Abstract
Cleavage of phosphodiester bonds by small ribonuclease mimics within different bulge-loops of RNA was investigated. Bulge-loops of different size (1–7 nt) and sequence composition were formed in a 3′ terminal fragment of influenza virus M2 RNA (96 nt) by hybridization of complementary oligodeoxynucleotides. Small bulges (up to 4 nt) were readily formed upon oligonucleotide hybridization, whereas hybridization of the RNA to the oligonucleotides designed to produce larger bulges resulted in formation of several alternative structures. A synthetic ribonuclease mimic displaying Pyr–Pu cleavage specificity cleaved CpA motifs located within bulges faster than similar motifs within the rest of the RNA. In the presence of 10 mM MgCl2, 75% of the cleavage products resulted from the attack of this motif. Thus, selective RNA cleavage at a single target phosphodiester bond was achieved by using bulge forming oligonucleotides and a small ribonuclease A mimic.
INTRODUCTION
Sensitivity of RNA phosphodiester bonds to cleaving reagents is determined by the nature of adjacent nucleosides and their involvement in secondary and tertiary interactions in the RNA structure (1–4). Recently, it was shown that base stacking strongly affects the reactivity of RNA phosphodiester bond towards cleavage (5,6). Cleavage of single-stranded RNA oligomers, as well as of RNA hairpin-loops and bulge-loops by imidazole buffer (7), metal ions (8–11), amines (12) or detergents (13) has been extensively investigated. Results of the studies evidence that phosphodiester bonds within small hairpin loops have enough conformational freedom to be cleaved by the in-line mechanism (14), however involvement of the loops in tertiary interactions can suppress their reactivity (15). Pb2+, Zn2+, Mg2+ and complexes of these cations cleave phosphodiester bonds within bulge loops (8,14,16) although less readily than they do within single-stranded oligoribonucleotides (8). Efficiency of cleavage by Pb2+ within bulges was shown to depend on the bulge sequence and length (14). Sensitivity of RNA bulges to non-metallated ribonuclease mimics has not been studied so far.
Sufficient conformational freedom and enhanced reactivity of some phosphodiester bonds in specific positions within the bulge-loops could be employed for achievement of increased cleavage by the catalytic groups delivered to these bonds by antisense oligonucleotides. Indeed, it was shown that cleavage of RNA by lanthanide (La3+ and Eu3+) complex conjugated to antisense oligonucleotide (17) or by transition metal complexes conjugated to 2′-O-methyl oligoribonucleotides (18) occurs more efficiently when the target sequence is located within a bulge loop. Thus, oligonucleotide binding, yielding highly reactive bulge loops, can thus be considered as an approach to induction of unique reactive sites in RNA, which can be selectively cleaved by small catalytic molecules. Activation of the target phosphodiester bond by interaction with acridine attached to addressing oligonucleotide was used to accomplish selective cleavage of RNA by lanthanide ions (19,20).
In the present work, we investigated cleavage of RNA bulge-loops of varying size and composition by imidazole and a small ribonuclease A mimic. We found that maximal reactivity of phosphodiester bonds is observed within Py–Pu motifs in the 4–7 nt long bulge-loops. RNA containing a single bulge loop of this structure can be selectively cleaved at this loop by a simple synthetic ribonuclease mimic.
MATERIALS AND METHODS
Ribonucleoside triphosphates, deoxyribonucleoside triphosphates and RNase A were from Sigma. [γ-32P]ATP with specific activity of ∼4000 Ci/mmol was from Biosan (Russia). Total tRNA from Escherichia coli used as a carrier to supplement labeled RNA was from State Research Centre of Virology and Biotechnology ‘Vector’ (Russia). RNase T1 and RNase T2 were from Boehringer Mannheim. Taq DNA polymerase was from Laboratory of Bioorganic Chemistry of Enzymes (this Institute). RNase H and T4 polynucleotide kinase were from Fermentas (Lithuania). RNase ONE, RNase U2 and ‘Large scale RNA production System Ribomax’ were from Promega (USA). GFX™ PCR DNA and Gel Band Purification Kit was purchased from Amersham (USA). Artificial ribonuclease ABL4C3 was synthesized in the Laboratory of Organic Synthesis (this Institute) by Dr D. Konevez.
Preparation of oligonucleotides
The following oligodeoxyribonucleotides (ONs) were synthesized by standard phosphoramidite chemistry and purified by successive ion-exchange and reverse-phase HPLC: ON1(0), GCACTCTGCTGTTCCT; ON2(1/−), CACTCTGC#GTTCCTTTC; ON3(1/+), CACTCTGC#AGTTCCTTTC; ON4(1/−), CAGCACTC#GCTGTTCC; ON5(2/−), CACTCTGC#TTCCTTTCG; ON6(2/+), CACTCTGC#ATTCCTTTCG; ON7(2/−), CAGCACTC#CTGTTCCTT; ON8(3/−), CACTCTGC#TCCTTTCGA; ON9(3/+), CACTCTGC# ATCCTTTCGA; ON10(3/−), AGCACTC#TGTTCCTTT; ON11(4/−), GCACTCTG#TCCTTTCGA; ON12(4/+), GCACTCTG#ATCCTTTCGA; ON13(4/−), AGCACTCT#TTCCTTTCG; ON14(4/−), CAGCACTC#GTTCCTTTC; ON15(5/−), AGCACTCT#TCCTTTCG; ON16(5/+), AGCACTCT#ATCCTTTCG; ON17(5/−), ACAGCAC# TGTTCCTTT; ON18(6/−), CAGCACTC#TCCTTTCGA; ON19(6/+), CAGCACTC#ATCCTTTCGA; ON20(6/−), ACAGCACT#TTCCTTTCG; ON21(7/−), ACAGCACT# TCCTTTCGA; ON22(7/+), ACAGCACT#ATCCTTTCGA; ON23(7/−), CACAGCAC#TTCCTTTCG; and ON24(7/−), CAGCACTC#CCTTTCGAT. ‘#’ in the sequences indicates the deletion sites where bulging of the RNA sequences occur upon hybridization with the oligonucleotides. The extra adenosines introduced opposite to the bulges are underlined.
Preparation of 216–312 fragment of influenza virus M2 RNA (M2-96 RNA)
M2-96 RNA was prepared by in vitro transcription using T7 RNA polymerase as described previously (21). DNA template corresponding to the nucleotides 216–312 of the influenza virus M2 RNA was prepared by PCR using the construct pSVK3M2 (bearing the M2 cDNA cloned into the KpnI and XhoI sites of the plasmid pSVK3) and forward 5′-ACAAGCTTTAATACGACTCACTCACTATAGGGCCTTCTACGGAAGGAGTACC-3′ and reverse 5′-CGAGACAAAATGACTGTCGTCAGC-3′ primers (T7 promotor sequence is underlined) (22). PCR product was purified using GFXTM PCR DNA and Gel Band Purification Kit. The in vitro transcription reaction was carried out using ‘Large scale RNA production System Ribomax’ according to the manufacturer's protocol. Secondary structure of the in vitro produced transcript was established using the data of probing with ribonucleases T1, T2, U2 and ONE as described previously (23).
RNA 5′ end labelling
Prior to 5′ end labelling, M2-96 transcript was dephosphorylated using bacterial alkaline phosphatase (BAP) (Fermentas) according to the described protocol (24). Briefly, the reaction mixture, containing 50 mM Tris–HCl (pH 8.5), 1 mM EDTA, 0.2% SDS, 2% formamide, 2.5 mM DTT, 10 μg of in vitro transcript of M2-96 RNA and 2 U of BAP in a total volume of 50 μl was incubated at 37°C for 1 h. BAP was added to the reaction mixture at 0 and 30 min of incubation time. The reaction was quenched by 1:1 phenol–chloroform (v/v) extraction followed by extraction of water phase with ethyl ester and ethanol precipitation.
Labelling of the 5′ end of the M2-96 transcript was performed with [γ-32P]ATP and T4 polynucleotide kinase under standard conditions (25). 32P-labeled RNA was isolated by electrophoresis in 12% denaturing polyacrylamide gel. RNA was visualized by autoradiography. 32P-labeled RNA was eluted from the gel with 300 μl of 0.3 M ammonium acetate, pH 6.0 and ethanol precipitated.
Hybridization of oligonucleotides with M2-96 RNA
Hybridization of oligonucleotides with in vitro transcript M2-96 RNA was studied using gel-mobility shift assay (26). Reaction mixtures (5 μl) containing 0.1 μM (0.1 μCi) 32P-labeled M2-96 RNA, 1 μM corresponding oligonucleotide, 50 mM Tris–HCl (pH 7.0), 0.1 mM EDTA, 200 mM KCl, 1 μg tRNA carrier were incubated at 37°C. Then, at definite times, 2.5 μl of loading buffer (20% Ficoll-400, 0.025% bromophenol blue and 0.025% xylene cyanol) was added and the mixtures were applied on running (20 V/cm) native 10% PAAG, with TBE as a running buffer, at 4°C. The gel was dried and analyzed using Molecular Imager FX (Bio-Rad). The total extent of RNA binding with oligonucleotide was determined as a ratio of radioactivity measured in the RNA/ON complex to the total radioactivity applied on the gel.
Effective association constant Kef
Hybridization of M2-96 RNA with oligonucleotides was performed under conditions of 10-fold oligonucleotide excess (0.1 μM of [32P]RNA and 1 μM oligonucleotide). The reaction mixtures were processed as described above. The effective association constants of RNA hybridization (Kef) were derived by minimizing the mean square deviation between the experimental data and theoretical curves obtained according to equation 1, using Origin 7.0 software.
| 1 |
where α is the extent of binding, k is the binding rate constant and [ON] the concentration of oligonucleotide.
Footprinting with RNase H
RNase H footprinting was performed as described previously (27). Briefly, the reaction mixture (10 μl) containing 0.1 μM (0.1 μCi) [5′-32P]M2-96 RNA, 50 mM HEPES (pH 7.5), 100 mM KCl, 0.05 mM EDTA, 10 mM MgCl2, 1 μg tRNA carrier and 1 μM oligonucleotide was incubated at 37°C for 1 h. Then 0.2 U of RNase H was added and the probe was incubated at 37°C for 15 min. Cleavage products were resolved on denaturing 18% PAAG. To identify the cleavage sites (here and in all cleavage experiments), the probes were run in parallel with ladders produced by RNase T1 (28) and 2 M imidazole buffer, pH 7.0 (5).
Probing of oligonucleotide–RNA complexes with 2 M imidazole buffer
Reaction mixture (20 μl) containing 0.1 μM (0.1 μCi) [5′-32P]M2-96 RNA, 2 M imidazole buffer (pH 7.0), 0.5 mM EDTA, 40 mM NaCl, 10 mM MgCl2, 2 μg tRNA carrier and 1 μM of corresponding oligonucleotide was incubated for 24 h at 20°C (29). Cleavage products were analyzed in 18% denaturing PAAG as described above.
RNase A cleavage
Reaction mixture (10 μl) contained 50 mM Tris–HCl (pH 7.0), 0.1 mM EDTA, 200 mM KCl, 1 μg tRNA carrier, 0.1 μM (0.1 μCi) [5′-32P]M2-96 RNA and 1 μM oligonucleotide. The mixture was incubated for 1 h at 37°C and then RNase A was added (concentration 10−5 mg/ml). The reaction was incubated for 1 min and quenched by adding 40 μl of tRNA carrier (0.05 μg/μl) and 50 μl of water-saturated phenol–chloroform. After phenol extraction, the cleavage products were precipitated with ethanol and analyzed by electrophoresis in denaturing 18% PAAG.
Cleavage assay with small artificial ribonuclease ABL4C3
To avoid any contamination of ABL4C3 (30) with ribonucleases, 2 mM solution of ABL4C3 was filtered through Centricon3 filters (Amicon, Dauvers, MA) as described in (31). Standard reaction mixture contained 0.1 μM (0.1 μCi) [5′-32P]M2-96 RNA, 50 mM Tris–HCl (pH 7.0), 0.1 mM EDTA, 200 mM KCl, 1 μg tRNA carrier and 1 μM oligonucleotide. In some experiments (see figure legends), the buffer was supplemented with 10 mM MgCl2. After 1 h of hybridization, compound ABL4C3 was added (final concentration 5 × 10−4 M) and the reaction mixture was incubated for 8 h. Cleavage products were analyzed using denaturing 18% PAAG. The gel was dried and analyzed using Molecular Imager FX. Total extent of RNA cleavage and extent of RNA cleavage at a specific site were determined as the ratio of radioactivity measured in the RNA fragment(s) to the total radioactivity applied on the gel.
RESULTS
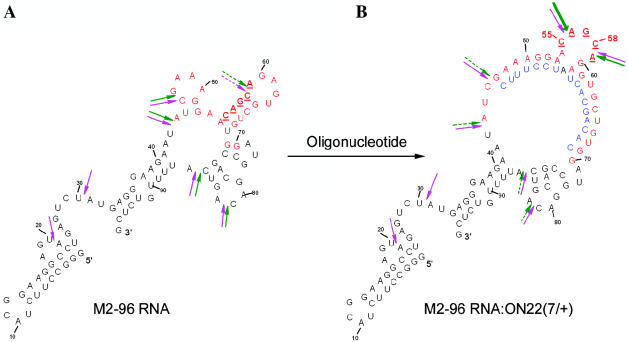
The model RNA used in our experiments is 96 nt long in vitro transcript of the 3′ end fragment of influenza virus M2 RNA (‘M2-96 RNA’) (Figure 1A). This RNA fragment retains the main elements (hairpins) of M2 RNA secondary structure and has the 5′ terminus convenient for labeling with 32P. Recently, we have found that CAGCA sequence (nucleotides 256–260) located in a stem-loop of M2 RNA is slightly sensitive to artificial ribonucleases ABLkC3 (31). We have chosen this sequence in M2-96 RNA (C55AGCA59 underlined in Figure 1) as a target to be placed in artificial bulge-loops. This sequence contains two Py-A motifs, which are known to display enhanced reactivity towards different RNA-cleaving compounds (32,33). We expected that this sequence would display even greater cleavage sensitivity by placing it into bulge-loop.
Figure 1.
Secondary structure of in vitro transcript of M2-96 RNA (A) and complex of M2-96 RNA with oligodeoxynucleotide ON22(7/+) resulting in bulge formation (B). RNA target and deoxy-ON are shown in red and blue, respectively. The arrows point to the cleavage sites induced in M2-96 RNA within the complex by artificial ribonuclease in the absence (pink) and in the presence (green) of 10 mM Mg2+. Solid and dashed arrows show strong and weak cleavage sites, respectively.
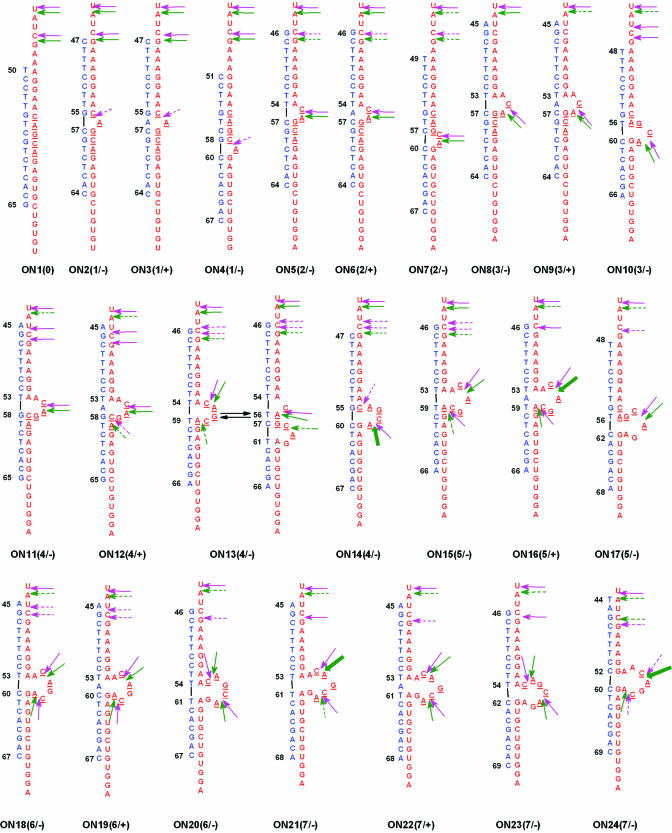
Sequences of oligodeoxyribonucleotides (15–18mers) were designed so that their hybridization with M2-96 RNA should result in formation of bulges containing up to 7 nt (Figures 1B and 2). The oligonucleotide sequences were selected to avoid self-complementarity and to have the shoulders of oligonucleotide providing similar affinities to their RNA complements. The oligonucleotides were named ONn(m/±), where n is oligonucleotide number, m corresponds to the size of the bulge formed upon the oligonucleotide hybridization, ‘+’ or ‘−’ indicates, if the oligonucleotide contains extra adenosine (mismatch) opposite to the bulge or not. Oligonucleotides also differed in the location of the target sequence within bulge (Figure 2). Oligonucleotide-induced bulge-loops contained either the C55–A56 or the C58–A59 target sequence or both. In the latter case [oligonucleotides ON18(6/−) and ON20 (6/−) and ON21(7/−), ON23(7/−) and ON24(7/−)], the sequences of the bulges differed by a one-nucleotide shift. Oligonucleotide ON1(0) forming a fully complementary complex with the sequence 50–65 of the RNA was used as a control.
Figure 2.
Sequences and structures of artificial bulge-loops in M2-96 RNA formed by hybridization with oligodeoxynucleotides. Red and blue letters correspond to M2-96 RNA and oligodeoxynucleotide sequences. Arrows show cleavage sites of RNA bound to the respective oligodeoxynucleotide by artificial ribonuclease in the absence (pink) or in the presence (green) of Mg2+. The arrow size (thickness) corresponds to cleavage intensities.
Hybridization assay
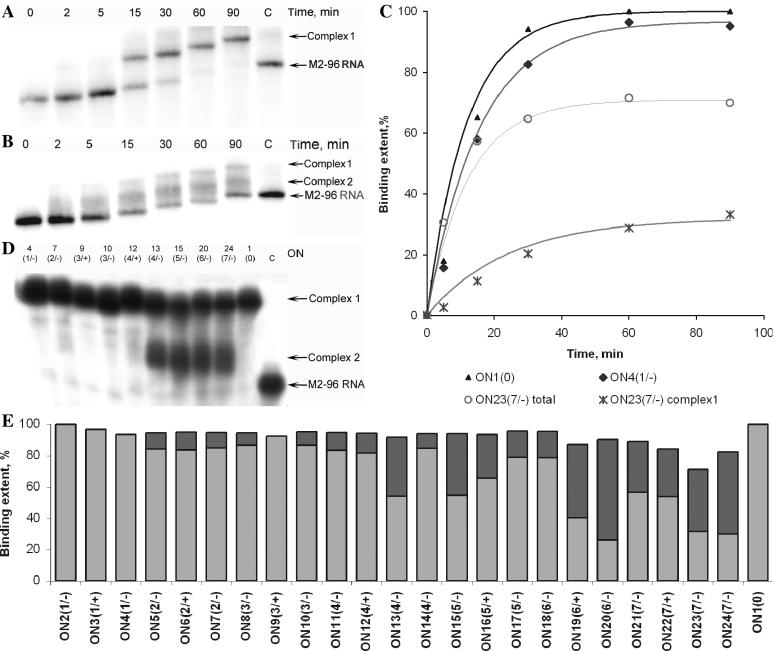
Gel-mobility shift assay was used to follow hybridization of oligonucleotides with M2-96 RNA at 37°C (Figure 3). All the oligonucleotides tested (at concentration 1 μM) formed complexes with the complementary sequence in M2-96 RNA. Figure 3A and B show time course of M2-96 RNA:ON complex formation for oligonucleotides ON4(1/−) and ON23(7/−), respectively. Oligonucleotide ON4(1/−) efficiently binds to M2-96 RNA. During the first minutes of incubation, M2-96 RNA rapidly forms a complex with ON4(1/−), which accumulates in the course of the following incubation. This complex, named complex 1, was suggested to correspond to the complementary duplex with a bulge-loop. In case of the oligonucleotide ON23(7/−) designed to form in the RNA the largest (7 nt) bulge (Figure 3B), a considerable portion of M2-96 RNA remained unbound even after 1.5 h of incubation. In contrast to ON4(1/−), ON23(7/−) formed two types of complexes with the RNA. The additional complex, named complex 2 has higher electrophoretic mobility as compared with the complex 1. Complex 2 could apparently be either an imperfect complex or the complex folded in some alternative structure. Complex 2 was formed faster than complex 1; after 1 h both complexes were present in similar amounts. This type of M2-96 RNA:ON23(7/−) hybridization was observed at different oligonucleotide concentrations (data not shown). The kinetic curve of M2-96 RNA:ON4(1/−) complex 1 formation reaches plateau (∼95%) after 45 min incubation similarly to the control oligonucleotide ON1(0) binding. Association constants (Kef) evaluated from these curves are 108 and 3.1 × 107 M−1 for ON1(0) and ON4(1/−), respectively. Extent of M2-96 RNA binding with ON23(7/−) reaches its plateau (complex 1, 28% and complex 2, 41%) after 1 h of incubation. The observed Kef for M2-96:ON23(7/−) complex 1 formation was estimated to be 106 M−1, which is considerably (one and two orders of magnitude) lower than the respective constants for ON4(1/−) and ON1(0).
Figure 3.
Hybridization of oligodeoxynucleotides with M2-96 RNA (gel-mobility shift assay). (A and B) Kinetics of ON4(1/−)(A) and ON23(7/−) (B) hybridization to M2-96 RNA. Lanes 0–90, M2-96 RNA was incubated in the presence of oligonucleotides for the indicated times (min). Lane C shows control, the M2-96 RNA in the absence of oligonucleotide. Positions of M2-96 RNA and the RNA:oligonucleotide complexes are given on the right. Gel-mobility shift assay was carried out as described in Materials and Methods. (C) Secondary plot of the data shown in (A and B) for ON4(1/−), ON23(7/−) and for ON1(0) [primary data for ON1(0) not shown]. Triangles are for ON1(0), diamonds for ON4(1/−), asterisks for the complex 1 formation with ON23(7/−) and circles correspond to the total RNA binding to oligonucleotide ON23(7/−) (complex 1 + complex 2). (D) Gel-mobility shift assay of M2-96 binding with oligonucleotides. Prior to analysis, M2-96 RNA (10−7 M) was incubated with respective oligonucleotide (1 μM) for 1 h at 37°C. Locations of M2-96 RNA and RNA:oligonucleotide complexes are shown on the right. C, control, M2-96 RNA. Oligonucleotide numbers are shown on the top. (E) Efficiencies of M2-96 RNA binding with oligonucleotides. Formations of complex 1 and complex 2 are shown with light grey and dark grey, respectively. Hybridization conditions are as in (D). Experimental error not exceeded 10%.
Figure 3D and E display the data on M2-96 RNA hybridization with oligonucleotides inducing bulges of different length. It is seen that after 1 h of incubation, 70–98% of M2-96 RNA is bound to oligonucleotides. ONs hybridization with M2-96 RNA is not affected by the presence of 10 mM MgCl2 (data not shown). Efficient binding (95 ± 5%) was observed for oligonucleotides forming bulges up to 6 nt long. In the case of ON21(7/−)–ON24(7/−), 70–90% of the RNA is involved in complex formation. Complex 1 for each of the oligonucleotides displayed an electrophoretic mobility similar to that of the complex formed with the fully complementary control oligonucleotide ON1(0) [Figure 3D, lane 1(0)]. Four oligonucleotides—ON2(1/−), ON3(1/+), ON4(1/−) and ON9(3/+) formed with M2-96 RNA only complex 1 (Figure 3E). In the case of the oligonucleotides forming bulges up to 3 nt [ON5(2/−), ON6(2/+), ON7(2/−), ON8(3/−) and ON10 (3/−)], the portion of complex 2 typically did not exceed 10% of the total. Yield of the complex 2 increased from 12 to 64% with the increase in bulge length from 4 to 7 nt (Figure 3E). Surprisingly, yield of the complex 2 was rather high (36%) in the case of ON13(4/−). This can be attributed to alternative bulge formation due to A/T base pairs flanking the bulge (Figure 2).
Probing the structure of M2-96:oligonucleotide complexes
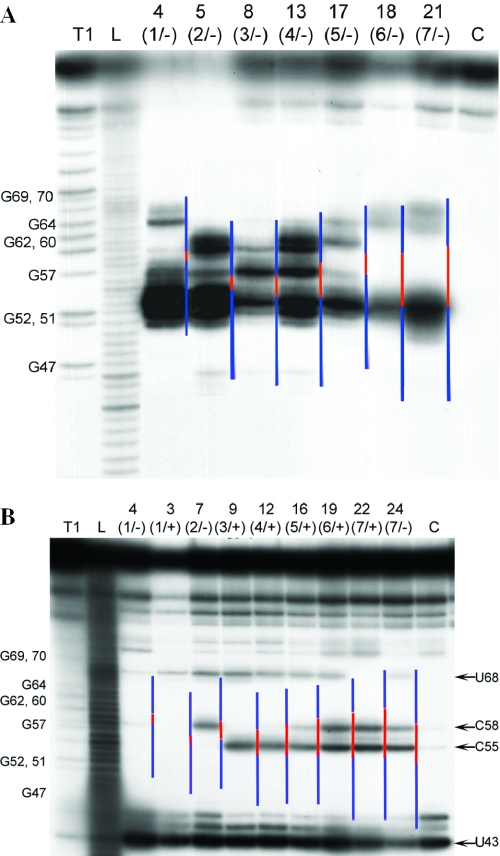
To characterize the structure of M2-96 RNA:ON complexes, we used RNase H (Figure 4A) allowing identification of nucleotides in M2-96 RNA involved in hybridization, additionally RNase A (Figure 4B) was used to map single-stranded regions and thereby the size of the bulges. As expected, RNase H cuts were gradually shifted to the 3′ end of RNA in accordance with the increasing length of a bulge, while intensities of the cuts gradually decreased. M2-96 RNA bound to oligonucleotides ON4(1/−), ON5(2/−), ON8(3/−) and ON13(4/−) (bulges up to 4 nt) was cleaved by RNase H within the complementary sequences. The most intensive cleavages occurred at G:C pairs. M2-96:ON17(5/−) complex was also cleaved within the entire complementary sequence, but the cuts in the 3′ end of the duplex were weak. In the complexes with 6–7-member bulges, formed by oligonucleotides [ON18(6/−) and ON21(7/−)], only three cuts were detected: one strong cut at position G52 and two weak cuts at G64 and G67, which is an evidence for poor hybridization of the 5′ part of oligonucleotides to their complement in the RNA. No cuts outside of the complementary sequences were observed except for the ON13(4/−), where strong cleavage at G57 occurred. This cleavage site can be attributed to alternative structure of the bulge, containing bulged C55 and C58AG60 sequence instead of C55AGC58 (Figure 2). Thus, RNase H footprinting clearly shows that in the case of oligonucleotides forming of 5–7-member bulges the 5′ part of oligonucleotides is loosely bound to the RNA.
Figure 4.
Probing of M2-96 RNA:oligodeoxynucleotide complexes with RNase H (A) and with RNase A (B). Radioautographs of denaturing 18% PAAG. Numbers of oligonucleotides are shown on the top. Lane L, imidazole ladder; lane T1, RNA cleavage with RNase T1 under denaturing conditions; and C, control, incubation of the RNA with RNase H or RNase A in the absence of oligodeoxynucleotide. Lines on the right-hand side of each lane indicate the position of the designed bulge (red) and double-stranded regions (blue). Cleavage conditions are described in Materials and Methods.
All the designed bulges contained CpA motifs that could be detected using RNase A as a probe. Incubation of M2-96 bound to oligonucleotides ON7(2/−), ON9(3/+), ON12(4/+), ON16(5/+), ON19(6/+), ON22(7/+) or ON24(7/−) with RNase A resulted in RNA cleavage at C55–A56 and/or C58–A59 sites, in accordance with the expected bulge structures (Figure 4B). Small 1-member bulges were not cleaved by RNase A. Besides, in the case of all tested oligonucleotides except for ON22(7/+) cleavage at U68−G69 next to 3′ end of the duplex was observed. In the native M2-96 RNA, this bond is resistant to cleavage with RNase A, since nucleotides U68 and G69 are located in a double-stranded region (Figure 1A). Upon RNA hybridization with oligonucleotides forming bulge up to 6 nt the RNA stem (nucleotides 54–58 and 64–68) unfolds, U68 becomes unpaired, and the phosphodiester bond U68–G69 is cleaved by RNase A. Complementary sequences of oligonucleotides ON22(7/+) and ON 24(7/−) include U68 and G69 and in complexes with these oligonucleotides this bond is protected from RNase A. This is the case for the ON22(7/+), whereas ON24(7/−) formed a ‘breathing’ complex with M2-96 RNA and did not prevent RNA cleavage at this particular site. To summarize, the probing provided direct evidences that the oligonucleotides tested bind to M2-96 RNA at their complementary sequences and form bulge-loops. In particular, oligonucleotides ON2–ON10, ON12 and ON14 form stable complexes with bulges up to 4 nt long, while in complexes with oligonucleotides designed to form longer bulges the 5′ sequence of the oligonucleotide is loosely bound to M2-96 RNA, thus resulting in several alternative structures of the complexes under these conditions.
Cleavage of RNA bulges with 2 M imidazole buffer and artificial ribonuclease ABL4C3
2 M imidazole buffer (pH 7.0) is known as an agent cleaving single-stranded sequences in RNA (29). Incubation of M2-96 RNA:ON complexes in 2 M imidazole buffer adjusted at pH 7.0 at 37°C for 24 h induce RNA cleavage within loops and within artificial bulges. From the data shown in Figure 5B, the bulge formation in M2-96 RNA upon binding of oligonucleotides ON2–ON14 is clearly seen, while no cleavage at sequences involved in DNA:RNA interaction is observed. These data indicate that these oligonucleotides are capable of strong binding with the RNA. In the case of oligonucleotides designed to form bulges longer than 4 nt (ON15–ON24), 2 M imidazole buffer cleaves phosphodiester bonds in the bulge at a rate similar to that of phosphodiester bonds that were expected to be hybridized. For oligonucleotides ON20(6/−), ON21(7/−) and ON24(7/−) imidazole cleavage pattern did not differ from the control, which indicates that RNA regions 54–58 and 64–68 complementary to the oligonucleotides remain partly unpaired. These data suggest that M2-96 RNA complexes with oligonucleotides ON17(5/−), ON18(6/−) and ON21(7/−), as detected with RNase H, are rather weak and/or form several alternative structures, which are in equilibrium. Probing of RNA:ON complexes with 2 M imidazole buffer clearly discriminates between stable complexes and the complexes with alternative structures caused by ‘breathing’ complexes.
Figure 5.
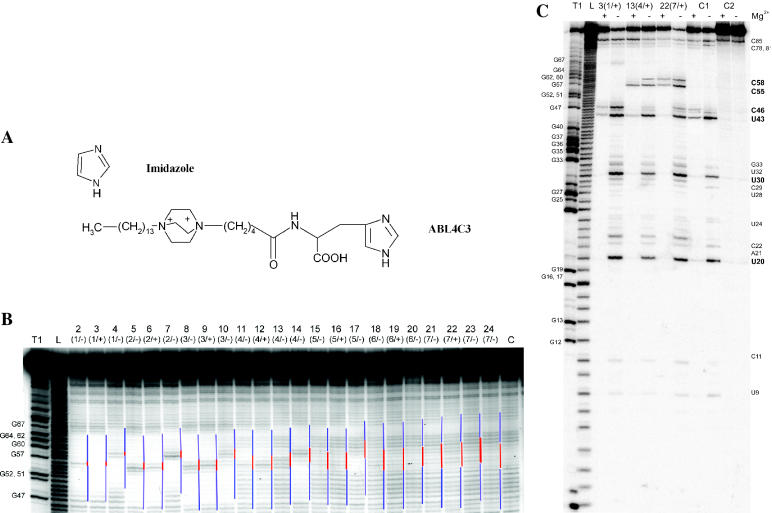
Cleavage of the oligonucleotide:M2-96 RNA complex with 2 M imidazole buffer and artificial ribonuclease ABL4C3. (A) Structure of imidazole and the artificial ribonuclease ABL4C3 (26). The two positive charges on N1 and N4 have the distance of two neighboring phosphodiester bonds and are the RNA-binding domain. (B) Cleavage of M2-96 RNA with imidazole buffer. Lane C, incubation of M2-96 RNA in 2 M imidazole buffer in the absence of oligonucleotide. Lane L, imidazole ladder; lane T1, RNA cleavage with RNase T1 under denaturing conditions. Lines on the right-hand side of each lane indicate the position of the designed bulge (red) and double-stranded regions (blue). (C) Cleavage of 1, 4 and 7-member bulges with ABL4C3 in the presence (+) and in the absence (−) of 10 mM MgCl2. Oligonucleotides are indicated on the top. Lane C1, cleavage of M2-96 RNA with ABL4C3 in the absence of oligonucleotide. Lane C2, incubation control. Cleave sites are indicated on the right, the major cuts are in boldface.
Artificial ribonuclease ABL4C3 is a conjugate of 1,4-diazabicyclo[2.2.2]octane substituted with a tetradecamethylene fragment at the bridge position, and histidine. ABL4C3 (Figure 4A) cleaves RNA preferentially at Py–Pu bonds located in single-stranded regions (31).
Complexes of M2-96 RNA with the oligonucleotides were incubated in the reaction buffer (see above) containing 0.5 mM ABL4C3 at 37°C for 8 h. Patterns of M2-96 RNA cleavage in the presence and in the absence of Mg2+ are shown in Figure 5C. In the absence of oligonucleotides and MgCl2, ABL4C3 cleaves M2-96 RNA at three major sites: U20–A21, U30–A31 and U43–A44 (Figure 5C, lane C1− and Figure 1A). Less intensive cleavages are observed at phosphodiester bonds after A21, C22, U24, U28, G33 and C46. To be noted, the same phosphodiester bonds are cleaved with RNase A (data not shown). Addition of 10 mM MgCl2 inhibits cleavage (Figure 5C, lane C1+). Cleavage at the sequences U20–A21, U30–A31, U43–A44 and C46–G47 occurs, although less efficient and cleavage of other sites is completely arrested. Apparently, this is the result of RNA structure stabilization by magnesium ions. In the presence of oligonucleotide ON3(1/+), the pattern of the RNA cleavage by ABL4C3 is similar to that induced by RNase A. No cleavage at the 1 nt bulges was observed. M2-96:ON13(4/+) complex containing 4-member artificial bulge is cleaved by ABL4C3 at the C55–A56 phosphodiester bond both in the presence and in the absence of magnesium. In the absence of Mg2+, the cleavage extent at the C55–A56 bond was comparable with the cleavage extent at U20–A21, U30–A31 and U43–A44 bonds, which remained unaffected by oligonucleotide binding and were the same as in the control (Figure 5C, lane C1−). The presence of MgCl2 drastically affected the cleavage pattern (Figures 5C and 2): the bulged C55–A56 bond became the major cleavage site (13%), while the phosphodiester bond U43–A44 was cleaved poorly (4%) and bonds U20–A21 and U30–A31 were not cleaved at all. In the complex with oligonucleotide ON22(7/+) forming a 7-member bulge, cleavage occurred at both C55–A56 and C58–A59 bonds. Total extents of RNA cleavage at these CpA bonds were 18% and 15% in the absence and in the presence of Mg2+, respectively, while total cleavage extent within the RNA was 80% and 20%, respectively. The selectivity (ratio of cleavage extent within the bulge to the total RNA cleavage) in the bulge in the absence and in the presence of MgCl2 equaled 0.22 and 0.75, respectively, assuming total cleavage extent as 1. Thus, selective RNA cleavage within the bulge is achieved in the presence of magnesium.
M2-96 cleavage with RNase A under similar conditions (in the presence and in the absence of magnesium) were performed to check if selective RNA cleavage within bulge-loops can be achieved by using natural enzyme (see Figure 2 in Supplementary Material). It turns out, that in the presence of Mg2+ M2-96 RNA cleavage within short bulges is suppressed, and in the longer bulges the phosphodiester bonds are cleaved with rate similar to the other loops of RNA (U6, U20, U30, U43).
Effect of bulge structure on the cleavage selectivity
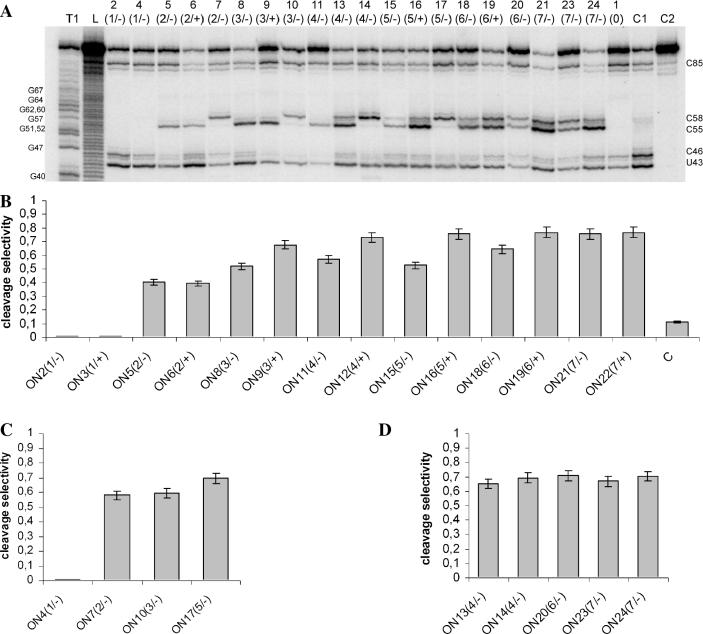
Figure 6A shows primary results of oligonucleotide:M2-96 RNA complex cleavage with ABL4C3 in the presence of 10 mM MgCl2. Under these conditions, only the bonds after U43, C46 and C85 were cleaved by the artificial ribonuclease; therefore, only the upper part of autoradiogram is shown. To compare the cleavage level with the bulge structures, the intensities of cleavage within the target sequence are marked in Figure 2 by green arrows. Minor cleavages at the target bonds C55–A56 and C58–A59 (cleavage selectivity was 0.12) were observed in the absence of oligonucleotide (Figure 6A, lane C1). In the presence of bulge-forming oligonucleotides, the intensity of U43–A44 bond cleavage is considerably lower as compared to the control (Figure 6A, lane C1). The cleavage intensities at the target sites are gradually increased with increasing bulge size. Similar to RNase A, compound ABL4C3 cleaved CpA motifs within the bulges of different size except for the 1-member bulges. The latter, C55 (or C58) is involved in base paring with oligonucleotide and the adenine (A56 or A59) is bulged out [Figure 6A, lanes 2(1/−) and 4(1/−); and Figure 2] and this combination is not cleavable by ABL4C3. In the symmetric situation, when C58 is bulged and A59 is base-paired [ON13(4/−), ON15(5/−) and ON16(5/+)], some cleavage with poor efficiency is observed. This is in accordance with data on RNA bulge loops cleavage by metal ions (16).
Figure 6.
Sensitivity of artificial RNA bulge-loops to cleavage with chemical ribonuclease ABL4C3 in the presence of 10 mM MgCl2. Products of M2-96 RNA:oligonucleotide duplex cleaved with compound ABL4C3 resolved in 18% denaturing PAAG (A). Oligonucleotide numbers referring to Figure 2 are indicated on the top. C1, cleavage of M2-96 RNA with ABL4C3 in the absence of oligonucleotide. C2, incubation control. Lane L, imidazole ladder; and lane T1, RNA cleavage with RNase T1 under denaturing conditions. The cleaved bonds in M2-96 RNA are indicated on the right. Selectivity of RNA cleavage within bulges calculated as a ratio of cleavage extent within bulge to total extent of RNA cleavage (B–D).
To identify features determining the bulge sensitivity to cleavage by ABL4C3, we investigated the influence of bulge length, location of CpA motifs in the bulge and of an extra adenine on the reaction (Figure 6B–D). Selectivity of cleavage within bulges was calculated as a ratio of cleavage extent within the bulge to total extent of RNA cleavage (Figure 6B–D). Both, in the presence and in the absence of Mg2+ (for primary data, see Supplementary Material), the intensities of RNA cleavage within bulges are higher than in the free, unhybridized M2-96 RNA (column C in Figure 6B corresponds to the lane C1 in Figure 6A). The selectivity of RNA cleavage within bulges is much higher when the reaction is supplied with Mg2+, except for the case of one member bulges (cf. Figures 5C and 6).
The data shown in Figure 6B indicate that in the case of oligonucleotides containing an extra adenine opposite to the bulge [ONn(m/+)] the cleavage selectivity is similar (2-member bulges) or reliably higher (longer bulges), than in the case of corresponding oligonucleotides lacking a mismatched adenine [ONn(m/−)]. It is worth noting that oligonucleotides with and without extra adenine display similar binding mode and binding efficiency (Figure 3E). Thus, the difference in cleavage selectivity can be attributed to more flexible structures of the bulge formed opposite to the extra adenine which results in better conditions for transesterification to occur.
Among short bulges, the highest selectivity (∼0.7) was achieved in a 4-member bulge [Figure 5C, ON13(4/+) and Figure 6B], containing C55–A56 bond in the apical position. This advantageous position of the CpA in the bulge containing 4 nt provide the same cleavage selectivity as observed in the case of long bulges containing two CpA motifs [ON16(5/+), ON19(6/+) and ON21(7/+)]. The total extents of RNA cleavage in the case of complexes with 6- and 7-member bulges are 1.5 times higher than those observed for the 4-member bulges: maximal cleavage extent (in the presence of magnesium) of 70% is observed for the M2-96:ON21(7/−) complex. Thus, maximal selectivity of cleavage achieved (0.70–0.75) in the 4-member and in the 7-member bulges corresponds to cleavage extents of 14 and 55%, respectively.
Selectivity of RNA cleavage within bulges containing only C58–A59 bond (Figure 6C) is higher than within the bulges of the same length containing C55–A56 bond (Figure 6B), but the total extent of RNA cleavage is lower. The selectivity of RNA cleavage within bulges containing both CpA motifs is shown in Figure 6B and D. The highest selectivities and the highest total cleavage extents are achieved in the case of 6- and 7-member bulges in which the C55A56 motif is located in the apical position. To summarize, efficient site-selective cleavage of M2-96 RNA by compound ABL4C3 is achieved in the presence of MgCl2 in the complexes with oligonucleotides forming bulges longer than 3 nt and containing CpA motif in the apical position.
DISCUSSION
We undertook the first systematic study of sensitivity of RNA bulges to cleavage with an imidazole-containing agent. Previously, cleavage of artificial bulges containing up to 4 nt with Me2+ has been investigated (15). It was shown that cleavages predominantly occurred at the phosphodiester bond on the 3′ side of the bulged nucleotide. We observed similar results upon cleavage of RNA:oligonucleotide complexes by 2 M imidazole buffer (see Figure 5A). Artificial ribonucleases cleaved a smaller set of phosphodiester bonds in the bulges and the cleavage extents are affected by the structure of the bulge. Our systematic screening of CpA motif location within bulges shows that bulges containing CpA motifs in the apical position are cleaved with the highest rate. The cleavage rates within small bulges are lower than in bulges longer than 4 nt. This observation is in contrast to the reported data for Me2+ ions, for which the 2 nt bulges are more preferential (34). On the other hand, similar results have been obtained upon cleavage of short hairpins by Zn2+ and Zn2+ complex (35). It was shown that phosphodiester bond in the apical position of a 5-member bulge was cleaved with the highest rate.
To achieve high rates of RNA cleavage and high reaction turnover, it is necessary that (i) the oligonucleotide should efficiently bind the RNA and the cleavage-sensitive complex should be formed, and (ii) the oligonucleotide would easily dissociate from the complex after cleavage took place and hybridize with the next RNA molecule. Prolonged incubation time (20 h) of RNA:ON complexes in the presence of 2 M imidazole buffer reveals that M2-96 RNA:oligonucleotide complexes, containing bulges up to 4 nt dissociate slowly, while complexes with longer bulges exist in equilibrium of hybridized (complex 1 and complex 2) and free forms. On the other hand, the long bulges display the highest cleavage selectivity in the presence of Mg2+. Thus, complexes containing longer bulges seem favorable for RNA targeting.
RNA stem-loop structures are well investigated and several algorithms for prediction of their stability have been developed (36–38), however experimental data on stabilities of RNA:DNA bulges are limited and rather inconsistent. It is known that bulge-loops change the structure of dsRNA by introducing a kink into RNA and into the regular structure of RNA:DNA helices, dependent on the length and structure of the bulge (39–41). Due to a variety of interactions in which the bulged bases participate, the geometry and stability of bulges cannot be predicted precisely from the thermodynamic data available. Thermodynamic evaluations predict that destabilization of a duplex produced by a bulge increases monotonously as bulge size increases (42–44), but in some cases the ΔG37 values are the same for bulges of different size. In our study, we observed a decrease of RNA:oligonucleotide complex stability and formation of ‘breathing’ complexes. We assume that this decrease in stability and therefore, lower proportion of the perfect complex in the mixture (complex 1) is due to the instability of long bulges rather than to the steric difficulties of RNA unfolding necessary for the complex formation, as all the oligonucleotides tested targeted the same M2-96 RNA region. According to the published data, the nearest nucleotide neighbors strongly influence stability of bulges: in some cases, sequences containing purine bases adjacent to the bulge stabilize it (45). Other authors (46) reported that average value for the bulges flanked by two purines, by two pyrimidines, and by one purine and a pyrimidine were the same within experimental error. Our results demonstrate that only the 4-member bulges were reliably less stable in case of A:T closing pairs, but for other bulges no correlation of bulge stability and bulge sequence was observed, probably because sequences of the bulges were approximately similar (see Figure 2).
RNA cleavage catalyzed by natural ribonucleases and some chemical compounds occurs via transesterification reaction, which involves nucleophilic attack of the 2′ oxygen on the adjacent phosphorus center (47–49). Transesterification is permitted only when the attacking 2′ oxygen nucleophile is positioned in line with the 5′ oxyanion leaving group, in such a way that the leaving group is located directly on the opposing side of the phosphorus center relative to the nucleophile. The in-line conformation of any phosphodiester bond linkage is characterized by defined values of torsion angles in the nucleotide and defined interatomic distance between the 2′ oxygen nucleophile and the phosphorus electrophile (50). Attempts were made to find the sites with near to in-line conformation within different RNAs because these sites can display high cleavage sensitivity and can be the site of spontaneous RNA cleavage. Good correlations between theoretical and empirical data were obtained only for short RNAs, but in the case of long RNA (longer than 40 nt) the accuracy was poor, which could be ascribed to the differences of RNA structure in crystal and in solution.
The crystallization data show that nucleotides within natural and artificial bulge-loops can be either stacked into RNA helix (51,52) or bulged out (53–56). This flexibility can help to assume the in-line conformation. This mechanism can be proposed for RNA cleavage in the absence of Mg2+. Addition of magnesium ions makes RNA structure more rigid (57) and stabilizes the bulge. This decreases spontaneous mobility of nucleotides and their feasibility of assuming in-line conformation. Using X-ray crystallography, it was shown that in the presence of 2 mM MgCl2, adenine in a bulge is near to the in-line conformation and the distance between 2′ oxygen nucleophile and the phosphorus electrophile is short enough for the transesterification reaction to occur (58). There is no data available for the conformation of other bases in the presence of Mg2+, but we can suggest that in our case some of the bulged phosphodiester bonds exist in advantageous conformation for the efficient cleavage and that Mg2+ ions clamp this state. At the same time, the C43–G44 motif remains single-stranded but conformation of the internucleotide bond becomes unfavorable for transesterification, and this situation is also anchored under these conditions. This combination results in the site-specific cleavage of the target sequence using non-covalently attached cleaver.
Site-selective RNA cleavage can be achieved by using conjugates of oligonucleotides with RNA cleaving agents. It is known that conjugates bearing RNA cleaving groups at 5′ or 3′ ends slowly dissociate from the complex after cleavage occurred, thereby leading to a low if any reaction turnover (17). This can be improved by placing the catalytic group in the middle of the oligonucleotide and by creating artificial bulges within target RNA (15,37); however, bulge-targeting conjugates remain to be designed (17,34).
Binary system can be considered as an alternative way of achieving site-selective RNA cleavage. In this system, the cleaver is free and specially designed oligonucleotides induce very sensitive phosphodiester bonds in the target RNA. In our study, the total cleavage extent of RNA reaches 71%, where 55% fall in the cleavage of CpA motifs within the artificial bulge. This is the first example of site-selective 96mer RNA cleavage by using a binary system consisting of an oligonucleotide and of a small RNase A mimic.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We acknowledge Dr Elena Kostenko for cloning of plasmid pSVK3M2. This work was supported by grants from The Welcome Trust (No. 063630), RAS program ‘Molecular and Cellular Biology’, ‘Science to medicine’, ‘Gene-targeted biologically active compounds as antiviral and anticancer drags’, RFBR (No. 02-04-48664), SB RAS (Interdisciplinary grant No. 50, and grant in Support of Young Scientists). I.L.K. was supported by the Leonhard-Euler Fellowship Program (A-04/00939) of the the German Academic Exchange Service DAAD. Funding to pay the Open Access publication charges for this article was provided by RAS program ‘Molecular and Cellular Biology’ and SB RAS Interdisciplinary grant no. 50.
REFERENCES
- 1.Hosaka H., Sakabe I., Sakamoto K., Yokoyama S., Takaku H. Sequence-specific cleavage of oligoribonucleotide capable of forming a stem and loop structure. J. Biol. Chem. 1994;269:20090–20094. [PubMed] [Google Scholar]
- 2.Bibillo A., Figlerowicz M., Ziomek K., Kierzek R. The nonenzymatic hydrolysis of oligoribonucleotides. VII. Structural elements affecting hydrolysis. Nucleosides Nucleotides Nucleic Acids. 2000;19:977–994. doi: 10.1080/15257770008033037. [DOI] [PubMed] [Google Scholar]
- 3.Kierzek R. Hydrolysis of oligoribonucleotides: influence of sequence and length. Nucleic Acids Res. 1992;20:5073–5077. doi: 10.1093/nar/20.19.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaukinen U., Lyytikainen S., Mikkola S., Lonnberg H. The reactivity of phosphodiester bonds within linear single-stranded oligoribonucleotides is strongly dependent on the base sequence. Nucleic Acids Res. 2002;30:468–474. doi: 10.1093/nar/30.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaukinen U., Venalainen T., Lonnberg H., Perakyla M. The base sequence dependent flexibility of linear single-stranded oligoribonucleotides correlates with the reactivity of the phosphodiester bond. Org. Biomol. Chem. 2003;1:2439–2447. doi: 10.1039/b302751a. [DOI] [PubMed] [Google Scholar]
- 6.Kaukinen U., Lonnberg H., Perakyla M. Stabilisation of the transition state of phosphodiester bond cleavage within linear single-stranded oligoribonucleotides. Org. Biomol. Chem. 2004;2:66–73. doi: 10.1039/b309828a. [DOI] [PubMed] [Google Scholar]
- 7.Vlassov A.V., Vlassov V.V., Uhlenbeck O.C. RNA hydrolysis catalyzed by imidazole as a reaction for studying the secondary structure of RNA and complexes of RNA with oligonucleotides. Dokl. Akad. Nauk. 1996;349:411–413. [PubMed] [Google Scholar]
- 8.Zagorowska I., Kuusela S., Lonnberg H. Metal ion-dependent hydrolysis of RNA phosphodiester bonds within hairpin loops. A comparative kinetic study on chimeric ribo/2′-O-methylribo oligonucleotides. Nucleic Acids Res. 1998;26:3392–3396. doi: 10.1093/nar/26.14.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrow J.R. Hydrolytic cleavage of RNA catalyzed by metal ion complexes. Met. Ions Biol. Syst. 1996;33:561–592. [PubMed] [Google Scholar]
- 10.Mikkola S., Kaukinen U., Lonnberg H. The effect of secondary structure on cleavage of the phosphodiester bonds of RNA. Cell Biochem. Biophys. 2001;34:95–119. doi: 10.1385/CBB:34:1:95. [DOI] [PubMed] [Google Scholar]
- 11.Pan T., Dichtl B., Uhlenbeck O.C. Properties of an in vitro selected Pb2+ cleavage motif. Biochemistry. 1994;33:9561–9565. doi: 10.1021/bi00198a023. [DOI] [PubMed] [Google Scholar]
- 12.Bibillo A., Figlerowicz M., Kierzek R. The non-enzymatic hydrolysis of oligoribonucleotides VI. The role of biogenic polyamines. Nucleic Acids Res. 1999;27:3931–3937. doi: 10.1093/nar/27.19.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riepe A., Beier H., Gross H.J. Enhancement of RNA self-cleavage by micellar catalysis. FEBS Lett. 1999;457:193–199. doi: 10.1016/s0014-5793(99)01038-8. [DOI] [PubMed] [Google Scholar]
- 14.Ciesiolka J., Michalowski D., Wrzesinski J., Krajewski J., Krzyzosiak W.J. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J. Mol. Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 15.Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V.V., Westhof E., Ebel J.P., Giege R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J. Mol. Biol. 1985;184:455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- 16.Husken D., Goodall G., Blommers M.J., Jahnke W., Hall J., Haner R., Moser H.E. Creating RNA bulges: cleavage of RNA in RNA/DNA duplexes by metal ion catalysis. Biochemistry. 1996;35:16591–16600. doi: 10.1021/bi961700c. [DOI] [PubMed] [Google Scholar]
- 17.Hall J., Husken D., Haner R. Towards artificial ribonucleases: the sequence-specific cleavage of RNA in a duplex. Nucleic Acids Res. 1996;24:3522–3526. doi: 10.1093/nar/24.18.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niittymaki T., Kaukinen U., Virta P., Mikkola S., Lonnberg H. Preparation of azacrown-functionalized 2′-O-methyl oligoribonucleotides, potential artificial RNases. Bioconjug. Chem. 2004;15:174–184. doi: 10.1021/bc034166b. [DOI] [PubMed] [Google Scholar]
- 19.Kuzuya A., Mizoguchi R., Morisawa F., Machida K., Komiyama M. Metal ion-induced site-selective RNA hydrolysis by use of acridine-bearing oligonucleotide as cofactor. J. Am. Chem. Soc. 2002;124:6887–6894. doi: 10.1021/ja025653p. [DOI] [PubMed] [Google Scholar]
- 20.Kuzuya A., Mizoguchi R., Sasayama T., Zhou J.M., Komiyama M. Selective activation of two sites in RNA by acridine-bearing oligonucleotides for clipping of designated RNA fragments. J. Am. Chem. Soc. 2004;126:1430–1436. doi: 10.1021/ja0389568. [DOI] [PubMed] [Google Scholar]
- 21.Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch E.M., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J.P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silberklang M., Gillum A.M., RajBhandary U.L. Use of in vitro 32P labelling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- 25.Silberklang M., Prochiantz A., Haenni A.L., Rajbhandary U.L. Studies on the sequence of the 3′-terminal region of turnip-yellow-mosaic-virus RNA. Eur. J. Biochem. 1977;72:465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- 26.Ceglarek J.A., Revzin A. Studies of DNA–protein interactions by gel electrophoresis. Electrophoresis. 1989;10:360–365. doi: 10.1002/elps.1150100514. [DOI] [PubMed] [Google Scholar]
- 27.Donis-Keller H. Site specific enzymatic cleavage of RNA. Nucleic Acids Res. 1979;7:179–192. doi: 10.1093/nar/7.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donis-Keller H., Maxam A.M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlassov V.V., Vlassov A.V. Cleavage of RNA by imidazole. In: Zenkova M.A., editor. Artificial Ribonucleases, Nucleic Acids and Molecular Biology. Vol. XIII. Heidelberg: Springer Verlag; 2004. pp. 49–88. [Google Scholar]
- 30.Konevets D.A., Bekk I.E., Sil'nikov V.N., Zenkova M.A., Shishkin G.V. Chemical ribonucleases. 3. Synthesis of organic catalysts of hydrolysis of phosphodiester bonds based on quaternary salts of 1,4-diazabicyclo(2.2.2)octane. Bioorg. Khim. 2000;26:852–861. [PubMed] [Google Scholar]
- 31.Zenkova M., Beloglazova N., Sil'nikov V., Vlassov V., Giege R. RNA cleavage by 1,4-diazabicyclo[2.2.2]octane-imidazole conjugates. Methods Enzymol. 2001;341:468–490. doi: 10.1016/s0076-6879(01)41171-2. [DOI] [PubMed] [Google Scholar]
- 32.Kierzek R. Nonenzymatic hydrolysis of oligoribonucleotides. Nucleic Acids Res. 1992;20:5079–5084. doi: 10.1093/nar/20.19.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beloglazova N., Vlassov A., Konevetc D., Sil'nikov V., Zenkova M., Giege R., Vlassov V. Mechanism and specificity of RNA cleavage by chemical ribonucleases. Nucleosides Nucleotides. 1999;18:1463–1565. doi: 10.1080/07328319908044753. [DOI] [PubMed] [Google Scholar]
- 34.Astrom H., Williams N.H., Stromberg R. Oligonucleotide based artificial nuclease (OBAN) systems. Bulge size dependence and positioning of catalytic group in cleavage of RNA-bulges. Org. Biomol. Chem. 2003;1:1461–1465. doi: 10.1039/b212216b. [DOI] [PubMed] [Google Scholar]
- 35.Kaukinen U., Bielecki L., Mikkola S., Adamiak W., Lonnberg H. The cleavage of phosphodiester bonds within small RNA bulges in the presence and absence of metal ion catalysts. J. Chem. Soc. Perkin 2. 2001;2:1024–1031. [Google Scholar]
- 36.Antao V.P., Lai S.Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antao V.P., Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dale T., Smith R., Serra M.J. A test of the model to predict unusually stable RNA hairpin loop stability. RNA. 2000;6:608–615. doi: 10.1017/s1355838200992495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhattacharyya A., Murchie A.I., Lilley D.M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 40.Zacharias M., Hagerman P.J. Bulge-induced bends in RNA: quantification by transient electric birefringence. J. Mol. Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 41.Luebke K.J., Tinoco I., Jr Sequence effects on RNA bulge-induced helix bending and a conserved five-nucleotide bulge from the group I introns. Biochemistry. 1996;35:11677–11684. doi: 10.1021/bi960914r. [DOI] [PubMed] [Google Scholar]
- 42.Delisi C., Crothers D.M. Prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 1971;68:2682–2685. doi: 10.1073/pnas.68.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gralla J., Crothers D.M. Free energy of imperfect nucleic acid helices. 3. Small internal loops resulting from mismatches. J. Mol. Biol. 1973;78:301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- 44.Longfellow C.E., Kierzek R., Turner D.H. Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry. 1990;29:278–285. doi: 10.1021/bi00453a038. [DOI] [PubMed] [Google Scholar]
- 45.LeBlanc D.A., Morden K.M. Thermodynamic characterization of deoxyribooligonucleotide duplexes containing bulges. Biochemistry. 1991;30:4042–4047. doi: 10.1021/bi00230a031. [DOI] [PubMed] [Google Scholar]
- 46.Znosko B.M., Silvestri S.B., Volkman H., Boswell B., Serra M.J. Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges. Biochemistry. 2002;41:10406–10417. doi: 10.1021/bi025781q. [DOI] [PubMed] [Google Scholar]
- 47.Raines R.T. Ribonuclease A. Chem. Rev. 1998;98:1045–1066. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 48.Oivanen M., Kuusela S., Lonnberg H. Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by Bronsted acids and bases. Chem. Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 49.Zhou D.M., Taira K. The hydrolysis of RNA: from theoretical calculations to the hammerhead ribozyme-mediated cleavage of RNA. Chem. Rev. 1998;98:991–1026. doi: 10.1021/cr9604292. [DOI] [PubMed] [Google Scholar]
- 50.Soukup G.A., Breaker R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borer P.N., Lin Y., Wang S., Roggenbuck M.W., Gott J.M., Uhlenbeck O.C., Pelczer I. Proton NMR and structural features of a 24-nucleotide RNA hairpin. Biochemistry. 1995;34:6488–6503. doi: 10.1021/bi00019a030. [DOI] [PubMed] [Google Scholar]
- 52.Smith J.S., Nikonowicz E.P. NMR structure and dynamics of an RNA motif common to the spliceosome branch-point helix and the RNA-binding site for phage GA coat protein. Biochemistry. 1998;37:13486–13498. doi: 10.1021/bi981558a. [DOI] [PubMed] [Google Scholar]
- 53.Ippolito J.A., Steitz T.A. The structure of the HIV-1 RRE high affinity rev binding site at 1.6 A resolution. J. Mol. Biol. 2000;295:711–717. doi: 10.1006/jmbi.1999.3405. [DOI] [PubMed] [Google Scholar]
- 54.Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Kundrot C.E., Cech T.R., Doudna J.A. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 55.Cate J.H., Gooding A.R., Podell E., Zhou K., Golden B.L., Szewczak A.A., Kundrot C.E., Cech T.R., Doudna J.A. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 56.van den Worm S.H., Stonehouse N.J., Valegard K., Murray J.B., Walton C., Fridborg K., Stockley P.G., Liljas L. Crystal structures of MS2 coat protein mutants in complex with wild-type RNA operator fragments. Nucleic Acids Res. 1998;26:1345–1351. doi: 10.1093/nar/26.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leroy J.L., Gueron M. Electrostatic effects in divalent ion binding to tRNA. Biopolymers. 1977;16:2429–2446. doi: 10.1002/bip.1977.360161108. [DOI] [PubMed] [Google Scholar]
- 58.Portmann S., Grimm S., Workman C., Usman N., Egli M. Crystal structures of an A-form duplex with single-adenosine bulges and a conformational basis for site-specific RNA self-cleavage. Chem. Biol. 1996;3:173–184. doi: 10.1016/s1074-5521(96)90260-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.