Abstract
Genetically modified (GM) crops are the fastest adopted commodities in the agribiotech industry. This market penetration should provide a sustainable basis for ensuring food supply for growing global populations. The successful completion of two decades of commercial GM crop production (1996–2015) is underscored by the increasing rate of adoption of genetic engineering technology by farmers worldwide. With the advent of introduction of multiple traits stacked together in GM crops for combined herbicide tolerance, insect resistance, drought tolerance or disease resistance, the requirement of reliable and sensitive detection methods for tracing and labeling genetically modified organisms in the food/feed chain has become increasingly important. In addition, several countries have established threshold levels for GM content which trigger legally binding labeling schemes. The labeling of GM crops is mandatory in many countries (such as China, EU, Russia, Australia, New Zealand, Brazil, Israel, Saudi Arabia, Korea, Chile, Philippines, Indonesia, Thailand), whereas in Canada, Hong Kong, USA, South Africa, and Argentina voluntary labeling schemes operate. The rapid adoption of GM crops has increased controversies, and mitigating these issues pertaining to the implementation of effective regulatory measures for the detection of GM crops is essential. DNA-based detection methods have been successfully employed, while the whole genome sequencing using next-generation sequencing (NGS) technologies provides an advanced means for detecting genetically modified organisms and foods/feeds in GM crops. This review article describes the current status of GM crop commercialization and discusses the benefits and shortcomings of common and advanced detection systems for GMs in foods and animal feeds.
Keywords: Genetically modified crops, Detection, Real-time PCR, ELISA, Next-generation sequencing, Transgenic
Introduction
The production of plants with improved quality traits such as disease resistance, prolonged shelf-life, and drought resistance by conventional breeding is extremely time consuming (Delaney 2015). However, the demand for food due to an ever-expanding global population and changes in eating habits has continually increased the demand for more productive food and feed crops. In fact, the provision of sufficient food to feed an estimated 9.7 billion people by 2050 (FAO 2017) and approximately 11.0 billion by 2100 (James 2015) is one of the major challenges of this century. Genetically modified (GM) crops provide an opportunity to increase food and feed production efficiently by generating plants with higher yields and greater nutritional benefits in reasonably short times (The Gaurdian 2016). GM crops offer the possibility of expanding the accessible gene pool for breeding by overcoming sexual incompatibilities between plants and providing the opportunity to use genes with beneficial traits and regulatory elements to express genes of prokaryotic or viral origin (Southgate et al. 1995).
GM crops contain novel genes (transgenes) with improved quality traits, such as herbicide tolerance, and allow the developmental process to be dramatically accelerated (Gressey 2013). GMOs (genetically modified organisms) enable the barrier of sexual incompatibility between plant species to be overcome and enormously increase the size of the available gene pool (Gressey 2013). GM crops have revolutionized agricultural commodities by allowing breeders to introduce specific genes from a wide variety of sources to produce more useful and productive crops (Tarafdar et al. 2014). The rapid adoption of GM crops within the agricultural sector has increased agricultural productivity, contributed to economic growth, and allowed food demand to be met (James 2015). The term “genetically modified organism” means an organism in which genetic material has been altered in a way that does not occur naturally by mating and/or natural recombination (Directive 2001/18/EC). GM crops are modified using recombinant DNA technology in three different ways, that is, transgenic, cisgenic, or intragenic. ‘Transgenic’ modification involves the insertion of foreign DNA from an unrelated genus or species. ‘Cisgenic’ involves the insertion of one or more gene of related species or from a crossable donor. The introduction of specific alleles/genes present in the gene pool, without any DNA sequence change, into new varieties is termed ‘cisgenesis’ (Schouten et al. 2006), and such processes accelerate the breeding of species with long reproduction cycles with no linkage drag. On the other hand, ‘intragenic’ modifications involve the use of genetic elements from other plants from the same sexually compatible gene pool and, thus, the coding regions of genes are combined with promoters and terminators of different genes from the same sexually compatible gene pool. Furthermore, there is a certain reluctance to accept GM foods created by transgenesis rather than cisgenics, as the latter process appears to be more natural. Introduction of the R1 gene, which provides resistance to late blight of potato, from wild-type potato (Solanum demissum) to cultivated potato (S. tuberosum) is a cisgenic process. However, the transfer of the Bt gene from the bacterium Bacillus thuringiensis to the cotton genome to produces pest-resistant cotton is an example of transgenesis (Wu et al. 2008). Genetically modified crops are exploited to develop the desired quality traits, such as drought, temperature, or salinity tolerance or disease resistance (Key et al. 2008). The Flavr Savr tomato was the first genetically modified crop developed by employing anti-sense technology and introduced to the market in 1994 (Bates et al. 2005), and since then more than 100 GMOs have been approved worldwide for use in commercial foods or feeds (http://www.gmo-compass.org, http://www.agbios.com/dbase.php) (Redenbaugh et al. 1992).
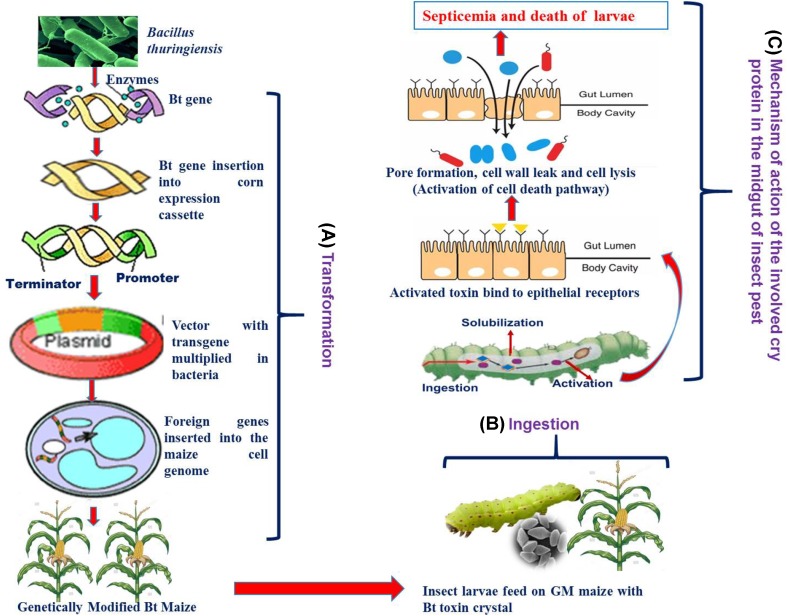
Genetically modified Bt corn carries gene variants of Cry proteins from the soil bacterium B. thuringiensis. These proteins, also known as Bt toxins, species-specifically kill important plant pests like insects of the orders Lepidoptera, Coleoptera, Diptera, and others if ingested (Trapero et al. 2016). B. thuringiensis is a Gram-positive spore-forming bacterium with entomopathogenic properties and a long history of safe use as a sprayable biopesticide (Trapero et al. 2016). Parasporally formed crystals are predominantly composed of one or more proteins (Cry and Cyt toxins) also called δ-endotoxin, which lyse epithelial cells of the insect midgut by inserting pores into the plasma membrane. Cry toxins are innocuous to humans, vertebrates, and plants and are completely biodegradable (Tabashnik et al. 2003). The effect of Bt maize on the life cycle of the insect is depicted in Fig. 1.
Fig. 1.
Schematic representation of the effect of Bt maize on the life cycle of insect. A Transformation of Bt gene in maize. B Ingestion of GM maiz expressing Bt gene by insect larvae. C Mechanism of action of the involved cry protein in the midgut of insect pest
The most widely accepted genetically modified traits in GM crops are herbicide tolerance and insect resistance. GM soybean, maize, canola, and cotton are the most common examples of these crops in the market (James 2014). Developing countries like India and China are the largest producers of genetically modified Bt cotton (Markoulatos et al. 2004; Trapero et al. 2016). The most common pest of cotton is the bollworm (corn earworm; Helicoverpa zea) and the next most common ones are the boll weevil and pink bollworms (Wu et al. 2008). Bt cotton (Gossypium hirsutum) MON531 lepidopteran-resistant variety expresses Cry1Ac in the cotton plant (Randhawa et al. 2014). In 1998, maize MON 810 was the first genetically modified crop approved for commercial cultivation in the EU (Randhawa et al. 2016), and this was followed in 2010 by the AMFLORA potato (also known as EH92-527-1), which has a high amylopectin content. AMFLORA received criticism due to the presence of the antibiotic resistance gene nptII and was withdrawn from the EU market by the European Commission in 2013 due to procedural errors during the approval process http://www.basf.com/group/corporate/en/products-andindustries/biotechnology/plant-biotechnology/amflora. Directive (2001/18/EC) addresses GMO Regulation 1829/2003 on genetically modified food and feed, and regulation 1830/2003 concerns the traceability and labeling of GMOs. The first generation of GM crops contained a single Bt gene (Cry1Ac, Cry1ab, etc.) and enhanced economic benefits to farmers by increasing yields and cost-effectiveness (Bawa and Anilakumar 2013).
Some cultivars of GM corn and cotton are ‘stacked’ events as they carry a transgene/foreign DNA for insect resistance (IR) and herbicide tolerance (HT) traits (Halpin 2005). Additionally, USDA-ERS (2013) claimed that 50% of GM crops are ‘stacked’ events in GM corn and cotton. Genetically modified crops of the first generation possess properties that are relevant only for agriculture praxis. In particular, herbicide- and insecticide-tolerant GM crops belong to this category. GM plants of the second generation differ, because embedded traits are intended to provide benefits to consumers and for industrial applications. A database of GM crop information is available in the link http://cera-gmc.org/index.php?action=gm_crop_database and is accessible to the public (Wu et al. 2014; Lin and Pan 2016). Third-generation GM crops carry a transgene construct that has not been used in other (known) GM crops and has undergone minimal recombination or modification (Holst-Jensen et al. 2012; Lin and Pan 2016). Third-generation GM crops have been successfully commercialized for recombinant vaccine production, producing industrial products, such as, monoclonal antibodies, vaccines, plastics and biofuel, and for bioremediation (Ma et al. 2003; Sticklen 2005; Conrad 2005; Key et al. 2008; James 2015).
Global production status of GM crops
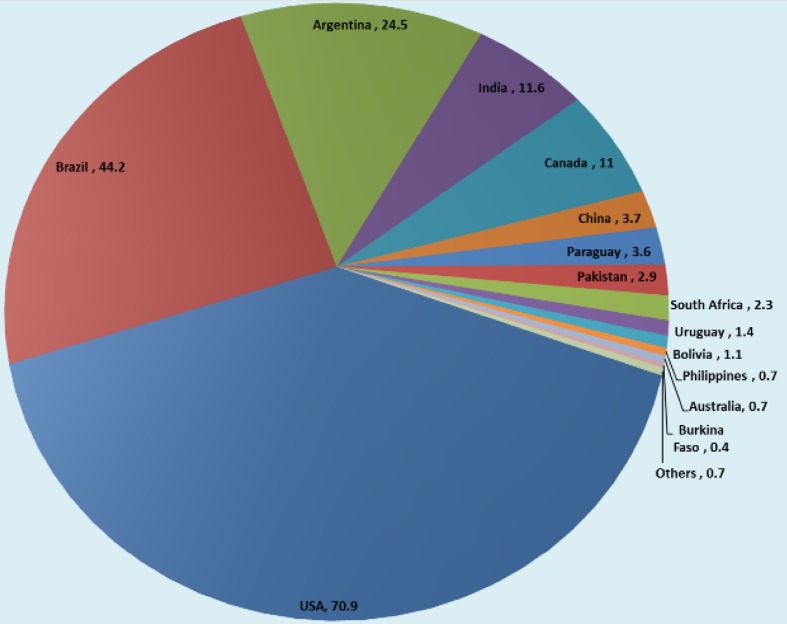
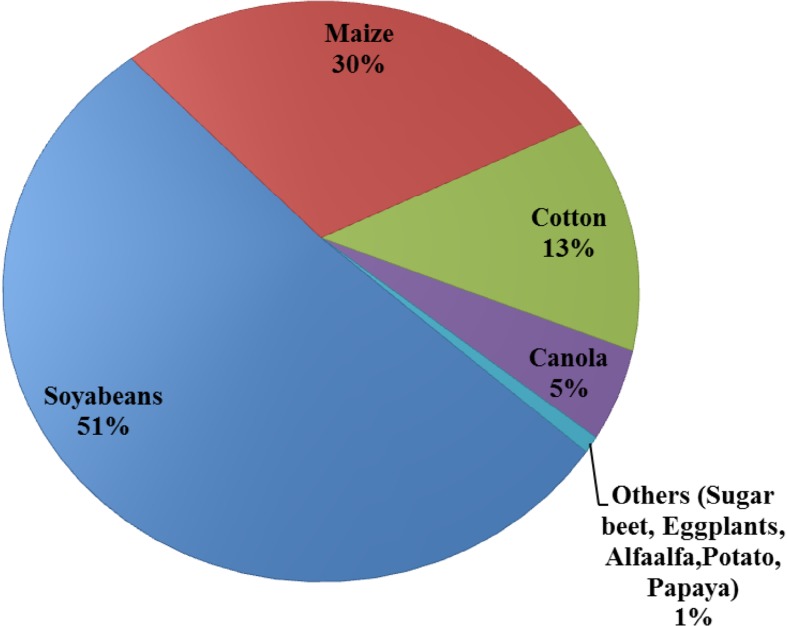
According to a recent release in ISAAA briefs (James 2015), the global production status of genetically modified crops increased by 100-fold between 1996 and 2015 from 1.7 to 179.7 million ha (1996–2015) (James 2015) (Fig. 2). Subsequently, there was a tremendous increase in the commercialization of GM crops (Fig. 3) at a rate unsurpassed during the history of modern agriculture (James 2015). Currently, USA is the world’s largest producer of GM crops with 70.9 million ha (39%), 90% of which is accounted for by maize, soybean, and cotton. Brazil is the second largest with 44.2 million ha (25% of the total global production) and planted stacked events (HT/IR) on a record 11.9 million ha. Argentina is the world’s third leading producer with 24.3 million ha. India has ranked fourth with 11.6 million ha of Bt cotton and has registered phenomenal growth in cotton production and topped the world with 95% resilient adoption rate. (James 2015). Canada has ranked fifth with 11.0 million ha (Fig. 4). Hence, the five major global GM crops are soybean, cotton, maize, and canola. In 2015, 82% (90.7 of 111 million ha) of the soybean planted were GM soybean strains, whereas GM cotton accounted for 68% (25.1 of 37 million ha) of global cotton production (Figs. 3, 4; James 2015). Of the 184 million hectares of maize planted, global 55.2 million ha (30%) was GM maize (Table 1). Moreover, herbicide-tolerant GM canola accounted for 25% of global planting (9 of 36 million ha) in 2015 (James 2015). The annual total for these four crops was 368 million ha, and 181.5 million ha (49%) comprised GM crops. According to a recent survey, the agronomic and economic benefits of GM crops are significant, as these benefits are dependent on the modified trait and geographical area (Klumper and Qaim 2014). High-yielding insect-resistant (IR) and herbicide-tolerant (HR) crops are greatly adopted by developing countries. Recently, genetically modified potato (Innate™) generation I with multi-trait resistance to black-spot bruising and browning was developed using RNA interference technology (Simplot Company) and successfully commercialized in 160 ha in the USA (James 2015). Innate™ II with a disease resistance trait for late blight of potato was subsequently approved. In terms of genetically modified animals, landmark approval of the first GM salmon was granted by FDA in 2012 for commercial food production and human consumption. The products are expected to be available in the US market by 2018 (James 2015).
Fig. 2.
The global areas of production of GM crops in 2015 (total. 181.5 million ha). Others include (Myanmar 0.3, and Mexico 0.2; Spain 0.1; Colombia 0.1; Sudan 0.1 and Honduras, Chile, Portugal, Cuba, Czech Republic, Romania, Slovakia, Costa Rica, Bangladesh less than 0.1 million ha).
Source: James 2015
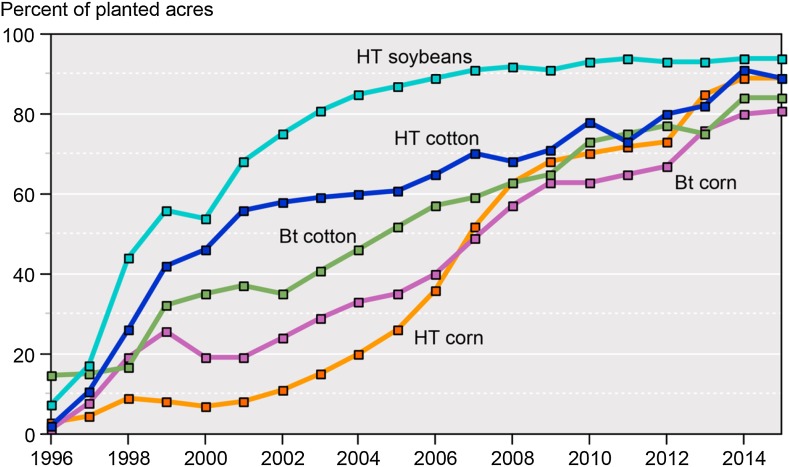
Fig. 3.
US statistics of Adoption of GM crops from 1996–2015 (Source: USDA, Economic Research Service using data from Fernandez-Cornejo and McBride (2002) for the year 1996–99 and USDA, National Agricultural Statistics Service, June Agricultural Survey for the years 2000–15; http://www.ers.usda.gov/data-products/adoption-of-genetically-engineered-crops-in-the-us/recent-trends-in-ge-adoption.aspx)
Fig. 4.
Global production of major GM and other GM crops and their percent statistics (James 2016)
Table 1.
The global area of production of GM crops in 2015.
Source: James 2015
| Rank | Country | Area (mil. ha) | GM crops |
|---|---|---|---|
| 1 | USA | 70.9 | Maize, soybean, cotton, canola, sugar beet, alfalfa, papaya, squash |
| 2 | Brazil | 44.2 | Maize, soybean, cotton |
| 3 | Argentina | 24.5 | Maize, soybean, cotton |
| 4 | India | 11.6 | Cotton |
| 5 | Canada | 11 | Canola, maize, soybean, sugar beet |
| 6 | China | 3.7 | Cotton, papaya, poplar, tomato, sweet pepper |
| 7 | Paraguay | 3.6 | Soybean, maize, cotton |
| 8 | Pakistan | 2.9 | Cotton |
| 9 | South Africa | 2.3 | Maize, soybean, cotton |
| 10 | Uruguay | 1.4 | Soybean, maize |
| 11 | Bolivia | 1.1 | Soybean |
| 12 | Philippines | 0.7 | Maize |
| 13 | Australia | 0.7 | Cotton, canola |
| 14 | Burkina Faso | 0.4 | Cotton |
| 15 | Myanmar | 0.3 | Cotton |
| 16 | Mexico | 0.2 | Cotton, soybean |
| 17 | Spain | 0.1 | Maize |
| 18 | Colombia | 0.1 | Cotton, maize |
| 19 | Sudan | 0.1 | Cotton |
| 20 | Honduras | <0.1 | Maize |
| 21 | Chile | <0.1 | Maize, soybean, canola |
| 22 | Portugal | <0.1 | Maize |
| 23 | Cuba | <0.1 | Maize |
| 24 | Czech Republic | <0.1 | Maize |
| 25 | Romania | <0.1 | Maize |
| 26 | Slovakia | <0.1 | Maize |
| 27 | Costa Rica | <0.1 | Cotton, soybean |
| 28 | Bangladesh | <0.1 | Brinjal, eggplant |
| Total | 181.5 |
Detection of genetically modified organisms in GM crops
GM crops are 100% genetically modified, that is, each cell of the transgenic plant contains a copy of the transgene insert. The detection and quantification of GM content in GM foods or feed is an important consideration before acceptance/commercialization (Pandey et al. 2010; Delaney 2015).
Biosafety measurements came into existence as ‘safety filter’ by testing and labeling of genetically modified organisms in GM produce for food/feed and, thus, minimized the potential risks to the environment and human health (Delaney 2015). GM labeling requirements for food products were introduced by the EU (Regulation (EC) 258/97) to determine whether products were recognized as “safe” for consumer uptake and commercialization. The traceability and contents of GMOs in GM crops and associated labeling systems are mandatory requirements in many countries (Australia, Brazil, Chile, China, EU, India, Indonesia, Israel, Japan, Philippines, Russia, South Korea, Taiwan, and Thailand), whereas in Argentina, Canada, and USA they are voluntary (Directive: EC/1829/2003; Delaney 2015).
The successful commercialization of GM crops is hampered by existing strict regulations and many countries have enacted legislation to regulate GMO and GMO-derived products (Wu et al. 2014; Fraiture et al. 2015a, b). Data on molecular characterization, data analysis of an inserted gene of interest, and location in the genome are regarded as important requirements by regulatory authorities. Hence, the labeling and traceability of GMOs in food/feed are huge contemporary issues with respect to the release of GM crops before commercialization, as evidenced by EU legislation (Miraglia et al. 2004; Yang et al. 2005a, b). The precautionary measures deployed in accordance with the Cartagena Protocol on Biosafety and Convention on Biological Diversity (Gruere and Rao 2007; Fraiture et al. 2015a, b) ensure adequate levels of protection during the transfer, handling, and proper labeling of GM crops (Gruere and Rao 2007). The enforcement of a ‘threshold value’ has created a demand for the development of reliable and accurate detection methods (Gressey 2013). The European Commission (EC) is responsible for establishing rules for the assessment and authorization of GM food labeling, traceability, and commercialization of GMOs in individual European Union (EU) member states (Mitchell 2003; Davidson 2010). The EU has strict regulations for the labeling of genetically modified organisms, GM foods and recombinant DNA crops, and those from genetic engineering technology with focused interest for the requirement of highly sensitive detection methods. Furthermore, the specificity of detection methods becomes more of an issue as the number of product types increases, mixed GMO products are devised, or different processing methods are introduced. To meet the demands of the regulatory labeling issues for GM crops, sensitive and reliable methods that utilize advanced protein techniques and DNA-based detection methods are required.
An acceptable GMO detection technique should:
Detect all known GMOs.
Provide quantitative data on the content and/or presence of GMOs.
Enable the testing of extensive ranges of food and agriculture produce.
Be reliable, reproducible, and produce minimum levels of false-positive results.
Be sensitive and accurate under all laboratory conditions.
Phenotype-based detection
The bioassays are used to detect the GM crops and analyze adverse environmental and health and safety issues. A bioassay is a complementary technique for providing analytical and biological information about herbicides and determining their phytotoxic effects (Marmiroli et al. 2000). These phenotypic bioassays provide essential information on soil–plant–herbicide relationships (Marmiroli et al. 2000) and on responses of plants to herbicides (Borem and Almeida 2011). Phenotype detection allows the presence of a trait particularly used for herbicide resistance (HR) and insect resistance (IR), such as of Cry1a protein or gene in GM maize for MON80100, MON801, MON802, MON809, and others, to be determined (Torres et al. 2003; Ladics et al. 2015). Herbicide (HR) bioassays are claimed to be accurate, cheap, and user friendly and are used by seed companies for quality assurance purposes. Currently, the herbicide-resistant bioassay is commercially available for soybean, maize, cotton, oilseed rape, etc. (Ladics et al. 2015). The efficiency of the bioassay is based on the number of total seed samples tested and germinated, and insect resistance (IR) is determined by counting insect larvae that die immediately after feeding the leaves of genetically modified GM crop. However, such bioassays are not available for the testing of processed foods, crushed grain, or for the identification of a single GM event.
DNA-based detection methods
Southern blotting
The molecular characterization of a single copy gene in the genome of a transformed sample or GM crop is performed by Southern blot hybridization (Southern 1975). SB is the most commonly used technique for the nucleic acid (DNA)-based detection of successful gene integration, copy number insertion, and incorporation of sequences from the transformation plasmid backbone. SB is a low-density screening method that employs a combination of few restriction enzymes and probes. Recently, Zastraw-Hayes et al. (2015) described the use of an innovative Southern-by-Sequencing approach, which provides sequence-level information about the DNA flanking sequences at insertion sites (used to characterize genomic locations), coupled with NGS to develop event-specific PCR assays for insertions (Zastraw-Hayes et al. 2015).
PCR-based detection
Molecular detection methods based on PCR are used to determine the presence of small quantities of foreign DNA/transgene inserts in genetically modified crops. Traditionally, the target DNA contains CaMV 35 constitutive promoter, NOS terminator, nptII (antibiotic resistance gene), and the Ti plasmid of Agrobacterium tumefaciens (Yang et al. 2013; Datukishvili et al. 2015; Randhawa et al. 2016). The increased production of GM crops harboring stacked events has limited the abilities of sensitive, user-friendly, cost-effective detection techniques to detect the presence of genetically modified organisms (Fu et al. 2015; Fraiture et al. 2015a, b). As a result, other techniques are also gaining importance with high thermal and chemical stability of DNA, apparent potential automation, and sensitivity, and thus the onset of sequence-based high-throughput technologies is trending (Zimmermann et al. 2000). To address the regulatory needs for GM crop labeling effectively, reliable analytical methods are urgently required for the detection and quantification of the existence of GM content in GM foods and feeds.
Event-specific PCR
Event-specific PCR is used to detect genetically modified organisms using trait-specific events, i.e., the junction sequences for integration between a transgene construct and the plant genome. The PCR assay technique used to amplify this DNA sequence is referred to as ‘transformation event’-specific PCR (Wu et al. 2014; Randhawa et al. 2016). The integration of a single transgene sequence into a plant genome is a random event and is highly unlikely to occur at the same genomic locus in two different GMOs, and this can be used to devise unambiguous detection assays specific for the transgenic construct and the integration site to be devised. The sequence information of a GMO is essentially required to detect legitimate transgenic events and a non-GM sample is needed as reference material (Wu et al. 2014). The P35S CaMV promoter and NOS terminator are the GMO screening targets most widely used for identification in the EU recommendation for commercial food and feed (Peano et al. 2005; Wu et al. 2014). An assay specific for the transgenic construct developed for the specific detection of maize Bt 11, MON810, GA21, and LY038 has been devised (Windels et al. 2003; Randhawa et al. 2016), and similarly Roundup Ready transformation-event-specific PCR for soybean (Lin and Pan 2016). A European Union database of 50 GM reference events for analysis based on the Compendium of Reference Methods for the GMO Analysis is available at GMOMETHODS and http://gmo-crl.jrc.ec.europa.eu/ (Randhawa et al. 2016; Guertler et al. 2013; Randhawa and Singh 2012; Wu et al. 2010). The event-specific PCR can potentially be utilized as a marker for screening of trait-specific known GMOs and their identification.
Construct-specific detection
Construct-specific PCR detection is based on two adjoining DNA sequences carrying the ‘transgene’ also resulting in GMOs containing the same construct. Construct-specific assays targeting ctp2-Cry2Ab2, ctp2-cp4epsps, P35S-cry1Ac, and P35S-UidA have been reported (Lee et al. 2007; Grohmann et al. 2009; Randhawa and Singh 2012; Chhabra et al. 2014; Randhawa et al. 2016). However, construct-specific PCR is unable to detect a single GMO event, and hence considered unreliable and simple for screening of GMOs in GM crop.
Multiplex PCR
Multiplex PCR is a variant of conventional PCR, which involves simultaneous amplification of multiple target gene sequences in a single reaction and saves time, effort, and cost. A multiplex PCR system developed for amplification of four target DNA sequences of (cp4-epsps, m-epsps, pat, bar, and ribulose bisphosphate carboxylase RBCL gene) for the simultaneous detection of GM soybean, maize, rice, and their products (Jinxia et al. 2011) and seven gene sequences containing the lec and zein genes (Kim et al. 2013; Huber et al. 2013; Randhawa et al. 2016) have been developed. Multiplex assays have been described for salinity- and drought-tolerant GM tomatoes, GM potatoes with better protein quality, and Bt crops including Bt brinjal, Bt okra, Bt potato, and Bt rice (Randhawa et al. 2016). However, the development of multiplex assays requires careful testing and validation.
Real-time PCR
Quantitative detection by real-time PCR is a method of choice for GM content detection in food or feed (Mano et al. 2009; Holst-Jensen 2013). The existence of threshold limits for GM contents in produce or crops is mandatory, and sensitive and accurate methods are needed. Real-time PCR coupled with fluorometric measurements of an internal probe during the reaction enables GMO contents or gene expression levels to be precisely quantified (Peano et al. 2005). Each test sequence includes the analysis of a full set of standards, to produce standard curves for determining GMO contents of unknowns (Randhawa et al. 2013, 2014). Moreover, tests for crops like wheat, maize, rapeseed, cotton, cassava, and brinjal have been successfully developed (Li et al. 2004; Lee et al. 2006; Wu et al. 2007; Beltran et al. 2009; Ballari et al. 2013).
LAMP PCR
The loop-mediated isothermal amplification (LAMP) method is a relatively simple and field-adaptable technique for sequence determination and offers a potential alternative to PCR (Lee et al. 2009). Target DNA is amplified under isothermal conditions using DNA polymerase (from Bacillus stearothermophilus) and amplification results in the formation of magnesium pyrophosphate. The LAMP method does not involve the use of a thermal cycler and is used to detect GMOs specifically and sensitively. Target event-specific sequences are available for transgenic MS8 and RF3 oilseed rape, transgenic element CaMV 35S, and NOS terminator. Moreover, a LAMP protocol combined with gel electrophoresis was developed for the detection of GM oil seed rape with a detection limit of 0.01%, and the detection of three GM rice events has been reported (Randhawa et al. 2014; Chen et al. 2012; Kiddle et al. 2012). In addition to this, a bioluminescent real-time reporter (BART) of LAMP, targeting the P35S, T-NOS, and Adh1 gene, was developed for the MON810 GM maize event (Kiddle et al. 2012), and event-specific assays have been developed for two soybean events, three GM rice events, and seven GM maize events (Chen et al. 2011, 2012; Guan et al. 2010). Similarly, event-specific visual and real-time LAMP assays have been described for the detection of two major commercialized Bt cotton events, MON531 and MON15985 (Randhawa et al. 2015). The portability, isothermal nature, and the lack of a need for a DNA extraction kit mean that real-time LAMP assays are well suited to GMO detection in the field (Randhawa et al. 2016).
Matrix
The matrix-based GMO screening approach is a combination of five target elements developed for 81 authorized/unauthorized GM events as determined by the European Union (Waiblinger et al. 2010). This matrix approach involves a stepwise process integrated with a decision support system (DSS) and a selection of target genetically modified organisms (GMOs) and their data analysis techniques (Randhawa et al. 2016). A GMOtrack algorithm has been developed http://kt.ijs.si/software/GMOtrack/GMOseek.html for the detection of GMO events in crop species and for potential target screening, and is freely available (Holst-Jensen et al. 2012). The matrix-based GMO screening approach provides a user-friendly, cost-effective means for detecting authorized or unauthorized GM plants (Kralj-Novak et al. 2009; James 2009; Waiblinger et al. 2010).
Protein-based detection methods
The detection of GMOs in plants is restricted to fresh/frozen foods or unprocessed food samples, because at high temperature proteins are denatured. GMO protein samples are resolved by one-dimensional SDS–gel electrophoresis. However, the differences between GMO and reference samples are not well resolved. Two-dimensional gel electrophoresis provides better resolution, but does not generally provide unequivocal identifications of transgene products, unless combined with immunological methods. The expression levels of genetically modified products in plants with strong constitutive promoter P35S (CaMV) expression will constitute only 0–2% of total soluble protein (Padgette et al. 1995). The concentration of a genetically modified organism (GMO) in plant tissues (>10 µg per tissue sample) means that given the limit of detection (LOD) of a typical protein, the presence of recombinant or genetically modified protein can only be detected in up to 1% of GMOs (Stave 2002). Immune-sensor techniques are mainly used to analyze serum and blood samples (Morgan et al. 1996), and the immunological methods described below depend on the availabilities of highly specific antibodies.
ELISA
PCR ELISA (enzyme-linked immunosorbent assay) offers a high-throughput approach suitable for automation. ELISA is based on the specific hybridization of immobilized, biotinylated PCR products (CaMV 35S Promoter) with digoxigenin-labeled internal probes suitable for colorimetric detection. ELISA is less time consuming than other detection techniques and can detect as little as 0.1 ng of amplicons in 2 h. ELISA has been successfully employed to detect the protein encoded by the cp4-epsps gene in Roundup Ready soybean (Rogan et al. 1999; Kamle and Ali 2013) and to detect Bt proteins like Cry1Ac and Cry1Ab (Vazquez-Padron et al. 2000). A monoclonal antibody-based sandwich ELISA immunoassay for Cry1Ac and Cry2Ab in cotton seeds/leaf samples has also been reported (Shan et al. 2007; Markoulatos et al. 2004; Kamle and Ali 2013). Various ELISA kits are commercially available for the detection of specific proteins of Cry1C, Cry3A, Cry2A, Cry9C, pat, and nptII (Anklam et al. 2002).
Western blot
Western blot is a highly sensitive qualitative technique for the detection of target proteins present below or above threshold levels in genetically modified samples, in particular, when applied to insoluble proteins and the detection limits vary between 0.1 and 1% in plant/seed samples, depending on protein expression levels (Markoulatos et al. 2004). Both ELISA and western blot techniques have been employed to detect Monsanto’s Roundup Ready soybean (Markoulatos et al. 2004).
Immunostrips
Immunostrip tests are important because they provide rapid, qualitative, and semi-quantitative information on GM proteins present and are provided as ready to use kits, which are useful for the initial screening of seed/grains. The immunostrips/dipsticks provide results in 5–10 min and are available for the detection of Cry1Ab, Cry1Ac, Cry2Ab, and Cp4-EPSPS (Fagan et al. 2001; Monsanto 1997; Kamle and Ali 2013), but not available for all commercialized GMOs. However, one important limitation of these tests is that all commercially available GMOs encode for a particular protein and, hence, are required to have a specific detection method. The second limitation of these kits is that their sensitivities lie within the range 0.1–1% and are highly inferior to PCR. It is noteworthy that ELISA and lateral flow (dipstick) detection techniques are trait specific and the same trait target proteins found in different GMOs contain the same construct (Fagan et al. 2001). Thus, antibody-based methods may not reliably differentiate GMOs precisely (e.g., maize Bt-176, Bt II, and Mon 810 contain the same cry protein). The glyphosate-resistant maize variety GA21 expresses a transgene EPSPS protein which contains few amino acids derived from the native plant (Nelson 2001). Besides this, the structures of both the native plant and transgenic EPSPS are so similar that the antibodies developed are not able to differentiate between these two (Patel 2002). Hence, no immune tests have been developed for the glyphosate-resistant maize GA21 event. Consequently, PCR-based detection methods are more reliable and accurate for GMO detection, but during the early stage of detection immunoassays or tests are also considered as suitable options.
Other detection methods
Chromatography
Chromatographic methods are being developed when the genetically modified organisms contain fatty acids, triglycerides, and certain chemicals that can only be detected suitably by qualitative ways. These analytical techniques were developed using high-performance liquid chromatography (HPLC) coupled with atmospheric pressure chemical ionization mass spectrometry (APCI-MS) in GM canola for the presence of triglycerides (Byrdwell et al. 2001; Jin et al. 2016). Quantification was performed with a flame ionizer detector (FID) coupled with HPLC and it precluded GM canola oils from containing an increased percentage of triglycerides for more oxidative stability in canola and soybean (Byrdwell et al. 2001).
Near infrared (NIR) spectroscopy
Near infrared (NIR) spectroscopy is used mainly for cereals and grains in elevators, usually for the non-destructive analysis of whole grains to determine the moisture, protein, oil, fiber, and starch contents. NIR is used to differentiate Roundup Ready soybean from conventional soybean (Agelet et al. 2013). In this technique, the spectral scans were obtained using Infratech 1220 spectrometer, where whole-grain samples flowed through a fixed path length for distinguishing Roundup Ready soybean from conventional soybean. The test is fast, cost-effective, takes less than a minute, and sample preparation is not needed because it uses whole kernels (about 300 g), which are dropped are dropped into measurement cells or flow-through system (Anklam et al. 2002). However, NIR’s major limitation is that it is unable to generate spectra for large sets of samples for identification purposes and changes in DNA or protein.
Biosensor-based detection
Biosensor is an analytical device which converts a biological element or response into electrical signals (Baeumner et al. 2003; Nakamura and Karube 2003). The biological element can be a complex structure, such as a tissue or organelle, or can be composed of isolated structures, such as a specific tissue or organelle, or it can be composed of isolated molecules, such as antibodies, enzymes, or nucleic acids (Minunni et al. 2001; Arugula et al. 2014). Biosensor-based detection is fast expanding for exploring high-throughput screening of GMO’s content for appropriate labeling for on-site import and commercialization. However, biosensors are not able to improve analysis under particular conditions and to accurately determine GMO contents in the produce (Nakamura and Karube 2003; Minunni et al. 2001; Arugula et al. 2014). Furthermore, biosensors are generally not convenient during the identification of GMO at the preliminary stage.
SPR (surface plasmon resonance)
SPR is a sensitive and selective screening method for the detection of GMOs and utilizes PCR and a commercially available surface plasmon resonance (SPR) affinity biosensor (Biacore XTM). In SPR-based detection, the target DNA sample is hybridized with SPR transduction using Biocore XTM to allow hybridization at the sensor surface using an immobilized probe (Sawata et al. 1999; Miyachi et al. 2000; Kneipp 2007). The immobilized probes are specific for the 35S promoter and NOS terminator sequences, which are characteristic of GMOs.
SPR imaging is particularly useful for single-stranded DNA. No labeling is required, but the technology is limited to the detection of single-stranded DNA samples only, which include target P35S and NOS-T (Nica et al. 2004), 35S from soy and maize (Mariotti et al. 2002), and GM maize (Feriotto et al. 2002).
SERS spectroscopy
SERS (surface enhanced Raman scattering) spectroscopy involves the interaction of light, molecules and metal nanostructures, which enhance Raman signals that can resolve structures down to the single molecule level. This is a flexible tool for biological analysis due to its excellent ability to detect a wide target biomolecules (Kneipp et al. 2002; Chen and Liu 2012; Arugula et al. 2014). An SERS-barcoded nanosensor was developed for the detection of Bt gene-transformed rice expressing insecticidal proteins. The barcode sensor designed by fabricating an SERS barcoded with a golden core for optical enhancement, a layer Raman reporter molecule absorbed onto the surface of the gold core for spectroscopic barcoding, and a silica shell to protect and the functionalization’s and conjugations of oligonucleotide strands targeting DNA strands. Two transgenes cry1Ab and cry1Ac for Bt rice (Bai et al. 2006) were used as a fusion gene sequence and compared with surface plasmonic spectroscopy for conventional rice to construct a specific SERS-based detection method. SERS spectra were finally used to decode the testing results. The detection assay showed good precision, accuracy, and sensitivity with a detection limit of 0.1 pg/ml (Leoni et al. 2011).
Advanced detection methods
DNA walking
The specific identification of unknown nucleotide sequences adjacent to known DNA regions in the given genome using specific primers for a known sequence is referred to as DNA walking, and the final product is further sequenced using Sanger sequencing technology (Spalinskas et al. 2013a). There are three categories of DNA walking as follows: (1) the restriction method involves digestion of genomic DNA using the desired restriction enzyme targeting sites close to the sequence of interest, such as the junction between the unknown and known sequences (Fraiture et al. 2015a, b). The obtained fragments are then self-circularized or ligated to a DNA cassette, named inverted PCR. (2) Extension based on a sequence-specific primer amplification method. The resulting ssDNA is ligated into either a DNA cassette or 3′ tail. This strategy has been successfully used on GM maize (MON810), rice (LLRICE62), soybean (A2704-12), rapeseed (T45), and cotton (LTCOTTON25) to characterize the transgene cassette and flanking regions (Spalinskas et al. 2013b; Fraiture et al. 2015a, b). (3) Primer-based methods combine (random or degenerate primers) to target specific primers using PCR strategies (Spalinskas et al. 2013a). The DNA walking method is of great value of importance for the detection of GMOs in processed and unprocessed food/feed. This approach involves two semi-nested PCRs to increase the yield and specificity for GMO targets, which is crucial when GMO is present at very low levels (Spalinskas et al. 2013b). This technique has been successfully implemented and adapted for routine GM crops analysis according to EU-regulatory guidelines for array of crops and is both rapid and sensitive. Therefore, regulatory elements such as promoter (P35S) and terminator (NOS) are commonly used as targets (Raymond et al. 2010; Yang et al. 2005a, b; Wang et al. 2011; Fraiture et al. 2015a, b; Ruttink et al. 2010; Kok et al. 2014). However, this approach is not preferred for the detection of unknown elements in GM crops.
Next-gen sequencing technologies
Molecular characterization of GMOs in GM crops is currently performed by Southern blot and PCR in combination with Sanger sequencing to target the precise locations of transgenes integrated into the host genome and to detect backbone sequences for transformation vectors. Recently, Kok et al. (2014) reported on the number of stacked traits used to develop GM crops. Increased sequencing throughput possibilities, at continuously decreasing costs, combined with NGS technologies have increased the reliability of genomic research in a diverse range of applications. NGS is the most efficient approach for the molecular detection of GMOs (Leoni et al. 2011; Guttikonda et al. 2016) and has been adapted for applied plant breeding techniques and the identification of small insertions and deletions in Arabidopsis thaliana (Chao et al. 2013) and Rice (Wahler et al. 2013). NGS has been demonstrated to have advantages over Southern blotting (Guttikonda et al. 2016). It also enables transcriptome profiling (Chu and Corey 2012) and detects altered expression profiles of transgene inserts and the relative abundances of pools of small regulatory RNAs (siRNA pools) in genetically modified crops and the sequenced data produced should be analyzed strictly under risk assessment (Ramon et al. 2014). Furthermore, cost-effective methods are developed to detect and analyze the GM crops and NGS coupled with whole genome sequencing (WGS) provides precise, specific results for the data analysis of GM crops. It has been revealed to provide a powerful means of characterizing GM crops having massive data developed for vip3Aa2 from MIR162 using Hiseq (Illumina) (Liang et al. 2014), Bt11 gene (454 system, Roche), Bt176 gene, LEC, P35S/CTP4, CP4-EPSPS, P35S, and T-NOS (454 system, Roche) (Song et al. 2014; Fraiture et al. 2015a, b). Consequently, the deployment of whole genome sequencing (WGS) technology for generating massive data can replace successfully the next-gen sequencing technology as the genomes of the majority of GM crops being considered for sequence-based analysis or under consideration in the near future.
Conclusion and future prospects
Twenty successful years of GM foods as commodity crops provides substantial multiple benefits to farmers concerning social, economic, and health benefits. Currently, a 100-fold increase in GM crop production has been recorded since their introduction in 1996. Rapid adoption of GM crops demands accurate and sensitive trait-specific assay detection methods of GMOs in GM crops before their commercial release. Time and effort should be devoted to on-farm trials to avoid potential risks. The appropriate regulatory framework with defined parameters should be employed to carefully assess the environmental and health risks associated with the use of GM crops. In addition to this, an array of sensitive and advanced sequence-based techniques and biosensor detection methods need to be explored for the detection of GMOs in GM crops. Recently, a transgenic cotton expressing Cry10Aa toxin conferring resistance to cotton boll weevil has been developed (Ribeiro et al. 2017). Similary, a label-free biosensor using single-walled carbon nanotube (SWCNT) for GMO detection was developed and it is sensitive and able to determine the GM food samples efficiently (Tam 2015). Consequently, despite the benefits of GM crops, these are often not carefully tested and adequately stored to avoid mixing of GM seeds with non-GM seed samples. Hence, developing countries, before releasing GM crops for commercialization, need to assess the impact and risk analysis of the GM crops and make their own strict regulatory framework and implementation policies.
Acknowledgements
The authors are thankful to their respective universities for the support. We are also grateful to the United States Department of Agriculture Economic and Research Service (Seth J. Wechsler) and Prof. Clive James and Mr. Clement Dionglay, Global Knowledge Center on Crop Biotechnology, ISAAA, South East Asia, Center, Khush Hall, IRRI, Los Banos, Laguna, Phillipines, for giving permission to include their data and figures in this review paper.
Author contributions
The authors M.K. and P.K. conceived, designed, edited, and wrote the manuscript. J.K.P. and V.K.B. edited the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Madhu Kamle and Pradeep Kumar contributed equally to this work.
Contributor Information
Pradeep Kumar, Email: pkbiotech@gmail.com.
Vivek K. Bajpai, Email: vbiotech04@gmail.com
References
- Agelet LE, Armstrong PR, Tallada JG, Hurburgh CR., Jr Differences between conventional and glyphosate tolerant soybeans and moisture effect in their discrimination by near infrared spectroscopy. Food Chem. 2013;141:1895–1901. doi: 10.1016/j.foodchem.2013.04.087. [DOI] [PubMed] [Google Scholar]
- Anklam E, Gadani F, Heinze P, Pijnenburg H, Eede VDG. Analytical methods for detection and determination of genetically modified organisms in agricultural crops and plant derived food products. Eur Food Res Technol. 2002;214:3–26. doi: 10.1007/s002170100415. [DOI] [Google Scholar]
- Arugula A, Zhang YY, Simonian AL. Biosensors as 21st century technology for detecting genetically modified organisms in food and feed. Anal Chem. 2014;86(1):119. doi: 10.1021/ac402898j. [DOI] [PubMed] [Google Scholar]
- Baeumner AJ, Cohen RN, Miksic V, Min JH. RNA biosensor for the rapid detection of viable Escherichia coli in drinking water. Bios Bioelectron. 2003;18(4):405–413. doi: 10.1016/S0956-5663(02)00162-8. [DOI] [PubMed] [Google Scholar]
- Bai Y, Jiang M, Cheng J, Wang D. Effects of Cry1Ab toxin on Propylea japonica (Thunberg) (Coleoptera: Coccinellidae) through its prey, Nilaparvata lugens Stål (Homoptera: Delphacidae), feeding on transgenic Bt rice. Environ Entomol. 2006;35:1130–1136. doi: 10.1603/0046-225X-35.4.1130. [DOI] [Google Scholar]
- Ballari RV, Martin A, Gowda LR. Detection and identification of genetically modified EE-1 brinjal (Solenum melongena) by single, multiplex and SYBR (®) real time PCR. J Sci Food Agric. 2013;93:340–347. doi: 10.1002/jsfa.5764. [DOI] [PubMed] [Google Scholar]
- Bates SL, Zhao JZ, Roush RT, Shelton AM. Insect resistance management in GM crops: past, present and future. Nat Biotech. 2005;23(1):57–62. doi: 10.1038/nbt1056. [DOI] [PubMed] [Google Scholar]
- Bawa AS, AnilaKumar KR. Genetically modified foods: safety, risks and public concerns—a review. J Food Sci Technol. 2013;50(6):1035–1046. doi: 10.1007/s13197-012-0899-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran J, Jaimes MH, Echeverry Y, Lopez LD, Duque MC, Tohme P. Quantitative analysis of transgenes in cassava plants using real-time PCR technology. In Vitro Cell Dev Biol Plant. 2009;45:48–56. doi: 10.1007/s11627-008-9159-5. [DOI] [Google Scholar]
- Borem A, Almeida GD. Plantas geneticamente modificadas: desafios e oportunidades para regiões tropicais. Visconde de Rio Branco: Suprema; 2011. p. 390. [Google Scholar]
- Byrdwell WC, Neff WE, List GR. Tryglyceride analysis of potential margarine base stocks by high-performance liquid chromatography with atmospheric pressure chemical ionization mass spectrometry and flame ionization detection. J Agric Food Chem. 2001;49(1):446–457. doi: 10.1021/jf0008801. [DOI] [PubMed] [Google Scholar]
- Chao Y, Ma L, Yang Y, Ju F, Zhang XX, Wu WM. Metagenomic analysis reveals significant changes of microbial compositions and protective functions during drinking water treatment. Sci Rep. 2013;3:35–50. doi: 10.1038/srep03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu Y. Ag-nanoparticle-modified single Ag nanowire for detection of melamine by surface-enhanced Raman spectroscopy. J Raman Spectr. 2012;43:986–991. doi: 10.1002/jrs.3137. [DOI] [Google Scholar]
- Chen L, Guo J, Wang Q, Kai G, Yang L. Development of the visual loop-mediated isothermal amplification assays for seven genetically modified maize events and their application in practical samples analysis. J Agric Food Chem. 2011;59:5914–5918. doi: 10.1021/jf200459s. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang X, Jin N, Zhou Y, Huang S, Miao Q, Zhu Q, Xu J. Endpoint visual detection of three genetically modified rice events by loop-mediated isothermal amplification. Int J Mol Sci. 2012;13:14421–14433. doi: 10.3390/ijms131114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Corey DR. RNA Sequencing: Platform Selection, Experimental Design, and Data Interpretation. Nucleic Acid Therap. 2012;22(4):271–274. doi: 10.1089/nat.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R, Randhawa GJ, Bhoge RK, Singh M. Qualitative and quantitative PCR-based detection methods for authorized genetically modified cotton events in India. J AOAC Int. 2014;97(5):1299–1309. doi: 10.5740/jaoacint.13-271. [DOI] [PubMed] [Google Scholar]
- Conrad U. Polymers from plants to develop biodegradable plastics. Trends Plant Sci. 2005;105:11–12. doi: 10.1016/j.tplants.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Cornejo JF, McBride WD (2002) Adoption of bioengineered crops. USDA Agricultural Economic Report No. (AER810) pp. 67
- Datukishvili N, Kutateladze T, Gabriadze I, Bitskinashvili K, Vishnepolsky B. New multiplex PCR methods for rapid screening of genetically modified organisms in foods. Front Microbiol. 2015;6:757. doi: 10.3389/fmicb.2015.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson J. GM plants: science, politics and EC regulations. Plant Sci. 2010;178:94–98. doi: 10.1016/j.plantsci.2009.12.005. [DOI] [Google Scholar]
- Delaney B. Safety assessment of foods from genetically modified crops in countries with developing economies. Food Chem Toxicol. 2015;86:132–143. doi: 10.1016/j.fct.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Directive 2001/18/EC of the European Parliament and of the Council of 12 March 2001 on the deliberate release into the environment of genetically modified organisms and repealing Council Directive 90/220/EEC-Commission Declaration. Off J L 106:0001–0039
- Fagan J, Schoel B, Haegert A, Moore J, Beeby J. Performance assessment under field conditions of a rapid immunological test for transgenic soybeans. Int J Food Sci Technol. 2001;36:1–11. doi: 10.1046/j.1365-2621.2001.00419.x. [DOI] [Google Scholar]
- FAO (2017) FAOSTAT Online database. http://faostat.fao.org/. Accessed Jan 2017
- Feriotto G, Borgatti M, Mischiati C, Bianchi N, Gambari R. Biosensor technology and surface plasmon resonance for real-time detection of genetically modified roundup ready soybean gene sequences. J Agric Food Chem. 2002;50(5):955–962. doi: 10.1021/jf0109773. [DOI] [PubMed] [Google Scholar]
- Fraiture MA, Herman P, Taverniers I, et al. Validation of a sensitive DNA walking strategy to characterize unauthorized GMOs using model food matrices mimicking common rice products. Food Chem. 2015;173:1259–1265. doi: 10.1016/j.foodchem.2014.09.148. [DOI] [PubMed] [Google Scholar]
- Fraiture MA, Herman P, Lefevre L, Taverniers I, De Loose M, Deforce D, Roosens NH. Integrated DNA walking system to characterize a broad spectrum of GMOs in food/feed matrices. BMC Biotech. 2015;15:76. doi: 10.1186/s12896-015-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Zhu P, Wang C, Huang K, Du Z, Tian W, Wang Q, Wang H, Xu W, Zhu S. A highly sensitive and specific method for the screening detection of genetically modified organisms based on digital PCR without pretreatment. Sci Rep. 2015;5(12715):1–10. doi: 10.1038/srep12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressey D. Transgenics: a new breed. Nature. 2013;497:27–29. doi: 10.1038/497027a. [DOI] [PubMed] [Google Scholar]
- Grohmann L, Brunen NC, Nemeth A, Waiblinger HU. Collaborative trial validation studies of real-time PCR-based GMO screening methods for detection of the bar gene and the ctp2-cp4epsps construct. J Agric Food Chem. 2009;57:8913–8920. doi: 10.1021/jf901598r. [DOI] [PubMed] [Google Scholar]
- Gruere GP, Rao SR. A review of international labelling policies of genetically modified food to evaluate India’s proposed rule. AgBioforum. 2007;10:51–64. [Google Scholar]
- Guan X, Guo J, Shen P, Yang L, Zhang D. Visual and rapid detection of two genetically modified soybean events using loop-mediated isothermal amplification method. Food Anal Methods. 2010;3(4):313–320. doi: 10.1007/s12161-010-9132-x. [DOI] [Google Scholar]
- Guardian (2016) https://www.theguardian.com/science/2016/nov/17/plants-genetically-modified-to-boost-photosynthesis-produce-greater-yields-study-shows. Accessed on 11.03.2017
- Guertler P, Huber I, Pecoraro S, Busch U. Development of an event-specific detection method for genetically modified rice Kefeng-6 by quantitative real-time PCR. J Verbraucherschutz Lebensmittelsicherheit. 2013;7(1):63–70. doi: 10.1007/s00003-011-0748-6. [DOI] [Google Scholar]
- Guttikonda SK, Marri P, Mammadov J, Ye L, Soe K, Richey K, et al. Molecular characterization of transgenic events using next generation sequencing approach. PLoS One. 2016;11(2):1–17. doi: 10.1371/journal.pone.0149515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin C. Gene stacking in transgenic plants—the challenge for 21st century plant biotechnology. Plant Biotech J. 2005;3:141–155. doi: 10.1111/j.1467-7652.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- Holst-Jensen A, Bertheau Y, de Loose M, Grohmann L, Hamels S, Hougs L. Detecting unauthorized genetically modified organisms (GMOs) and derived materials. Biotechnol Adv. 2012;30(6):1318–1335. doi: 10.1016/j.biotechadv.2012.01.024. [DOI] [PubMed] [Google Scholar]
- Holst-Jensen A (2013) Real-time PCR analysis of genetically modified organisms. In: Rodriguez-Lazaro D (ed) Real time PCR in food science, Caister Academic, Norfolk
- Huber I, Block D, Sebah F, Debode D, Morisset L, Grohmann G, Berben D, Stebih M, Milavec J, Zel U. Busch Development and validation of duplex, triplex and and pentaplex real-time PCR screening assays for the detection of genetically modified organisms in food and feed. J Agric Food Chem. 2013;61:10293–10301. doi: 10.1021/jf402448y. [DOI] [PubMed] [Google Scholar]
- James C (2009) Global status of commercialized Biotech/GM crops. ISAAA Brief 41, Ithaca
- James C (2014) ISAAA released the global status of commercialized Biotech/GM crops. ISAAA Brief 49, Ithaca
- James C (2015) Global status of commercialized Biotech/GM crops. ISAAA Breifs 51
- James C (2016) Global Status of Commercialized Biotech/GM Crops: 2016. ISAAA Brief No. 52, International Service for the Acquisition of Agri-biotech Applications, Ithaca, NY
- Jin C, Viidanoja J, Zhang Y, Ikonen E, Root A, Romanczyk M, Manheim J, Eric Dziekonski, Kenttamma HI. Comparison of atmospheric pressure chemical ionization and field ionization mass spectrometry for the analysis of large saturated hydrocarbons. Anal Chem. 2016;88(21):10592–10598. doi: 10.1021/acs.analchem.6b02789. [DOI] [PubMed] [Google Scholar]
- Jinxia A, Qingzhang L, Xuejun G, Yanbo Y, Lu L, Zhang M. A multiplex nested PCR assay for the simultaneous detection of genetically modified soybean, maize and rice in highly processed products. Food Cont. 2011;22(10):1617–1623. doi: 10.1016/j.foodcont.2011.03.018. [DOI] [Google Scholar]
- Kamle S, Ali S. Genetically modified crops: detection strategies and biosafety issues. Gene. 2013;522:123–132. doi: 10.1016/j.gene.2013.03.107. [DOI] [PubMed] [Google Scholar]
- Key S, Ma JKC, Drake PM. Genetically modified plants and human health. J Royal Soc Med. 2008;101(6):290–298. doi: 10.1258/jrsm.2008.070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiddle G, Hardinge P, Buttigieg N, Gandelman O, Pereira C, McElgunn CJ, Rizzoli M, Jackson R, Appleton N, Moore C, Tisi LC, Murray JAH. GMO detection using a bioluminescent real time reporter (BART) of loop mediated amplification (LAMP) suitable for field use. BMC Biotechnol. 2012;12:15. doi: 10.1186/1472-6750-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim EH, Yea MC, Kim HY. Validation of A multiplex PCR detection kit for screening of herbicide-tolerant genes in genetically modified crops. J Kor Soc Appl Biol Chem. 2013;56(2):251–254. doi: 10.1007/s13765-012-3229-4. [DOI] [Google Scholar]
- Klumper W, Qaim M. A meta-analysis of the impacts of genetically modified crops. Plos One. 2014;9(11):16–29. doi: 10.1371/journal.pone.0111629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneipp K. Surface enhanced Raman scattering. Phys Today. 2007;60:40–42. doi: 10.1063/1.2812122. [DOI] [Google Scholar]
- Kneipp K, Haka AS, Kneipp H. Surface-enhanced raman spectroscopy in single living cells using gold nanoparticles. Appl Spectrosc. 2002;56:150–154. doi: 10.1366/0003702021954557. [DOI] [Google Scholar]
- Kok EJ, Pedersen J, Onori R, Sowa S, Schauzu M, De SA. Plants with stacked genetically modified events: to assess or not to assess? Trends Biotechnol. 2014;32:70–73. doi: 10.1016/j.tibtech.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Kralj-Novak P, Gruden K, Morisset D, Lavrac N, Stebih D, Rotter A, et al. GMOtrack: generator of cost-effective GMO testing strategies. J AOAC Int. 2009;92:1739–1746. [PubMed] [Google Scholar]
- Ladics GS, Bartholomaeus A, Bregitzer P, Doerrer NG, Gray A, Holzhauser T, et al. Genetic basis and detection of unintended effects in genetically modified crop plants. Transgenic Res. 2015;24:587–603. doi: 10.1007/s11248-015-9867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kang SH, Park YH, Min DM, Kim YM. Quantitative analysis of two genetically modified maize lines by real-time PCR. J Microbiol Biotechnol. 2006;16:205–211. [Google Scholar]
- Lee SH, Kim JK, Yi BY. Detection methods for biotech cotton MON 15985 and MON 88913 by PCR. J Agric Food Chem. 2007;55(9):3351–3357. doi: 10.1021/jf070036b. [DOI] [PubMed] [Google Scholar]
- Lee D, La Mura M, Allnutt TR, Powell W. Detection of genetically modified organisms (GMOs) using isothermal amplification of target DNA sequences. BMC Biotech. 2009;9:7–9. doi: 10.1186/1472-6750-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni C, Volpicella M, De Leo F, Gallerani R, Ceci LR. Genome walking in eukaryotes. FEBS J. 2011;278:3953–3977. doi: 10.1111/j.1742-4658.2011.08307.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Hansen JL, Liu Y, Zemetra RS, Berger PH. Using real-time PCR to determine transgene copy number in wheat. Plant Mol Biol Rep. 2004;22:179–188. doi: 10.1007/BF02772725. [DOI] [Google Scholar]
- Liang C, Van Dijk JP, Scholtens IMJ, et al. Detecting authorized and unauthorized genetically modified organisms containing vip3A by real-time PCR and next-generation sequencing. Anal Bioanal Chem. 2014;406(11):2603–2611. doi: 10.1007/s00216-014-7667-1. [DOI] [PubMed] [Google Scholar]
- Lin C-H, Pan TM. Perspectives on genetically modified crops and food detection. J Food Drug Anal. 2016;24:1–8. doi: 10.1016/j.jfda.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JKC, Drake PMW, Christou P. The production of recombinant pharmaceutical proteins in plants. Nature. 2003;4:794–805. doi: 10.1038/nrg1177. [DOI] [PubMed] [Google Scholar]
- Mano J, Shigemitsu N, Futo S, Akiyama H, Teshima R, Hino A, Furui S, Kitta K. Real-time PCR array as a universal platform for the detection of genetically modified crops and its application in identifying unapproved genetically modified crops in Japan. J Agric Food Chem. 2009;57:26–37. doi: 10.1021/jf802551h. [DOI] [PubMed] [Google Scholar]
- Mariotti E, Minunni M, Mascini M. Surface plasmon resonance biosensor for genetically modified organism’s detection. Anal Chim Acta. 2002;453:165–172. doi: 10.1016/S0003-2670(01)01458-1. [DOI] [Google Scholar]
- Markoulatos P, Siafakas N, Papathoma A, Nerantzis E, Betzios B, Dourtoglou VM. Qualitative and quantitative detection of protein and genetic traits in genetically modified food. Food Rev Int. 2004;20:275–296. doi: 10.1081/FRI-200029418. [DOI] [Google Scholar]
- Marmiroli N, Agrimonti C, Visioli G, Colauzzi M, Guarda G, Zuppini A. Silencing of G1-1 and A2-1 genes. Effects on general plant phenotype and on tuber dormancy in Solanum tuberosum L. Potato Res. 2000;43:313–323. doi: 10.1007/BF02360537. [DOI] [Google Scholar]
- Minunni M, Tombelli S, Pratesi S, Mascini M, Piatti P, Bogani P, Buiatti M. A piezoelectric affinity biosensor for genetically modified organisms (GMOs) detection. Anal Lett. 2001;34:825–840. doi: 10.1081/AL-100103595. [DOI] [Google Scholar]
- Miraglia M, Berdal K, Brera C, Corbisier P, Holst-Jensen A, Kok EJ, Marvin HJ, Schimmel H, Rentsch J, Van Rie JP, Zagon J. Detection and traceability of genetically modified organisms in the food production chain. Food Chem Toxicol. 2004;42:1157–1180. doi: 10.1016/j.fct.2004.02.018. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Europe angers US with strict GM labeling. Nat Biotechnol. 2003;21:6. doi: 10.1038/nbt0103-6a. [DOI] [PubMed] [Google Scholar]
- Miyachi H, Yano K, Ikebukuro K, Kono M, Hoshina S, Karube I. Application of chimeric RNA–DNA oligonucleotides to the detection of pathogenic microorganisms using surface plasmon resonance. Anal Chim Acta. 2000;407:1–10. doi: 10.1016/S0003-2670(99)00779-5. [DOI] [Google Scholar]
- Monsanto Corn/Tolerance to the herbicide glyphosate (GA21) (1997) Petition submitted, August 20. FDA biotechnology notification files number 51
- Morgan CL, Newman DJ, Price CP. Immunosensors: technology and opportunities in laboratory medicine. Clin Chem. 1996;42:193–209. [PubMed] [Google Scholar]
- Nakamura H, Karube I. Current research activity in biosensors. Anal Bioanal Chem. 2003;377(3):446–468. doi: 10.1007/s00216-003-1947-5. [DOI] [PubMed] [Google Scholar]
- Nelson GC. Traits and techniques of GMOs. In: Nelson GC, editor. Genetically modified organisms in agriculture. San Diego: Academic Press; 2001. pp. 7–13. [Google Scholar]
- Nica AG, Mascini M, Ciucu AA (2004) DNA-based biosensor for detection of genetically-modified organisms. Analele Universit ăŃii din Bucure ş ti–Chimie, Anul XIII (serie nouă), vol I-II, pp 85–94
- Padgette SR, Kolacz KH, Delannay X, Re DB, LaVallee BJ, Tinius CN, Rhodes WK, Otero YI, Barry GF, Eichholz DA, Peschke VM, Nida DL, Taylor NB, Kishore GM. Development, identification, and characterization of a glyphosate-tolerant soybean line. Crop Sci. 1995;35:1451–1461. doi: 10.2135/cropsci1995.0011183X003500050032x. [DOI] [Google Scholar]
- Pandey A, Kamle M, Yadav LP, Muthukumar M, Kumar P, Gupta V, Ashfaque M, Pandey BK. Genetically modified food: its uses, prospects and safety assessment. Biotechnol. 2010;9(8):448–458. [Google Scholar]
- Patel PD. Biosensors for measurement of analyte implicated in food safety: a review. Trends Anal Chem. 2002;21:96–115. doi: 10.1016/S0165-9936(01)00136-4. [DOI] [Google Scholar]
- Peano C, Bordoni R, Gulli M, Mezzelani A, Samson MC, De Bellis G, Marmiroli N. Multiplex polymerase chain reaction and ligation detection reaction/universal assay technology for the traceability of genetically modified organisms in foods. Anal Biochem. 2005;346:90–100. doi: 10.1016/j.ab.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Ramon M, Devos Y, Lanzoni A, Liu Y, Gomes A, Gennaro Waigmann A. RNAi based GM plants: food for thought for risk assessors. Plant Biotechnol J. 2014;12(9):1271–1273. doi: 10.1111/pbi.12305. [DOI] [PubMed] [Google Scholar]
- Randhawa GJ, Singh M. Multiplex, construct-specific and real-time PCR-based analytical methods for Bt rice with cry1Ac gene. J AOAC Int. 2012;95(1):186–194. doi: 10.5740/jaoacint.10-429. [DOI] [PubMed] [Google Scholar]
- Randhawa GJ, Singh M, Morisset D, Sood P, Zel J. Loop mediated isothermal amplification: rapid visual and real-time methods for detection of genetically modified crops. J Agric Food Chem. 2013;61:11338–11346. doi: 10.1021/jf4030085. [DOI] [PubMed] [Google Scholar]
- Randhawa GJ, Morisset D, Singh M, Zel J. GMO matrix: a cost-effective approach for screening unauthorized genetically modified events in India. Food Control. 2014;38:124–129. doi: 10.1016/j.foodcont.2013.10.013. [DOI] [Google Scholar]
- Randhawa GJ, Chhabra R, Bhoge RK, Singh M. Visual and real-time event-specific loop-mediated isothermal amplification based detection assays for Bt cotton events MON531 and MON15985. J AOAC Int. 2015;98(5):1207–1214. doi: 10.5740/jaoacint.14-269. [DOI] [PubMed] [Google Scholar]
- Randhawa G, Singh M, Sood P. DNA based methods for the detection of genetically modified events in food supply and chain. Curr Sci. 2016;110(6):1000–1009. doi: 10.18520/cs/v110/i6/1000-1009. [DOI] [Google Scholar]
- Raymond P, Gendron L, Khalf M. Detection and identification of multiple genetically modified events using DNA insert fingerprinting. Anal Bioanal Chem. 2010;396(6):2091–2102. doi: 10.1007/s00216-009-3295-6. [DOI] [PubMed] [Google Scholar]
- Redenbaugh K, Hiatt B, Martineau B, Kramer M, Sheehy R, Sanders R, Houck C, Emlay D. Safety assessment of genetically engineered fruits and vegetables: a case study of the Flavr Savr tomato. Boca Raton: CRC Press; 1992. p. 288. [Google Scholar]
- Ribeiro TP, Arraes FB, Lourenco-Tessutti IT, Silva MS, Lisei-de-Sa ME, Lucena WA, Macedo LL, Lima JN, Santos Amorim RM, Artico S, Alves-Ferreira M, Mattar Silva MC, Grossi-de-Sa MF. Transgenic cotton expressing Cry10Aa toxin confers high resistance to the cotton boll weevil. Plant Biotechnol J. 2017;15:1–13. doi: 10.1111/pbi.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan GJ, Dudin YA, Lee TC. Immunodiagnostic methods for detection of 5-enolpyruvylshikimate-3-phosphate synthase in Roundup Ready® soybeans. Food Control. 1999;10:407–414. doi: 10.1016/S0956-7135(99)00083-3. [DOI] [Google Scholar]
- Ruttink T, Demeyer R, Van Gulck E. Molecular tool box for the identification of unknown genetically modified organisms. Anal Bioanal Chem. 2010;396(6):2073–2089. doi: 10.1007/s00216-009-3287-6. [DOI] [PubMed] [Google Scholar]
- Sawata S, Kai E, Ikebukuro K, Iida T, Honda T, Karube I. Application of peptide nucleic acid to the direct detection of deoxyribonucleic acid amplified by polymerase chain reaction. Biosens Bioelectron. 1999;14(4):397–404. doi: 10.1016/S0956-5663(99)00018-4. [DOI] [PubMed] [Google Scholar]
- Schouten HJ, Krens FA, Jacobsen E. Do cisgenics plants warrantless stringent oversight? Nat Biotechnol. 2006;24:753. doi: 10.1038/nbt0706-753. [DOI] [PubMed] [Google Scholar]
- Shan G, Embrey SK, Schaffer BW. A highly specific enzyme-linked immunosorbent assay for the detection of Cry1Ac insecticidal crystal protein in transgenic Wide Strike cotton. J Agric Food Chem. 2007;55:5974–5979. doi: 10.1021/jf070664t. [DOI] [PubMed] [Google Scholar]
- Song Q, Wei G, Zhou G. Analysis of genetically modified organisms by pyrosequencing on a portable photodiode-based bioluminescence sequencer. Food Chem. 2014;154:78–83. doi: 10.1016/j.foodchem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/S0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southgate EM, Davey MR, Power JB, Merchant R. Factors affecting the genetic engineering of plants by microprojectile bombardment. Biotechnol Adv. 1995;13:631–657. doi: 10.1016/0734-9750(95)02008-X. [DOI] [PubMed] [Google Scholar]
- Spalinskas R, Van den Bulcke M, Milcamps A. Efficient retrieval of recombinant sequences of GM plants by Cauliflower Mosaic Virus 35S promoter-based bidirectional LT-RADE. Eur Food Res Technol. 2013;237(6):1025–1031. doi: 10.1007/s00217-013-2078-7. [DOI] [Google Scholar]
- Spalinskas RM, den Bulcke Van, Vanden Eede G, Milcamps A. LT-RADE: an efficient user-friendly genome walking method applied to the molecular characterization of the insertion site of genetically modified maize MON810 and riceLLRICE62. Food Anal Method. 2013;6(2):705–713. doi: 10.1007/s12161-012-9438-y. [DOI] [Google Scholar]
- Stave JW. Protein immunoassay methods for detection of biotech crops: applications, limitations and practical considerations. J AOAC Int. 2002;85:780–786. [PubMed] [Google Scholar]
- Sticklen M. Plant genetic engineering to improve biomass characteristics for biofuels. Curr Opin Biotechnol. 2005;17:315–319. doi: 10.1016/j.copbio.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Tabashnik BE, Carrière Y, Dennehy TJ, Morin S, Sisterson MS, Roush RT, Shelton AM, Zhao JZ. Insect resistance to transgenic Bt crops: lessons from the laboratory and field. J Econ Entomol. 2003;96:1031–1038. doi: 10.1093/jee/96.4.1031. [DOI] [PubMed] [Google Scholar]
- Tam PD. Genetically modified organism (GMO) detection by biosensor based on SWCNT material. Curr Appl Phys. 2015;15:397–401. doi: 10.1016/j.cap.2015.01.017. [DOI] [Google Scholar]
- Tarafdar A, Kamle M, Prakash A, Padaria JC. Transgenic plants: issues and prospects. In: Govil JN, Anandakumar P, editors. Plant biotechnology. USA: Studium Press LLC Publisher; 2014. pp. 337–375. [Google Scholar]
- Torres AC, Nascimento WM, Paiva SAV, Aragao FAS. Bioassay for detection of transgenic soybean seeds tolerant to glyphosate. Pesq Agropec Bras Brasília. 2003;38:1053–1057. doi: 10.1590/S0100-204X2003000900005. [DOI] [Google Scholar]
- Trapero C, Wilson IW, Stiller WN, Wilson LJ. Wilson enhancing integrated pest management in GM cotton systems using host plant resistance. Front Plant Sci. 2016;7:500. doi: 10.3389/fpls.2016.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA-ERS (2013) http://www.ers.usda.gov/data-products/adoption-of-genetically-engineered-crops-in-the-us/recent-trends-in-ge-adoption.aspx
- Vazquez-Padron RI, et al. Cry1Ac protoxin from Bacillus thuringiensis sp. Kurstaki HD-73 binds to surface proteins in the mouse small intestine. Biochem Biophys Res Commun. 2000;271:54–58. doi: 10.1006/bbrc.2000.2584. [DOI] [PubMed] [Google Scholar]
- Wahler D, Wahler L, Schauser J, Bendiek L, Grohmann Next-generation sequencing as a tool for detailed molecular characterization of genomic insertions and flanking regions in genetically modified plants: a pilot study using a Rice event unauthorized in the EU. Food Anal Method. 2013;6:1718–1727. doi: 10.1007/s12161-013-9673-x. [DOI] [Google Scholar]
- Waiblinger HU, Grohmann L, Mankertz J, Engelbert D, Pietsch K. A practical approach to screen for authorised and unauthorised genetically modified plants. Anal Bioanal Chem. 2010;396(6):2065–2072. doi: 10.1007/s00216-009-3173-2. [DOI] [PubMed] [Google Scholar]
- Wang WX, Zhu TH, Lai FX, Fu Q. Event-specific qualitative and quantitative detection of transgenic rice Kefeng 6 by characterization of the transgene flanking sequence. Eur Food Res Technol. 2011;232(2):297–305. doi: 10.1007/s00217-010-1389-1. [DOI] [Google Scholar]
- Windels P, Bertrand S, Depicker A, Moens W, Bockstaele E, De Loose M. Qualitative and event-specific PCR real-time detection methods for Star Link maize. Eur Food Res Technol. 2003;216:259–263. doi: 10.1007/s00217-002-0652-5. [DOI] [Google Scholar]
- Wu Y, Wu G, Xiao L, Lu C. Event-specific qualitative and quantitative PCR detection methods for transgenic rapeseed hybrids MS1 × RF1 and MS1 × RF2. J Agric Food Chem. 2007;55:8380–8389. doi: 10.1021/jf0717337. [DOI] [PubMed] [Google Scholar]
- Wu KM, Lu YH, Feng HQ, Jiang YY, Zhao JZ. Suppression of cotton bollworm in multiple crops in china in areas with Bt toxin-containing cotton. Science. 2008;321:1676–1678. doi: 10.1126/science.1160550. [DOI] [PubMed] [Google Scholar]
- Wu G, Wu Y, Nie S, Zhang L, Xiao L, Cao Y, Lu C. Real-time PCR method for detection of the transgenic rice event TT51-1. Food Chem. 2010;119:417–422. doi: 10.1016/j.foodchem.2009.08.031. [DOI] [Google Scholar]
- Wu Y, Wang Y, Li J, Wei Li Zhang L, Li Y, Li X, Li J, Zhu L, Wu G. Development of a general method for detection and quantification of the P35S promoter based on assessment of existing methods. Sci Rep. 2014;4:7358. doi: 10.1038/srep07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu S, Pan A, Zhang K, Wang Z, Zhou Z, Zhang D. Event specific qualitative and quantitative polymerase chain reaction detection of genetically modified MON863 maize based on the 5′-transgene integration sequence. J Agric Food Chem. 2005;53(24):9312–9318. doi: 10.1021/jf051782o. [DOI] [PubMed] [Google Scholar]
- Yang L, Xu S, Pan A, Zhang K, Wang Z, Zhou Z, Zhang D. Event specific qualitative and quantitative polymerase chain reaction detection of genetically modified MON863 maize based on the 5′-transgene integration sequence. J Agr Food chem. 2005;53(24):9312–9318. doi: 10.1021/jf051782o. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu F, Xua H, Aguilar ZP, Niu R, Yuan Y, Sun J, You X, Lai W, Xiong Y, Wan C, Wei H. Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable Salmonella typhimurium, Escherichia coli O157: H7 and Listeria monocytogenes in food products. Food Microbiol. 2013;34:418–424. doi: 10.1016/j.fm.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Zastraw-Hayes GM, Lin H, Sgmund AL, Hoffman JL, Alarcon CM, Hayes KR, Richmond TA, Jeddeloh JA, May GD, Beatty MK. Southern-by-sequencing: a robust screening approach for molecular characterization of genetically modified crops. Plant Genome. 2015;8(1):1–15. doi: 10.3835/plantgenome2014.08.0037. [DOI] [PubMed] [Google Scholar]
- Zimmermann A, Luthy J, Pauli U. Event specific transgene detection in Bt11 corn by quantitative PCR at the integration site. LWT Food Sci Technol. 2000;33:210–216. doi: 10.1006/fstl.2000.0637. [DOI] [Google Scholar]