Abstract
This study examines the extraction and hydrolysis of proteins using single factor and Box–Behnken Design (BBD). From single factor tests, optimised extraction parameters were 1% alkali concentration, 40 °C temperature, 60 min time, and 1:20 solid to alkali ratio. Under these conditions; 924.31 mg/g of total protein was obtained from Limonia acidissima (L acidissima). The maximum degree of hydrolysis was 39.82% at pH 2, enzyme to substrate ratio 2.5% (w/w), and hydrolysis time was 42.41 min using BBD design. L acidissima seed protein hydrolysate showed 32.94% DPPH and 88.18% of ABTS activity at concentration of 100 µg/ml and 1 mg/ml, respectively. Reducing power of 0.16 and metal chelating activity of 87.39% was obtained from 5 mg/ml protein hydrolysates. This implied that L acidissima seed protein hydrolysate could be utilised in protein rich product or as protein supplements.
Keywords: L acidissima seed, Protein hydrolysate, BBD, Antioxidant activity
Introduction
The origin for L acidissima is India and Ceylon. Fruit is used in the preparation of chutneys and jelly and jam (Morton 1987). It has got various medicinal properties such as anti-tumor (Saima et al. 2000), antimicrobial activity (Rahman and Gray 2002), anti-inflammatory, antipyretic and analgesic activity (Ahamed et al. 2008), antioxidant activity (Sonawane and Arya 2013, 2015) and anti-diabetic (Patel et al. 2012). There are recent reports which indicate that defatted L acidissima seed is rich in protein (49.51%) and also like Na, Mn, Mg, K, Cu, Fe, and Zn (Sonawane et al. 2016).
The digestion of various proteins benefits in liberation of bio-functional peptides (including animal and plant sources). The bio-functional peptides which are present in an inactive form in their parent proteins are released during gastrointestinal digestion or food processing (Matsui et al. 2002). The bio-functional activities of peptides depend on structure, composition, and sequence. In recent years, an extensive literature has also encouraged on bioactive peptides, with a view as functional ingredients pointed at fitness (Ryan et al. 2011).
The ambitions of present study were to boost extraction and hydrolysis using response surface methodology (RSM)—BBD which examine as high degree of protein hydrolysate, as well as to evaluate its potential as an antioxidant agent.
Materials and methods
L acidissima was acquired from a local market in one lot (APMC market, Navi Mumbai, India). The outer hard shell of L acidissima fruit was removed followed by scooping of pulp from shell was carried out. The seeds were detached from pulp by washing under running water. Furthermore, overnight drying was carried at 45 ± 2 °C and stored at −20 °C to prevent spoilage and to maintain uniformity in the entire research. The analytical grade chemicals were used in experiment.
Preparation of defatted L acidissima seed flours
The flour mill was cleaned and L acidissima seeds were ground to obtain a fine powder. The fat from the seed flour sample was removed by soaking flour in petroleum ether with stirring for 6 h at 37 ± 1 °C. The solvent was decanted and the process was repeated thrice with fresh solvent. After removal of solvent, flour was air-dried at room temperature and ground to pass through 40 mesh sieve. Further, these flours were packed into airtight polyethylene bags and stored at 4 °C until the extractions were done (at least 2 weeks) (Kanbargi et al. 2016).
Optimization of extraction parameter for defatted L acidissima seed protein
The defatted L acidissima seed flour was suspended in alkali extraction. Further, this were incubated at elected time, temperature and solid-to-alkali ratio to enhance protein extraction. The resulting slurry was centrifuged (12,000 rpm for 60 min at 4 °C) after incubation. The supernatant was analysed for protein content using Bradford assay (1976) and expressed as mg protein per gram of flour sample. Optimization parameters obtained by single factor experiments are reported in Table 1. The supernatant was further precipitated at pH 4 and washed thrice with distilled water, followed by drying in a vacuum oven at 40 °C for 16 h.
Table 1.
Evaluation of parameters used for protein extraction optimization from defatted L acidissima seed flour
| Variable parameter (range) | Constant parameters | |||
|---|---|---|---|---|
| Alkali optimization | Alkali concentration | Temperature | Time | Solid to liquid ratio |
| Alkali concentration (0.2–1.2%) | – | 40 | 60 | 1:40 |
| Temperature (20–50 °C) | O.C | – | 60 | 1:40 |
| Time (0–150 min) | O.C | O.C | – | 1:40 |
| Solid to liquid ratio | O.C | O.C | O.C | – |
O.C optimized parameter
Enzymatic hydrolysis of L acidissima seed protein using one factor
Six proteases (papain, pepsin, protease, pancreatin, trypsin, and chymotrypsin) were screened (data is not shown) was selected to boost optimal condition for hydrolysis to improve degree of hydrolysis. L acidissima seed proteins 1% (w/v) were suspended in distilled water and subjected to enzymatic hydrolysis with pepsin were optimized. The pH for hydrolysis ranged between 2 and 3, enzyme to substrate ratio (Es) 0.5–3% (w/w) and hydrolysis time varied from 30 to 180 min. During hydrolysis, pH was maintained and the reaction was carried out at 37 °C. The hydrolysate kept in a water bath at 100 °C for 10 min to deactivate enzymes. The supernatant centrifuged at 10,000 rpm for 20 min and subjected to degree of hydrolysis (DH) as mentioned below.
Degree of hydrolysis
The DH was determined following TCA method (Hoyle and Merritt, 1994) with slight modifications. In the first part, 500 μl of hydrolyzed protein samples were mixed with the equal amount, i.e. 500 μl of 20% TCA solution. After incubation of 30 min, supernatant were centrifuged at 10,000 rpm and protein content was determined by the method of Lowry, Rosebrough, Farr, and Randal (1951). Finally, DH was determined using following equation:
Experimental design
A three-level-three-factor BBD (Design-Expert1 6 software, Stat-Ease Inc., Minneapolis, USA) was employed to predict the optimal hydrolysis conditions of L acidissima using pepsin with 17 experiments (Box and Draper 1987) (Table 2). The three levels of the variables: pH (2.0, 2.2 and 2.4), E/S (1.5, 2.0 and 2.5%) and time (30, 60 and 90 min) were selected for enzymatic hydrolysis using pepsin. The DH was used as response to measure hydrolysis (Table 2).
Table 2.
Enzymatic hydrolysis experimental design for L acidissima seed protein
| Run | pH | ES | Time | DH |
|---|---|---|---|---|
| 1 | 2 | 1.5 | 60 | 36.55 ± 1.17 |
| 2 | 2.4 | 1.5 | 60 | 28.97 ± 3.99 |
| 3 | 2 | 2.5 | 60 | 40.31 ± 6.34 |
| 4 | 2.4 | 2.5 | 60 | 32.94 ± 5.45 |
| 5 | 2 | 2 | 30 | 25.95 ± 0.84 |
| 6 | 2.4 | 2 | 30 | 10.55 ± 0.38 |
| 7 | 2 | 2 | 90 | 21.11 ± 0.46 |
| 8 | 2.4 | 2 | 90 | 17.60 ± 0.32 |
| 9 | 2.2 | 1.5 | 30 | 12.11 ± 0.98 |
| 10 | 2.2 | 2.5 | 30 | 16.94 ± 1.71 |
| 11 | 2.2 | 1.5 | 90 | 17.35 ± 1.43 |
| 12 | 2.2 | 2.5 | 90 | 19.77 ± 1.60 |
| 13 | 2.2 | 2 | 60 | 32.20 ± 2.40 |
| 14 | 2.2 | 2 | 60 | 32.16 ± 4.23 |
| 15 | 2.2 | 2 | 60 | 32.03 ± 1.66 |
| 16 | 2.2 | 2 | 60 | 33.45 ± 0.58 |
| 17 | 2.2 | 2 | 60 | 33.10 ± 0.87 |
The experimental data obtained were fitted by the following regression equation:
| 1 |
where Y is DH (predicted response), β 0, β 1, β 2, β 3… β 33 were the regression coefficients, X 1-H, X 2-E/S ratio and X 3-time.
Assessment of molecular weight (MW) of peptides
The MW of peptides formed during hydrolysis was determined by SDS-PAGE. For electrophoresis, 30% acrylamide- bis-acrylamide solution mixture was used for the preparation of 4% stacking gel and 15% resolving gel. 10–20 μl of sample mixed with the sample buffer was loaded into the well. Gel was run at 60 V for ~90–120 min for the visualization of bands, silver staining was done.
Antioxidant studies of L acidissima seed protein hydrolysate
The optimal conditions provided by the BBD were used to produce protein hydrolysate from L acidissima. The supernatant was collected by centrifuging (10,000 rpm for 20 min at 4 °C). Furthermore, supernatant (neutralized) was dried in vacuum oven at 40 °C until protein hydrolysates were in dried form.
ABTS assay
Antioxidant activity of protein hydrolysate was verified using ABTS method (Re et al. 1999). The 7 mM ABTS and 2.45 mM potassium persulfate was mixed and incubated at 37 °C for 16–17 h. The OD was set at 0.7 (±0.02) by diluting with 0.01 M phosphate buffer saline at 734 nm. The ABTS cations 260 µl (absorbance of 0.700 ± 0.02) mixed to the 40 µl of the hydrolyzed sample and absorbance was measured at 734 nm immediately after 6 min.
DPPH activity
DPPH free radicals were used to assess activity of protein hydrolysate using Sahreen et al. (2010) method with slight modification. The sample (40 µl) mixed with 120 µl of methanol which further mixed with 40 µl of DPPH (0.15 mM) prepared in methanol. After 15 min incubation in dark room OD was taken at 517 nm.
Determination of ferric reducing antioxidant power (FRAP)
FRAP assay was developed by Benzie and Strain (1996). The mixture for FRAP reagent preparation were tenfold acetate buffers: onefold TPTZ: onefold FeCl3. The reaction mixture 200 µl of FRAP reagent was added to 20 µl of the sample and absorbance was recorded at 593 nm after incubation of 15 min at 37 °C.
Metal chelation activity
Metal chelating activity described by Decker and Welch (1990) was used to determine the activity with small modifications. In chelation test, protein hydrolysate (50 µl) sample was added into 10 µl of FeCl2 (2 mM) and then it was mixed with 250 µl of double distilled water. Furthermore, 20 µl of ferrozine solution (5 mM) was mixed to the reaction by shaking for 2 min. The color reduction was recorded by measuring the absorbance at 562 nm after 10 min of incubation. The chelating activity was calculated as a percentage using
Statistical analysis
All experiments were performed in triplicate and were expressed as a mean ± standard deviation. The SPSS (Statistical Package for Social Sciences) for Windows version (16.0) was used to analyze the data (SPSS Inc., Chicago, IL). Statistical significance was considered at p < 0.05.
Results and discussion
Extraction and optimization of process parameter for protein from defatted L acidissima seed
Extraction of protein depends upon the alkali concentration, a buffer to sample ratio, pH, temperature, and time (Wani et al. 2006).
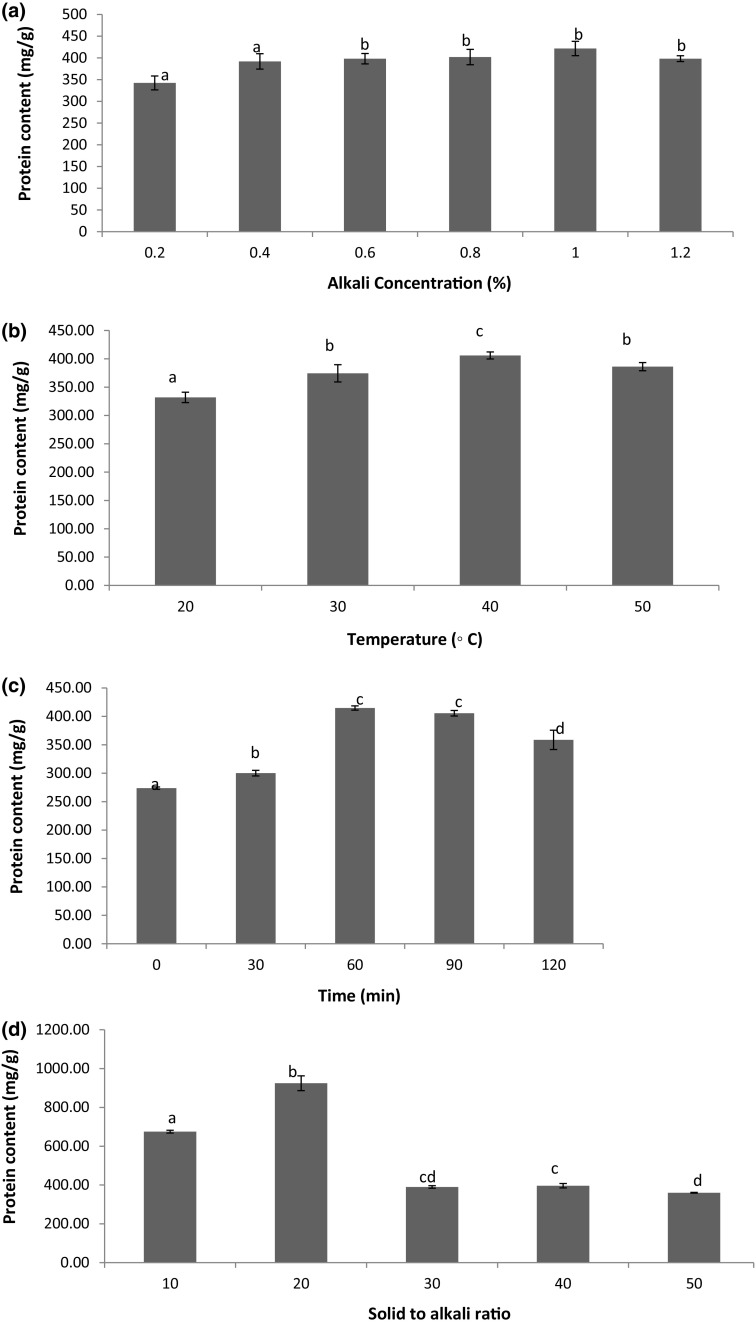
Effect of various alkali concentrations on protein content is signified in Fig. 1a). From figure it can be seen that as concentration increased up to 0.4%; insignificant difference in protein content of L acidissima seed was observed. Further, as the concentration of alkali increased; an increase in protein content up to 1% was observed followed by a significant decrease in the concentration. Thus, an optimal concentration for extraction was considered as 1% alkali. This could be due to fact that acidic proteins are soluble in alkali solution thus causing an increase in protein content with respect to increased alkali concentration (Zhang et al. 2009). However, further decrease in protein concentration was observed and this could be due to the denaturation of proteins.
Fig. 1.
Effect of a alkali concentration; b Temperature; c Time; d solid to alkali ratio on protein content of defatted L acidissima seed flours (Duncan test)
Temperature is a key factor which affects protein content during extraction (Fig. 1b). As the temperature increased from 20 to 40 °C; significant improvement in protein concentration (331.94–405.88 mg/g) was observed; however, significant decrease was seen at 50 °C. Hence, 40 °C was considered as an optimum temperature in the experiment. A similar kind of study was performed for grape seed protein (Lv et al. 2011). Deng et al. (2014) also observed a decrease in protein content of kiwifruit seeds with increased temperature (above 35 °C) due to the denaturation of proteins.
The influence of time on protein extraction of L acidissima is shown in Fig. 1c). As time increased from 10 to 60 min; significant increase in protein content, i.e. from 273.80 to 414.77 mg/g was observed followed by a significant decrease. The increase in the protein concentration due to the longer time protein dissolves. Thus, an optimum time of 60 min was finalised for protein extraction. The plausible explanation for such observation is given by Lv et al. (2011). They observed 30 min as the total time of extraction of protein later it was stable due to the protein solubility equilibrium.
The effect of solid to alkali ratio is represented in Fig. 1d. Initially at 1:20 ratio; significantly high protein content, i.e., 924.31 mg/g was recorded and as further ratio increased from 30 to 50 ml/g, it decreased 390.17–360.19 mg/g. Hence, 1:20 (solid to alkali ratio) was selected as optimal to increase protein content in extraction. A similar solid/solvent ratio was obtained by Lv et al. (2011), during grape seeds protein extraction.
From the single factor test, optimum extraction parameters were obtained as following alkali concentration 1%, temperature 40 °C, time 60 min, and solid to alkali ratio 1:20. Under this condition total protein content was 924.31 mg/g achieved form L acidissima. This may be due to the high alkali, which is responsible to inhibit the common activity of protease and causes ionization of phenolic compounds and also prevent the forming hydrogen bonding with protein (Loomis and Battaile 1966; Hochstrasser et al. 1988).
Optimization of process parameter for hydrolysis of protein from defatted L acidissima seed protein
The DH is a useful tool to understand the effect of pH, an enzyme to substrate ratio, and time on hydrolysis of L acidissima protein. In protein hydrolysates, PCL (peptide chain length) is produced with desired functional properties which explain that higher the DH value corresponds to low formation of PCL or inverse (Adler-Nissen 1986).
Effect of pH on hydrolysis of protein was observed and depicted in Fig. 2a. As the pH increased from 2 to 3, the degree of hydrolysis was decreased. There was no significant difference observed after pH 2.2 (p < 0.05). Hence, pH range of 2–2.4 was selected for further optimization study.
Fig. 2.

Effect of a pH; b Enzyme to substrate (%); c Time (min); on hydrolysis of protein from defatted L acidissima seed protein (Tukey test)
As enzyme concentration increased from 0.5 to 3% (w/w, enzyme/substrate); DH was increased (Fig. 2b). However, 2–3% was selected for further optimization.
Effect of time on DH is represented in Fig. 2c. Initially high DH was observed, i.e., up to 60 min, later, it was in stationary phase up to 150 min (which showed no significant difference between 60 and 150 min) and further it significantly increased due to the longer extraction time. Hence, 30–90 min ranges were selected for optimization study. Thus, these results reconfirmed that hydrolysis is a function of time and enzyme concentration.
Response surface methodology
The interaction of hydrolysis parameters: pH (X 1), ES (X 2) and time (X 3) having impact on DH of L acidissima seed protein, were studied by employing a BBD design. The single factor helps in the designing experiment on the basis of DH as response. Table 2 shows that the experimental outcomes obtained by design. The various conditions of software DH was varied in between 10.55 and 40.31%.
Model fitting
In our experiment, pepsin was most suitable enzyme which produces peptides with a high DH.
Furthermore, efforts were made to investigate interaction of pH, E/S ratio and time on the DH using BBD. The results are shown in Table 2. The best explanatory model equation fitted into Eq. (1) is given in Eq. (2) which corresponds to DH (%),
| 2 |
The significance of the coefficient evaluated using analysis of variance (ANOVA) and presented in Table 3. Quadratic model was significant (p < 0.05) and in coded variables is given in Eq. (2). F value and p value was used in the determination of each coefficient. If F value is greater than p value, then corresponding variables would be more significant (Atkinson and Donev 1992).
Table 3.
ANOVA for Response Surface Quadratic Model
| Source | Sum of squares | df | Mean square | F value | p value prob >F | |
|---|---|---|---|---|---|---|
| Model | 1305.686 | 9 | 145.0762 | 127.866 | <0.0001 | Significant |
| X 1-pH | 143.3144 | 1 | 143.3144 | 126.3132 | <0.0001 | |
| X 2-Es | 28.01546 | 1 | 28.01546 | 24.69202 | 0.0016 | |
| X 3-time | 13.29038 | 1 | 13.29,038 | 11.71376 | 0.0111 | |
| X 1 X 2 | 0.011,495 | 1 | 0.011,495 | 0.010131 | 0.9226 | |
| X 1 X 3 | 35.35,149 | 1 | 35.35,149 | 31.15778 | 0.0008 | |
| X 2 X 3 | 1.484,312 | 1 | 1.484,312 | 1.30823 | 0.2903 | |
| 20.01717 | 1 | 20.01717 | 17.64256 | 0.0040 | ||
| 0.024382 | 1 | 0.024382 | 0.021489 | 0.8876 | ||
| 1073.634 | 1 | 1073.634 | 946.2703 | <0.0001 | ||
| Residual | 7.94217 | 7 | 1.134596 | |||
| Lack of fit | 6.294256 | 3 | 2.098085 | 5.092704 | 0.0749 | Not significant |
| Pure error | 1.647914 | 4 | 0.411979 | |||
| Cor total | 1313.628 | 16 | ||||
| R-squared | 0.993954 | |||||
| Adj R-squared | 0.986181 | |||||
| Pred R-squared | 0.921376 | |||||
| C.V. % | 4.086604 |
The DH was a linear term of pH (X 1), an enzyme to substrate ratio (X 2) and time (X 3) followed by a quadratic term of pH(), and time () and interaction terms X 1 X 3. On the other hand, quadratic terms of the enzyme to substrate ratio (), and other interaction terms of X 1 X 2 and X 2 X 3 were not significant (p > 0.05).
The lack of fit test dealings the failure of the model (insignificant p-value of 0.0749) which indicates data in the experiment fitted well and acceptable to describe observed data. The results are presented in Table 3. The R 2 = 0.9940 indicated that the fitted model can explain 99.40% of the variation in the data. Also high degree of correlation was observed from Adj R 2 (0.9862) and Pred R 2 (0.9214). This model had a CV (coefficient of variation) of 4.09% suggesting a good precision and high reliability of the experiment achieved.
Interpretation of response surface model
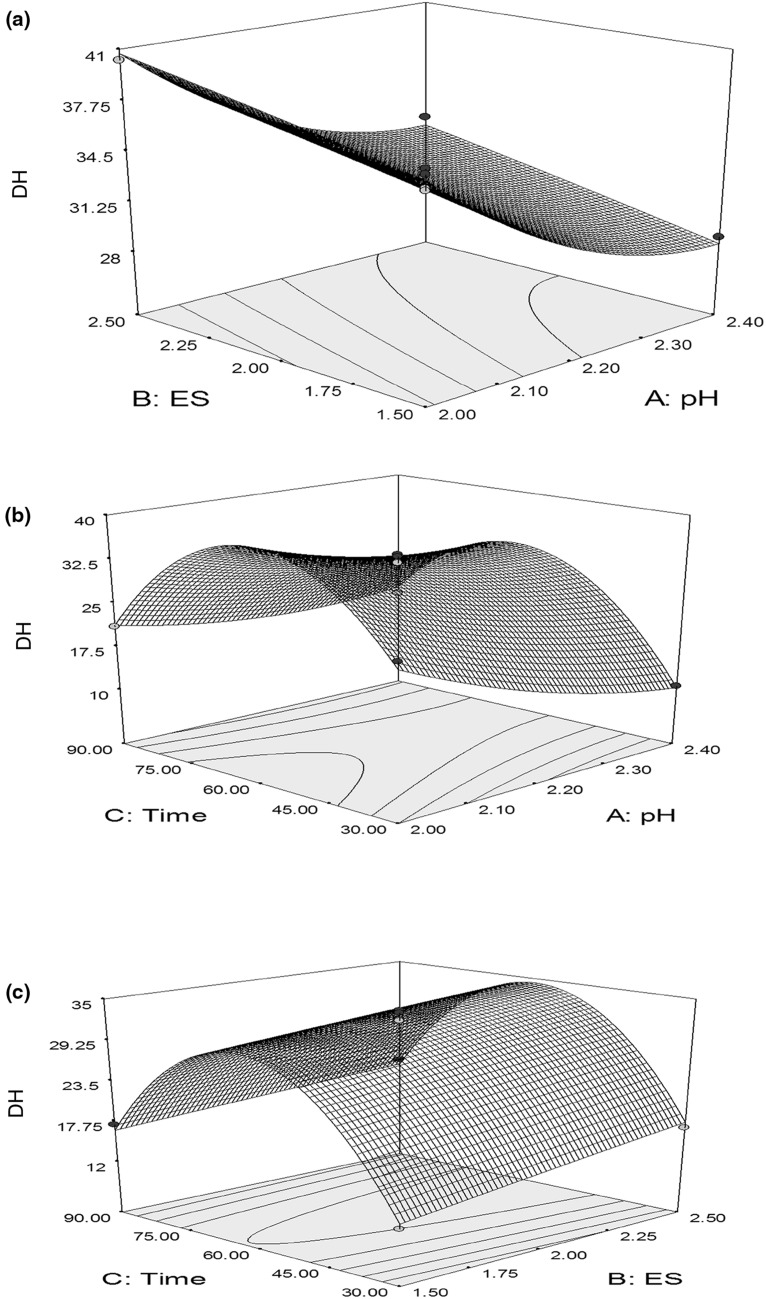
The optimum values of the variables within the given ranges are shown by graphical representation of the regression equation. This can be presented by three-dimensional response surfaces and contour plots of the models. The interaction between E/S and pH on the DH of the hydrolysates of L acidissima seed protein at any constant hydrolysis time was depicted in Fig. 3a. As the concentration of enzyme was increased; DH was also increased. Whereas at increased pH; decreased DH was observed. At pH 2; as concentration of enzyme increased; increased DH was observed. Similar trends were observed at high pH 2.4. However, at low (1.5%) and high (2.5%) concentration of enzyme, if pH was varied; decreased order in DH was noted.
Fig. 3.
Response surface plot: effect of a Es and pH b time and pH c ES and time on degree of hydrolysis of protein from defatted L acidissima seed
The interaction between time and pH on the DH of the hydrolysates of L acidissima seed protein at any constant Es is presented in Fig. 3b. Initially, as hydrolysis time increases DH increase and start decrease after a certain time (75 min), decrease in the DH observed as increase in the pH. Whereas in Fig. 3c noted that DH increased up to certain time and further it slightly decreased. But as enzyme concentration increased DH increased.
Validation of experiments
Confirmation of experiments was accomplished to suitability in the experiment. The optimal conditions for the predicted target degree of hydrolysis 36.59% corresponded to pH 2, an enzyme to substrate ratio 2.5% (w/w), and hydrolysis time 42.41 min. The experimental value of 39.82 ± 4.30 (%) was found close to predicted value. Hence, the model is valid for optimizing the process of hydrolysate from L acidissima seed protein.
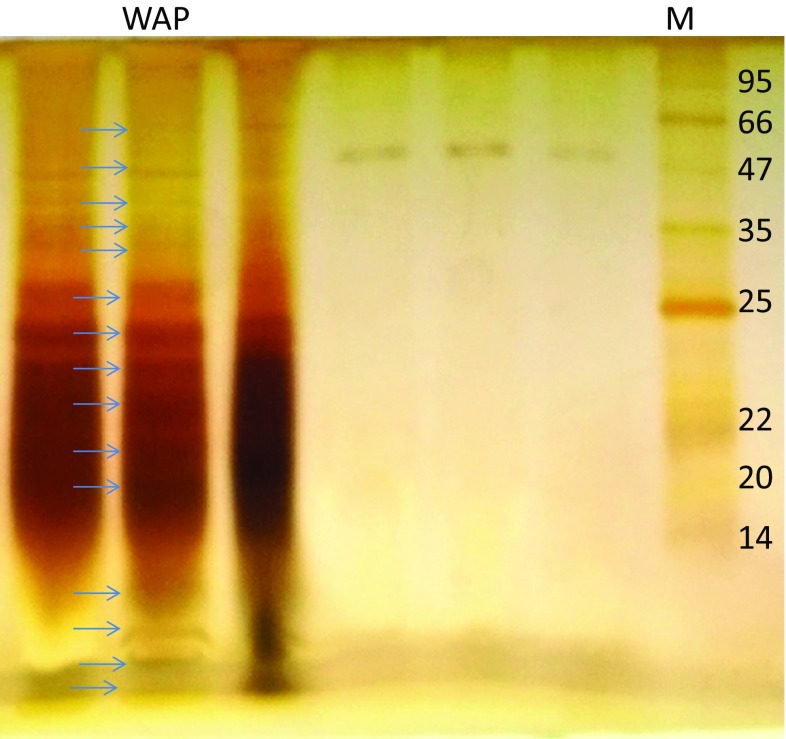
Distribution of molecular weight in L acidissima seed protein hydrolysates
Polypeptide band pattern was observed in L acidissima seed protein hydrolysates (Fig. 4). L acidissima seed protein hydrolysates composed of 15 protein bands with a molecular weight between 95 and 14 and below 14 kDa, low molecular weight peptide band pattern was also observed below 14 kDa. The degree of hydrolysis of L acidissima seed protein (39.82%) produces a range of different peptide bands. Suetsuna and Nakano (2000) documented that pepsin is help in hydrolysis of proteins.
Fig. 4.
SDS page of L acidissima seed protein hydrolysates
Antioxidant activity of L acidissima seed protein hydrolysates
It is a widely suggested to perform more than two assays for estimation of antioxidant activity due to the possible irregularity of different radicals which are measured by DPPH, FRAP and ABTS assays. The DPPH and ABTS assay evaluate the scavenging capacity, and FRAP assay was used to reflect the ferric-reducing activity. The secondary antioxidant potential was measured by MCA (metal chelating assay) and demonstrates inhibitory effect on generation of radicals.
Scavenging activity by DPPH
Radical scavenging activity is measured by reduction in absorbance. Table 4 represents the scavenging activity of the L acidissima seed protein hydrolysate at various dosages. The L acidissima seed protein hydrolysates have DPPH radical scavenging activity at a dosage of 20–100 µg/ml, whereas Trolox at 2–10 µg/ml (Table 4). The protein hydrolysate 100 µg/ml showed 32.94% inhibition which was closed to 6 µg/ml of Trolox. This is a disadvantage of DPPH scavenging activity assay in the case of protein, DPPH is only soluble in methanol or ethanol solution and when the concentration of protein was increased, precipitation occurred, which is a big hurdle to measure antioxidant activity of protein by DPPH. This may be due to the hydrophilic nature of hydrolysate in which possibility of accumulation of shorter peptides and amino acids. The increased polarity of the low-MW protein hydrolysate makes them more difficult to react with hydrophobic (Zhu et al. 2008).
Table 4.
Radical scavenging activity and reducing capacity of L acidissima seed peptides
| Peptides (µg/ml) | Scavenging activity of DPPH (%) | Trolox (µg/ml) | Scavenging activity of DPPH (%) | Peptides (mg/ml) | Scavenging activity of ABTS (%) | Trolox (µg/ml) | Scavenging activity of ABTS (%) |
|---|---|---|---|---|---|---|---|
| 20 | 10.14 ± 0.32 | 2 | 11.9 | 0.2 | 57.09 ± 5.12 | 5 | 12.0 |
| 40 | 15.02 ± 0.48 | 4 | 24.8 | 0.4 | 74.06 ± 1.19 | 10 | 22.3 |
| 60 | 16.73 ± 0.54 | 6 | 35.4 | 0.6 | 81.48 ± 0.85 | 15 | 38.8 |
| 80 | 22.66 ± 0.73 | 8 | 48.0 | 0.8 | 85.00 ± 0.58 | 20 | 46.2 |
| 100 | 32.94 ± 1.05 | 10 | 58.5 | 1 | 88.18 ± 0.71 | 25 | 51.6 |
| Reducing capacity of wood apple seed peptides | |||||||
|---|---|---|---|---|---|---|---|
| Peptides (mg/ml) | FRAP Abs593 nm | Trolox (µM/ml) | FRAP Abs593 nm | Peptides (mg/ml) | Metal chelating activity (%) | Trolox (µg/ml) | Metal chelating activity (%) |
| 1 | 0.03 ± 0.02 | 10 | 0.220 | 1 | 51.91 ± 1.19 | 0.1 | 3.67 |
| 2 | 0.05 ± 0.01 | 20 | 0.426 | 2 | 67.23 ± 1.55 | 0.2 | 14.69 |
| 3 | 0.07 ± 0.01 | 30 | 0.635 | 3 | 80.65 ± 1.86 | 0.3 | 16.33 |
| 4 | 0.09 ± 0.01 | 40 | 0.809 | 4 | 81.73 ± 1.88 | 0.4 | 21.41 |
| 5 | 0.16 ± 0.01 | 50 | 0.986 | 5 | 87.39 ± 2.01 | 0.5 | 30.83 |
Results were expressed as the mean ± SD of three determinations
Scavenging activity by ABTS
Antioxidant activity lipophilic and hydrophilic molecules analysed using ABTS decolourization assay that determined by measuring absorbance at 734 nm in which reaction of hydrogen donating antioxidants with the ABTS radical. ABTS scavenging activity by protein hydrolysate was increased as the concentration of peptides increased (Table 4). 0.2 mg/ml of protein hydrolysate showed 57.09% of scavenging activity which was close to 25 µg/ml of Trolox that is 51.6%. The difference in the inhibition of DPPH and ABTS radical by L acidissima seed protein hydrolysates was observed. This may be due to the DPPH assay performed in organic media, while the ABTS assay in aqueous media. The contradictory phenomenon between the activities is due to the differences of solubility and diffusivity of radicals and peptides in the reaction system (Arnao 2000).
FRAP
A compound shows its potential antioxidant activity by reducing capacity that may serve as a marker of antioxidant power. The reducing agents in tested samples results in the development of the complex tripiridiltriazine (TPTZ)–Fe(II), which is monitored at 595 nm. As the concentration of L acidissima protein hydrolaste (1–5 mg/ml) increased; the absorbance was increased (0.03–0.16) at 593 which were comparatively low to Trolox (Table 4). At 5 mg/ml of protein hydrolysate concentration; the absorbance value of 0.16 was observed that was corresponding to 0.22 absorbance (at concentration of 10 µM/ml of Trolox). An increased absorbance was noted as the concentration of protein hydrolysates and Trolox increased. This increase could be due to the formation of peptide cleavages which resulted into improved hydrogen ions availability during the course of reaction (Kong and Xiong 2006).
Metal chelating ability
The ferrous ion chelating ability of L acidissima protein hydrolysates is shown in Table 4. The high chelating effect was observed at a high concentration of hydrolysates that is 1–5 mg/ml. 87.30% of chelating activity was showed by 5 mg/ml of hydrolysate. It is well-known that the side chains of the acidic and basic amino acids plays an important role in chelating metal ions (Saiga et al. 2003).
Antioxidant activity obtained from different assay showed variation due to reaction mechanism such as single electron transfer (SET) which corresponds to FRAP and hydrogen atom transfer (HAT) mechanism corresponds to ABTS assay (Prior et al. 2005).
Antioxidant capacity of proteins and their hydrolysates comes from their ability to inactivate reactive oxygen species (ROS), quench free radicals, chelate prooxidative transition metals, and donate electron/hydrogen (Elias et al. 2008). The amino acid composition and sequence, the size and the configuration (Chen et al. 1996; Kong et al. 2008) and hydrophobicity (Sarmadi and Ismail 2010) having influence on the antioxidant properties of proteins and their hydrolysates. Xie et al. (2008) reported amino acids, such as Tyr, Lys, Arg, Gly, Leu, and His is playing key role for antioxidant activity.
Conclusions
In summary, the study of single factors test employed in the extraction and hydrolysis of protein from L acidissima seed. The predicted profile showed that the optimum conditions for protein hydrolysis were a degree of hydrolysis 39.82% corresponded to pH 2, an enzyme to substrate ratio 2.5% (w/w), and hydrolysis time was 42.41 min. Through the confirmative test, model can be widely used for hydrolysis of protein. This investigation confirms that L acidissima seed protein hydrolysate could be used as a valuable source of antioxidative peptides. This may forecast their future application into functional food formulations and/or dietary supplements.
Acknowledgements
The research is funded by University Grant Commission (UGC), Government of India. We are also thankful to Advance Enzyme, Mumbai, India for providing enzymes as gift samples during this study.
Compliance with ethical standards
Conflicts of interest
We do not have any conflicts of interest.
References
- Adler-Nissen J. Enzymic hydrolysis of food proteins. London: Elsevier Applied Science; 1986. [Google Scholar]
- Ahamed SM, Swamy SK, Jayaverra KN, Rao JV, Kumar S. Anti inflammatory, antipyretic and analgesic activity of methanolic extract of Feronia limonia. Pharmacol. 2008;3:852–857. [Google Scholar]
- Arnao MB. Some methodological problems in the determination of antioxidant activity using chromogen radicals: a practical case. Trends Food Sci Technol. 2000;11:419–421. doi: 10.1016/S0924-2244(01)00027-9. [DOI] [Google Scholar]
- Atkinson AC, Donev AN. Optimum experimental designs. Oxford: Clarendon Press; 1992. [Google Scholar]
- Benzie IFF, Strain JJ. The ferric reducing ability of plasma as a measure of “antioxidant power”: the FRAP assay. Anal Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- Box GEP, Draper NR. Empirical model-building and response surfaces. New York: Wiley; 1987. p. 249. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen HM, Muramoto K, Yamauchi F, Nokihara K. Antioxidant activity of designed peptides based on the antioxidative peptide isolated from digests of a soybean protein. J Agricul Food Chem. 1996;44(9):2619–2623. doi: 10.1021/jf950833m. [DOI] [Google Scholar]
- Decker EA, Welch B. Role of ferritin as a lipid oxidation catalyst in muscle food. J Agricul Food Chem. 1990;38:674–677. doi: 10.1021/jf00093a019. [DOI] [Google Scholar]
- Deng J, Sun T, Cao W, Fan D, Cheng N, Wang B, Gao H, Yang H. Extraction optimization and functional properties of proteins from kiwi fruit (Actinidia chinensis Planch.) Seeds. Int J Food Prop. 2014;17(7):1612–1625. doi: 10.1080/10942912.2013.772197. [DOI] [Google Scholar]
- Elias RJ, Kellerby SS, Decker EA. Antioxidant activity of proteins and peptides. Crit Rev Food Sci Nutr. 2008;48(5):430–441. doi: 10.1080/10408390701425615. [DOI] [PubMed] [Google Scholar]
- Hochstrasser DF, Harrington MG, Hochstrasser AC, Miller MJ, Merril CR. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem. 1988;173(2):424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- Hoyle NT, Merritt JOHN. Quality of fish protein hydrolysates from herring (Clupea harengus) J Food Sci. 1994;59(1):76–79. doi: 10.1111/j.1365-2621.1994.tb06901.x. [DOI] [Google Scholar]
- Kanbargi KD, Sonawane SK, Arya SS. Functional and antioxidant activity of Ziziphus jujube seed protein hydrolysates. Food Measure. 2016;10:226–235. doi: 10.1007/s11694-015-9297-5. [DOI] [Google Scholar]
- Kong BH, Xiong YL. Antioxidant activity of zein hydrolysate in liposome system and the possible mode of action. J Agricul Food Chem. 2006;54:6059–6068. doi: 10.1021/jf060632q. [DOI] [PubMed] [Google Scholar]
- Kong XZ, Zhou HM, Hua YF. Preparation and antioxidant activity of wheat gluten hydrolysates (WGHs) using ultrafiltration membranes. J Sci Food Agricul. 2008;88(5):920–926. doi: 10.1002/jsfa.3172. [DOI] [Google Scholar]
- Loomis WD, Battaile J. Plant phenolic compounds and the isolation of plant enzymes. Phytochem. 1966;5(3):423–438. doi: 10.1016/S0031-9422(00)82157-3. [DOI] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randal RJ. Protein analysis with Folin-phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lv C, Jia X, Li M, Yang J, Zhao G. Optimization of extraction process of crude protein from grape seeds by RSM. Food Sci Technol Int. 2011;17(5):437–445. [Google Scholar]
- Matsui T, Yukiyoshi A, Doi S, Sugimoto H, Yamada H, Matsumoto K. Gastrointestinal enzyme production of bioactive peptides from royal jelly protein and their antihypertensive ability in SHR. J Nut Biochem. 2002;13:80–86. doi: 10.1016/S0955-2863(01)00198-X. [DOI] [PubMed] [Google Scholar]
- Morton JF (1987) Wood-Apple. In: Fruits of warm climates, Flare Books, Miami, Florida, p 190–191
- Patel DK, Kumar R, Laloo D, Hemalatha S. Diabetes mellitus: an overview on its pharmacological aspects and reported medicinal plants having ant diabetic activity. Asian Pac J Tropical Biomed. 2012;2(5):411–420. doi: 10.1016/S2221-1691(12)60067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior RL, Wu X, Schaich K. Standardized method for the determination of antioxidant capacities and phenolics in foods and dietary supplements. J Agricul Food Chem. 2005;53:4290–4302. doi: 10.1021/jf0502698. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Gray AI. Antimicrobial constituents from the stem bark of Feronia limonia. Phytochem. 2002;59(1):73–77. doi: 10.1016/S0031-9422(01)00423-X. [DOI] [PubMed] [Google Scholar]
- Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic Biol Med. 1999;26:1231–1237. doi: 10.1016/S0891-5849(98)00315-3. [DOI] [PubMed] [Google Scholar]
- Ryan JT, Ross RP, Bolton D, Fitzgerald GF, Stanton C. Bioactive peptides from muscle sources. Meat Fish. 2011;3:756–791. doi: 10.3390/nu3090765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahreen S, Khan M, Khan R. Evaluation of antioxidant activities of various solvent extracts of Carissa opaca fruits. Food Chem. 2010;122:1205–1211. doi: 10.1016/j.foodchem.2010.03.120. [DOI] [Google Scholar]
- Saiga A, Tanabe S, Nishimura T. Antioxidant activity of peptides obtained from porcine myofibrillar proteins by protease treatment. J Agricul Food Chem. 2003;51:3661–3667. doi: 10.1021/jf021156g. [DOI] [PubMed] [Google Scholar]
- Saima Y, Das AK, Sarkar KK, Sen AK, Sur P. An antitumor pectic polysaccharide from Feronia limonia. Int J Biol Macromolec. 2000;27(5):333–335. doi: 10.1016/S0141-8130(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Sarmadi BH, Ismail A. Antioxidative peptides from food proteins. Peptides. 2010;31:1949–1956. doi: 10.1016/j.peptides.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Sonawane S, Arya SS. Antioxidant activity of jambhul, L acidissima, ambadi and ambat chukka: an indigenous lesser known fruits and vegetables of India. Adv Food Sci Technol. 2013;5(3):270–275. [Google Scholar]
- Sonawane SK, Arya SS. Effect of drying and storage on bioactive components of jambhul and wood apple. J Food Sci Technol. 2015;52:2833–2841. doi: 10.1007/s13197-014-1321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane SK, Bagul MB, LeBlanc JG, Arya SS. Nutritional, functional, thermal and structural characteristics of Citrullus lanatus and Limonia acidissima seed flours. Food Measu. 2016;10(1):72–79. doi: 10.1007/s11694-015-9278-8. [DOI] [Google Scholar]
- Suetsuna K, Nakano T. Identification of an antihypertensive peptide from peptic digest of wakame. J Nut Biochem. 2000;11:450–454. doi: 10.1016/S0955-2863(00)00110-8. [DOI] [PubMed] [Google Scholar]
- Wani AA, Sogi DS, Grover L, Saxena DC. Effect of temperature, alkali concentration, mixing time and meal/solvent ratio on the extraction of watermelon seed proteins—a response surface approach. Biosyst Eng. 2006;94:67–73. doi: 10.1016/j.biosystemseng.2006.02.004. [DOI] [Google Scholar]
- Xie Z, Huang J, Xu X, Jin Z. Antioxidant activity of peptides isolated from alfalfa leaf protein hydrolysate. Food Chem. 2008;111:370–376. doi: 10.1016/j.foodchem.2008.03.078. [DOI] [PubMed] [Google Scholar]
- Zhang M, Gao J, Yang H. Functional properties of 7 s globulin extracted from cowpea vicilins. Cereal Chem. 2009;3:261–266. doi: 10.1094/CCHEM-86-3-0261. [DOI] [Google Scholar]
- Zhu LJ, Chen J, Tang XY, Xiong YL. Reducing, radical scavenging, and chelation properties of in vitro digests of alcalase treated zein hydrolysate. J Agricul Food Chem. 2008;56:2714–2721. doi: 10.1021/jf703697e. [DOI] [PubMed] [Google Scholar]