Abstract
DNA mismatch repair in Escherichia coli has been shown to be involved in two distinct processes: mutation avoidance, which removes potential mutations arising as replication errors, and antirecombination which prevents recombination between related, but not identical (homeologous), DNA sequences. We show that cells with the mutSΔ800 mutation (which removes the C-terminal 53 amino acids of MutS) on a multicopy plasmid are proficient for mutation avoidance. In interspecies genetic crosses, however, recipients with the mutSΔ800 mutation show increased recombination by up to 280-fold relative to mutS+. The MutSΔ800 protein binds to O6-methylguanine mismatches but not to intrastrand platinated GG cross-links, explaining why dam bacteria with the mutSΔ800 mutation are resistant to cisplatin, but not MNNG, toxicity. The results indicate that the C-terminal end of MutS is necessary for antirecombination and cisplatin sensitization, but less significant for mutation avoidance. The inability of MutSΔ800 to form tetramers may indicate that these are the active form of MutS.
INTRODUCTION
The Dam-directed mismatch repair (MMR) system of Escherichia coli removes potential mutations arising as replication errors (mutation avoidance) (1–3). Correction of biosynthetic errors by MMR occurs directly after replication, just behind the replication fork, where the parental DNA strand is fully methylated at GATC (dam) sequences and the newly synthesized strand is not yet methylated. When base mispairs arise in such hemi-methylated DNA, they are bound initially by the MutS protein which subsequently recruits MutL and MutH to form a ternary complex. Incision by activated MutH occurs on the unmethylated strand at a nearby GATC sequence, followed by excision, in either the 3′ or 5′ direction, of the mismatched base pair and surrounding sequence. Re-synthesis by the replicative polymerase, DNA polymerase III, restores the correct nucleotide sequence and the resulting nick is sealed by DNA ligase to complete the repair process. Subsequently, the repaired DNA strand is methylated at GATC sequences by Dam methyltransferase and this methylation prevents further MMR action. This model has both genetic and biochemical support including a mutator phenotype associated with mutS mutants (1,2).
MMR also plays a role in preventing recombination between related, but not identical (homeologous), DNA sequences (anti-recombination) (4). Genetic crosses between E.coli and Salmonella enterica Serovar Typhimurium are sterile unless the recipient contains mutations in the mutS or mutL MMR genes. This result suggests that MMR either impedes or actively reverses recombination intermediates or destroys them, with the former having some experimental support (5–7). On a biochemical level, MutS and MutL block RecA-mediated strand exchange between the fd and M13 genomes, which are 3% divergent, but not between M13–M13 genomes (8). In the homeologous reaction, MutS has a greater effect than MutL, which is effective only in combination with MutS. The intimate connection between mutation avoidance and antirecombination is shown by the phenotype of E.coli mutS mutations which are defective in both these processes (4). A survey of a large number of E.coli mutS mutants failed to detect any with only one of the phenotypes (9). Similarly, we have been unable to find a mutant with only one of these phenotypes among a collection of dominant-negative alleles (10).
E.coli dam mutants are more sensitive to the cytotoxic action of cisplatin and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) than wild type (11,12). Mutations in genes that incapacitate MMR (mutS, mutL or mutH) in a dam background confer a level of resistance to these agents similar to that of the wild type. This result indicates that inappropriate MMR on chemically modified DNA can sensitize dam cells to these cytotoxic agents (11–13). MMR in wild-type cells is restricted to one strand in the hemimethylated region behind the replication fork because MutH endonuclease cannot use fully methylated DNA as a substrate. In dam mutants, however, MMR can occur on either strand anywhere on the chromosome because MutH can use unmethylated DNA as a substrate. It has been proposed that MMR-induced single-strand breaks or gaps can be converted to double-strand breaks (DSBs) either by MutH action at the same GATC sequence on the opposite strand (14) or by replication fork collapse (15,16). The DSBs so formed require recombination for their repair thereby making recombination essential for viability (15). As a result of these DSBs, recombination capacity is limiting (15) and when dam cells are exposed to cisplatin a large number of DSBs accumulate (16). MMR is required for the formation of cisplatin-induced DSBs (16) suggesting that they probably occur at cisplatin lesions undergoing MMR at either unreplicated lesions or those formed with mismatches after the action of translesion polymerases. The accumulation of DSBs in dam mutants, which have limited recombination capacity, and the reduced ability of MutS to prevent RecA-mediated strand transfer of platinated DNA during recombinational repair (17) provides a plausible model to explain why MMR sensitizes dam cells to cisplatin.
To explain MMR sensitization of E.coli dam mutants by MNNG, the ‘futile cycling’ model was proposed (11). In this model, the replicative polymerase inserts either a T or C opposite template O6-methylguanine (O6-meG). Since neither of these bases is considered a ‘good’ match by the MMR system, a futile cycle of insertion and removal ensues preventing progression of the replication fork. Such futile cycling in dam mutants can also occur at sites away from the replication fork as a consequence of MMR recognition and MutH incision on unmethylated DNA at O6-meG–C base pairs to promote DSB formation (13).
The MutS protein binds specifically to heteroduplex DNA containing base mismatches (18) or with greatest affinity to small insertion/deletion loops (IDLs) (19). It also binds platinated GG intrastrand cross-links (20) and O6-meG mispaired with either C or T (21). The crystal structure of E.coli MutS bound to an oligonucleotide with a G–T mismatch has been solved using a derivative of the MutS protein, MutSΔ800, which lacks the C-terminal 53 amino acids (22). The MutSΔ800 mutant crystallizes as a dimer and retains the ability to bind DNA and ATP, just as full-length MutS does (22). Further analysis into the properties of the MutS and MutSΔ800 proteins by equilibrium sedimentation and gel filtration show that MutS dimers can assemble into higher order oligomeric structures, while the MutSΔ800 mutant is restricted to dimer formation only (23). The ability of MutS to form tetramers suggests this oligomeric state is important in MutS function. A similar conclusion was reached with the MutS protein from Thermus species (24,25) for which a crystal structure is also available (26).
In this report, we show that the C-terminal end of MutS is critical for antirecombination and cisplatin sensitization, but less significant for mutation avoidance.
MATERIALS AND METHODS
Strains, plasmids and media
Strains GM3819 (Δdam-16::Kan), GM4799 (mutS458::mTn10Kan), GM5550 (recA56 mutS458::mTn10Kan) and GM5556 (Δdam-16::Kan mutS215::Tn10) are derivatives of AB1157 (thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1). AB259 (HfrH) was obtained from E.A. Adelberg (Yale University). Salmonella strains SA536 and SA977 were obtained from the Salmonella Genetic Stock Centre, University of Calgary, Canada. Plasmids with the mutSΔC800 (22) (referred to as mutSΔ800 in this paper) and mutSΔ680 (27) were derived from pMQ372 (27) carrying the mutS+ gene. Full descriptions of strains and plasmids can be found at http://users.umassmed.edu/martin.marinus/dstrains.html. Bacteriological media have been described previously (15,28) except for the MacConkey Agar Base (Difco) medium which was supplemented with 50 g/l galactose. Ampicillin, rifampicin and streptomycin were included in media, when required, at 100 μg/ml.
Estimation of mutant frequency, conjugational crosses and cytotoxicity
Spontaneous mutant frequencies were measured as described elsewhere (28,29) and conjugation experiments were performed as described previously (15) except that mating mixtures were incubated for 1 h at 37°C for homologous crosses and 3 h for the homeologous crosses. Cell survival after exposure to cisplatin or MNNG was measured as described previously (11,29).
Proteins and DNA
MutS protein was purchased from USB and dialyzed before use against 20 mM KPO4, pH 7.4, 0.1 mM EDTA, 1 mM PMSF and 10 mM beta-mercaptoethanol. We found that heteroduplex binding (see below) and ATPase specific activity (data not shown) were the same as that for the MutSΔ800 protein. MutL protein was a gift from F. Lopez de Saro and M. O'Donnell (Rockefeller University). MutSΔ800 was purified from strain GM7854 [pmutSΔ800/BL21(lambdaDE3)]. Briefly, the cells were grown at 37°C to an OD600 of 0.8 and shifted to 26°C for induction with 1 mM IPTG. The cells were grown for an additional 3 h at 26°C before harvesting. The cells were lysed using a Microfluidizer (Microfluidics Corp.) and then centrifuged at 27 000 g for 30 min. The supernatant was treated with streptomycin sulfate and ammonium sulfate as described previously (18). The resulting fraction was loaded onto a heparin agarose column (Amersham Pharmacia) and eluted with a linear gradient of 100–400 mM KCl. The fractions containing MutS were subsequently loaded onto a ceramic hydroxylapatite column (Sigma) and eluted using a linear concentration gradient of 20–120 mM KCl. This produced a single MutS peak eluting at 50–70 mM KCl. Fractions containing MutSΔ800 were pooled, concentrated using a Centriprep column (Millipore) and frozen at −75°C. The protein was at least 95% pure as determined by SDS–PAGE analysis. Protein concentration was determined by ninhydrin analysis (30).
M13mp18 DNA RFI and single-stranded circular DNA forms were from New England Biolabs (NEB). The RFI form was digested with DraIII and BglII restriction endonucleases (NEB) to remove lac operon DNA. Single-stranded circular DNA of bacteriophage fd was prepared by growing a culture of strain AB259 to about 1 × 108 cells, adding fd at an m.o.i of 1 and allowing the culture to grow for an additional 3–4 h. The culture was centrifuged to remove bacteria, and 20% polyethyleneglycol 8000 and 2.5 M NaCl solution were added to the supernatant (1:3) and left on ice for 30 min. The mixture was centrifuged at 9500 g for 10 min and the phage pellet resuspended in TE buffer. The DNA was phenol extracted, ethanol precipitated and its concentration determined by UV spectroscopy.
Heteroduplex construction, electrophoretic mobility shift assay and RecA strand exchange assay
Heteroduplex DNA with a single IDL and labeled with P32 was constructed and analyzed by electrophoretic mobility shift assay as described previously (19,27). The oligonucleotide 5′-ATACGGAAGTTAAAGTO6meGCGGATCATCTCTAG-3′ (Sigma-Genosys) was annealed with its complementary sequence to form either O6meG–C or O6meG–T base pairs. A DNA oligonucleotide provided by Jennifer Robbins and J.M. Essigmann (MIT), designated Xlink (5′-CCT CTC CTT GGT CTT CTC CTC TCC-3′) contains a cisplatin cross-link between the two guanines and was annealed to its complementary sequence to form either GG–CC or GG–CT mismatches. The platinated and methylated oligonucleotides were not radiolabeled. The P32-labeled DNA and the methylated and platinated DNA, which were stained with Vistra Green (Amersham-Pharmacia) for 1 h, were quantitated using a Fuji Phosphorimager. RecA-mediated strand transfer was performed and measured as described except that M13 and fd molecules were used in place of the phiX174 molecules used previously (17).
RESULTS
Spontaneous mutation frequency of wild-type and mutS strains
Strain GM4799, a mutS null mutant (27), was transformed with plasmids bearing the mutS+, mutSΔ680 and mutSΔ800 genes. The mutSΔ680 and mutSΔ800 designations indicate where the 853 amino acid MutS protein was truncated. The mutSΔ680 mutation has a null mutator phenotype and the MutSΔ680 protein is unable to form oligomers and has reduced ability to interact with MutL (27). It is used in experiments presented here as the null mutS control. The transformed strains were tested for reversion of the argE3 and galK2 markers of the host strain as well as resistance to rifampicin. The results in Table 1 and Figure 1 show that the GM4799 strain with the mutSΔ800 or mutS+ plasmids did not display a mutator phenotype, but that GM4799 with mutSΔ680 plasmid did. The result with rifampicin resistance for the mutSΔ800 and mutS+ strains confirmed that obtained previously by Lamers et al. (22) and Biswas et al. (31). The reversion to Arg+ and Gal+ papillae formation was marginally higher in the GM4799 with the mutSΔ800 plasmid culture compared to mutS+, but both these values were much lower than in the mutSΔ680 plasmid control strain. We conclude that the mutSΔ800 allele does not confer a mutator phenotype to the cell.
Table 1.
Spontaneous mutant frequencies
| Plasmid allele | Arg+ | RifR |
|---|---|---|
| MutS+ | 4 | 1 |
| mutSΔ680 | 66 | 72 |
| mutSΔ800 | 8 | 1 |
Cultures of strain GM4799 (mutS null mutation) with the indicated plasmid allele were plated on media selective for rifampicin-resistance or arginine prototrophy. The numbers represent mutant colonies per 108 cells plated.
Figure 1.

Gal reversion assay. Dark Gal+ revertants are shown on a background of white Gal− colonies. From left to right: the GM4799 strain (mutS null mutation) with plasmids bearing mutS+, mutSΔ680 and mutSΔ800, respectively.
Interspecies crosses
Strain GM4799 with each of the mutS plasmids was used as a recipient in conjugal crosses with E.coli (homologous) or S.enterica Serovar Typhimurium (homeologous) donors. With the E.coli donor, AB259, recombinants were formed after 60 min of mating, at the same frequency with each GM4799 strain indicating no effect of the mutS alleles on homologous recombination (Table 2). With the Salmonella donors SA536 or SA977, however, the GM4799 recipient with the mutSΔ800 plasmid formed recombinants 283- and 100-fold higher than the wild-type recipient, respectively. These values, however, are about half that for the GM4799 strain with the mutSΔ680 plasmid indicating that the mutSΔ800 allele does not completely relieve the inhibition produced by a fully proficient MMR system. The level of homeologous recombination was the same with either strain GM4799 or GM4799 containing the mutSΔ680 plasmid (data not shown).
Table 2.
Yield of recombinants in homologous and homeologous crosses
| Donor | Plasmid in recipient | Recipient | Selected marker(s) | Frequency | Fold increase |
|---|---|---|---|---|---|
| AB259 | mutS+ | Rec+ | Thr+ Leu+ | 2.8 × 105 | — |
| AB259 | mutSΔ680 | Rec+ | Thr+ Leu+ | 2.5 × 105 | 1 |
| AB259 | mutSΔ800 | Rec+ | Thr+ Leu+ | 2.0 × 105 | 1 |
| SA536 | mutS+ | Rec+ | Arg+ | 120 | 1 |
| SA536 | mutSΔ680 | Rec+ | Arg+ | 59 000 | 492 |
| SA536 | mutSΔ800 | Rec+ | Arg+ | 34 000 | 283 |
| SA977 | mutS+ | Rec+ | Arg+ | 35 | 1 |
| SA977 | mutSΔ680 | Rec+ | Arg+ | 10 665 | 296 |
| SA977 | mutSΔ800 | Rec+ | Arg+ | 3410 | 100 |
| SA536 | mutS+ | recA56 | Arg+ | 30 | 1 |
| SA536 | mutSΔ680 | recA56 | Arg+ | 80 | 2.7 |
| SA536 | mutSΔ800 | recA56 | Arg+ | 60 | 2 |
| SA977 | mutS+ | recA56 | Arg+ | 8 | 1 |
| SA977 | mutSΔ680 | recA56 | Arg+ | 120 | 15 |
| SA977 | mutSΔ800 | recA56 | Arg+ | 85 | 10 |
Donor and recipient (GM4799 and GM5550) cultures (at 1–2 × 108 cells per ml) were mated for 60 min (AB259) or 3 h (SA strains). The numbers in the table represent the Thr+ Leu+ [StrR] or Arg+ [StrR] recombinants in 50 μl of undiluted mating mixture.
To ensure that the results of the crosses described above were due to recombination, we repeated the crosses with a recombination-deficient (recA56) derivative of GM4799, GM5550. With the E.coli donor, AB259, no recombinants were detected in any of the crosses with the GM5550 recipients (data not shown). With the Salmonella donors, there was more than a 99% decrease in recombinant formation in the GM5550 strains with the mutSΔ800 and mutSΔ680 plasmids. We conclude that the mutSΔ800 mutation significantly reduces antirecombination in interspecies crosses in a recA-dependent manner.
Sensitivity to cytotoxic agents
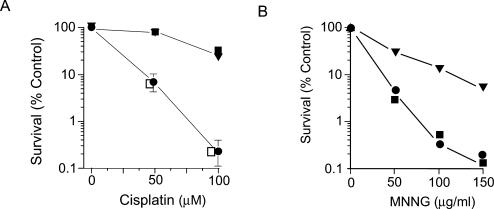
E.coli dam mutants are more sensitive to cisplatin and MNNG than wild type (11,12). Figure 2A shows that a dam mutS strain, GM5556, carrying the mutS+ plasmid is more sensitive to cisplatin than the same strain bearing the mutSΔ680 (null mutation) plasmid or the mutSΔ800 plasmid, indicating that the mutSΔ800 gene product is unable to promote MMR sensitization. Figure 2A also shows that the survival of GM3819, a dam mutS+ strain, with the mutSΔ800 plasmid is the same as with a mutS+ plasmid. This result indicates that the mutSΔ800 allele in multicopy does not have a dominant-negative phenotype in wild-type (mutS+) cells.
Figure 2.
Survival of cells exposed to cisplatin and MNNG. Cell survival of strain GM5556 (Δdam-16::Kan mutS::Tn10) containing plasmids with mutS+ (closed circles), mutSΔ680 (inverted triangles), and mutSΔ800 (closed squares) after exposure to cisplatin (A) and MNNG (B). The open squares in (A) represent the survival of GM3819 (Δdam-16::Kan mutS+) with the mutSΔ800 plasmid.
Figure 2B shows that when exposed to MNNG, the GM5556 strain with the mutS+ plasmid is sensitive but that with the mutSΔ680 plasmid is resistant. In contrast to the results with cisplatin, GM5556 with the mutSΔ800 plasmid has the same survival as the strain with the mutS+ plasmid.
The simplest interpretation of the data in Figure 2 is that while the MutSΔ800 protein is unable to recognize diguanyl platinated cross-links, it can recognize O6-meG mismatches with enough efficiency to produce the same phenotype in cells as MutS.
MutS and MutSΔ800 binding to platinated oligonucleotides
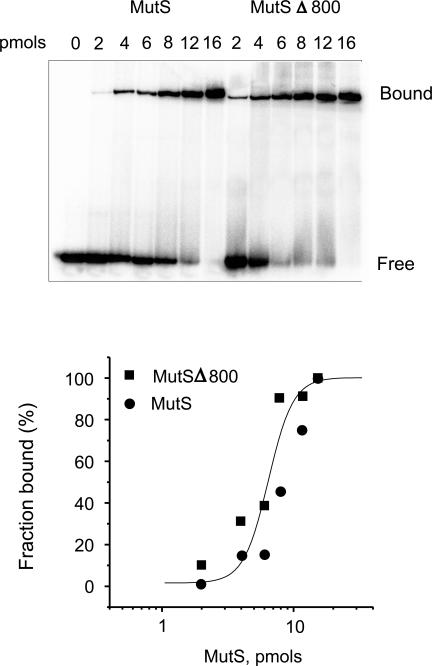
To investigate if MutS and MutSΔ800 have altered binding to cisplatin cross-links, we tested the ability of both proteins to bind to homoduplex, heteroduplex and platinated DNA substrates in an electrophoretic mobility shift assay. The data in Figure 3 show that MutSΔ800 has approximately the same affinity for P32-labeled heteroduplex DNA with a single base IDL as MutS. There was no significant binding of these proteins to a fully base-paired homoduplex substrate under the same experimental conditions (data not shown).
Figure 3.
Binding isotherm of MutS and MutSΔ800 to heteroduplex DNA. The upper panel shows the binding of MutS and MutSΔ800 to a P32-labeled heteroduplex DNA with a single base IDL. The heteroduplex concentration was 0.6 pmol. The data are shown in the lower panel as a binding isotherm.
The substrates for MutS binding to platinated DNA included unplatinated homoduplex DNA, unplatinated heteroduplex DNA with a G–T mismatch, and derivatives of each of these with a single intrastrand diguanyl cross-link (Figure 4). The unplatinated homoduplex acts as a negative control for the diguanyl cross-link paired with CC, and is considered to be a biologically relevant adduct produced from the reaction of cisplatin and DNA. The GG–CT substrate acts as a positive control ensuring that MutS can bind to mismatched DNA in this sequence context. The mobility shifts shown in Figure 4 were monitored by fluorescence after staining with VistraGreen. As expected, there was no detectable band shifting with MutS and MutSΔ800 to unplatinated homoduplex DNA at the concentrations used for specific binding (Figure 4A, lanes 2–5). In contrast to the results in Figure 3, where MutS and MutSΔ800 bind the IDL with near equivalent affinity, we did detect a difference in binding between MutS and MutSΔ800 to the unplatinated heteroduplex containing a G–T mismatch (Figure 4B, lanes 2–5). The addition of 4 μM of MutS produced a discernible retardation of the oligonucleotide (lane 3) while, at the same concentration, MutSΔ800 did not (lane 5). At this concentration, MutS forms two distinct bands (lane 3), only one of which is present with MutSΔ800 (lane 5) and may indicate that MutS first binds as a dimer and subsequently is converted to a tetramer. With 8 μM, the extent of heteroduplex binding by MutSΔ800 was about 80% that of MutS (lanes 2 and 4).
Figure 4.
Binding of MutS and MutSΔ800 to modified DNA. (A) The binding of MutS and MutSΔ800 to unplatinated (lanes 1–5) and platinated homoduplex (lanes 6–10) DNA. No enzyme (lanes 1 and 6); MutS, 8 μM (lanes 2 and 7) and 4 μM (lanes 3 and 8); and MutSΔ800, 8 μM (lanes 4 and 9) and 4 μM (lanes 5 and 10). The DNA was visualized by staining the gel with Vistra Green. (B) The binding of MutS and MutSΔ800 to unplatinated (lanes 1–5) and platinated heteroduplex (lanes 6–10). Protein additions are the same as in (A). (C) The binding of MutS and MutSΔ800 to O6-meG–C (lanes 1–4) and O6-meG–T (lanes 5–8) DNA. MutS, 8 μM (lanes 1 and 5) and 16 μM (lanes 2 and 6); and MutSΔ800, 8 μM (lanes 3 and 7) and 16 μM (lanes 4 and 8).
There was a significant difference in binding of MutS and MutSΔ800 to DNA with a GG cross-link opposite two C residues (Figure 4A, lanes 6–10). In fact, no binding of MutSΔ800 to this DNA was observed (Figure 4A, lanes 9 and 10) although MutS, at the same concentration, did produce a considerable band shift (Figure 4A, lanes 7 and 8). On the other hand, the DNA with a GG cross-link opposite CT residues, a product of translesion polymerase synthesis, was bound by MutSΔ800 to about the same extent as MutS at 4 nM but less so at 8 nM due to partial shifting (Figure 4B, lanes 6–10). With this substrate, the MutS and MutSΔ800 upper bands are at about the same location. This could be due to MutSΔ800 recognizing this substrate as a G–T mismatch and the presence of the platinated guanines or that MutSΔ800 arrays on this substrate to a greater extent than MutS.
The results in Figure 4C show that both MutS and MutSΔ800 were able to bind to a duplex DNA with a single O6-meG–C (lanes 1–4) or O6-meG–T (lanes 5–8). There was no binding to homoduplex DNA at the concentrations used (data not shown). The difference in the binding affinities of MutSΔ800 for the types of heteroduplexes used in Figures 3 and 4 are relevant because the affinity of these proteins for the same substrate can differ due to sequence context or the type of mismatch or the affinity for base mismatches versus a one base IDL.
MutSΔ800, therefore, has impaired specific binding of platinated intrastrand GG–CC cross-links, but not intrastrand GG–CT cross-links or a single base IDL or O6-meG–C or O6-meG–T mismatches. Since MutS can form tetramers while MutSΔ800 cannot, this suggests that the ability to form tetramers could be necessary for efficient MutS processing of the biologically relevant platinated intrastrand cross-link.
RecA-mediated strand exchange in vitro
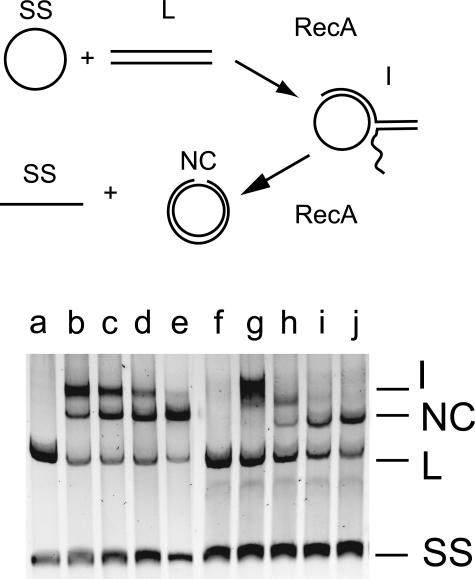
In this assay, the ability of MutS to inhibit RecA-mediated strand transfer between M13–M13 (homologous) and M13-fd (homeologous) phage DNA molecules is measured (8). Figure 5 shows the results of a typical assay. For the M13–M13 homologous substrates (lanes a–e), nicked circle (NC) product and intermediate (I) bands are visible after 5 min (lane b) and these increase and decrease, respectively, during the reaction (lanes c–e). The homeologous M13-fd reaction (lanes f–j), is clearly slower, with no nicked circle product visible at 5 min (lane g) and a lower yield (72%) at 90 min (lane j). The graph in Figure 6 shows that the addition of 100 nM MutS or MutSΔ800 has no effect on the rate or yield of the reaction using the homologous M13–M13 substrates. Addition of 25 or 100 nM MutS, however, reduces exchange between homeologous M13-fd substrates by 48% and 98%, respectively (Figure 6A). When MutSΔ800 is added, the corresponding percentages were 22% and 58%, respectively (Figure 6B).
Figure 5.
RecA-catalyzed strand transfer. The upper panel shows a schematic diagram of the reaction. Single-strand (SS) circular DNA reacts with linear (L) duplex to form intermediate (I) structures which are converted into nicked-circle (NC) products. The fluorograph shows the results from the homologous M13–M13 (lanes a–e) and the homeologous M13-fd (lanes f–j) reactions. Samples were removed at time 0 (lanes a and f), 5 min (lanes b and g), 15 min (lanes c and h), 45 min (lanes d and i) and 90 min (lanes e and j).
Figure 6.
Kinetics of MutS inhibition of RecA-mediated strand exchange. The graphs show the effect of varying concentrations of MutS (A) and MutSΔ800 (B) on RecA strand exchange using homologous (M13–M13) and homeologous (M13-fd). Closed circles, M13–M13; crosses, M13–M13 plus 100 nM protein; closed squares, M13-fd; open squares, M13-fd plus 25 nM protein; open circles, M13-fd plus 100 nM protein; and inverted triangles, M13-fd plus 25 nM protein plus 40 nM MutL.
Worth et al. (8) showed that MutL stimulates MutS binding to M13-fd homeologous duplexes when the latter is at a sub-optimal concentration. In the experiment shown in Figure 6, 25 nM MutS or MutSΔ800 only partially inhibits RecA-catalyzed strand transfer. Inclusion of MutL, at 15 or 40 nM to these reactions, however, completely abolishes RecA transfer activity (Figure 6) indicating that MutL dimers can interact with MutS or MutSΔ800. MutL by itself had no effect on the rate or product yield in the reaction. We showed previously that there was no binding of MutL alone to platinated cross-linked DNA (17).
DISCUSSION
The results described in this report show that MutS and MutSΔ800 proteins, produced from multicopy plasmids, impart different properties to cells. Although cells with the MutSΔ800 or MutS protein are not mutators (Table 1 and Figure 1), the MutSΔ800-containing cells are deficient in antirecombination (Table 2) and not sensitized to cisplatin cytotoxicity (Figure 2A). This is the first demonstration of a separation of function phenotype for an E.coli mutS mutation. We propose that the C-terminal domain of MutS is required for efficient antirecombination and cisplatin sensitization, but not mutation avoidance or MNNG sensitization. Given that MutSΔ800 can only form monomers and dimers, we postulate that the inability to form tetrameric MutS could be responsible for this phenotype. The data reported here also suggests that although MutS and MutSΔ800 may bind base mismatches as dimers, conversion to the terameric form may be required for subsequent reactions in mismatch repair.
The results we have obtained in this report are best explained by the findings of Bjornson et al. (23) who have argued that the MutS tetramer is likely the active form of MutS in mutation avoidance. To support this conclusion, they found that MutSΔ800 protein bound to mismatched DNA less efficiently than MutS. Our results also indicate that MutSΔ800 does have reduced affinity for GG–CT base mismatches (Figure 4B) but not with the one base IDL (Figure 3) or O6-meG mismatches (Figure 4C). Given that MutSΔ800 has reduced affinity for some base mismatches, we propose that the lack of a mutator phenotype in cells over-expressing MutSΔ800 (Table 1 and Figure 1) can be explained by the MutS dimer being able to cope with the few mismatches generated behind the replication fork. The atomic structure of MutSΔ800 indicates that mismatch binding does occur. In interspecies crosses, however, the much larger number of mismatches overwhelms the MutSΔ800 protein's reduced mismatch binding and processing ability, thereby leading to reduced antirecombination as shown in Table 2. An essential part of our proposal, therefore, is that the cell's response with dimeric MutS depends on the number of mismatches involved; if there are few, overproduced dimeric MutS can substitute for tetrameric MutS at its normal cellular concentration. The data in Figures 5 and 6, showing a greater amount of M13-fd heteroduplex formation by MutSΔ800 compared to MutS, is consistent with this model. The combination of MutL and MutSΔ800 is as effective as MutS and MutL where the level of mismatching is 3%. We predict that at higher levels of mismatching, such as the 17% in E.coli–Salmonella crosses, MutSΔ800 would be much less effective than MutS, even in the presence of MutL.
Bjornson et al. (23) also showed that MutH-induced incision of heteroduplex DNA, in the presence of MutL, was reduced by MutSΔ800. This observation can also help to explain the response of dam mutants with the over-expressed MutSΔ800 protein to cisplatin and MNNG. For cells exposed to MNNG, there is sufficient binding of MutSΔ800 to O6-meG mismatches to provoke a sensitization response even though there is reduced MutH-induced incision (Figure 2B). For dam cells expressing MutSΔ800 and exposed to cisplatin, however, the reduced binding of MutSΔ800 to intrastrand diguanyl-cisplatin cross-links and the reduced MutH-induced incision activity allow the cell to repair, by recombination, the few DSBs that might be formed (Figure 2B).
The model proposed above regarding the number of mismatched base pairs and the ability of over-expressed MutSΔ800 to deal with them makes the strong prediction that when MutSΔ800 is expressed from a single-copy gene, the dam cells containing it would have different responses for spontaneous mutagenesis and resistance to cisplatin and MNNG compared to expression from a multicopy plasmid. Experiments to test this prediction are in progress.
Surprisingly, MutSΔ800 when expressed in a wild-type background does not display a dominant negative phenotype with regard to cisplatin sensitivity (Figure 2). This result appears to rule out the possibility that MutS, which is at a low cellular concentration, could be titrated out by the plasmid expressed MutSΔ800, leading to a dominant negative phenotype. We have not yet quantitated the protein levels of the plasmid expressed MutSΔ800 versus the chromosomally expressed copy of MutS. If the MutSΔ800 protein is less stable than MutS, then the ratio of the two proteins may be such that active mixed multimers are formed allowing for near-full functionality of the protein (Figure 2), which is absent in a population containing only the MutSΔ800.
The results in Figure 4 indicate that the C-terminal end of MutS is required for binding to platinated GG–CC cross-links, which could also indicate a requirement for MutS tertamerization, in that the dimeric MutSΔ800 cannot recognize the lesion, whereas the MutS does. At present, it is not known how this cross-link opposite CC differs in structure from that opposite CT to allow efficient recognition by MutS, but it is possible that binding to a platinated lesion requires critical contacts with residues residing in the C-terminus of MutS, offering an alternative to the tetramerization requirement. Furthermore, deletion of the C-terminus may disrupt the binding of MutS to low-affinity lesions, such as the intrastrand GG cross-links (10- to 40-fold lower than a G–T mismatch (20,23), but not to a high affinity one base IDL (Figure 3). Either explanation is supported by the survival results for cells with the mutSΔ800 plasmid exposed to cisplatin (Figure 2). In addition to these alternatives to tetramerization, we cannot rule out a critical loss of interaction(s) between MutSΔ800 and other proteins, such as the beta-clamp (32) which can ultimately affect the efficiency of recognition or repair processes.
That MutS can bind to platinated GG–CC cross-links suggests that MMR-induced DSBs can occur in unreplicated DNA of dam mutants at these sequences, as well as cross-links opposite CT bases, which can be formed by translesion polymerases (33,34). Platinated GG–CC cross-links can also be formed during recombinational repair of DSBs and these are bound by MutS which can block further branch migration (17), thereby preventing DSB repair. The ability of MutS to bind these cross-links can explain the observed cisplatin sensitivity of the dam mutS+ strain in Figure 2A. In contrast, the decreased binding of MutSΔ800 to such lesions reduces the number of mismatch repair-induced DSBs and thereby promotes survival of the dam mutSΔ800 strain to cisplatin (Figure 2A).
Finally, we note that mammalian cell lines also show sensitivity to cisplatin and MNNG and MMR-deficient lines derived from them are resistant to both compounds (35,36), although whether cisplatin resistance is due specifically to MMR-deficiency has recently been challenged (37,38). Cisplatin-resistant cells isolated from patients treated with this drug have also been shown to be deficient in MMR (39). Like the bacterial MutS protein, the human MutS-alpha counterpart also binds to cisplatin intrastrand cross-links and O6-meG mismatches (40). The various models proposed to explain mismatch repair-dependent drug-resistance in mammalian cells have been reviewed in (41). The data reported here with E.coli MutSΔ800 have no counterpart in eukaryotic systems, thereby preventing selection of one model over another.
Acknowledgments
We thank Meindert Lamers and Titia Sixma for the mutSΔC800 plasmid; Kenneth Sanderson for the Salmonella donor strains; Jennifer Robbins for construction and purification of the cross-linked platinated oligonucleotide; Dianne Schwarz for assistance with the radiolabeled band shifts; F. Lopez de Saro and M. O'Donnell for the gift of MutL protein and Mary Munson for invaluable help and guidance during protein purification. This work was supported by grant GM63790 from the National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health.
REFERENCES
- 1.Modrich P., Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 2.Schofield M.J., Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 3.Marti T.M., Kunz C., Fleck O. DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol. 2002;191:28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 4.Rayssiguier C., Thaler D.S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 5.Westmoreland J., Porter G., Radman M., Resnick M.A. Highly mismatched molecules resembling recombination intermediates efficiently transform mismatch repair proficient Escherichia coli. Genetics. 1997;145:29–38. doi: 10.1093/genetics/145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harfe B.D., Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 7.Fabisiewicz A., Worth L., Jr Escherichia coli MutS,L modulate RuvAB-dependent branch migration between diverged DNA. J. Biol. Chem. 2001;276:9413–9420. doi: 10.1074/jbc.M005176200. [DOI] [PubMed] [Google Scholar]
- 8.Worth L., Clark S.R.M., Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc. Natl Acad. Sci. USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junop M.S., Yang W., Funchain P., Clendenin W., Miller J.H. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair (Amst.) 2003;2:387–405. doi: 10.1016/s1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- 10.Wu T.H., Marinus M.G. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karran P., Marinus M.G. Mismatch correction at O6-methylguanine residues in E.coli DNA. Nature. 1982;296:868–869. doi: 10.1038/296868a0. [DOI] [PubMed] [Google Scholar]
- 12.Fram R.J., Cusick P.S., Wilson J.M., Marinus M.G. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol. Pharmacol. 1985;28:51–55. [PubMed] [Google Scholar]
- 13.Marinus M.G. Dr Jekyll and Mr Hyde: how the MutSLH repair system kills the cell. In: Higgins N.P., editor. The Bacterial Chromosome. Washington, DC: ASM Press; 2005. pp. 413–430. [Google Scholar]
- 14.Au K.G., Welsh K., Modrich P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 15.Marinus M.G. Recombination is essential for viability of an Escherichia coli dam (DNA adenine methyltransferase) mutant. J. Bacteriol. 2000;182:463–468. doi: 10.1128/jb.182.2.463-468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowosielska A., Marinus M.G. Cisplatin induces DNA double-strand break formation in Escherichia coli dam mutants. DNA Repair (Amst.) 2005 doi: 10.1016/j.dnarep.2005.03.006. in press. [DOI] [PubMed] [Google Scholar]
- 17.Calmann M.A., Marinus M.G. MutS inhibits RecA-mediated strand exchange with platinated DNA substrates. Proc. Natl Acad. Sci. USA. 2004;101:14174–14179. doi: 10.1073/pnas.0406104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su S.S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl Acad. Sci. USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker B.O., Marinus M.G. Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl Acad. Sci. USA. 1992;89:1730–1734. doi: 10.1073/pnas.89.5.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourrier L., Brooks P., Malinge J.M. Binding discrimination of MutS to a set of lesions and compound lesions (base damage and mismatch) reveals its potential role as a cisplatin-damaged DNA sensing protein. J. Biol. Chem. 2003;278:21267–21275. doi: 10.1074/jbc.M301390200. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen L.J., Samson L. The Escherichia coli MutS DNA mismatch binding protein specifically binds O(6)-methylguanine DNA lesions. Carcinogenesis. 1996;17:2085–2088. doi: 10.1093/carcin/17.9.2085. [DOI] [PubMed] [Google Scholar]
- 22.Lamers M.H., Perrakis A., Enzlin J.H., Winterwerp H.H., de Wind N., Sixma T.K. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 23.Bjornson K.P., Blackwell L.J., Sage H., Baitinger C., Allen D., Modrich P. Assembly and molecular activities of the MutS tetramer. J. Biol. Chem. 2003;278:34667–34673. doi: 10.1074/jbc.M305513200. [DOI] [PubMed] [Google Scholar]
- 24.Takamatsu S., Kato R., Kuramitsu S. Mismatch DNA recognition protein from an extremely thermophilic bacterium, Thermus thermophilus HB8. Nucleic Acids Res. 1996;24:640–647. doi: 10.1093/nar/24.4.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas I., Ban C., Fleming K.G., Qin J., Lary J.W., Yphantis D.A., Yang W., Hsieh P. Oligomerization of a MutS mismatch repair protein from Thermus aquaticus. J. Biol. Chem. 1999;274:23673–23678. doi: 10.1074/jbc.274.33.23673. [DOI] [PubMed] [Google Scholar]
- 26.Obmolova G., Ban C., Hsieh P., Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 27.Wu T.H., Marinus M.G. Deletion mutation analysis of the mutS gene in Escherichia coli. J. Biol. Chem. 1999;274:5948–5952. doi: 10.1074/jbc.274.9.5948. [DOI] [PubMed] [Google Scholar]
- 28.Marinus M.G., Morris N.R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J. Mol. Biol. 1974;85:309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- 29.Nowosielska A., Calmann M.A., Zdraveski Z., Essigmann J.M., Marinus M.G. Spontaneous and cisplatin-induced recombination in Escherichia coli. DNA Repair (Amst.) 2004;3:719–728. doi: 10.1016/j.dnarep.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Rosen H. A modified ninhydrin colorimetric analysis for amino acids. Arch. Biochim. Biophys. 1957;67:10–15. doi: 10.1016/0003-9861(57)90241-2. [DOI] [PubMed] [Google Scholar]
- 31.Biswas I., Obmolova G., Takahashi M., Herr A., Newman M.A., Yang W., Hsieh P. Disruption of the helix–u-turn–helix motif of MutS protein: loss of subunit dimerization, mismatch binding and ATP hydrolysis. J. Mol. Biol. 2001;305:805–816. doi: 10.1006/jmbi.2000.4367. [DOI] [PubMed] [Google Scholar]
- 32.Lopez de Saro F.J., O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl Acad. Sci. USA. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaisman A., Chaney S.G. The efficiency and fidelity of translesion synthesis past cisplatin and oxaliplatin GpG adducts by human DNA polymerase beta. J. Biol. Chem. 2000;275:13017–13025. doi: 10.1074/jbc.275.17.13017. [DOI] [PubMed] [Google Scholar]
- 34.Vaisman A., Masutani C., Hanaoka F., Chaney S.G. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry. 2000;39:4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 35.Branch P., Masson M., Aquilina G., Bignami M., Karran P. Spontaneous development of drug resistance: mismatch repair and p53 defects in resistance to cisplatin in human tumor cells. Oncogene. 2000;19:3138–3145. doi: 10.1038/sj.onc.1203668. [DOI] [PubMed] [Google Scholar]
- 36.Kartalou M., Essigmann J.M. Mechanisms of resistance to cisplatin. Mutat. Res. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 37.Massey A., Offman J., Macpherson P., Karran P. DNA mismatch repair and acquired cisplatin resistance in E. coli and human ovarian carcinoma cells. DNA Repair (Amst.) 2003;2:73–89. doi: 10.1016/s1568-7864(02)00187-8. [DOI] [PubMed] [Google Scholar]
- 38.Claij N., Te Riele H. Msh2 deficiency does not contribute to cisplatin resistance in mouse embryonic stem cells. Oncogene. 2004;23:260–266. doi: 10.1038/sj.onc.1207015. [DOI] [PubMed] [Google Scholar]
- 39.Fink D., Nebel S., Aebi S., Zheng H., Cenni B., Nehme A., Christen R.D., Howell S.B. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 40.Duckett D.R., Drummond J.T., Murchie A.I., Reardon J.T., Sancar A., Lilley D.M., Modrich P. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc. Natl Acad. Sci. USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stojic L., Brun R., Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst.) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]