Abstract
The ends of linear chromosomes are capped by protein–DNA complexes termed telomeres. Telomere repeat binding factors 1 and 2 (TRF1 and TRF2) bind specifically to duplex telomeric DNA and are critical components of functional telomeres. Consequences of telomere dysfunction include genomic instability, cellular apoptosis or senescence and organismal aging. Mild oxidative stress induces increased erosion and loss of telomeric DNA in human fibroblasts. We performed binding assays to determine whether oxidative DNA damage in telomeric DNA alters the binding activity of TRF1 and TRF2 proteins. Here, we report that a single 8-oxo-guanine lesion in a defined telomeric substrate reduced the percentage of bound TRF1 and TRF2 proteins by at least 50%, compared with undamaged telomeric DNA. More dramatic effects on TRF1 and TRF2 binding were observed with multiple 8-oxo-guanine lesions in the tandem telomeric repeats. Binding was likewise disrupted when certain intermediates of base excision repair were present within the telomeric tract, namely abasic sites or single nucleotide gaps. These studies indicate that oxidative DNA damage may exert deleterious effects on telomeres by disrupting the association of telomere-maintenance proteins TRF1 and TRF2.
INTRODUCTION
The ends of linear chromosomes are capped by protein–DNA complexes termed telomeres. These structures protect the chromosome ends and prevent them from being recognized as DNA double strand breaks. Telomere dysfunction results as a consequence of the gradual loss of telomeric DNA that occurs during cellular proliferation in the absence of telomerase, or upon the loss of critical telomere-maintenance proteins (1). Cellular consequences of telomere dysfunction include telomere end fusions and genomic instability, apoptosis or senescence [reviewed in (2)]. Increasing evidence indicates that DNA damage may contribute directly to the loss of telomeric DNA and function.
Numerous studies have reported increased erosion and loss of telomeric DNA in human fibroblasts after mild oxidative stress-induced by hyperoxia, mitochondrial dysfunction, arsenic or UVA irradiation (3–7). In these studies the antioxidant treatment prevented telomere attrition. Consistent with this, high expression of the antioxidant enzyme, extracellular superoxide dismutase, was associated with decreased telomere erosion rates and increased cellular lifespan in human fibroblast cell lines (8). The exact mechanism of oxidation-induced telomere erosion is unknown. Oxidizing and alkylating agents induce a higher density of single strand breaks (SSBs) in telomeric DNA, compared with minisatellites and the bulk genome (9), and provoke erosion of the 3′ telomeric single strand tail (10). Telomeric DNA is also highly susceptible to oxidative lesion formation in vitro (4). However, the consequences of DNA lesions in telomeric DNA on structure and function are unknown. These studies suggest that the telomeres are particularly sensitive to oxidative stress, and that DNA damage in telomeres may contribute to telomere erosion.
Human telomeres consist of 5–15 kb of TTAGGG tandem repeats and terminate in a 3′ single strand tail. This tail is proposed to loop back and invade the telomeric duplex tract resulting in a large t-loop that protects the chromosomal ends (11). This structure is formed and maintained by protein complexes that associate with the telomeric end. Human telomere repeat binding factors (TRF) 1 and 2 bind duplex (TTAGGG)n DNA, and regulate telomere length and access of the 3′ tail (2). Defects in TRF2 induce loss of the 3′ tail, telomere end fusions and either apoptosis or senescence even though telomeres are not critically short (1). TRF1 acts in telomere length homeostasis, and may have protective roles since Trf1 deletion in mice causes embryonic lethality (12) and telomere end fusions (13). In addition, Trf1 deficient ES cells display decreased levels of TRF2 at the chromosome ends (13). TRF1 and TRF2 bind to human telomeric DNA directly with exquisite sequence specificity, and are critical for recruiting other proteins to duplex telomeres that function in proper telomere maintenance and capping, including TIN2, RAP1, POT1 and Ku [reviewed in (2)]. Indeed maintenance of the precise telomeric sequence was found to be critical for proper function (14) and is presumably required for the association of TRF1 and TRF2. In total, loss of the DNA or protein components of the telomeres can have severe cellular consequences.
The consequences of DNA lesions in the telomeres on structure and function are unknown. Of the lesions induced by oxidative stress, 8-dihydro-2′-deoxyguanine (8oxoG) is one of the most abundant. While it does not block replication by DNA polymerase, it is highly mutagenic and could alter telomeric sequence (15). 8oxoG is primarily repaired through base excision repair (BER) [reviewed in (16)]. In BER, a modified base is removed, the DNA is incised at the abasic site, a polymerase incorporates one or a few nucleotides at the breaks, the displaced residues are removed and the nick is sealed. We hypothesize that oxidative lesions and/or repair intermediates in telomeric DNA may interfere directly with the recognition by the TRF1 and TRF2 proteins. Here, we observed that the presence of a single or multiple 8oxoG lesions in defined telomeric substrates disrupted binding by TRF1 and TRF2 proteins to varying degrees. The presence of BER intermediates, namely a single nucleotide gap and abasic lesions, within the telomeric tracts also inhibited TRF1 and TRF2 interaction with the substrate. Our studies indicate that efficient repair of DNA damage and modified bases in the telomeres are critical for the association of telomere-maintenance proteins.
MATERIALS AND METHODS
Proteins
Restriction enzymes and T4 polynucleotide kinase (PNK) were from New England BioLabs. Recombinant histidine-tagged human TRF1 and TRF2 proteins were purified using a baculovirus/insect cell expression system as described previously (17). The baculovirus constructs for TRF2 and TRF1 were generously provided by Dr Titia de Lange (Rockefeller University, New York, NY). The recombinant GST-tagged TRF2ΔN mutant lacking the N-terminal domain was generated by PCR using the gene encoding human TRF2 as template. The PCR primers used were 5′-TTGGATCCGAGGCACGGCTGGAAGAG and 5′-CGGAATTCGTTTCAGTTCATGCCAAGTC (Midland Certified Reagents Co.) The amplified fragment (encoding amino acids 45–501) was purified and cloned into the BamHI and EcoRI sites of the pGEX-6p-2 vector (Amersham Pharmacia). The expression and purification of the GST–TRF2ΔN protein, in Escherichia coli, was as described previously (18). Recombinant human APE1 protein was purified as described previously (19).
Substrate preparation
Oligonucleotides in Table 1 were from Midland Certified Reagents Co. or Integrated DNA Technologies. 5′ end labeling of oligonucleotides was achieved with [γ-32P]ATP (3000 Ci/mmol) and T4 PNK. To construct substrates containing site-specific 8oxoG (X) or tetrahydrofuran (F) lesions, the first 11 oligonucleotides (Table 1) were 5′ end labeled and then annealed to the template oligonucleotide (TPL) in a 1:2 molar ratio. To construct the nicked and one nucleotide gap substrates, oligonucleotides NIK or GAP (Table 1), respectively, were 5′ end labeled and then annealed with DS to TPL in a 1:2:2 ratio. Annealing reactions were in 50 mM LiCl, to prevent the formation of G-quadruplex DNA, and incubated for 95°C for 5 min and then cooled to room temperature.
Table 1.
Oligonucleotides used in substrate preparations
| Sequence (5′→3′) | |
|---|---|
| CTRL | GTGGATCCGTACTTAGGGTTAGGGTTAACACGAATTCGA |
| 8oxoG1 | GTGGATCCGTACTTAGXGTTAGGGTTAACACGAATTCGA |
| 8oxoG2 | GTGGATCCGTACTTAGXXTTAGGGTTAACACGAATTCGA |
| 8oxoG3 | GTGGATCCGTACTTAXXXTTAGGGTTAACACGAATTCGA |
| 8oxoG1-1 | GTGGATCCGTACTTAGXGTTAGXGTTAACACGAATTCGA |
| AP-1a | GTGGATCCGTACTTAFGGTTAGGGTTAACACGAATTCGA |
| AP-1b | GTGGATCCGTACTTAGFGTTAGGGTTAACACGAATTCGA |
| AP-1c | GTGGATCCGTACTTAGGFTTAGGGTTAACACGAATTCGA |
| AP-2a | GTGGATCCGTACTTAGGFTTAFGGTTAACACGAATTCGA |
| AP-2b | GTGGATCCGTACTTAFGGTTAGGFTTAACACGAATTCGA |
| AP-3 | GTGGATCCGTACTTAFFFTTAGGGTTAACACGAATTCGA |
| NIK | GTGGATCCGTACTTAGG |
| GAP | GTGGATCCGTACTTAG |
| DS | GTTAGGGTTAACACGAATTCGA |
| TPL | TCGAATTCGTGTTAACCCTAACCCTAAGTACGGATCCAC |
| MXT | GTGGATCCGTACGATGGTTTTAGGTGAACACGAATTCGA |
| MXB | TCGAATTCGTGTTCACCTAAAACCATCGTACGGATCCAC |
Telomeric repeats are underlined.
DNA binding assays
Reactions (10 μl) for TRF1 and the TRF2ΔN mutant were performed in TEL buffer (20 mM HEPES, pH 7.9, 150 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 5% glycerol, 0.1% NP-40 and 5% β-casein) with substrate and protein amounts as indicated in the figure legends. The reactions were incubated at 4°C for 20 min, followed by the addition of 2 μl of the dye (0.25% bromophenol blue), and were loaded on a 5% native polyacrylamide gel (37.5:1). Electrophoresis was at 4°C and 200 V for 1.5 h (TRF1) or 2.5 h (ΔN-TRF2) in 1× TAE buffer. Reactions were visualized by Phorphorimager analysis (Molecular Dynamics). The percentage bound was calculated as described previously (20), and was corrected for background in the no enzyme control.
Binding reactions for full-length TRF2 (10 μl) were performed in TRF2 binding buffer (40 mM Tris, pH 8.0, 12 mM MgCl2, 5 mM DTT, 1 mM ATP and 100 μg/ml BSA) with substrate and protein amounts as indicated in figure legends. The reactions were incubated at 4°C for 20 min, followed by the addition of 5 μl 3× dye (0.25% bromophenol blue, 10% ficoll) and were loaded on a 1% agarose gel (Seakem GTG, BioWhittaker Molecular Applications). Electrophoresis was at 4°C and 100 V for 4 h in 0.5× TBE buffer. Gels were dried onto Hybond-XL membranes (Amersham), followed by Phorphorimager analysis. The percentage of bound substrate was calculated as described previously (20) for 100 and 200 nM TRF2 (lack of detectable products at the lower TRF2 concentrations for some substrates precluded reliable quantification) (Figure 5). Values were corrected for background in the no enzyme control, and represent the mean and standard deviation from at least three independent experiments.
Figure 5.
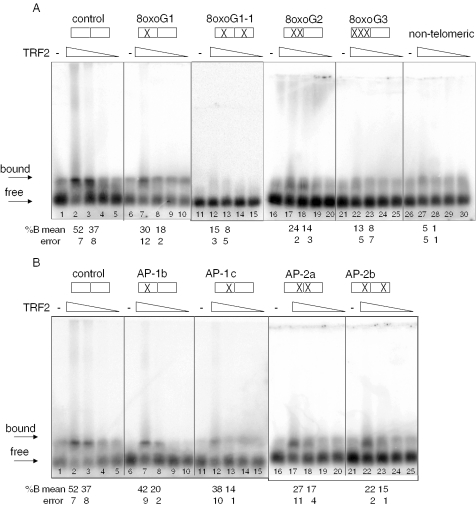
8oxoG and AP lesions disrupt TRF2 recognition of telomeric DNA. Boxes indicate the GGG runs of the tandem telomeric repeats, and (X) marks the position of 8oxoG (A) or abasic (B) lesion(s) within the G3 runs (see Table 1). Substrates (2.5 nM) were incubated alone (−) or together with decreasing concentrations of TRF2 (200, 100, 50 or 25 nM) for 20 min at 4°C in TRF2 binding buffer. Reactions were run on 1% agarose gels. The arrow indicates the unbound substrate. % Bound was determined as described in ‘Materials and Methods’. Values represent the mean and error from two to four independent experiments.
APE1 incision assay
For the incision reactions (10 μl), substrate AP-1a/TPL (1 nM) and TRF1 (amounts indicated in the figure legend) were pre-incubated at 4°C for 20 min in TEL buffer, followed by the addition of APE1 (28 pM). The reactions were incubated at 37°C for 3 min, followed by the addition of 5 μl stop buffer [95% (v/v) formamide, 20 mM EDTA, 0.1% (w/v) bromophenol blue and xylene cyanol]. The reactions were heated at 95°C for 10 min and loaded on an 18% denaturing polyacrylamide (19:1) gel. Electrophoresis was conducted at 250 V for 1.5 h in 1× TBE buffer.
RESULTS
TRF1 binding is inhibited by single or multiples 8oxoG lesions
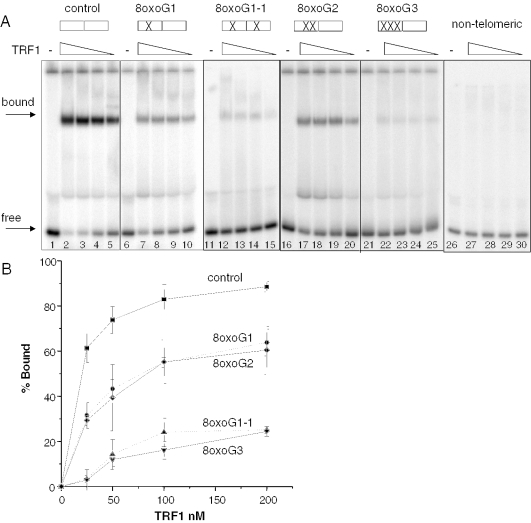
TRF1 binds predominantly to telomeric DNA as a homo-dimer, and each TRF1 monomer binds to a 5′-TAGGGTTR-3′ minimal consensus sequence (21). To determine whether oxidative lesions interfere with TRF1 binding, we used 39 bp DNA substrates that contained two tandem, overlapping monomer-binding sites, in which guanine residues were replaced with site-specific 8oxoG lesions (Table 1). Electrophoresis mobility shift assays (EMSAs) were performed with purified human recombinant TRF1 to obtain binding curves. Migration of the telomeric substrates was retarded as a function of TRF1 concentration (Figure 1A, lanes 1–25), whereas the non-telomeric substrate was not shifted (lanes 26–30), attesting to the specificity of TRF1 for human telomeric DNA. The binding curve began to plateau at ∼50 nM TRF1 for the undamaged control substrate (Figure 1B), thus, all comparisons are made at 25 nM TRF1 which falls within the linear range of the curve. Since the central G of a telomeric tract was previously found to be the most susceptible to oxidation in vitro (7), this G was replaced with an 8oxoG residue. The lesion disrupted TRF1 binding by 50%, relative to the control (61 ± 6% bound reduced to 31 ± 6%) (Figure 1A, compare lanes 5 and 10, and Figure 1B). The presence of a single 8oxoG in each telomere repeat (monomer-binding site) dramatically decreased the percentage of bound substrate 18-fold to barely detectable levels (Figure 1A, lane 15). However, when two 8oxoGs were present within a single telomere repeat, TRF1 binding was decreased only 2-fold, relative to the control, to 29 ± 2% bound (Figure 1A, lane 20). Therefore, the presence of one or two 8oxoGs in a single telomere repeat had similar effects on TRF1 binding. In contrast, substitution of all three tandem guanines with 8oxoGs dramatically disrupted TRF1 binding and the percentage of bound substrate was decreased 21-fold, relative to the control (Figure 1A, lane 25). In summary, a single oxidized guanine in each monomer-binding site, or the saturation of a single site with oxidized guanines, most strongly altered TRF1 binding.
Figure 1.
8oxoG inhibits TRF1 binding to telomeric DNA. Boxes indicate the GGG runs of the tandem telomeric repeats, and (X) marks the position of the 8oxoG lesion(s) within the G3 runs (see Table 1). Substrates (2.5 nM) were incubated alone (−) or together with decreasing concentrations of purified TRF1 (200, 100, 50 or 25 nM) (lanes 2–5, 7–10, and 12–15, 17–20, 22–25 and 27–30) for 20 min at 4°C in TEL buffer. The reactions were run on 5% acrylamide native gels and representative phosphorimager scans are shown in (A). Arrows indicate bound or free substrate. The percentage bound was determined as described in ‘Materials and Methods’ and plotted against TRF1 concentration as shown in (B) Square, control; circle, 8oxoG1; diamond, 8oxoG2; triangle, 8oxoG1-1; and inverted triangle, 8oxoG3. Values and error bars represent the mean and SD from three independent experiments.
Single nucleotide gaps disrupt TRF1
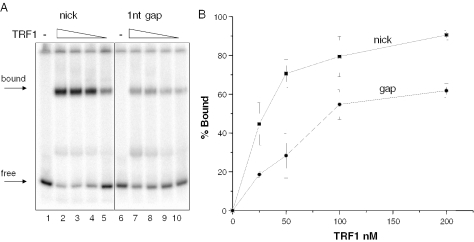
Telomeric DNA was reported to contain a higher frequency of SSBs compared with the bulk genome after cellular treatments with oxidative and alkylating agents (9). These breaks may represent direct damage-induced SSBs or indirect BER intermediates, including single nucleotide gaps that exist prior to nucleotide incorporation by DNA polymerase β (22). TRF1 binding was not significantly altered by the presence of a single strand nick within the telomeric tract (Figure 2, lanes 1–5). However, we cannot rule out the possibility that common chemical modifications at the termini of an SSB (i.e. a 3′-phosphate or phosphoglycolate) (16) might alter TRF1 binding. The presence of a single nucleotide gap within the telomeric tract reduced TRF1 binding ∼3-fold (18 ± 6.4% bound) (Figure 2, lane 10), relative to the control (61 ± 6% bound) (Figures 1A, lane 5 and Figure 1B) at 25 nM TRF1. Thus, the loss of a single nucleotide in the telomeric tract disrupted TRF1 binding, while a simple nick did not.
Figure 2.
TRF1 binding is inhibited by a single nucleotide gap in telomeric DNA. Substrates containing either a nick or single nucleotide gap (gap) within the tandem telomeric repeats were constructed as described in ‘Materials and Methods’. Substrates (2.5 nM) were incubated alone (−) or together with decreasing concentrations of TRF1 (200, 100, 50 or 25 nM) (lanes 2–5 and 7–10) for 20 min at 4°C in TEL buffer. The reactions were run on 5% acrylamide native gels and representative phosphorimager scans are shown in (A). Arrows indicate bound or free substrate. The percentage bound was determined as described in ‘Materials and Methods’ and plotted against TRF1 concentration as shown in (B). Square, nick and circle, single nucleotide gap.
TRF1 recognition is inhibited by abasic lesions
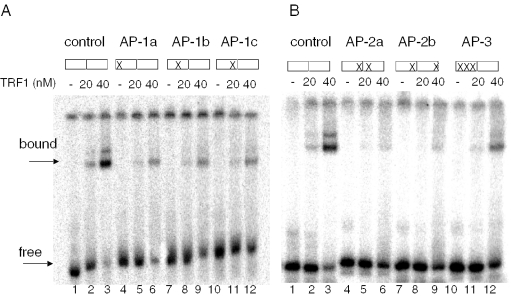
Another important intermediate in BER is the apurinic or apyrimidinic (AP) site. These lesions result from the removal of a damaged base by a DNA glycosylase that initiates BER, but also arise spontaneously ∼10 000 times per cell per day (23). We tested substrates in which various guanine residues within the telomeric tract were replaced with a tetrahydrofuran, an AP site analog, for binding by TRF1 (Table 1). The presence of a single AP residue decreased TRF1 (20 nM) binding 2- to 4-fold (lanes 4–12), compared with the undamaged control (lanes 1–3), regardless of which residue in the G run was replaced with an AP site (Figure 3A) (Table 2). However, the presence of a single AP residue in each telomeric repeat inhibited TRF1 binding even more dramatically (6- to 7-fold) (Figure 3B, lanes 1–9) (Table 2). When the guanines in a single telomere repeat were saturated with AP lesions, binding was decreased 4-fold relative to the control (Figure 3B, lanes 10–12) (Table 2). Thus, the presence of multiple abasic lesions within the telomeric tracts appeared to be less disruptive to TRF1 binding than multiple 8oxoG lesions. Nevertheless, these experiments indicate that the loss of a single or multiple bases within a telomeric tract results in disruption of TRF1 interactions with the substrate.
Figure 3.
Abasic sites in telomeric DNA inhibit TRF1 binding. Effects of single (A) or multiple (B) abasic sites in the telomeric DNA. Boxes indicate the GGG runs of the tandem telomeric repeats, and (X) marks the position of the abasic lesion(s) within the G3 runs (see Table 1). Substrates (1 nM) were incubated alone (lanes 1, 4, 7 and 10) or together with increasing concentrations of TRF1 (20 or 40 nM) (lanes 2–3, 5–6, 8–9 and 11–12) for 20 min at 4°C in TEL buffer. Reactions were run on 5% acrylamide native gels. Arrows indicate the position of bound and unbound substrate.
Table 2.
Relative TRF1 binding activity with AP-containing telomeric substrates
| Substrate | 20 nM TRF1 | 40 nM TRF1 |
|---|---|---|
| Control | 1 | 1 |
| AP-1a | 0.45 ± 0.11 | 0.41 ± 0.24 |
| AP-1b | 0.38 ± 0.12 | 0.41 ± 0.01 |
| AP-1c | 0.24 ± 0.03 | 0.43 ± 0.08 |
| AP-2a | 0.17 ± 0.00 | 0.27 ± 0.04 |
| AP-2b | 0.14 ± 0.03 | 0.30 ± 0.06 |
| AP-3 | 0.23 ± 0.07 | 0.47 ± 0.18 |
Values were normalized to the control and represent the mean and error from two independent binding assays.
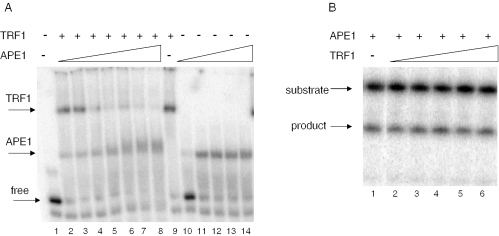
Although the presence of AP residues in telomeric DNA decreased TRF1 binding, interaction with the DNA substrate was not completely abolished, raising the possibility that TRF1 could interfere with repair proteins. To test this we examined the binding of TRF1 and APE1 simultaneously to a telomeric substrate containing an AP residue (AP-1a, Table 1). APE1 is the major human repair endonuclease for AP sites (24). The majority of the substrate (1 nM) was bound by a large excess of TRF1 (200 nM) alone (Figure 4A, lane 9). However, the intensity of the band representing bound TRF1 decreased, and the amount of APE1–DNA complex increased as a function of APE1 concentration (Figure 4A, lanes 2–8). An APE1–DNA complex was apparent even at the lowest APE1 concentration (11 nM) (lane 2), and the TRF1–DNA complex was largely absent at near equal molar APE1 and TRF1 levels (Figure 4A, lane 5). A super shifted complex was not detected in any of the reactions (although some smearing was observed), indicating that APE1 and TRF1 did not form a stable co-complex on the substrate. Consistent with these results, we observed that TRF1 did not interfere with the catalytic activity of APE1. When APE1 was incubated with the AP-1a substrate, the enzyme incised the DNA strand 5′ to the AP residue resulting in a 15 nt product (Figure 4B, lane 1). The incision activity of APE1 was unaffected even in the presence of up to a 7000-fold molar excess of TRF1 (Figure 4B, lane 6). Our results indicate a higher affinity of APE1 for the AP-containing telomeric DNA, than TRF1, and suggest that even large excess of TRF1 is unlikely to interfere with the processing of damaged telomeric DNA by APE1.
Figure 4.
APE1 is active on abasic site-containing telomeric substrates that are pre-bound with TRF1. (A) APE1 binding to TRF1 pre-bound telomeric DNA. The AP-1a substrate (1 nM) (Table 1) was incubated with either TRF1 (200 nM) alone (lane 9) or together with increasing concentrations of APE1 (11, 28, 140, 700, 2800 nM, 11 μM and 28 μM) (lanes 2–8), or with APE1 alone (11, 140, 700, 2800 nM and 11 μM) (lanes 10–14) for 20 min at 4°C in TEL buffer. Reactions were initiated by the addition of APE1 and were run on 5% native gels. Arrows indicate the position of bound (TRF1 or APE1) and free substrate. (B) APE1 activity in the presence of TRF1. The AP-1a substrate (1 nM) was incubated with either APE1 (28 pM) alone (lane 1) or with increasing concentrations of TRF1 (1, 5, 20, 80 and 200 nM) (lanes 2–6) at 37°C for 3 min in TEL Buffer. Reactions were run on an 18% acrylamide denaturing gel.
TRF2 binding to telomeric DNA is disrupted by oxidative and abasic DNA lesions
Although both TRF1 and TRF2 bind telomeric duplex DNA, their roles in telomere maintenance differ. The TRF2-DNA binding domain is 56% identical to TRF1 (25), however, the loss of TRF2 binding to telomeres has different cellular consequences, compared with TRF1 loss. Expression of a TRF2 mutant lacking the N-terminus and the C-terminal DNA binding domain was found to induce loss of telomeric 3′ tails and telomere end fusions, whereas, a similar TRF1 truncated mutant did not (1,26). These proteins also differ biochemically in that TRF2, but not TRF1, was observed to promote t-loop formation of model telomeric ends in vitro (27,28). Therefore, we performed EMSA with TRF2 to determine the effects of oxidized lesions on TRF2 binding. As observed by us and others, TRF2–DNA complexes failed to shift into acrylamide gels (20,29). However, previous reports indicated successful detection of TRF2–DNA complexes migrating into agarose gels (30,31). Yet, using the reaction buffers described in these prior reports, or those used here for TRF1 (Figures 1–3), we detected significant binding of TRF2 to the non-telomeric substrate via the agarose gel assay, indicating a lack of specificity (data not shown). This probably resulted from the basic N-terminus of TRF2 making non-specific contacts with the negatively charged DNA backbone. Thus, we used a reaction buffer for which we previously observed TRF2 complex formation specifically with a (TTAGGG)4 containing substrate, but not with an equivalent substrate in which the sequence was scrambled (20). We also increased the Mg2+ concentration to further inhibit non-specific interactions between the TRF2 N-terminus and the DNA phosphate backbone. Under these conditions, the addition of TRF2 shifted the migration of the telomeric substrate into the agarose gel (Figure 5A, lanes 1–5), but did not significantly shift the non-telomeric DNA (lanes 26–30).
Following development of the binding assay, we next tested TRF2 binding to the substrates containing site-specific 8oxoGs or the abasic analog residue (Table 1). When a single telomeric repeat contained either one or two tandem 8oxoGs, the percentage of TRF2-bound products decreased by nearly 50%, from 37 ± 8% bound to 18 ± 2% and 14 ± 3% bound, respectively, at 100 nM TRF2 (Figure 5A, lanes 3, 8 and 18). The presence of a single 8oxoG residue in each telomeric repeat decreased the percentage of bound substrate to barely detectable levels above background (8 ± 5% bound at 100 nM TRF2) (Figure 5A, lanes 11–15). Likewise, saturation of the guanine residues in a single telomere repeat with 8oxoGs dramatically decreased TRF2 binding to negligible levels (Figure 5A, lanes 21–25). Overall, the data indicate that the specific binding of full-length TRF2 to telomeric DNA is disrupted by the presence of oxidized guanines in the telomeric tract.
Next, we tested the ability of abasic DNA to alter TRF2 binding using the substrates with defined AP sites (Table 1). The replacement of the second or third guanine residue with an AP residue decreased TRF2 (100 nM) binding 1.8- and 2.6-fold, respectively, compared with the control (Figure 5B, lanes 3, 8 and 23). The presence of a single AP site in each telomeric repeat also decreased binding ∼2-fold, to 17 ± 4 and 15 ± 1% bound, compared with the control (37 ± 8% bound) at 100 nM TRF2 (Figure 5B, lane 3, 18 and 23). Therefore, the loss of a single or multiple bases within a telomeric tract results in disruption of TRF2 interactions with the substrate, similar to TRF1.
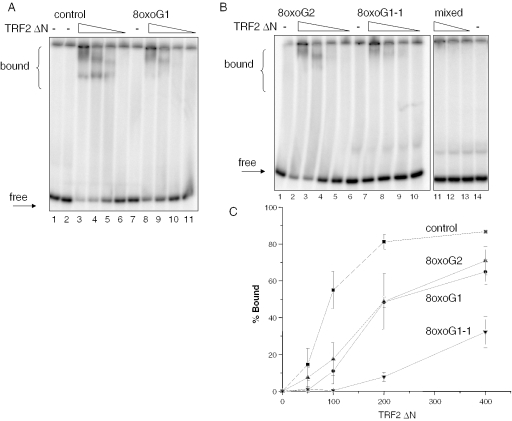
To further confirm the 8oxoG inhibitory effect on TRF2 recognition and to obtain binding curve, we performed gel-shift assays under similar conditions as for TRF1, but with a TRF2 mutant that lacks the N-terminal domain (TRF2ΔN). This mutant bound specifically to the telomeric control substrate, but not to the non-telomeric substrate (Figure 6A, lane 1–6 and Figure 6B, lanes 11–14), further supporting the notion that the N-terminal domain is responsible for the non-specific DNA interactions. The binding curve began to plateau at ∼200 nM TRF2ΔN for the undamaged control substrate (18 nM) (Figure 1C). Therefore the comparisons with the damaged substrates are performed at 100 nM TRF2ΔN, which falls within the linear portion of the curve. A single 8oxoG (G1) or multiples 8oxoGs (G2) within a single telomere repeat decreased TRF2ΔN binding 5- to 3-fold to an average of 11 and 17% bound, respectively, compared with the control (55% bound) (Figure 6). At this protein amount, the presence of a single 8oxoG lesion in each telomeric repeat (oxoG1-1) reduced binding to negligible levels (Figure 6B, lane 9). For this substrate, binding was only detected at the higher TRF2ΔN levels (200–400 nM), but was considerably reduced (up to 10-fold) compared with the control (Figure 6C). Therefore, 8oxoG is probably interfering with specific contacts between the DNA binding domains of TRF1 and TRF2 and the telomeric DNA. These results precisely mimic those using the full-length TRF2 protein (see above).
Figure 6.
A TRF2 mutant lacking the N-terminus binds specifically to telomeric DNA and is inhibited by 8oxoG. Effects of single (A) or multiple (B) 8oxoG lesions within G runs of telomeric DNA. Reaction conditions and substrates were as in Figure 1. ‘mixed’ denotes non-telomeric sequence. Substrates (18 nM) were incubated alone or together with decreasing concentrations of TRF2 (400, 200, 100 or 50 nM) for 20 min at 4°C in TEL reaction buffer. The reactions were run on 5% acrylamide native gels. Arrows indicate bound or free substrate. (C) Percentage bound was determined as described in ‘Materials and Methods’ and plotted against TRF2ΔN concentration. Square, control; circle, 8oxoG1; triangle, 8oxoG2; inverted triangle, 8oxoG1-1. Values and error bars represent the mean and SD from three independent experiments.
DISCUSSION
In this study, we tested the effects of site-specific 8oxoG lesions and the presence of BER intermediates within telomeric tracts, on the binding activity of the critical telomere-maintenance proteins TRF1 and TRF2. We observed that a single 8oxoG lesion resulted in at least a 50% reduction in the amount of TRF1- or TRF2-bound substrate. When each telomere repeat, or monomer-binding site, contained an 8oxoG lesion, the amount of TRF1- or TRF2-bound substrate was decreased to barely detectable levels. TRF1 binding was largely unaffected by the presence of a single strand nick within the telomeric tract, but binding was reduced 3-fold by the presence of a single nucleotide gap. We also tested the effect of removing a base, rather than an entire nucleotide, using site-specific tetrahydrofuran residues (an abasic site analog). Similar to 8oxoG lesions, we found an AP site inhibited TRF1 and TRF2 binding ∼50%, regardless of which guanine in the telomeric tract was replaced. These studies indicate that oxidative stress and oxidative DNA damage in particular may exert deleterious effects on telomeres through inhibition of TRF telomeric protein binding.
Structural data show that the presence of an 8oxoG-C base pair in B-form DNA imparts no gross distortion (32), suggesting that the disruption of TRF1 and TRF2 binding is due to the disruption of specific contacts required for the recognition of telomeric sequence. TRF1 and TRF2 belong to a family of eukaryotic TRFs that contain conserved domains which resemble the DNA binding motif of the c-Myb transcription activator family (25). TRF1 and TRF2 each contain a single myb-like domain in the C-terminus and bind to telomeric DNA as a homo-dimer [reviewed in (2)]. The solution structure of the TRF1 myb-like domain complexed with DNA containing the TTAGGGTTA sequence revealed specific molecular contacts between the DNA bases and TRF1 amino acids (25). A helix-turn-helix (HTH) motif lies in the DNA major groove, and amino acids in the helix contact the TAGG tandem bases and the complementary C residues of the 2nd and 3rd Gs in the (G)3 run (25). The stretch of Gs is essential for high-affinity binding by a protein fragment with the TRF1 myb-domain (21,25). Here, we report that modification of G residues in the telomeric tract disrupted binding by the full-length TRF1 and TRF2 proteins. Conversion of any of the G bases to an abasic site decreased binding by both full-length TRF1 and TRF2 (Table 2) (Figures 3 and 5B), with alteration of the 3rd G having the strongest effect. The loss of the 3rd guanine base may alter the position of the paired C, and which may disrupt fundamental contacts between this C and the TRF1 amino acids. 8oxoG substitution at the central G may similarly disrupt specific amino acid contacts with this G, but may also alter interactions with its complementary C residue (Figure 1). We have demonstrated that the loss of contacts with guanine bases (tetrahydrofuran substitution or nucleotide loss) or the chemical modification of guanine residues (oxidation), disrupt TRF1 and TRF2 interactions with telomeric DNA.
In general we detected that the presence of a lesion in both telomeric repeats inhibited TRF1 and TRF2 binding more strongly than the presence of two lesions within a single repeat (Figures 1, 3 and 5–6) (Table 2). Previous reports indicate that binding via both myb-domains of a TRF1 dimer cooperatively increases the thermal stability and affinity of the protein–DNA complex, compared with complexes in which only one myb-domain is engaged (21). Consistent with this, we found that disruption of both myb-DNA binding sites more dramatically inhibited TRF1 and TRF2 binding, compared with alteration of a single site (Figures 1, 3 and 5–6) (Table 2). The spatial distance between the two myb-DNA binding sites does not alter the affinity of TRF1 homo-dimers for the substrate (21). This is consistent with biochemical studies that support roles for TRF1 and TRF2 in remodeling telomeric ends into complex structures via mediating interaction or bridging between different regions of the telomeric stretch (27,33). Therefore, 8oxoG or abasic DNA lesions need not occur in adjacent telomeric repeats to influence TRF1 and TRF2 binding, and subsequent modeling of the telomeric end into higher-order structures. This implies, distant lesions can disrupt the higher-order restructuring of the telomeric DNA by preventing TRF binding. Nevertheless, evidence indicates that lesions are formed in close proximity (within 10–20 bp) as a consequence of ionizing radiation [(34), reviewed in (35)]. These clustered lesions contain oxidized bases, abasic sites and SSBs on the same and/or opposite strands.
Telomere length has been found to correlate with sensitivity to some DNA damaging agents. Transgenic telomerase knock-out mice with shortened telomeres, are hypersensitive to alkylating agents (36), IR (37), doxorubicin (38) and arsenic-induced oxidative stress (6) compared with non-transgenic controls. Consistent with this, telomerase expression decreased the sensitivity of cultured fibroblasts with short telomeres to IR, bleomycin, hydrogen peroxide and etoposide by elongating the telomeres (39). Reasons for the hypersensitivity are unknown, but shorter telomeres have less bound TRF1 and TRF2 (17,26). Based on our findings here, we propose that shorter telomeres, with already lower levels of bound TRFs, may be more sensitive to further loss of TRF proteins upon oxidative damage. Consistent with this, the over expression of TRF2 or telomerase, protected cultured cardiomyocytes from oxidative stress-induced apoptosis (40). We found increased binding of TRF2 to the oxidized substrates at the higher TRF2 concentrations (Figures 5 and 6).
Our findings highlight the importance of repairing DNA damage at the telomeric end, since damage may result in the loss of critical telomere-maintenance proteins. One mechanism of oxidative stress-induced telomere loss has been proposed to be blocking of the replication fork in the telomeres by the presence of unrepaired nucleotides or bases (4). Blocked and/or aborted replication may lead to strand breaks and the loss of telomere repeats, and the subsequent loss of associated TRF1 and TRF2 proteins. This is a likely scenario for blocking lesions such as thymine glycol, but 8ooxoG does not significantly block or impede DNA polymerases (15). Rather, we propose that 8oxoG may interfere with proper telomere function by directly inhibiting TRF1 and TRF2 association, or by causing mutagenesis if unrepaired. Loss of telomere-associated TRF2 has been found to induce telomere dysfunction, including telomere end fusions, even in the absence of telomere shortening (1). The binding of the TRF proteins to the telomere repeat sequence is probably a critical step of the maintenance process. In summary, oxidative stress may cause the dissociation of telomeric TRF1 and TRF2 via a combination of the loss of telomeric DNA repeats and the inhibitory effects of DNA lesions on TRF1 and TRF2 binding. Further studies are required to determine the direct consequences of oxidative DNA lesions in the telomeric DNA on telomere structure and function.
Acknowledgments
We thank Drs Nadja Souza-Pinto and Heng Kuan Wong for critical reading of the manuscript, and Lale Dawut for technical support. Funding to pay the Open Access publication charges for this article was provided by NIH.
REFERENCES
- 1.van Steensel B., Smogorzewska A., De Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 2.De Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 3.Tchirkov A., Lansdorp P.M. Role of oxidative stress in telomere shortening in cultured fibroblasts from normal individuals and patients with ataxia-telangiectasia. Hum. Mol. Genet. 2003;12:227–232. doi: 10.1093/hmg/ddg023. [DOI] [PubMed] [Google Scholar]
- 4.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., Trimarchi J.R., Smith P.J., Keefe D.L. Mitochondrial dysfunction leads to telomere attrition and genomic instability. Aging Cell. 2002;1:40–46. doi: 10.1046/j.1474-9728.2002.00004.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu L., Trimarchi J.R., Navarro P., Blasco M.A., Keefe D.L. Oxidative stress contributes to arsenic-induced telomere attrition, chromosome instability, and apoptosis. J. Biol. Chem. 2003;278:31998–32004. doi: 10.1074/jbc.M303553200. [DOI] [PubMed] [Google Scholar]
- 7.Oikawa S., Kawanishi S. Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 1999;453:365–368. doi: 10.1016/s0014-5793(99)00748-6. [DOI] [PubMed] [Google Scholar]
- 8.Serra V., von Zglinicki T., Lorenz M., Saretzki G. Extracellular superoxide dismutase is a major antioxidant in human fibroblasts and slows telomere shortening. J. Biol. Chem. 2003;278:6824–6830. doi: 10.1074/jbc.M207939200. [DOI] [PubMed] [Google Scholar]
- 9.Petersen S., Saretzki G., von Zglinicki T. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp. Cell Res. 1998;239:152–160. doi: 10.1006/excr.1997.3893. [DOI] [PubMed] [Google Scholar]
- 10.Stewart S.A., Ben Porath I., Carey V.J., O'Connor B.F., Hahn W.C., Weinberg R.A. Erosion of the telomeric single-strand overhang at replicative senescence. Nature Genet. 2003;33:492–496. doi: 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- 11.Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., De Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 12.Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., De Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 2003;23:6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwano T., Tachibana M., Reth M., Shinkai Y. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- 14.Hanish J.P., Yanowitz J.L., De Lange T. Stringent sequence requirements for the formation of human telomeres. Proc. Natl Acad. Sci. USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 16.Wilson D.M., III, Sofinowski T.M., McNeill D.R. Repair mechanisms for oxidative DNA damage. Front. Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- 17.Opresko P.L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M.M., Bohr V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 18.von Kobbe C., Harrigan J.A., May A., Opresko P.L., Dawut L., Cheng W.H., Bohr V.A. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol. Cell. Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erzberger J.P., Barsky D., Scharer O.D., Colvin M.E., Wilson D.M., III Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 21.Bianchi A., Stansel R.M., Fairall L., Griffith J.D., Rhodes D., De Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson S.H., Kunkel T.A. Passing the baton in base excision repair. Nature Struct. Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 23.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 24.Wilson D.M., III, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa T., Okamura H., Nagadoi A., Konig P., Rhodes D., Nishimura Y. Solution structure of a telomeric DNA complex of human TRF1. Structure. 2001;9:1237–1251. doi: 10.1016/s0969-2126(01)00688-8. [DOI] [PubMed] [Google Scholar]
- 26.Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., De Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stansel M., Lange T., Griffith J.D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimura S.H., Maruyama H., Ishikawa F., Ohki R., Takeyasu K. Molecular mechanisms of DNA end-loop formation by TRF2. Genes Cells. 2004;9:205–218. doi: 10.1111/j.1356-9597.2004.00719.x. [DOI] [PubMed] [Google Scholar]
- 29.Machwe A., Xiao L., Orren D.K. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 30.Li B., Oestreich S., De Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 31.Dantzer F., Giraud-Panis M.J., Jaco I., Ame J.C., Schultz I., Blasco M., Koering C.E., Gilson E., Menissier-de Murcia J., de Murcia G., et al. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol. Cell. Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipscomb L.A., Peek M.E., Morningstar M.L., Verghis S.M., Miller E.M., Rich A., Essigmann J.M., Williams L.D. X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl Acad. Sci. USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchi A., Smith S., Chong L., Elias P., De Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutherland B.M., Bennett P.V., Sidorkina O., Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dianov G.L., O'Neill P., Goodhead D.T. Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. Bioessays. 2001;23:745–749. doi: 10.1002/bies.1104. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Suarez E., Goytisolo F.A., Flores J.M., Blasco M.A. Telomere dysfunction results in enhanced organismal sensitivity to the alkylating agent N-methyl-N-nitrosourea. Cancer Res. 2003;63:7047–7050. [PubMed] [Google Scholar]
- 37.Wong K.K., Chang S., Weiler S.R., Ganesan S., Chaudhuri J., Zhu C., Artandi S.E., Rudolph K.L., Gottlieb G.J., Chin L., et al. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nature Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- 38.Lee K.H., Rudolph K.L., Ju Y.J., Greenberg R.A., Cannizzaro L., Chin L., Weiler S.R., DePinho R.A. Telomere dysfunction alters the chemotherapeutic profile of transformed cells. Proc. Natl Acad. Sci. USA. 2001;98:3381–3386. doi: 10.1073/pnas.051629198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubio M.A., Davalos A.R., Campisi J. Telomere length mediates the effects of telomerase on the cellular response to genotoxic stress. Exp. Cell Res. 2004;298:17–27. doi: 10.1016/j.yexcr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Oh H., Wang S.C., Prahash A., Sano M., Moravec C.S., Taffet G.E., Michael L.H., Youker K.A., Entman M.L., Schneider M.D. Telomere attrition and Chk2 activation in human heart failure. Proc. Natl Acad. Sci. USA. 2003;100:5378–5383. doi: 10.1073/pnas.0836098100. [DOI] [PMC free article] [PubMed] [Google Scholar]