002;19:627–636. doi: 10.1080/07391102.2002.10506769. [DOI] [PubMed] [Google Scholar]
- 31.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1991;1:117–125. [PMC free article] [PubMed] [Google Scholar]
- 34.Fouillot N., Rossignol J.M. Translational stop codons in the precore sequence of hepatitis B virus pre-C RNA allow translation reinitiation at downstream AUGs. J. Gen. Virol. 1996;77:1123–1127. doi: 10.1099/0022-1317-77-6-1123. [DOI] [PubMed] [Google Scholar]
- 35.Hwang W.L., Su T.S. The encapsidation signal of hepatitis B virus facilitates preC AUG recognition resulting in inefficient translation of the downstream genes. J. Gen. Virol. 1999;80:1769–1776. doi: 10.1099/0022-1317-80-7-1769. [DOI] [PubMed] [Google Scholar]
- 36.Hackett P.B., Petersen R.B., Hensel C.H., Albericio F., Gunderson S.I., Palmenberg A.C., Barany G. Synthesis in vitro of a seven amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J. Mol. Biol. 1986;190:45–57. doi: 10.1016/0022-2836(86)90074-4. [DOI] [PubMed] [Google Scholar]
- 37.Nassal M., Junker-Niepmann M., Schaller H. Translational inactivation of RNA function: discrimination against a subset of genomic transcripts during HBV nucleocapsid assembly. Cell. 1990;63:1357–1363. doi: 10.1016/0092-8674(90)90431-d. [DOI] [PubMed] [Google Scholar]
- 38.Sen N., Cao F., Tavis J.E. Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting. J. Virol. 2004;78:11751–11757. doi: 10.1128/JVI.78.21.11751-11757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryabova L., Park H.S., Hohn T. Control of translation reinitiation on the cauliflower mosaic virus (CaMV) polycistronic RNA. Biochem. Soc. Trans. 2004;32:592–596. doi: 10.1042/BST0320592. [DOI] [PubMed] [Google Scholar]