Abstract
Ribozymes, RNA molecules that catalyze the cleavage of RNA substrates, provide an interesting alternative to the RNA interference (RNAi) approach to gene inactivation, especially given the fact that RNAi seems to trigger an immunological response. Unfortunately, the limited substrate specificity of ribozymes is considered to be a significant hurdle in their development as molecular tools. Here, we report the molecular engineering of a ribozyme possessing a new biosensor module that switches the cleavage activity from ‘off’ (a ‘safety lock’) to ‘on’ solely in the presence of the appropriate RNA target substrate. Both proof-of-concept and the mechanism of action of this man-made riboswitch are demonstrated using hepatitis delta virus ribozymes that cleave RNA transcripts derived from the hepatitis B and C viruses. To our knowledge, this is the first report of a ribozyme bearing a target-dependent module that is activated by its RNA substrate, an arrangement which greatly diminishes non-specific effects. This new approach provides a highly specific and improved tool with significant potential for application in the fields of both functional genomics and gene therapy.
INTRODUCTION
The ability of ribozymes (RNA enzymes) to catalyze the cleavage of RNA substrates makes them attractive potential molecular tools [reviewed in (1,2)]. Moreover, ribozymes are an interesting alternative to the RNA interference (RNAi) approach to gene inactivation as the use of RNAi seems to trigger an immunological response (3–5). However, the use of ribozymes in this regard is limited by several factors, including: delivery problems, effective concentration in vivo and cellular stability (1,2). In addition, considerable effort has been directed toward the improvement of substrate specificity of ribozymes. For example, the approach of systematic enrichment of ligands by exponential evolution has resulted in the evolution and selection of allosteric ribozymes, which exhibit an activity that is both controllable and sensitive (6–13). Inactive allosteric RNAs are activated by the binding of effectors that cause conformational transitions yielding active ribozymes. This is well illustrated by allosteric ribozymes for which the cleavage activity is regulated by either small biosensor molecules (e.g. ATP) or proteins (e.g. Tat) (8,13). However, the in vivo potential of allosteric ribozymes might be limited because they involve a third species (i.e. the independent effector), which complicates the approach. Furthermore, the presence of a regulatory module does not improve the substrate specificity of these ribozymes in terms of avoiding the cleavage of an inappropriate target. In other words, none of these ribozymes is specifically activated by its own substrate, which should be considered ideal for greatly diminishing any non-specific effects.
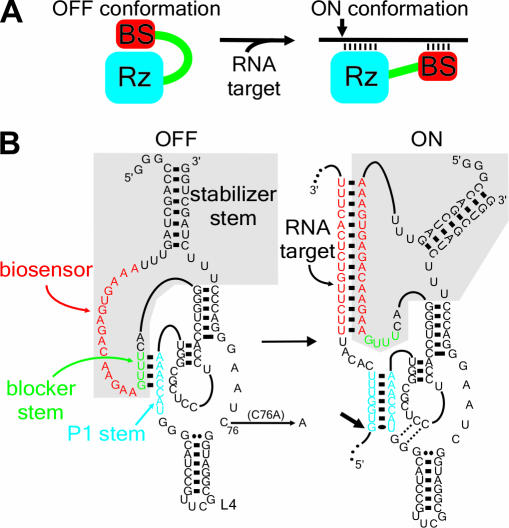
With the goal of generating highly specific ribozymes that could be regulated by the presence/absence of their target substrates, we started out with the concept that a ribozyme should be linked to a target-dependent module that acts as a biosensor (Figure 1A). In the absence of the target, the ribozyme should be inactive (‘off’, ‘safety lock’ conformation), while in the presence of the desired substrate the biosensor should recognize it and activate (turn on) the ribozyme's cleavage activity. Accordingly, a rational design led to a ribozyme controlled by a novel specific on/off adapter (SOFA). The original delta ribozyme (δRz) derived from the hepatitis delta virus was used as a suitable model (14,15). Substrate recognition of this ribozyme is based solely on the formation of the P1 stem, which necessarily requires 7 bp (16). The SOFA includes three sequence segments: a blocker, a biosensor (BS) and a stabilizer (Figure 1B, gray section). In the absence of the target, the blocker forms an intramolecular stem with the P1 strand, resulting in no cleavage. Upon the addition of the target, the biosensor anneals with the substrate, thereby releasing the P1 strand so that it can subsequently hybridize with the substrate and initiating formation of the active conformation. Thus, the target has two simultaneous roles: one as an activator and the other as a substrate. The biosensor acts as a riboswitch regulating the catalytic activity. This study presents both the proof-of-concept and the first characterization of the molecular mechanism of action of this new man-made riboswitch.
Figure 1.
The concept of the SOFA-ribozyme. (A) Schematic representation of both the ‘off’ and ‘on’ conformations of the SOFA-ribozyme. The ribozyme (Rz) and the biosensor (BS) are in blue and red, respectively. The small arrow indicates the cleavage site. (B) Secondary structure and nucleotide sequence of SOFA+-δRz-303 in both the ‘off’ and the ‘on’ conformations. The gray section indicates the SOFA module. The P1 stem of the ribozyme and the biosensor are in blue and red, respectively. The blocker sequence is represented in green. The C76A mutation that produces an inactive ribozyme version is indicated. The numbering system is according to (15).
MATERIALS AND METHODS
HBV, HCV and ribozyme DNA constructs
The hepatitis B virus (HBV) 1190 nt fragment was excised from pCHT9/3091 (17) using SacI and EcoRI, and then subcloned into pBlueScript SK, generating pHBV-1190. The shorter HBV 44 nt substrate was produced using a PCR-based strategy with T7 sense primer (5′-TTAATACGACTCACTATAGGG-3′) and antisense primer (5′-CTTCCAAAAGTGAGACAAGAAATGTGAAACCACAAGAGTTGCCCTATAGTGAGTCGTATTAA-3′). The plasmid pHCVA was obtained by cloning the 1348 nt HCV 5′ sequence from pHCV-1b (18) into the HindIII/BamHI sites of pcDNA3 (Invitrogen). The original δ ribozymes were constructed as described previously (19). SOFA+/−-ribozymes were constructed using a PCR-based strategy that included two complementary and overlapping oligonucleotides. Briefly, two DNA oligonucleotides were used: the antisense oligonucleotide [5′-CCAGCTAGAAAGGGTCCCTTAGCCATCCGCGAACGGATGCCCA(N)6P1ACCGCGAGGAGGTGGACCCTG(N)4-BL-3′, where N is A, C, G or T and BL indicates blocker sequence]; and, the sense primer [5′-TTAATACGACTCACTATAGGGCCAGCTAGTT-T(N)7–20-BS(N)4-BLCAGGGTCCACC-3′] that permitted incorporation of the T7 RNA promoter. It is important to note that the (N) segments were varied to correspond to specific designed target sequences. The PCR products were purified by phenol:chloroform extraction, precipitated with ethanol and dissolved in water. In vitro transcriptions and purifications of the ribozymes were then performed as described below. A similar strategy was used to build the different versions of ribozyme. All SOFA−-ribozymes possess the same biosensor sequence, i.e. 5′-GAACATCG GTATCAC-3′.
For the in vivo experiments, the open reading frame (ORF) of the HBV C gene was amplified from pCH9T/3091 (17) using two oligonucleotides, a sense (5′-TATCTAAAGCTAGCTTCATGTCCTACTGTTCAAGCCTCC-3′) and an antisense (5′-TAGTGAAACTCGAGAATAAAGCCCAGTAAAGTTCCCA-3′) primer. The PCR product was cloned into the NheI and XhoI restriction enzyme sites of the pIND vector (Invitrogen), yielding the pINDHBVC vector. For the cloning of the ribozymes, we first constructed an expression cassette using two oligonucleotides composed (5′→3′) by the KpnI and EcoRV restriction sites followed by a poly(A) signal. This cassette was subsequently inserted into HindIII/XhoI-linearized pcDNA3. Second, the PCR products corresponding to the minigenes of the different versions of the ribozyme described above were reamplified using two oligonucleotides (sense, 5′-ATCCATCGGGTACCGGGCCAGTTAGTTT-3′; and antisense, 5′-CCAGCTAGAAAGGGTCCCTTAGCCATCCGCG-3′) that permitted the introduction of a KpnI restriction site at the 5′ end and also created a blunt 3′ end. The DNA products were then cloned in KpnI/EcoRV-linearized pcDNA3. The resulting plasmids were named pmδRz-X, where X represents the cleavage position of the ribozyme.
RNA synthesis
Both ribozymes and substrates were synthesized by run-off transcription from PCR products, HindIII-linearized plasmid pHBV-1190 and XbaI-linearized plasmid pHCVA templates. Run-off transcriptions were performed and the resulting transcripts purified as described previously (19). Subsequently, the substrates were dephosphorylated and 5′ end-labeled with 32P as described previously (19).
Ribozyme cleavage assays
In all reactions, the ribozyme and substrate were preincubated together at 37°C for 2–5 min, and subsequently magnesium was added in order to initiate the cleavage. Changing the order of the addition of the components did not modify the results. Except when indicated, all reactions were performed under single turnover conditions ([Rz] > [S]) using 1 μM ribozyme and trace amounts of either internally 32P-labeled or 32P-5′-end-labeled substrates at 37°C in a final volume of 10 μl containing 50 mM Tris–HCl, pH 7.5 and 10 mM MgCl2. For the time-course experiments, aliquots (0.8 μl) were removed at various times up to 3 h and were quenched by the addition of ice-cold formamide dye buffer (5 μl of 97% formamide, 10 mM EDTA, pH 8.0), electrophoresed on 6% PAGE gel and the gels analyzed with a radioanalytic scanner (PhosphorImager). For the multiple turnover reactions, the assays were performed at 56°C with an excess of substrate over ribozyme (10 μM versus 1 μM, respectively). Cleavage reactions for the mechanism analysis were carried out either with or without 5 μM of a facilitator (FCO, 5′-AAAGTGAGACAAGAA-3′), biosensor stem (BSO, 5′-TTCTTGTCTCACTTT-3′) and an unrelated (UNO, 5′-CCCAATACCACATCA-3′) oligodeoxynucleotide. The oligodeoxynucleotides were always mixed, for 5 min at 37°C, first with the RNA component that it bound to, and the reactions then initiated by the addition of the other RNA component (e.g. the substrate and FCO were preincubated together, and then the ribozyme was added to the mixture). Cleavage assays with the pools of mixed substrates were performed with trace amounts of radiolabeled substrates (50 000 c.p.m.), non-labeled RNA substrates (2 μM) and SOFA-ribozymes (500 nM), except in the case of the original ribozyme (WT, 2 μM). The different components were mixed simultaneously, and the reactions were then incubated for 2 h, analyzed on denaturing 10% PAGE gels and visualized by PhosphorImager.
Cell culture, DNA transfections and northern blot hybridization
HEK 293 EcR cells (Human Embryonic Kidney) were grown in DMEM (Sigma) supplemented with 10% fetal bovine serum (Wisent) and 0.4 mg/ml of zeocine (Invitrogen) at 37°C under a humidified 5% CO2 atmosphere. The cells (2 × 105) were co-transfected with a total of 2 μg of DNA using LipofectAMINE as per the manufacturer's instructions (Invitrogen). The pINDHBVC and pmδRz-X vectors were present in a ratio of 1:19. The transfected cells were induced 24 h later by adding ponasterone A to a final concentration of 1.1 μM, and total RNA extracted ∼44 h post-transfection using TriReagent (BioShop Canada Inc., Burlington). Northern blot analyses of RNA samples (10 μg) were performed as described previously (20). The HBV C probe was obtained by cloning the PCR product of the C gene amplified above into pBlueScript (SK) vector in the inverse orientation and then performing run-off transcription using the T7 RNA polymerase in the presence of [α-32P]UTP. The β-actin RNA probe used was prepared using the Strip-EZ™ RNA T7/T3 kit (Ambion). All hybridizations were carried out for 16–18 h at 65°C, and the membranes were exposed to PhosphorImager screens for 2–24 h.
RESULTS
Rational design of the SOFA-ribozyme
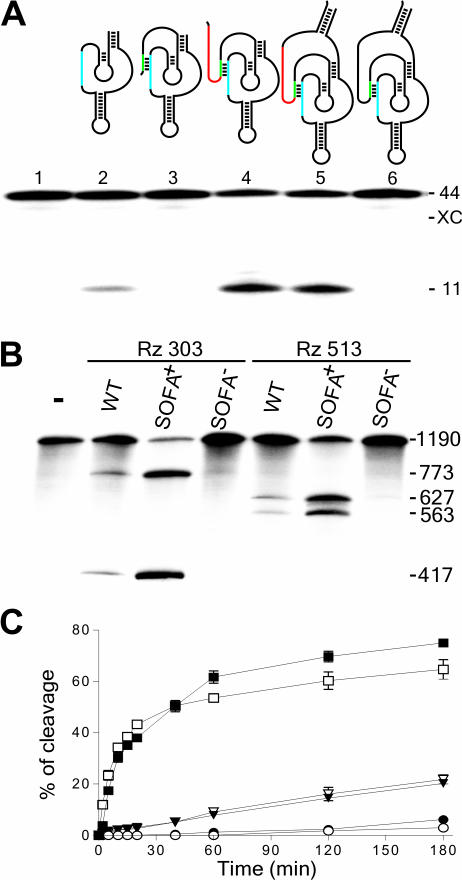
The molecular engineering of the specific ‘on/off’ adapter module followed several steps (Figure 2A). First, a blocker sequence that forms an intramolecular stem with the catalytic core of the ribozyme (i.e. the P1 strand region) was added at the 5′ extremity (Figure 2A, lane 3). The resulting molecule exhibited no cleavage of a small substrate 44 nt in size derived from the HBV RNA (for sequences, see Materials and Methods). The blocker acts as a ‘safety lock’ even though the nucleotide sequences of the δ ribozyme portions in both molecules were identical. Several mutant blocker sequences, either shorter ones or those producing mismatches, were constructed. All exhibited at least a residual cleavage activity, indicating that mutated blocker sequences do not prevent completely the ribozyme activity (data not shown). Second, the ribozyme was further extended at its 5′ end by the addition of a biosensor sequence of 15 nt complementary to the substrate. The resulting construct exhibited a cleavage activity several times higher than that of the original ribozyme (Figure 2A, compare lane 4 with lane 2). In this case, the substrate sequence bound by the biosensor was arbitrarily fixed 5 nt downstream of the last nucleotide participating in the formation of the P1 stem with the ribozyme. The region inserted between the P1 stem and the biosensor stem on the RNA substrate was named the spacer. Third, the ribozyme was further extended at both the 5′ and 3′ ends in order to permit the ends to be joined in a terminal stem (Figure 2A, lane 5). This last addition was included based on the observations revealing that the terminal P2 stem of the original δ ribozyme provides an outstanding stability to this RNA species (21). The presence of the stabilizer did not influence the ribozyme's level of cleavage. Both the biosensor and the blocker sequences function independently of the presence/absence of the stabilizer. Together, the three segments (blocker, biosensor and stabilizer) constitute the SOFA module. When the full version of the SOFA-ribozyme harbors a non-specific biosensor sequence (i.e. one unrelated to the substrate), the resulting construct exhibited no cleavage activity whatsoever (Figure 2A, lane 6). This SOFA−-ribozyme remains locked in its inactive conformation (‘off’), in contrast to its SOFA+-ribozyme counterpart that switched into the ‘on’ conformation.
Figure 2.
Rational design and proof-of-concept of the SOFA-ribozyme. (A) Schematic representation and typical autoradiogram of a denaturing 6% PAGE gel used to analyze the cleavage reactions of various versions of ribozyme. The P1 stem of the ribozyme (Rz), the blocker and the biosensor (BS) are in blue, green and red, respectively. The length of the bands, in nucleotides, is shown adjacent to the gel. The control (−) was performed in the absence of ribozyme (lane1), while lane 2 is in the presence of the original δRz. Lanes 3–5 are ribozymes progressively extended by the addition of the blocker (lane 3), the biosensor (lane 4) and the stabilizer (lane 5). Lane 6 is a SOFA−-ribozyme that consists of a ribozyme harboring an inappropriate biosensor sequence. (B) Typical autoradiogram of a denaturing 6% PAGE gel used to analyzing the cleavage reaction of the HBV-derived target by the original- (WT), SOFA-δRz-303 and -513 ribozymes. The incubation time was 3 h at 37°C. The length of the bands in nucleotides is shown adjacent to the gel. The control (−) was performed in the absence of ribozyme, while SOFA+ and SOFA− indicates SOFA harboring either the appropriate or inappropriate biosensor sequences, respectively. (C) Graphical representations of time courses for the cleavage reactions of SOFA+ (squares), SOFA− (circles) and the original (inversed triangles) ribozyme versions of δRz-303 (filled symbols) and δRz-513 (open symbols). Incubations were performed at 37°C and aliquots were recovered at 0, 2, 5, 10, 15, 20, 40, 60, 120 and 180 min.
The SOFA concept is not simply an extension of the complementary sequence. If this was the case, the SOFA− version would have to cleave to the same extent as the original ribozyme, but this is not what is observed. An original feature of the SOFA is the control of the ribozyme's activity by the blocking sequence that acts as a ‘safety lock’ preventing folding into an active structure, even in the presence of the appropriate P1 targeting site on the RNA substrate. Furthermore, the activation of the SOFA module does not require the presence of a third species as has been reported for other attempts at increasing ribozyme specificity (6,7,10,12,13).
Proof-of-concept of the SOFA-ribozyme
The proof-of-concept of the SOFA-ribozyme was performed using two accessible sites of the HBV RNA that have previously been selected for ribozyme cleavage (Rz-303 and Rz-513; for the sequences see Supplementary Figure 1) (19). These ribozymes inefficiently cleave an HBV-derived RNA of 1190 nt (Figure 2B, ∼15%), whereas the corresponding SOFA-ribozymes (SOFA+) possessing a biosensor exhibited a drastically improved level of cleavage at both sites (∼75%). SOFA−-ribozymes bearing a biosensor sequence unrelated to the target sequence were found to be barely active. These ribozymes remain locked by the blocker sequence in an inactive conformation. Time-course experiments reinforced the conclusion that appending of a specific SOFA significantly contributed to enhancing the level of cleavage activity (Figure 2C). Moreover, similar conclusions were reached for several other SOFA-ribozymes, albeit to different degrees (data not shown). Variation in the levels of cleavage might be due to several features, including differences in the binding efficiencies and the rate constants of the SOFA structural transitions (‘on/off’ conformations).
Characterization of the SOFA-ribozyme
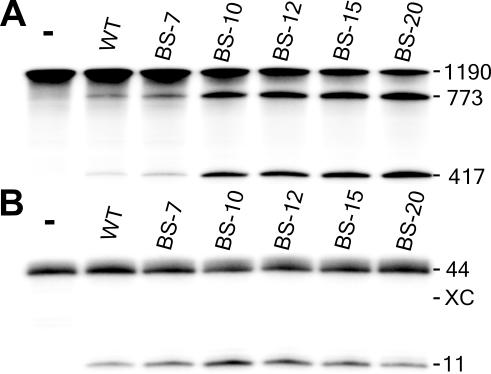
Cleavage activity was investigated using SOFA+-δRz-303 possessing biosensor sequences of various lengths. Under single turnover conditions, the cleavage levels increased in proportion to the size of the biosensor (Figure 3A): the longer the base-paired segment, the better the binding of the ribozyme to the substrate and the higher level of cleavage. Kinetic analyses permitted the determination of second order rate constants (Table 1, kcat/KM). The ribozymes SOFA+-δRz-303-BS10 and -BS15 had kcat/KM values of 1.5 × 105 min−1 M−1 and 2.3 × 105 min−1 M−1, respectively. These values are 16- and 25-fold higher than that of the original ribozyme (i.e. 9.3 × 103 min−1 M−1). These improvements are due to smaller KM and higher kcat values (3- and 8-fold each, respectively), probably representing a better formation of the active ribozyme–substrate complex that results in less dissociation and more cleavage. Either single or double mutations in the biosensor sequence of SOFA+-δRz-303-BS10, which produces a mismatch between the substrate and the ribozyme, were sufficient to cause a 25-fold reduction in the kcat/KM value (Supplementary Figure 2). The ribozyme SOFA−-δRz-303-BS10 had a kcat/KM value 2239 times smaller than that of its active counterpart. This observation is in good agreement with the idea that the blocker sequence acts to efficiently prevent the adoption of the active structure. Under multiple turnover conditions, a smaller HBV-derived substrate was used, since the length of the latter affects the turnover rate of the ribozyme (22). In this case, the level of cleavage increased in proportion to the length of the biosensor, up to 10 bp, at which point it decreased with increasing length (Figure 3B). Elongation of the biosensor stimulates the cleavage activity up to the point where, most probably, product release becomes rate limiting. Interestingly, in this specific experiment, the SOFA+-δRz-303-BS-10 performed four turnovers (i.e. since the concentration of substrate is 10-fold higher than that of the ribozyme, the cleavage of 40% substrate corresponds to four turnovers), while the original ribozyme completed only one. The observation of several turnovers indicated that the product release is not the rate-limiting step for the SOFA-ribozyme. A SOFA-ribozyme in which the cytosine in position 76 was mutated to adenosine was also synthesized (Figure 1B, SOFA+-δRzC76A-303). This mutation yields a SOFA−-ribozyme that exhibited the same binding ability, but is completely devoid of any of cleavage activity (data not shown). This result demonstrates that the biosensor is not directly implicated in the cleavage reaction; rather, it acts as a facilitator resulting in an increased level of cleavage. More importantly, the SOFA-ribozyme meets the classical criteria of an enzyme (e.g. it exhibits turnover) (23).
Figure 3.
Characterization of SOFA+-δRz-303. (A) Autoradiogram of a denaturing 6% PAGE gel showing the cleavage assays of the HBV-derived substrate by SOFA+-δRz-303 bearing biosensor stems of various lengths (BS-X, where X indicates the length of the stem). The reactions were performed under single turnover conditions at 37°C for 3 h. (B) Autoradiogram of a 20% PAGE gel showing the cleavage assays of the 44 nt HBV-derived substrate by a SOFA+-δRz-303 bearing a biosensor stem of various lengths. The reactions were performed at 37°C for 2 h under multiple turnover conditions. XC indicates the position of the xylene cyanol dye. For (A) and (B), the length of the bands in nucleotides is shown adjacent to the gel. The control (−) was performed in the absence of ribozyme.
Table 1.
Kinetic analysis of various versions of SOFA-δRz-303 under single turnover conditions
| Ribozymes | kcat (min−1) | KM (nM) | kcat/Km (min−1 M−1) |
|---|---|---|---|
| δRz-303 | 0.007 ± 0.001 | 750 ± 100 | 9.3 × 103 |
| SOFA+-δRz-303-BS15 | 0.053 ± 0.008 | 226 ± 30 | 2.3 × 105 |
| SOFA−-δRz-303-BS15 | 0.00008 ± 0.00002 | 600 ± 150 | 1.3 × 102 |
| SOFA+-δRz-303-BS10 | 0.023 ± 0.003 | 150 ± 20 | 1.5 × 105 |
| SOFA−-δRz-303-BS10 | 0.00004 ± 0.00001 | 600 ± 150 | 6.7 × 101 |
Overview of the molecular mechanism of a SOFA-ribozyme
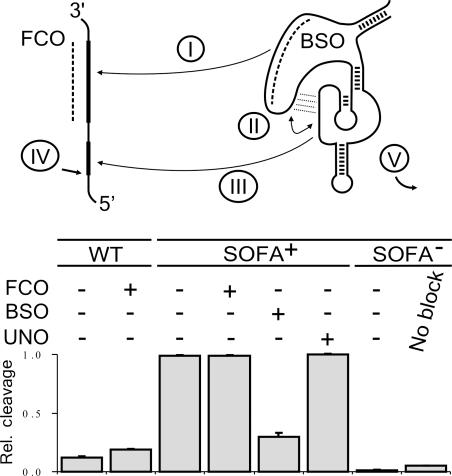
The mechanism of action of both the biosensor and blocker sequences was investigated using an oligonucleotide competition approach coupled with mutated ribozymes (Figure 4). The addition of an oligonucleotide possessing the same sequence as the biosensor (FCO) slightly increased the level of cleavage of the original Rz-303. This oligonucleotide acts as a facilitator that renders the binding site more accessible to the catalytic region of the ribozyme (24,25). In contrast, the presence of the FCO did not alter the level of cleavage of SOFA+-δRz-303 after 3 h of incubation; whereas, it required more time to reach the same cleavage level (data not shown). One possibility is that the binding of both the biosensor and the P1 sequence favorably competes with the FCO for the substrate. When the experiment was repeated using an oligonucleotide complementary to the biosensor sequence (BSO), the cleavage activity of the SOFA+-ribozyme was drastically decreased. Conversely, the presence of an oligonucleotide having an unrelated sequence (UNO) did not modify the cleavage level. Finally, the contribution of the blocker sequence was assessed using SOFA−-δRz-303. This ‘off’-version, which lacks the appropriate biosensor, but possesses the appropriate P1 stem, barely demonstrated a detectable level of cleavage. In contrast, a mutant lacking the blocker sequence exhibited a higher cleavage activity than did the SOFA−-ribozyme. Hence, the blocker plays its role by preventing the formation of the P1 stem in the absence of the appropriate biosensor. This requirement increases the specificity of the SOFA because it can only be activated by the desired substrate. Thus, the binding of the biosensor to the substrate is the first step as there is no cleavage activity if the biosensor stays in the locked conformation.
Figure 4.
Analysis of the mechanism of action of the SOFA-δ ribozyme. The upper panel shows the proposed sequential interactions between the δRz and the substrate. The roman numerals identify the steps of the mechanism. Dashed lines indicate oligonucleotide binding to either the substrate (i.e. FCO acting as facilitator) or the biosensor (BSO). The lower panel is a histogram showing the relative cleavage efficiencies as calculated from two independent sets of experiments using the original- (WT), SOFA+- and SOFA−-δRz-303 ribozymes incubated either with or without the FCO, BSO and unrelated (UNO) oligonucleotides.
Improved substrate specificity
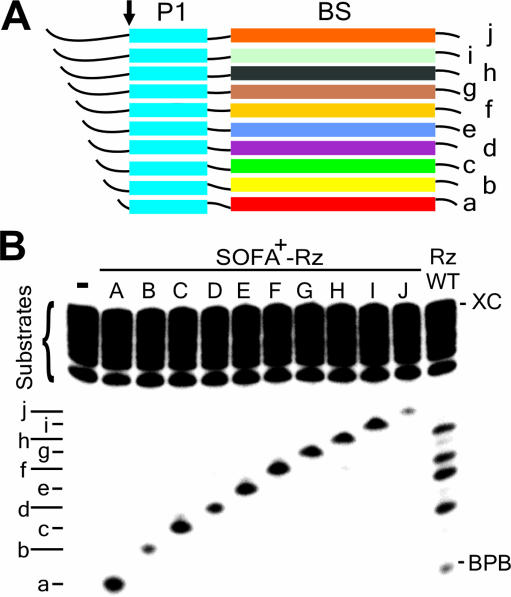
The specificity of a ribozyme can commonly be defined by its ability to discriminate between similar RNA substrates (23). In order to illustrate the gain in terms of substrate specificity, two distinct experiments were performed. First, 10 substrates were designed, each possessing an identical P1 binding sequence coupled to a completely different biosensor binding sequence. The substrates were successively extended at their 5′ ends by at least 2 nt in order to provide them with different electrophoretic mobilities (Figure 5A). When all of the substrates were incubated together with a given SOFA+-ribozyme, only the substrate harboring the relevant requirements, in terms of sequence recognition by the biosensor of that particular ribozyme, was cleaved (Figure 5B). The lower cleavage levels observed for substrates ‘b’ and ‘j’ were indicative of the influence of the biosensor's sequence identity. This experiment reveals that a ribozyme, activated by the proper substrate, does not cleave other substrates via a trans-cleavage mechanism. Conversely, the original ribozyme did not make this discrimination (lane WT), and all substrates were cleaved, although at different levels. This study shows the advantage of the ‘safety lock’ concept. Any non-specific cleavage was observed for the SOFA-ribozyme as compared with the original ribozyme results. Kinetic experiments were performed using an excess of ribozyme, thereby permitting determination of kobs that are near the kcat. Overall, the kobs of the SOFA-ribozymes cleaving the perfectly complementary substrates were at least three orders of magnitude higher than that of the other substrates (data not shown). Thus, each ribozyme cut efficiently only its perfectly complementary substrate.
Figure 5.
Trans-cleavage analysis of various SOFA-δ ribozymes. (A) Schematic representation of the HBV target substrates. All the ribozymes and substrates have a similar P1 stem sequences (in blue), but differ in their biosensor sequences (in various colors). The small arrow indicates the cleavage site (for the sequences, see Supplementary Table 1). (B) Cleavage assays of a pool of ten 5′ end-labeled substrates (a–j) by either a specific SOFA-δ ribozyme (named A–J), or the original ribozyme (WT). The reactions were performed at 37°C for 3 h. BPB and XC indicate the positions of the bromophenol blue and xylene cyanol dyes, respectively. The control (−) was performed in the absence of ribozyme.
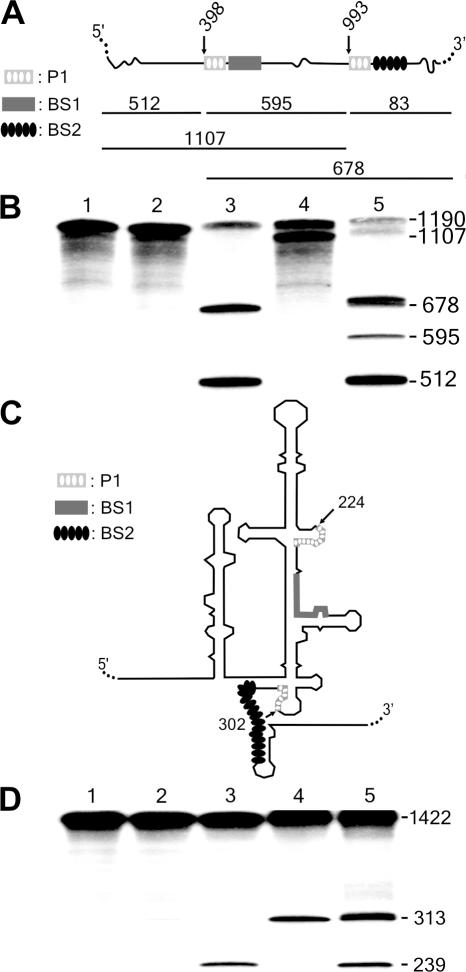
In a second experiment, we attempted to selectively cleave sites of long RNA molecules that included identical P1 binding sequences, but different biosensor sequences. A sequence 7 nt long was found to be present twice in the HBV fragment (Figure 6A and B, i.e. cleavage at positions 398 and 993). In addition, the classical δRzs did not permit the detection of cleavage at either of these sites, most likely because they were embedded in complex structures. This result is in agreement with a previous study that showed that these regions of HBV were inaccessible to ribozyme cleavage via three different protocols (19). In contrast, SOFA-δ ribozymes exhibited an efficient and specific cleavage at these sites (i.e. without any interference between the sites). This corroborates the power of the SOFA module, and points out that it enables the cleavage of a substrate uncleavable by the original ribozyme. Similar results were obtained when targeting a repeated sequence (i.e. the cleavage sites at positions 224 and 302) within the highly structured internal ribosome entry site (IRES) of the HCV (26,27). The wild-type ribozyme cleaved the HCV-derived transcripts at both sites (positions 224 and 302), although inefficiently. The SOFA+ versions produced a good level of specific cleavage (Figure 6C and D). More importantly, the position of the cleavages was dictated by the identity of the biosensor sequences. This implies that no cis-cleavage was observed in these studies. Clearly, these experiments establish the high substrate specificity of the SOFA module, in addition to illustrating its character as a facilitator (i.e. unwinding the secondary structure in the neighborhood of the target site). Thus, the accessibility of target sites became a lesser hurdle than is for classical ribozymes (20).
Figure 6.
Cis-cleavage analysis of various SOFA-δ ribozymes. (A and C) Schematic representation of the HBV and HCV target substrates, respectively. For these experiments, the sequence of the P1 stem is identical at each site, but the biosensor sequences are different (see the legend on the left of the Figure). (B) Autoradiogram of the cleavage assays of the 1190 nt HBV-derived target by SOFA-ribozymes cleaving at either position 398 or 993. Lane 1 is the control performed in the absence of ribozyme. Lane 2 is the reaction performed in the presence of the original δRz that cleaves at both sites (positions 398 and 993). Lanes 3 and 4 are the reactions performed with the SOFA+-δRz-398 and −993 ribozymes, respectively. Finally, lane 5 is the reaction performed in the presence of both the SOFA+-δRz-398 and -993. The reactions were performed at 37°C for 3 h. (D) Autoradiogram of the cleavage assays of a 1422 nt HCV-derived target by SOFA-δ ribozymes cleaving at either position 224 or 302 of the IRES. Lane 1 is the control performed in the absence of ribozyme. Lane 2 is the reaction performed in the presence of the original δRz that cleaves at both sites (positions 224 and 302). Lanes 3 and 4 are the reactions performed with the SOFA+-δRz-224 and -302 ribozymes, respectively. Finally, lane 5 is the reaction performed in the presence of both the SOFA+-δRz-224 and -302. The reactions were performed at 37°C for 3 h. All of the target site sequences of these substrates are presented in Supplementary Figure 1. The expected cleavage fragments are shown with their corresponding sizes in nt.
Activity of SOFA-ribozymes in cellular environment
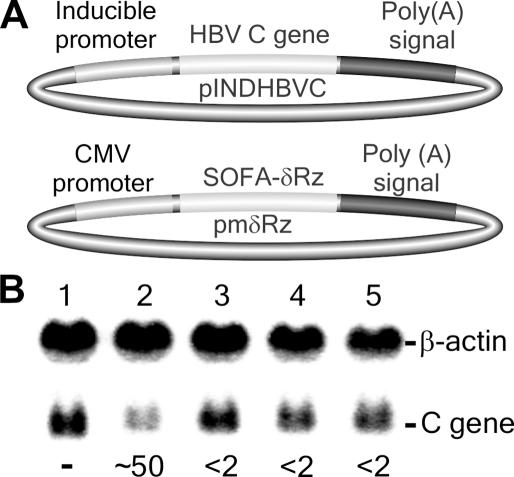
In order to demonstrate the potential of SOFA-ribozymes as molecular tools for gene-inactivation systems, a preliminary experiment targeting an HBV-derived transcript was performed in cell culture (Figure 7). First, the ORF of the HBV C gene was cloned into the inducible pIND vector (Figure 7A, upper part). Using HEK-293 EcR cells, which stably express the ecdysone receptor thereby permitting the inductor ponasterone A to enter the cell, it is possible to control the expression of the target RNA species. Second, several ribozymes were cloned downstream of the cytomegalovirus (CMV) promoter of a modified version of the pcDNA3 vector (Figure 7A, lower part; see also Materials and Methods). This modified version should permit the in vivo transcription by RNA polymerase II, and ensure localization of the ribozymes in the cytoplasm. Both vectors were co-transfected, and the expression of the HBV transcripts induced. The cells were then harvested and the RNA extracted and analyzed by northern blot hybridization. Figure 7B shows a typical autoradiogram. In the presence of SOFA+-δRz-303, an ∼50% reduction in the level of HBV C gene was observed (compare lanes 1 and 2). This is a reduction of several folds due to the presence of the SOFA+-δRz-303, as compared with the corresponding original ribozyme (data not shown). Conversely, no significant reduction was observed in the presence of SOFA+-δRzC76A-303 (<2%), an inactive mutant which is the ideal control for the presence of any antisense effect (20). Similarly, no decrease was observed in the presence of either SOFA−-δRz-303 or SOFA−-δRzC76A-303 (Figure 7B, lanes 3 and 4), illustrating the necessity of the appropriate biosensor for a ribozyme to be active, and supporting the ‘safety lock’ role played by the blocker sequence. Together, these data confirm the great potential of the target-dependent module for significantly improving ribozyme specificity in a cellular environment.
Figure 7.
Activity of SOFA-ribozymes in a cellular environment. (A) Schematic representation of the expression vectors for both the HBV C target gene (upper part) and the ribozymes (lower part). (B) Autoradiogram of northern blot hybridization. The transfection control was performed using pmδRz devoid of any ribozyme gene (lane 1). Lanes 2 and 3 are RNA samples extracted from HEK-293 EcR cultured cells expressing SOFA+-δRz-303 and SOFA+-δRzC76A-303, respectively. Lanes 3 and 4 are RNA samples extracted from cultured cells expressing SOFA−-δRz-303 and SOFA−-δRzC76A-303Rz, respectively. β-actin was used as a control for normalizing the results. The data below the autoradiogram correspond to the percentage of reduction of the C mRNA as compared with the control lane 1. Note that two independent sets of data were used to determine average values.
DISCUSSION
The concept of a target-dependent module provides for a new generation of biosensorized ribozymes possessing significantly improved substrate specificities and efficiencies. The ‘on’ conformation implies that a ribozyme with a greater affinity for its substrate subsequently cleaves them faster. Meanwhile, the ‘off’ conformation prevents the cleavage of an inappropriate target, acting as a ‘safety lock’. The design of the specific ‘on/off’ adapter was influenced by several factors. First, it is reminiscent of the human immune system, specifically the cytotoxic T lymphocyte's activation mechanism (28). The T lymphocytes bind specific cell surface molecules, which in turn dictate the T cell's responses (i.e. binding activates the T cells which in turn kill the target cell). In the same way, the SOFA-ribozyme hybridizes to the RNA target (the activator) and specifically cleaves it. Second, the biosensor's mechanism is reminiscent of that of an oligonucleotide acting as facilitator for ribozyme cleavage (24,25). However, the linkage of the biosensor directly to the ribozyme permitted a significant gain, in terms of cleavage activity, as compared with the use of two distinct molecules. As the activation is mediated by the substrate, this might constitute an important advantage for further use in a cellular environment because it implies that these are no limitations in terms of timing and localization for any additional components. Third, the blocker stem was influenced by the TRAP strategy (for targeted ribozyme-attenuated probe), in which a 3′ terminal attenuator anneals to conserved bases in the catalytic core to form the ‘off’ state of a hammerhead ribozyme (7). The blocker sequence of the SOFA module also inactivates the cleavage activity of the ribozyme by binding a sequence that is part of the catalytic core. Finally, the idea of a stabilizer stem that places the 3′ end of the SOFA module in a double-stranded region originated from the previous demonstration that the P2 stem of the δRz, which plays the same role in the wild-type ribozyme, provides an outstanding stability to this RNA species (21). However, it is important to note that the biosensor and blocker sequences function independently of the presence of the stabilizer domain. A version of the SOFA-ribozyme lacking the stabilizer sequence cleaved the HBV transcripts to the same extent as the original version (Figure 2A). Clearly, the SOFA module is the fruit of a rational design. Using the SELEX approach, it would have been impossible to develop this kind of module for a ribozyme.
The action of the SOFA module involves two key mechanisms: the ‘safety lock’ action of the blocker sequence and the activation of the ribozyme. Briefly, the SOFA module enables the ribozyme to only cleave the desired target sequence. Even with the presence of an appropriate P1 stem sequence in the substrate, if the sequence complementary to the biosensor is absent, no cleavage was observed (Figure 5). The ribozyme is blocked in an ‘off’ conformation by the blocker. Consequently, all tested SOFA−-ribozymes (i.e. those possessing an unrelated biosensor, but a good P1 ribozyme sequence) were representative of the ‘off’ conformation (e.g. Figures 2, 4, 5 and 7, ‘safety lock’). In other words, the ribozyme is always in an inactive conformation, except if the biosensor is triggered. A biosensor has to bind its complementary sequence in order to unlock a SOFA module, thereby permitting the folding of the catalytic core into an ‘on’ conformation. The action of the biosensor constitutes the second crucial mechanism associated with the SOFA action. Both the blocker and the biosensor contribute to the increased substrate specificity of the ribozyme's cleavage, although differently. Overall the presence of the biosensor increased the kcat/KM values at least one order of magnitude over that of the original ribozyme (see Table 1). In the case of the blocker, which affects the basal level of activity, we observed a decrease in the kcat/KM value of a SOFA−-ribozyme of two orders of magnitude (i.e. probably up to three orders of magnitude according to the kobs analyses for many other tested ribozymes) (L. J. Bergeron and J. P. Perreault, unpublished data). The latter effect is the most important one in terms of numbers because it contributes to the prevention of the cleavage of erroneous substrates. Together, the blocker and the biosensor of the SOFA concept have lead to the designing of efficient ribozymes that turnover and possess an improved substrate specificity.
The important gain in terms of substrate specificity was illustrated by several experiments. The experiment performed with the pool of 10 substrates having similar P1 stem sequences, but different complementary sequences for biosensor binding, showed that a SOFA+-ribozyme cleaved only the appropriate substrate without interference from the other substrates via a trans-cleavage mechanism (Figure 5). Moreover, this experiment showed that, even in the presence of the appropriate cleavage site on each substrate, the blocker mechanism permits the ribozyme to stay in the ‘off’ conformation until it is activated by the proper substrate. This was not the case with the original ribozyme that cleaved all substrates, although at different levels. The same conclusions were reached from the targeting of similar cis-cleavage sequences harboring different binding sequences for the biosensor in both the HBV and HCV transcripts (Figure 6). In this case, the ribozymes exhibited cleavage at the site proximal to the biosensors, but never to that of the distal site. This shows that the distance between the cleavage site and the binding sequence of the biosensor on a substrate is important. In this regard, further characterization should establish the optimal distance between the biosensor and the cleavage site.
The SOFA-module provides the ribozyme with improved substrate specificity. However, is this sufficient to target only one RNA species in a cellular environment? Considering only the base pairs formed during the two binding steps between an optimal SOFA-δ ribozyme and its substrate (7 bp for P1 stem binding + 10 bp for the biosensor stem), this means that 1 site should exist per 1.7 × 1010 bases (417). Since the human genome is composed of 3 × 109 bp, of which only ∼5% constitutes mRNA (i.e. 1.5 × 108 bases), this is more than sufficient to feel secure in terms of substrate specificity. However, this is true only if all nucleotides contribute fully to the specificity, meaning that no mismatches can be tolerated, which is not necessarily the case. Some experiments have to be performed in order to address this question. Although the probability of finding two sites appears to be negligible, if it does occur the SOFA-ribozyme will distinguish between the two. Other features also contribute favorably to the specificity. For example, the nucleotides 5′ of the cleavage site have been shown to contribute as an external determinant that defines the ability of a substrate to be cleaved (29,30).
Here, we have included initial data showing the inhibition of the expression of the HBV C gene. In this example, the expression of the SOFA-ribozymes was under the control of a polymerase II promoter (CMV) present in a pcDNA3 derived vector. It is interesting to note that a catalyst exhibiting a (biologically speaking) relatively short rate constant (see Figure 2B, half-life near 15 min) has a significant effect on the steady-state level of a target mRNA (Figure 7B, near 50% reduction in HBV C mRNA). Implicit in this data is the issue that target mRNA half-life is an important consideration or limitation, as only mRNAs with relatively long half-lives (>30 min) would be expected to be significantly down-regulated by such catalysts. However, selection of more active ribozymes will allow targeting of a wide range of mRNA targets. Other attempts at investigating the potential use of the SOFA-ribozyme as RNA molecular tools are currently underway. These experiments include use of various targets (i.e. endogenous mRNAs and viral RNAs), different approaches, such as RNA transfection and the generation of stably transfected cells for the production of the SOFA-ribozyme, as well as in several biological systems, such as prokaryotes and eukaryotes. Other research perspectives involve characterizing in intimate detail all features of the SOFA module, as well as the kinetics of its mechanism of action. Additionally, it will be of interest to verify whether or not the concept of the SOFA module offers some flexibility in its design. For example, the SOFA module was also introduced within the L4 loop (Figure 1B; data not shown). The blocker sequence of this later SOFA-module interacted with the sequence of the J4/2 junction of the ribozyme. The SOFA+ version exhibited a relatively high cleavage activity, while the SOFA− did not cleave the substrate. Clearly, this indicates that the SOFA module is malleable and can be easily adapted, since its function is based on the simple and rational hybridization potential of component parts.
To our knowledge, this is the first report of a ribozyme of an endonuclease-type that harbors a ‘safety lock’ target-dependent module that is activated by an RNA substrate and subsequently cleaves this molecule. More specifically, this new ribozyme concept is a single molecule, which can be considered as a clear advantage over other technologies (e.g. allosteric ribozymes). Finally, this new approach provides an improved tool with significant potential in both functional genomics and gene therapy. Furthermore, this concept can be substantially extended to other RNA drug-based molecules.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the technical assistance of D. Lévesque, F. Bolduc and B. Provencher-Dionne, and Gilles Robichaud and S. Abou Elela for critical reading of the original version. This work was supported by grants from the Canadian Institute of Health Research (CIHR) to J.P.P. The RNA group is supported by grants from the CIHR. L.J.B. was the recipient of a pre-doctoral fellowship from CIHR. J.P.P. is an investigator from the CIHR. Funding to pay the Open Access publication charges for this article was provided by CIHR grant number MOP-44002.
REFERENCES
- 1.Lewin A.S., Hauswirth W.W. Ribozyme gene therapy: applications for molecular medicine. Trends Mol. Med. 2001;7:221–228. doi: 10.1016/s1471-4914(01)01965-7. [DOI] [PubMed] [Google Scholar]
- 2.Michienzi A., Rossi J.J. Intracellular applications of ribozymes. Methods Enzymol. 2001;341:581–596. doi: 10.1016/s0076-6879(01)41178-5. [DOI] [PubMed] [Google Scholar]
- 3.Pebernard S., Iggo R.D. Determinants of interferon-stimulated gene induction by RNAi vectors. Differentiation. 2004;72:103–111. doi: 10.1111/j.1432-0436.2004.07202001.x. [DOI] [PubMed] [Google Scholar]
- 4.Carlson C.B., Stephens O.M., Beal P.A. Recognition of double-stranded RNA by proteins and small molecules. Biopolymers. 2003;70:86–102. doi: 10.1002/bip.10413. [DOI] [PubMed] [Google Scholar]
- 5.Sledz C.A., Holko M., de Veer M.J., Silverman R.H., Williams B.R. Activation of the interferon system by short-interfering RNAs. Nature Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 6.Breaker R.R. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 7.Burke D.H., Ozerova N.D.S., Nilsen-Hamilton M. Allosteric hammerhead ribozyme TRAPs. Biochemistry. 2002;41:6588–6594. doi: 10.1021/bi0201522. [DOI] [PubMed] [Google Scholar]
- 8.Wang D.Y., Sen D. Rationally designed allosteric variants of hammerhead ribozymes responsive to the HIV-1 Tat protein. Comb. Chem. High Throughput Screen. 2002;5:301–312. doi: 10.2174/1386207023330273. [DOI] [PubMed] [Google Scholar]
- 9.Wang D.Y., Lai B.H.Y., Feldman A.R., Sen D. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 2002;30:1735–1742. doi: 10.1093/nar/30.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson M.P., Ellington A.D. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 11.Soukup G.A., Breaker R.R. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuwabara T., Warashina M., Tanabe T., Tani K., Asano S., Taira K. A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell. 1998;2:617–627. doi: 10.1016/s1097-2765(00)80160-4. [DOI] [PubMed] [Google Scholar]
- 13.Tang J., Breaker R.R. Rational design of allosteric ribozymes. Chem. Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 14.Bergeron L.J., Ouellet J., Perreault J.P. Ribozyme-based gene-inactivation systems require a fine comprehension of their substrate specificities; the case of delta ribozyme. Curr. Med. Chem. 2003;10:2589–2597. doi: 10.2174/0929867033456486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih I.H., Been M.D. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 16.Nishikawa F., Roy M., Fauzi H., Nishikawa S. Detailed analysis of stem I and its 5′ and 3′ neighbor regions in the trans-acting HDV ribozyme. Nucleic Acids Res. 1999;27:403–410. doi: 10.1093/nar/27.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nassal M. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alaoui-Ismaili M.H., Gervais C., Brunette S., Gouin G., Hamel M., Rando R.F., Bedard J. A novel high throughput screening assay for HCV NS3 helicase activity. Antiviral Res. 2000;46:181–193. doi: 10.1016/s0166-3542(00)00085-1. [DOI] [PubMed] [Google Scholar]
- 19.Bergeron L.J., Perreault J.P. Development and comparison of procedures for the selection of delta ribozyme cleavage sites within the hepatitis B virus. Nucleic Acids Res. 2002;30:4682–4691. doi: 10.1093/nar/gkf598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Anjou F., Bergeron L.J., Larbi N.B., Fournier I., Salzet M., Perreault J.P., Day R. Silencing of SPC2 expression using an engineered δ ribozyme in the mouse βTC-3 endocrine cell line. J. Biol. Chem. 2004;279:14232–14239. doi: 10.1074/jbc.M310632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lévesque D., Choufani S., Perreault J.P. Delta ribozyme benefits from a good stability in vitro that becomes outstanding in vivo. RNA. 2002;8:464–477. doi: 10.1017/s1355838202020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy G., Ananvoranich S., Perreault J.P. Delta ribozyme has the ability to cleave in trans an mRNA. Nucleic Acids Res. 1999;27:942–948. doi: 10.1093/nar/27.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York, NY: W.H. Freeman and Company; 1999. Chapters 3, 10 and 13. [Google Scholar]
- 24.Jankowsky E., Schwenzer B. Oligonucleotide facilitators may inhibit or activate a hammerhead ribozyme. Nucleic Acids Res. 1996;24:423–429. doi: 10.1093/nar/24.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankowsky E., Schwenzer B. Efficient improvement of hammerhead ribozyme mediated cleavage of long substrates by oligonucleotide facilitators. Biochemistry. 1996;35:15313–15321. doi: 10.1021/bi961397f. [DOI] [PubMed] [Google Scholar]
- 26.Odreman-Macchioli F., Baralle F.E., Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J. Biol. Chem. 2001;276:41648–41655. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- 27.Kieft J.S., Zhou K., Jubin R., Murray M.G., Lau J.Y., Doudna J.A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 28.Janeway C.A., Travers P. Immunobiologie. Paris, France: DeBoeck Université; 1997. Chapter 7. [Google Scholar]
- 29.Deschênes P., Lafontaine D.A., Charland S., Perreault J.P. Nucleotides −1 to −4 of hepatitis delta ribozyme substrate increase the specificity of ribozyme cleavage. Antisense Nucleic Acid Drug Dev. 2000;10:53–61. doi: 10.1089/oli.1.2000.10.53. [DOI] [PubMed] [Google Scholar]
- 30.Perrotta A.T., Been M.D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.